Abstract
Mycobacterium avium subspecies paratuberculosis (MAP), the causative agent of Johne's disease in cattle and sheep, has unique iron requirements in that it is mycobactin-dependent for cultivation in vitro. The iron-dependent regulator (IdeR) is a well-characterized global regulator responsible for maintaining iron homeostasis in Mycobacterium tuberculosis (MTB). We identified an orthologous segment in the MAP genome, MAP2827, with >93 % amino acid identity to MTB IdeR. Electrophoretic mobility shift assays and DNase protection assays confirmed that MAP2827 binds the 19 bp consensus motif (iron box) on the MAP genome. Sequencing of MAP2827 from multiple isolates revealed a non-synonymous change (R91G) exclusive to sheep strains. Reporter gene assays and quantitative real-time RT-PCR assays in two diverse MAP strains and in an ideR deletion mutant of M. smegmatis (mc2155) suggested that both sheep MAP IdeR (sIdeR) and cattle MAP IdeR (cIdeR) repress mbtB transcription at high iron concentrations and relieve repression at low iron concentrations. On the other hand, bfrA (an iron storage gene) was upregulated by cIdeR when presented with MTB or the cattle MAP bfrA promoter, and was downregulated by sIdeR in the presence of MTB, or sheep or cattle MAP bfrA promoters, at high iron concentrations. The differential iron regulatory mechanisms between IdeR-regulated genes across strains may contribute to the differential growth or pathogenic characteristics of sheep and cattle MAP strains. Taken together, our study provides a possible reason for mycobactin dependency and suggests strong implications in the differential iron acquisition and storage mechanisms in MAP.
INTRODUCTION
Iron is a cofactor in several enzymic reactions due to its wide redox potential (De Voss et al., 1999; Wandersman & Delepelaire, 2004). Iron plays a central role in enzymic reactions involved in electron transport, nucleic acid synthesis and oxidative stress defence. Therefore, iron is critical for most living systems. The iron-dependent regulator (IdeR) of Mycobacterium tuberculosis (MTB) has been shown to regulate a repertoire of genes in response to iron concentration (Rodriguez et al., 2002). Comparative evaluation of mycobacterial genomes has revealed that ideR is present in all sequenced mycobacteria (Yellaboina et al., 2006). Structural and functional roles of IdeR have been extensively studied in MTB (Pohl et al., 1999; Rodriguez & Smith, 2003; Wisedchaisri et al., 2004, 2007). In the presence of iron, IdeR binds to a 19 bp promoter sequence called the ‘iron box’ and represses the transcription of genes involved in iron acquisition, while activating iron-storage genes. Inactivation of ideR is possible only in the presence of a second suppressor mutation, suggesting that ideR is an essential gene in MTB (Rodriguez et al., 2002). This rare mutant of ideR showed deregulated siderophore synthesis and increased sensitivity to oxidative stress. Thus it is clear that iron-concentration-induced gene regulation is essential for the survival of pathogenic mycobacteria.
Mycobacterium avium subsp. paratuberculosis (MAP) is very well known for its unique iron requirements for in vitro growth. Mycobactin dependency in laboratory-based culture systems is now well-established (Wheeler & Hanks, 1965). We recently demonstrated that despite mycobactin dependency in vitro, MAP upregulates mycobactin (mbt) synthesis genes inside bovine macrophages (Zhu et al., 2008). Similarly, proteomic (MAP grown in iron-sufficient and iron-limited conditions) analysis has also revealed that MAP regulates expression of an iron storage (bfrA) gene (H. K. Janagama & S. Sreevatsan; unpublished). These findings suggest that MAP employs a network of genes that are deployed to maintain iron homeostasis. IdeR, a transcriptional factor, monitors global iron regulation in MTB. Currently, there is a lack of understanding of iron regulation in MAP. In the current study, we characterized the IdeR of MAP and its functional role in iron-dependent gene regulation.
METHODS
Bacterial strains, DNA manipulations and protein expression.
MAP strains were grown in Middlebrook 7H9 supplemented with OADC enrichment medium and mycobactin J. M. smegmatis was grown in Luria–Bertani (LB) medium. Antibiotics (μg ml−1: kanamycin, 20; hygromycin, 100; streptomycin, 20; spectinomycin, 75) were added when necessary. Competent E. coli BL21(DE3) (Novagen) and E. coli TOP10F cells (Invitrogen) were grown in LB medium. Minimal medium was prepared as described previously (Rodriguez et al., 2002). DNA modifying enzymes were purchased from New England Biolabs. The predicted ORF of MAP2827 from the MAP K-10 genome was amplified using primers 2827NF (5′-GGAATTCCATATGATGAACGACCTGGTTGACACC-3′) and 2827BR (5′-CGCGGATCCTCAGACCTTTTCGACCTTGA-3′), which carried NdeI and BamHI restriction sites (underlined) at the 5′ end and cloned into predigested (NdeI and BamHI) vector pET-16b (Novagen). Proper insertion and orientation of MAP2827 into pET-16b was verified by sequencing. Competent E. coli BL21(DE3) cells (Novagen) were transformed with pET-16b carrying MAP2827 and induced with 1 mM IPTG for 4 h to over express MAP2827. The expressed MAP2827 with an N-terminal his-tag was purified using Ni-NTA columns (Qiagen). Recombinant MAP2827 was also obtained as a maltose-binding protein fusion protein (Bannantine et al., 2004).
Computational prediction of IdeR-regulated genes.
MTB IdeR regulates gene expression by binding to a 19 bp consensus sequence, termed the iron box, in gene promoter regions (Gold et al., 2001). The Prokaryotic Database of Gene Regulation (prodoric) is a knowledge base that encompasses molecular networks of prokaryotes, such as transcriptional regulation (www.prodoric.de) (Munch et al., 2003). Virtual footprint, a software tool in prodoric, is used for identification of the transcriptional factor binding sites in a bacterial genome. Using this software option, the iron box sequence (TWAGGTWAGSCTWACCTWA; where W=A/T and S=G/C) was queried in the MAP genome (Li et al., 2005) to retrieve similar sequences in the intergenic regions. Searches were allowed for five mismatches in the consensus sequence and were limited to −300 to +100 bases of each predicted ORF as reported by Gold et al. (2001).
Electrophoretic mobility shift assay (EMSA).
Physical binding of cattle MAP2827 to the promoter sequences of cattle mbtB and bfrA was carried out by EMSA. Promoter sequences containing the putative iron box of MAP bfrA or mbtB were amplified using 5′ biotin-labelled primers via PCR. Purified amplification products were used in DNA protein interaction studies. Binding reactions (Gold et al., 2001) were performed for 30 min in the presence or absence of 200 μM NiSO4 containing 20 fm DNA and 250 ng protein, and the complexes were resolved in a 10 % non-denaturing polyacrylamide gel. Following gel electrophoresis, the complexes were transferred onto a nylon membrane and detected using a chemiluminescence-based nucleic acid detection kit (Pierce).
DNase protection assay.
A DNase footprinting assay was performed to map the physical binding region of MAP2827 on the putative promoter sequence as described by Merighi et al. (2006) and Zianni et al. (2006). Cattle MAP mbtB or bfrA promoters carrying the predicted iron box sequence were cloned into pSM128 (see below). A 5′ FAM-labelled forward primer (which had the priming sites 100 bp upstream of the cloned segment) and a 5′ HEX-labelled reverse primer (which had the priming sites 100 bp downstream of the cloned segment) were used to amplify MAP promoters from pSM128. Binding of MAP2827 to the probe was performed as described for EMSA. Following DNA–protein interaction, complexes were digested partially with DNase I. A parallel partial DNase I digestion of the DNA probe alone was also performed. Digested DNA and DNA–protein complexes were purified individually to remove proteins and other salts contained in the reaction buffer. Purified fragments were resolved in a fragment analyser (Applied Biosystems 3130xl) and chromatograms were analysed using GeneMapper software (Applied Biosystems).
Reporter gene assays.
Functional activity of mbtB and bfrA promoters was assessed using a cattle MAP strain (MAP 1018) and an M. smegmatis ideR null mutant, SM3 (Dussurget et al., 1996). Predicted MAP promoter sequences containing the iron box were amplified using primers carrying ScaI restriction sites. Amplified products were restriction-digested with ScaI and ligated into a pre-digested (ScaI) promoterless integrative plasmid, pSM128 (Dussurget et al., 1999). Proper orientation of sequences in the plasmid was verified by sequencing. MAP 1018 was transformed with pSM128 carrying MAP mbtB or bfrA promoters. ORFs MAP2827 from MAP K-10 (cattle strain) or MAP 7565 (sheep strain) were amplified via PCR using primers that carried restriction sites for BamHI and HindIII. Amplified products were double-digested with BamHI and HindIII and ligated into a pre-digested (BamHI and HindIII) expression plasmid, pSM417. Proper orientation and ligation of MAP2827 into pSM417 was verified by sequencing. SM3 was transformed both with pSM128 carrying MAP promoter sequences and pSM417 carrying MAP2827. The recombinant MAP 1018 (carrying MAP mbtB and bfrA promoter–lacZ fusions) and M. smegmatis (carrying MAP mbtB or bfrA promoter–lacZ fusions and MAP2827) strains were cultured in minimal medium supplemented with 50 μM (high iron) or 1 μM FeCl3 (low iron) and analysed for β-galactosidase activity (Rodriguez et al., 1999), according to the manufacturer's recommendations (Promega; cat#E2000). Iron concentrations were selected based on the fact that a concentration above 25 μM is sufficient to activate the repressor activity of IdeR (G. M. Rodriguez & others, unpublished).
Real-time Q-RT-PCR assays.
Transcription of mbtB and bfrA was measured using real-time Q-RT-PCR. MAP 1018 (cattle strain) and MAP 7565 (sheep strain) were grown to mid-exponential growth phase (OD600=1.0) in MB7H9 medium containing OADC and mycobactin J. Bacterial cultures were washed five times in 1× PBS, resuspended in minimal medium containing no iron or 50 μM iron, and gene expression was allowed to continue for 3 h. RNA isolation was performed as described by Zhu et al. (2008). RNA was treated with DNase I (Ambion) and Q-RT-PCR was performed using QuantiFast SYBR Green mix (Qiagen) and gene specific primers in a LightCycler 480 (Roche). Gene expression values of mbtB and bfrA were normalized to 16s rRNA and reported as fold change (low iron/high iron).
Intracellular and in vitro MAP gene expression profiling.
Human primary CD14-positive monocytes were obtained from Lonza. Monocytes were differentiated into macrophages by the addition of recombinant human granulocyte macrophage colony-stimulating factor (R&D Systems). Following differentiation for 3 days in Teflon wells (Savillex Corporation), 2×106 macrophages were seeded into 25 cm2 tissue culture flasks. Macrophages were infected with cattle or human MAP strains at an m.o.i. of 20. RNA isolation, cDNA synthesis was performed as described by Wu et al. (2007) and Zhu et al. (2008). Briefly, first strand cDNA was synthesized from total RNA isolated from infected macrophages or MAP grown in broth cultures using primers that carried a defined 5′ end and a random nonamer at the 3′ end. MAP targets specifically expressed inside macrophages were obtained via hybridization to biotin-labelled MAP genomic DNA. Following second strand cDNA synthesis and PCR amplification, MAP cDNA obtained either from macrophages or broth cultures was individually hybridized onto MAP K-10 microarrays. MAP genomic DNA (gDNA) was used as reference in each hybridization (Faucher et al., 2006). Array normalizations and data analysis were performed as described by Paustian et al. (2008). Following normalization of gDNA and cDNA signals, fluorescence intensities were log2 transformed and reported as fold change. Genomic DNA-based normalization of expression data has been described by Talaat et al. (2002).
Statistical analysis.
All experiments were independently repeated at least three times in triplicate. Means of Miller units from reporter gene assays were compared across strains and treatments using a multiple range t-test with Bonferroni's correction.
RESULTS
MAP2827 shows sequence-specific physical binding to the promoter regions of mbtB and bfrA
The annotated version of MAP IdeR (MAP2827c) does not share amino acid identity with other mycobacterial IdeR proteins. Nucleotide sequence alignment of MAP2827c and MTB IdeR revealed 86 % similarity, suggesting that MAP IdeR may be transcribed in a different ORF in the same genomic location. A search for ORFs in all orientations revealed MAP2827 as a possible ORF encoding MAP IdeR.
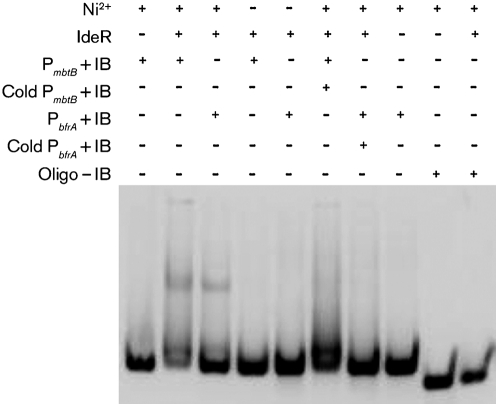
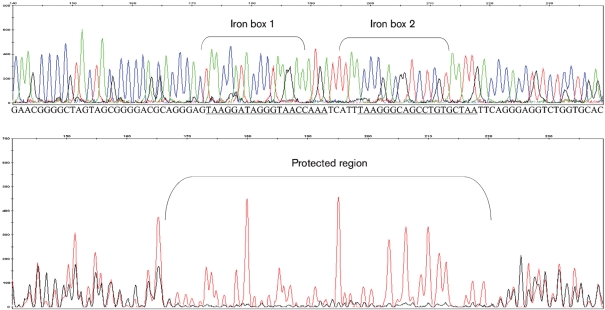
Computational predictions identified 24 genes that may be regulated by IdeR (Supplementary Table S1). We focused our analyses on proteins that serve two distinct functions – an iron acquisition protein, MbtB, and an iron storage protein, BfrA. We first tested the ability of MAP IdeR to bind to the predicted promoter regions using EMSA. Our results show that IdeR bound to the promoters of mbtB and bfrA in presence of Ni+2 (Fig. 1). Competition for the binding sites on IdeR using excess unlabelled promoter DNA abrogated the gel shift. Furthermore, there was no binding of IdeR to a divergent probe that carried no iron box sequence. The DNA sequence bound by IdeR was identified by DNase protection assays using fluorescently labelled intergenic regions of mbtB or bfrA containing the suspected iron box sequence. Results showed that IdeR recognizes and protects the iron box sequence on the MAP mbtB and bfrA promoters (Fig. 2). This demonstrates that MAP IdeR is a metal-dependent DNA-binding protein that interacts with a specific sequence in the promoter regions of two iron-regulated genes and is likely to function as a transcriptional regulator in MAP.
Fig. 1.
Electrophoretic mobility shift assay to demonstrate specificity of MAP2827 protein with the predicted promoters. Recombinant MAP2827 was reacted with mbtB and bfrA promoters for this assay. MAP IdeR reacts with the promoters only in presence of nickel (second and third lanes from left). P, Promoter; +/− IB, presence/absence of iron box; cold, unlabelled DNA; oligo, non-specific DNA.
Fig. 2.
DNase footprint assay showing the binding of the MAP2827 protein to the iron box sequence located on the MAP mbtB promoter. Red peaks in the electropherogram represent DNA alone and black peaks represent DNA pre-incubated with MAP2827 (bottom panel). Both reactions were partially digested with DNase I and analysed by capillary electrophoresis in a genetic analyser (Applied Biosystems 3130xl). The top panel shows an electropherogram of the promoter sequence with the terminal bases identified (iron box sequence is underlined). The protected region is indicated which overlaps the iron box sequence. Note the difference in the heights of black peaks (lost completely) relative to red peaks, indicating protection from DNase digestion.
Sequence analysis of the MAP ideR ORF and promoter regions of putative IdeR-regulated genes reveals polymorphisms between sheep and cattle MAP strains
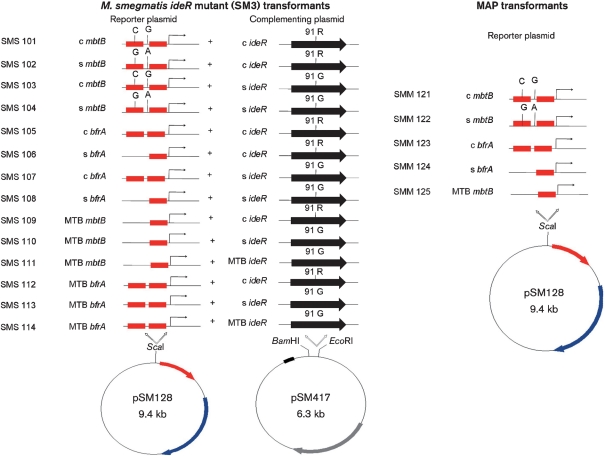
To establish that MAP2827/ideR was conserved across all genotypes of MAP, ideR and the mbtB and bfrA promoters of putative IdeR-regulated genes from cattle, human, bison, and sheep MAP isolates were sequenced. Data showed that ideR was conserved across all cattle (1018, K-10, 9123, 9142, 9245), bison (7560) and human (M4, M5, M6) isolates studied, but the sheep isolates (7565, 467, 394) showed a single nucleotide polymorphism that led to an amino acid change at residue 91 (R91G) (Supplementary Fig. S1). Similarly sequence analysis of the mbtB and bfrA promoter regions of IdeR transcribed genes identified polymorphisms exclusively in sheep MAP strains (Fig. 3). Of the two iron boxes present within the bfrA promoter, the box distal to the bfrA start site in sheep MAP isolates is deleted. Similarly, the mbtB promoter of sheep MAP isolates carries two single nucleotide polymorphisms (C-87G and G-71A relative to the ATG site of mbtA) compared to cattle MAP isolates. The C to G polymorphism is present within the iron box, whereas the G to A polymorphism is present outside the iron box.
Fig. 3.
Genotypes of SM3 constructs used to transform M. smegmatis and MAP transformants used in reporter assays. The polymorphisms present in the promoter regions of MAP mbtB and bfrA promoters are highlighted. Red boxes, iron boxes; black arrow, ideR ORF; c, cattle; s, sheep.
Functional assays reveal divergent control of promoters by sheep and cattle MAP IdeR
Transcriptional regulation of IdeR derived from cattle (cIdeR) and sheep (sIdeR) MAP strains was studied using promoter fusions to lacZ in an IdeR deletion mutant of M. smegmatis (SM3), and in intact cattle and sheep MAP strains. Controls used were dual empty vector (SM3 transformed with pSM128 and pSM417 without inserts), promoter alone (SM3 transformed with pSM128 carrying a promoter without an iron box sequence), iron box control (SM3 transformed with pSM128 carrying a promoter with an iron box sequence) and ideR alone (SM3 transformed with pSM417 carrying ideR of cattle or sheep MAP variant) grown in high- or low-iron medium.
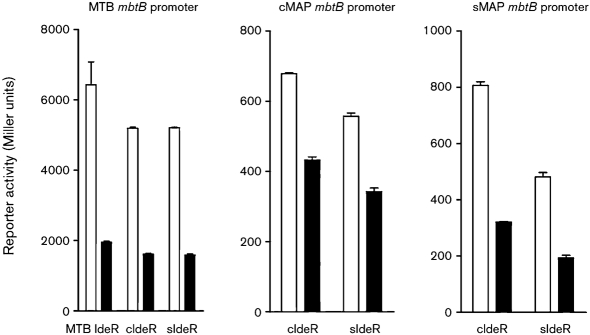
First, control experiments in an ideR-deleted M. smegmatis (SM3) showed that cIdeR or sIdeR repressed transcription of MTB mbtB under high-iron conditions, similar to MTB IdeR (Fig. 4). Since sequence analysis of ideR and the promoter regions of IdeR-regulated genes revealed polymorphisms between cattle and sheep MAP strains, we tested structure–function associations by swapping IdeR and promoters between cattle and sheep MAP variants, and measured transcription of the reporter gene in SM3. Results suggested that irrespective of polymorphisms in the promoter segments, mbtB transcription was repressed under iron-rich conditions by cIdeR and sIdeR (Fig. 4). These findings were statistically significant (P<0.05).
Fig. 4.
Reporter activity of mbtB promoter fusions of MTB mbtB, cattle MAP mbtB or sheep MAP mbtB to lacZ driven by cIdeR, sIdeR or MTB IdeR. Under iron-rich conditions, cIdeR, sIdeR and MTB IdeR repressed transcription of mbtB. Open bars, transcription in iron-depleted medium; filled bars, transcription in iron-rich medium. Means of three independent experiments, each performed in triplicate, are shown.
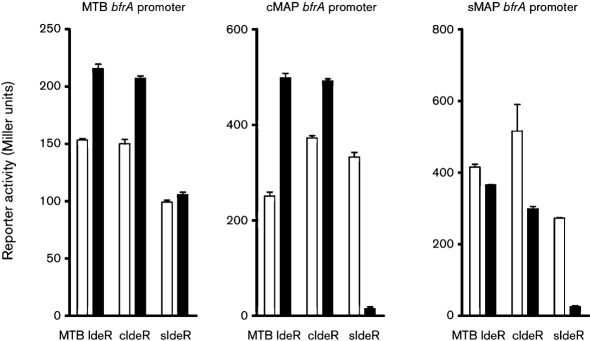
We then compared the activities of MTB (control), cIdeR and sIdeR on bfrA promoter binding and subsequent transcription. MTB IdeR and cIdeR showed identical activities on binding to cattle MAP or MTB bfrA promoters under iron-rich conditions (Fig. 5). In contrast, IdeRs from MTB, and sheep and cattle strains of MAP downregulated transcription in iron-excess growth medium in the presence of the sheep bfrA promoter (Fig. 5).
Fig. 5.
Reporter activity of bfrA promoter fusions of MTB bfrA, cattle MAP bfrA or sheep MAP bfrA to lacZ driven by cIdeR, sIdeR or MTB IdeR. Under iron-rich conditions, cIdeR, sIdeR and MTB IdeR showed repressed transcription with the sheep bfrA promoter. Open bars, transcription in iron-depleted medium; closed bars, transcription in iron-rich medium. Means of three independent experiments, each performed in triplicate, are shown.
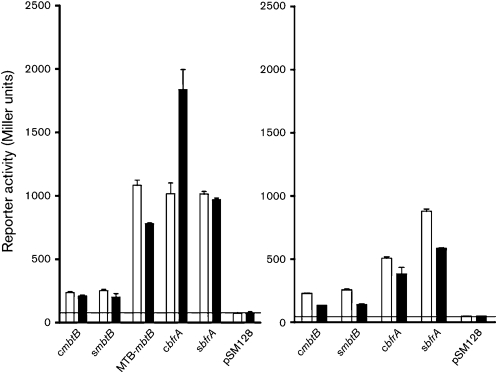
Second, these effects were studied in a native MAP background – we attempted to create an ideR deletion mutant of MAP and failed. Therefore, to address if the promoters are active in an MAP genetic background, we transformed cattle and sheep MAP strains with bfrA or mbtB promoters and measured reporter activity. Results suggested that both mbtB and bfrA promoters are active in MAP (Fig. 6). In the presence of excess iron, both cIdeR and sIdeR repressed mbtB transcription, whereas only sIdeR repressed transcription via the bfrA promoter.
Fig. 6.
Reporter activity of mbtB and bfrA promoter fusions to lacZ in two MAP strains. The line indicates the background reporter activity of the empty vector (pSM128). Both mbtB and bfrA are active in MAP. The activity of MAP mbtB promoters is lower in comparison to that of MTB mbtB. The bfrA promoter from a sheep MAP strain showed consistently lower activity under high-iron conditions, as shown in the SM3 background. Open bars, transcription in iron-depleted medium; closed bars, transcription in iron-rich medium; c, cattle; s, sheep. Means of three independent experiments, each performed in triplicate, are shown.
Since reporter gene assays suggested differential promoter activities and downstream gene expression between cattle and sheep MAP strains, we measured transcription of mbtB and bfrA in cattle and sheep MAP strains using Q-RT-PCR under high- and low-iron concentrations. Consistent with reporter gene assay data in SM3 and MAP backgrounds, the results showed that mbtB was repressed under low-iron growth conditions by cattle and sheep MAP strains, whereas bfrA transcription was derepressed only in the sheep MAP strain (Table 1).
Table 1.
Iron-concentration-dependent expression of mbtB and bfrA as measured by Q-RT-PCR in cattle and sheep strains of MAP
| Gene | Fold change (IE/ID)* |
|---|---|
| Derepressed by IdeR and iron | |
| Cattle bfrA | 2.63 |
| Repressed by IdeR and iron | |
| Sheep bfrA | 0.13 |
| Cattle mbtB | 0.40 |
| Sheep mbtB | 0.17 |
*Fold change for each target gene is calculated as described in Methods and presented as the ratio of expression under iron excess (IE) to that under iron depletion (ID). A fold change of 1 is considered as no change in gene expression in response to iron, whereas a fold change greater than or less than 1 is considered as derepressed or repressed, respectively, in response to iron.
Next, we performed microarray-based gene expression analysis on a cattle strain to confirm both functional assay data and computational predictions. Several bioinformatically predicted IdeR-regulated genes were transcribed in macrophages (iron-depleted environment) or in nutrient-rich broth cultures (Table 2). As expected, mbtA was downregulated whereas bfrA was upregulated in the nutrient-rich broth cultures in a cattle MAP strain. mbtE was upregulated in human macrophages. Expression patterns of bfrA in broth cultures (iron-rich environment) and mbtE in macrophages (iron-deficient) suggest that MAP regulates expression of these genes in an iron-dependent fashion.
Table 2.
Putative IdeR-regulated genes expressed in primary human macrophages or in broth cultures
| ORF ID | Predicted function/gene name | Fold change* | |
|---|---|---|---|
| In vivo | In vitro | ||
| MAP0024c | Hypothetical protein | – | −5.14 |
| MAP0025 | Acyl carrier protein | – | −8.20 |
| MAP0913c | fadE13 (fatty acid metabolism) | – | −7.64 |
| MAP1559c | Transcriptional repressor | 1.61 | 3.38 |
| MAP1560 | Hypothetical protein | – | −7.42 |
| MAP1595 | bfrA | – | 9.11 |
| MAP1762c | Iron permease FTR1 family protein | – | −7.42 |
| MAP2178 | mbtA | – | −7.94 |
| MAP2206 | Putative permease protein | −4.73 | −3.18 |
| MAP3778 | ATPase, AAA family protein | 2.18 | −2.44 |
| MAP2173c | mbtE | 2.05 | – |
*Fold change expressed as normalized log2 values; – represents <1.5-fold change for that target.
DISCUSSION
MAP is unique in its iron requirements owing to its mycobactin dependency in vitro. However, to date there is no comprehensive understanding either of iron metabolic pathways, in general, or the regulation of the mycobactin operon in MAP. In MTB, the iron-dependent regulator IdeR is the principal regulator of iron metabolism (Rodriguez & Smith, 2003). We report here that MAP2827 carries IdeR function and regulates mbtB and bfrA gene expression. Selection of mbtB and bfrA is based on their immediate roles in iron acquisition and iron storage, respectively.
IdeR controls expression of genes involved in iron acquisition and iron storage via iron box sequence recognition and binding. Computational screening of the MAP genome for iron box motifs identified 24 genes associated with diverse functions in iron metabolism. Transcriptional analysis confirmed expression of a subset of these genes in MAP upon macrophage infection (iron-restricted conditions) and/or broth cultures (iron-rich media). Expression data of bfrA in broth cultures (iron-rich) and mbtE inside macrophages (iron-deficient) clearly suggests that MAP regulates expression of these genes in an iron-dependent fashion. mbtE is a part of the mycobactin synthesis operon, a 10 gene cluster (mbtA–J), and the encoded protein carries a peptidyl carrier protein domain involved in mycobactin biosynthesis. The predicted iron box is located between mbtB and mbtA. These findings suggest that MAP IdeR regulates at least some of the genes computationally predicted to be in the IdeR regulon in this study. Genome mining to identify an IdeR homologue in the MAP genome revealed that the complementary sequence of the annotated ideR shared substantial amino acid similarities with the IdeR family of proteins. Sequence analysis of ideR (MAP2827) of diverse MAP strains revealed that the ideR ORF and IdeR-regulated promoters exhibit polymorphisms that could lead to functional variation between cattle and sheep strains.
It is believed that M. avium encompasses a diverse group of subspecies (Turenne et al., 2008) that are host-specific, and that this specialization occurred after diversification. Molecular epidemiological studies, comparative genomic hybridizations and host–pathogen interaction studies suggest that MAP is genotypically and phenotypically diverse (Motiwala et al., 2006a, b; Paustian et al., 2008). It is also now well-established that sheep MAP strains are extremely slow-growing organisms in routine laboratory cultures relative to cattle MAP strains. Sequence analysis of the ideR ORF and the promoters of IdeR-regulated genes demonstrated polymorphisms between cattle and sheep MAP strains. Functional studies with two IdeR-regulated genes, mbtB (iron acquisition gene) and bfrA (iron storage gene), revealed variations in bfrA regulation under adequate iron conditions. This variation may partially explain their in vitro phenotype in that the sheep strain may have a defective iron storage pathway leading to iron toxicity when excess iron is provided in culture media. More directed studies on iron storage mechanisms in sheep strains would be needed to fully address this contention.
It is counterintuitive for sheep strains to repress transcription of an iron storage gene under high iron conditions that are also expected to occur in an early phagosome environment (Wagner et al., 2005, 2006). Whether differential iron storage/regulation between sheep and cattle MAP strains exists or not would be better addressed when the genome sequence of a sheep strain of MAP is available for analysis.
Transcriptional start point assays have identified two promoter sites (Plow and Phigh) in the MTB bfrA intergenic region (Gold et al., 2001). It has been proposed that in the presence of iron, bfrA transcription is either downregulated (via Plow) or upregulated (via Phigh). Our data suggested that cIdeR in the presence of the cattle MAP bfrA promoter upregulated transcription in the presence of iron, probably behaving as Phigh. On the other hand, sIdeR or cIdeR from MAP or MTB IdeR downregulated bfrA transcription in the presence of a sheep MAP promoter under high iron conditions, suggesting Plow activity. This may be due to polymorphisms identified in the bfrA promoter of sheep MAP strains. Thus it is possible that there are alternate promoters for cattle and sheep bfrA. We are currently addressing these issues by transcriptional start point analysis in MAP.
The current study was aimed at determining if ideR of MAP encodes a functional protein. Our results show that MAP2827 controls transcription of genes involved in iron regulation. Analysis of the whole regulon of IdeR is underway and is expected to provide an understanding of iron regulation in MAP.
Acknowledgments
This work was supported in part by a USDA-NRI grant (2005-35204-16106) and Johne's Disease Integrated Program (USDA-CSREES 2008-55620-18710) awarded to S. S. The work was also supported by an NIH grant (AI 44856) awarded to I. S. We would like to thank the Microbial and Plant Genomics Institute, Biomedical Genomics Center and Computational Genetics Laboratory at the University of Minnesota for providing resources and services to perform these studies.
Abbreviations
EMSA, electrophoretic mobility shift assay
MAP, Mycobacterium avium subsp. paratuberculosis
MTB, Mycobacterium tuberculosis
cIdeR/sIdeR, cattle/sheep MAP IdeR
Footnotes
A multiple sequence alignment of MAP2827c, MAP2827, MAV3604 and RV2711 and a table of prodoric-predicted putative MAP IdeR regulated genes are available with the online version of this paper.
References
- Bannantine, J. P., Hansen, J. K., Paustian, M. L., Amonsin, A., Li, L. L., Stabel, J. R. & Kapur, V. (2004). Expression and immunogenicity of proteins encoded by sequences specific to Mycobacterium avium subsp. paratuberculosis. J Clin Microbiol 42, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Voss, J. J., Rutter, K., Schroeder, B. G. & Barry, C. E., III (1999). Iron acquisition and metabolism by mycobacteria. J Bacteriol 181, 4443–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussurget, O., Rodriguez, M. & Smith, I. (1996). An ideR mutant of Mycobacterium smegmatis has derepressed siderophore production and an altered oxidative-stress response. Mol Microbiol 22, 535–544. [DOI] [PubMed] [Google Scholar]
- Dussurget, O., Timm, J., Gomez, M., Gold, B., Yu, S., Sabol, S. Z., Holmes, R. K., Jacobs, W. R., Jr & Smith, I. (1999). Transcriptional control of the iron-responsive fxbA gene by the mycobacterial regulator IdeR. J Bacteriol 181, 3402–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher, S. P., Porwollik, S., Dozois, C. M., McClelland, M. & Daigle, F. (2006). Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc Natl Acad Sci U S A 103, 1906–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold, B., Rodriguez, G. M., Marras, S. A., Pentecost, M. & Smith, I. (2001). The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol Microbiol 42, 851–865. [DOI] [PubMed] [Google Scholar]
- Li, L., Bannantine, J. P., Zhang, Q., Amonsin, A., May, B. J., Alt, D., Banerji, N., Kanjilal, S. & Kapur, V. (2005). The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc Natl Acad Sci U S A 102, 12344–12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merighi, M., Majerczak, D. R., Zianni, M., Tessanne, K. & Coplin, D. L. (2006). Molecular characterization of Pantoea stewartii subsp. stewartii HrpY, a conserved response regulator of the Hrp type III secretion system, and its interaction with the hrpS promoter. J Bacteriol 188, 5089–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiwala, A. S., Janagama, H. K., Paustian, M. L., Zhu, X., Bannantine, J. P., Kapur, V. & Sreevatsan, S. (2006a). Comparative transcriptional analysis of human macrophages exposed to animal and human isolates of Mycobacterium avium subspecies paratuberculosis with diverse genotypes. Infect Immun 74, 6046–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiwala, A. S., Li, L., Kapur, V. & Sreevatsan, S. (2006b). Current understanding of the genetic diversity of Mycobacterium avium subsp. paratuberculosis. Microbes Infect 8, 1406–1418. [DOI] [PubMed] [Google Scholar]
- Munch, R., Hiller, K., Barg, H., Heldt, D., Linz, S., Wingender, E. & Jahn, D. (2003). prodoric: prokaryotic database of gene regulation. Nucleic Acids Res 31, 266–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paustian, M. L., Zhu, X., Sreevatsan, S., Robbe-Austerman, S., Kapur, V. & Bannantine, J. P. (2008). Comparative genomic analysis of Mycobacterium avium subspecies obtained from multiple host species. BMC Genomics 9, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl, E., Holmes, R. K. & Hol, W. G. (1999). Crystal structure of the iron-dependent regulator (IdeR) from Mycobacterium tuberculosis shows both metal binding sites fully occupied. J Mol Biol 285, 1145–1156. [DOI] [PubMed] [Google Scholar]
- Rodriguez, G. M. & Smith, I. (2003). Mechanisms of iron regulation in mycobacteria: role in physiology and virulence. Mol Microbiol 47, 1485–1494. [DOI] [PubMed] [Google Scholar]
- Rodriguez, G. M., Gold, B., Gomez, M., Dussurget, O. & Smith, I. (1999). Identification and characterization of two divergently transcribed iron regulated genes in Mycobacterium tuberculosis. Tuber Lung Dis 79, 287–298. [DOI] [PubMed] [Google Scholar]
- Rodriguez, G. M., Voskuil, M. I., Gold, B., Schoolnik, G. K. & Smith, I. (2002). ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun 70, 3371–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaat, A. M., Howard, S. T., Hale, W. T., Lyons, R., Garner, H. & Johnston, S. A. (2002). Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res 30, e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turenne, C. Y., Collins, D. M., Alexander, D. C. & Behr, M. A. (2008). Mycobacterium avium subsp. paratuberculosis and M. avium subsp. avium are independently evolved pathogenic clones of a much broader group of M. avium organisms. J Bacteriol 190, 2479–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D., Maser, J., Lai, B., Cai, Z., Barry, C. E., III, Honer Zu Bentrup, K., Russell, D. G. & Bermudez, L. E. (2005). Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J Immunol 174, 1491–1500. [DOI] [PubMed] [Google Scholar]
- Wagner, D., Maser, J., Moric, I., Vogt, S., Kern, W. V. & Bermudez, L. E. (2006). Elemental analysis of the Mycobacterium avium phagosome in Balb/c mouse macrophages. Biochem Biophys Res Commun 344, 1346–1351. [DOI] [PubMed] [Google Scholar]
- Wandersman, C. & Delepelaire, P. (2004). Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol 58, 611–647. [DOI] [PubMed] [Google Scholar]
- Wheeler, W. C. & Hanks, J. H. (1965). Utilization of external growth factors by intracellular microbes: Mycobacterium paratuberculosis and wood pigeon mycobacteria. J Bacteriol 89, 889–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisedchaisri, G., Holmes, R. K. & Hol, W. G. (2004). Crystal structure of an IdeR–DNA complex reveals a conformational change in activated IdeR for base-specific interactions. J Mol Biol 342, 1155–1169. [DOI] [PubMed] [Google Scholar]
- Wisedchaisri, G., Chou, C. J., Wu, M., Roach, C., Rice, A. E., Holmes, R. K., Beeson, C. & Hol, W. G. (2007). Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis. Biochemistry 46, 436–447. [DOI] [PubMed] [Google Scholar]
- Wu, C. W., Schmoller, S. K., Shin, S. J. & Talaat, A. M. (2007). Defining the stressome of Mycobacterium avium subsp. paratuberculosis in vitro and in naturally infected cows. J Bacteriol 189, 7877–7886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellaboina, S., Ranjan, S., Vindal, V. & Ranjan, A. (2006). Comparative analysis of iron regulated genes in mycobacteria. FEBS Lett 580, 2567–2576. [DOI] [PubMed] [Google Scholar]
- Zhu, X., Tu, Z. J., Coussens, P. M., Kapur, V., Janagama, H., Naser, S. & Sreevatsan, S. (2008). Transcriptional analysis of diverse strains Mycobacterium avium subspecies paratuberculosis in primary bovine monocyte derived macrophages. Microbes Infect 10, 1274–1282. [DOI] [PubMed] [Google Scholar]
- Zianni, M., Tessanne, K., Merighi, M., Laguna, R. & Tabita, F. R. (2006). Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J Biomol Tech 17, 103–113. [PMC free article] [PubMed] [Google Scholar]