Abstract
Krüppel-like factor 8 (KLF8) regulates critical cellular processes including cell cycle progression, transformation, epithelial-to-mesenchymal transition, migration and invasion by either repressing or activating target gene promoters. As a repressor, KLF8 recruits the CtBP co-repressor via its PVDLS repression motif. However, how KLF8 acts as an activator has not been determined. Here we report the identification of both the KLF8 activation domain and associated co-activators. By site-directed mutagenesis and cyclin D1 promoter reporter assays using both mouse fibroblasts and human epithelial cells, we determined that deletion of residues 100–260 or mutation of Q118-Q248 abolished KLF8 transactivity. This transactivity was dramatically reduced in p300−/−, CBP−/− or PCAF−/− cells and could be restored by re-expressing p300 or PCAF, but not CBP. Co-immunoprecipitation analyses demonstrated that KLF8 interacted with these co-activators whereas the Q118N-Q248N mutant did not. Chromatin immunoprecipitation experiments showed that KLF8 promoted histone acetylation at the promoter whereas the Q118N-Q248N mutant had a dramatic loss of this function. Western blotting revealed that unlike wild-type KLF8 the Q118N-Q248N was no longer able to upregulate cyclin D1 protein level. BrdU incorporation assays showed that the Q118N-Q248N mutant also lost the ability to promote DNA synthesis. Taken together, these results identified the KLF8 activation domain located between residues 101–260 where the well-conserved Q118 and Q248 are essential for recruiting p300 and PCAF to activate target gene transcription.
Keywords: krüppel-like factor 8, transcription activation, p300, creb-binding protein, p300/CBP associated factor, cyclin D1
Introduction
KLF8 is a Krüppel-like transcription factor (KLF) family member that plays a critical role in the regulation of important cellular processes including cell cycle progression,1–4 oncogenic transformation,5 epithelial-to-mesenchymal transition, migration and invasion.6 The expression and cellular function of KLF8 is positively regulated by cell signaling molecules such as Src and PI3K downstream of FAK1,2,7,8 and transcriptional activators including Sp1,2 and KLF1,9 and negatively regulated by transcriptional repression by KLF3,9 and post-translational sumoylation.3 KLF8 can function as either a transcriptional repressor3,6,10–12 or activator,1,3,4 by binding to GT-box (CACCC) promoter sequence via its C-terminal three C2H2 zinc fingers that are highly conserved among KLFs.13
As a transcriptional repressor similar to the KLF3 and KLF12, KLF8 recruits the C-terminal binding protein (CtBP) co-repressor to its N-terminal PVDLS repression motif10 to repress promoters of target genes such as γ-globin,10–12 KLF4,3 and E-cadherin.6 As a transcriptional activator, KLF8 has been found to directly bind and activate the cyclin D1 gene promoter,1,3 however KLF8 activation domain(s) or motif(s), and associated co-activator(s) remain unidentified.
p300, Creb-binding protein (CBP) and p300/CBP associated factor (PCAF) are transcription co-activators of the histone acetyltransferase (HAT) family.14 These co-activators can be recruited to a target gene promoter by a transcription activator. This process requires an activation domain of the transcription factor, which are usually rich in acidic aspartate and glutamate (D and E), proline (P) and/or glutamine (Q) amino acids (aa).15–17 At the gene promoter, these co-activators catalyze lysine acetylation on either histone tails,14 the transcription activator18 and/or the basal transcription machinery components.19 These reactions help unmask the promoter DNA from the nucleosomes, stabilize the transcription factor, enhance its interaction with DNA or protein partners, and/or promote transcription initiation and elongation. As a result, the transcription of the target gene is activated. Recent studies have suggested that these co-activators can mediate transcriptional activation by a number of KLF proteins including KLF1,20,21 KLF4,22 KLF6,23 and KLF13.24,25
In this study we determined the activation domain of KLF8 and the associated co-activators. We demonstrate that the activation domain is located between aa 101 and 260 where the well-conserved Q118 and Q248 residues make up its core. We show that recruitment of p300 and PCAF by this domain to target gene promoters is critical for KLF8 to regulate transcriptional activation and its associated cellular processes.
Results
Mutation of regions Q114-Q118 and Q245-Q248 causes a significant decrease in KLF8-mediated transcriptional activation
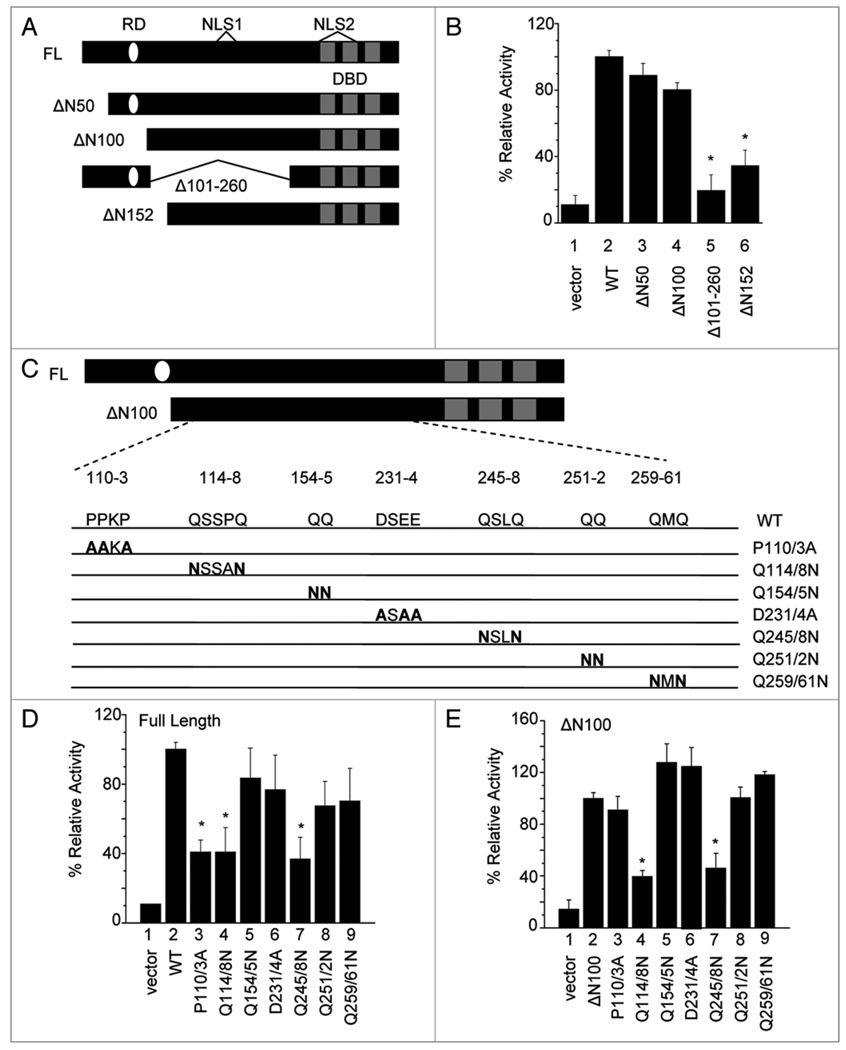
KLF8 was initially reported as a transcription repressor and contains a CtBP-binding repression motif (i.e., 86PVDLS90).10 We have previously shown that deletion of this repression motif (ΔN100) partially decreases the ability of KLF8 to mediate the activation of cyclin D1 gene promoter, suggesting that the repressor function of KLF8 is partially required for the activation of cyclin D1 transcription.1,3 However, we have demonstrated that this deletion mutant still dramatically activates the cyclin D1 promoter and that KLF8 directly binds the promoter in vivo.1,3 These observations suggest that the maximal activation requires both the repressor and activator functions of KLF8,1,3 and that an unknown activation domain or motif is located within the region between aa 101 and the C-terminal zinc finger domains of KLF8. To identify the precise activation domain, we first deleted this 101–260 aa region (Fig. 1A, Δ101–260) and tested the requirement of this region for the activation of the cyclin D1 gene promoter using promoter luciferase reporter assays. We found that this deletion did not disrupt the ability of KLF8 to bind to the cyclin D1 promoter (Suppl. Fig. 3) but resulted in a dramatic decrease in cyclin D1 promoter activity (Fig. 1B and compare column 5 with 2). This result suggests that the KLF8 activation domain or motif is located within the aa 101–260 region. To narrow down the activation domain region we performed stepwise N-terminal truncations to remove an additional 50 aa from this region. We found that the ΔN152 mutation caused a further decrease in promoter activity compared to the ΔN100 mutant (Fig. 1B and compare column 6 with 4). This result suggests that the region between aa 100–152 may contain critical amino acids for KLF8 mediated transcriptional activation. However, further truncation mutants failed to localize to the nucleus4 and therefore could not be used for these experiments.
Figure 1.
Mutation of regions Q114-Q118 and Q245–248 causes a significant decrease in cyclin D1 promoter activity. (A) Schematic diagram of KLF8 deletion mutants. The three zinc finger DNA binding domain (DBD), two nuclear localization signals (NLS) and the repression domain (RD) are depicted. (B) Luciferase activity was measured in NIH3T3 cells co-transfected with the cyclin D1 promoter reporter and the KLF8 deletion mutants. The empty vector and wild type KLF8 (WT) or a KLF8 construct lacking its repression domain (ΔN100) were used as negative and positive controls, respectively. After 16 h, cells were harvested for luciferase assays as described in Materials and Methods. Results were normalized to WT. (C) Schematic diagram of point mutations generated in the context of either full length KLF8 (FL) or ΔN100. Prolines (P) or aspartates (D) were mutated to alanines (A) and glutamines (Q) were mutated to asparagines (N). (D and E) NIH3T3 cells were co-transfected with the cyclin D1 promoter reporter and the point mutants in the context of either full length (D) or ΔN100 (E). Luciferase assays were performed similarly as described in (B). Results were normalized to either WT (D) or ΔN100 (E). Equal expression of the KLF8 constructs is shown in Supplemental Figure 1. All experiments were done in triplicate and the results represent mean ± S.E. of at least three experiments. *p < 0.05 compared to WT (B and D) or ΔN100 (E).
As a result, we decided to generate multiple point mutations within the aa 101–260 region using PCR generated site-directed mutagenesis to disrupt prolines, glutamines and/or acidic amino acids that are potential key components15–17 of the activation domain (Fig. 1C). We found that the Q114/8N and Q245/8N mutations, in both full-length and ΔN100 contexts caused a significant decrease in cyclin D1 promoter activity (Fig. 1D and E, compare columns 4 or 7 with 2). Interestingly, the P110/3A mutation caused a partial inhibition of the promoter activity only in the full length, but not ΔN100 context (Fig. 1D and E, compare column 3 with 2). These results suggest that amino acids within the regions Q114-Q118 and Q245-Q248 are essential for KLF8 mediated promoter activation.
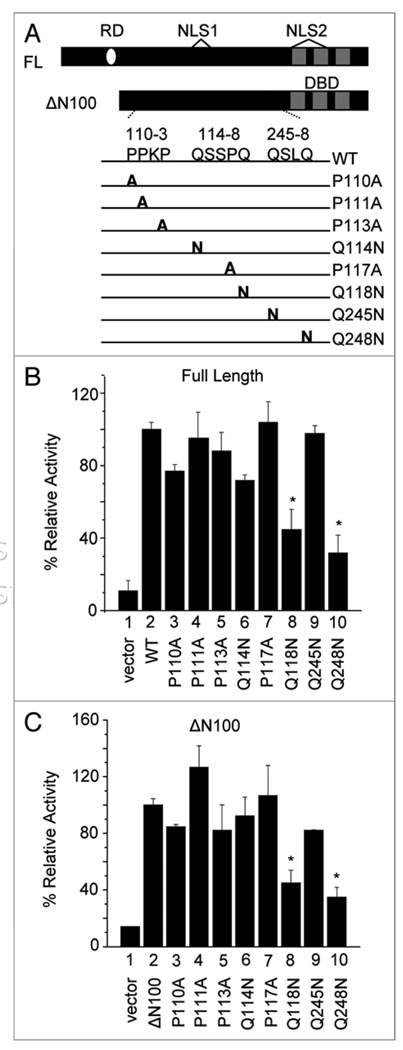
Amino acids Q118 and Q248 are core residues of the KLF8 activation domain
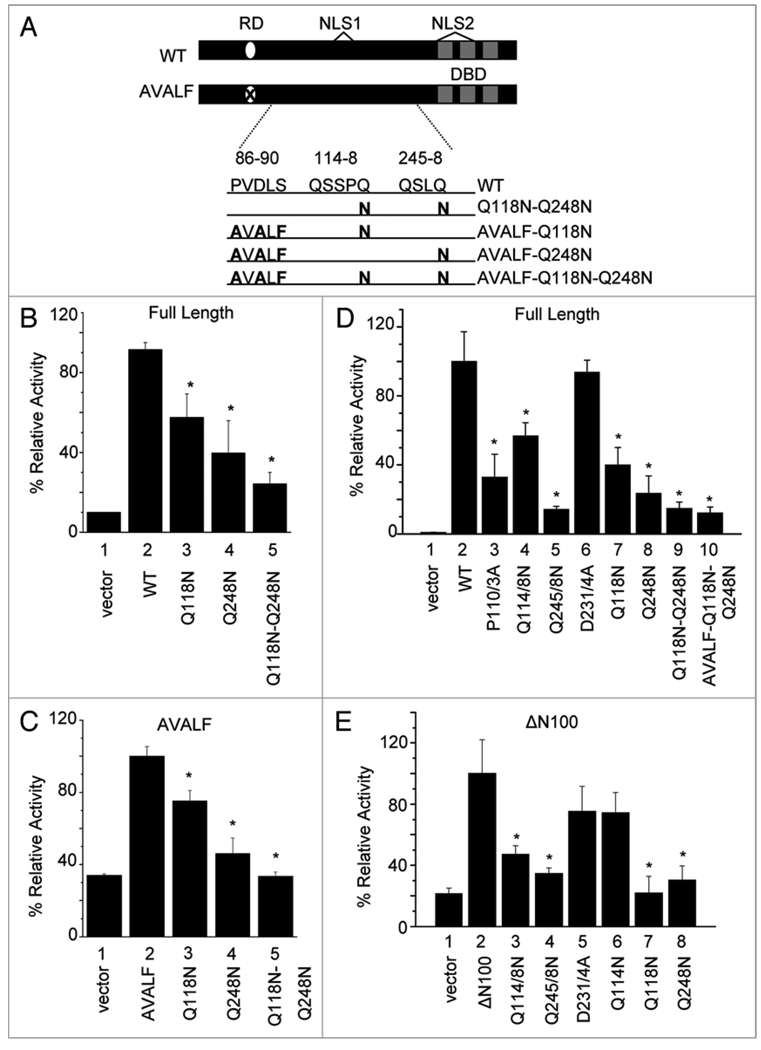
To further probe for the activation domain residues, we mutated each glutamine or proline individually within the 110–113, 114–118 and 245–248 aa regions. We found that the Q118N and Q248N mutations caused significant decreases in cyclin D1 promoter activity in both the full length and ΔN100 context, while other mutations did not (Fig. 2B and C, compare columns 8 and 10 with 2). Moreover, the Q118N–Q248N double mutation caused an even greater decrease in activity compared to Q118N or Q248N alone (Fig. 3B and compare column 5 with 3 and 4). Finally, a complete decrease in cyclin D1 promoter activity was seen when the Q118N–Q248N mutation was combined with the disruption of the repression motif (PVDLS→AVALF) (Fig. 3C and compare column 5 with 1). Similar results were obtained using the T80 human ovarian surface epithelial cells suggesting that this result is not cell type specific (Fig. 3D and E). Notably, these two residues are well conserved among KLF8 orthologs but not paralogs (Suppl. Fig. 4). These results suggest that the critical Q118 and Q248 make up the core of the KLF8 activation domain.
Figure 2.
Amino acids Q118 and Q248 are essential for KLF8 activation of cyclin D1 gene promoter. (A) Schematic diagram of single point mutations generated in either KLF8 full length or ΔN100 context. (B and C) the cyclin D1 gene promoter reporter luciferase assays were performed similarly as described in Figure 1. Equal expression of the KLF8 constructs is shown in Supplemental Figure 1. *p < 0.05 compared to WT (B) or ΔN100 (C).
Figure 3.
Q118 and Q248 are two critical residues required for KLF8 activation of cyclin D1 gene promoter. (A) Schematic diagram of Q118N, Q248N and/or Q118N-Q248N mutants in the context of full length KLF8 (WT) or with disruption of the repression motif (PVDLS to AVALF). (B–E) NIH3T3 (B and C) or T80 (D and E) cells were co-transfected with the cyclin D1 gene promoter reporter and the indicated constructs. Luciferase assays were performed similarly as described in Figure 1. Equal expression of the KLF8 constructs is shown in Supplemental Figure 1. *p < 0.05 compared to WT (B and D), AVALF (C), or ΔN100 (E).
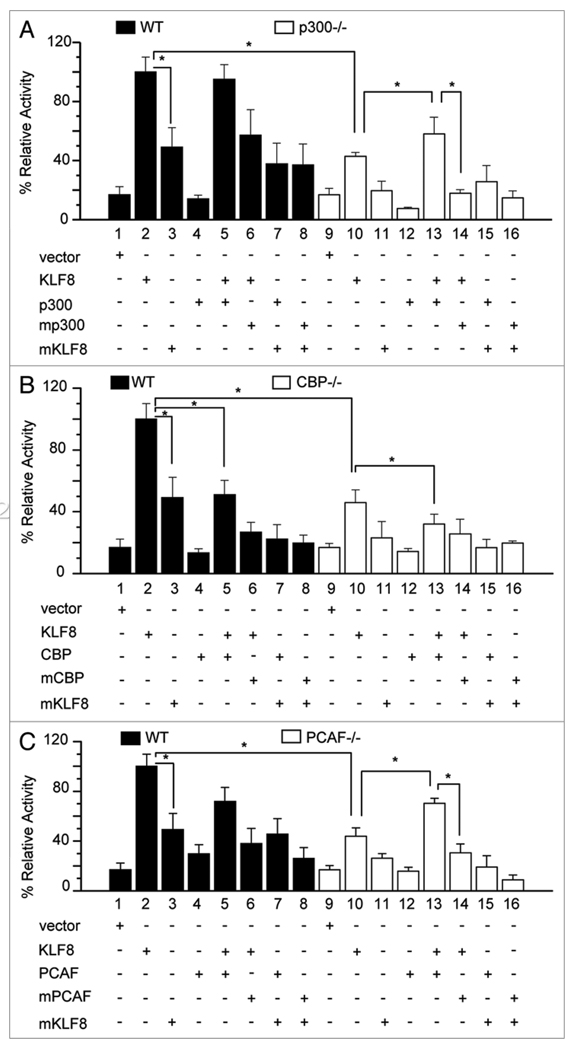
KLF8 requires the HAT domains of p300 and PCAF but not CBP to activate the cyclin D1 gene promoter
Several other KLF family members have been shown to require these co-activators to activate target gene promoters.20–25 To test if KLF8 also requires any of these co-activators to activate the cyclin D1 gene promoter, mouse embryonic fibroblast (MEF) cells either wild type (WT) or null for p300, PCAF or CBP, were used for luciferase assays. We found that KLF8 alone was not able to activate the cyclin D1 gene promoter in the null cells to equivalent levels as in the WT cells (Fig. 4A – C and compare column 10 with 2). In contrast, when p300 or PCAF was added back into the null cells, KLF8 promoted a significant increase in the promoter activity (Fig. 4A and C, compare column 13 with 10). However, their HAT defective mutants (ΔHAT) failed to do so (Fig. 4A and C, compare column 14 with 13). Surprisingly, the addition of CBP reduced the KLF8-mediated promoter activation both in the WT and CBP−/− backgrounds (Fig. 4B, compare column 5 with 2, or 13 with 10) independent of the HAT domain (Fig. 4B and compare columns 6 with 5 and 14 with 13). As expected, the Q118N–Q248N KLF8 mutant (mKLF8) lost the ability to fully activate the promoter in both WT and null cells (Fig. 4A – C and compare column 3 with 2, and 11 with 10). This loss of function could not be rescued by over- or re-expression of p300, CBP or PCAF (Fig. 4A – C, compare column 7 with 3, and 15 with 11). These results suggest that p300 and PCAF play a primary role in transcriptional activation by KLF8, presumably by catalyzing protein acetylation, and that KLF8 may depend on the Q118 and Q248 residues for recruiting the co-activators to the promoter.
Figure 4.
KLF8 requires p300 and PCAF to activate the cyclin D1 promoter in a HAT dependent manner. (A) Mouse embryonic fibroblasts (MEF) null for p300 (p300−/−) were co-transfected with the cyclin D1 promoter reporter plus the empty vector, KLF8, Q118N-Q248N-KLF8 (mKLF8), p300, or p300ΔHAT (mp300) in the indicated combinations. Luciferase assays were performed and cyclin D1 gene promoter activity levels were normalized to levels in KLF8-transfected wild type MEF (WT) cells. (B and C) Similar experiments were performed using CBP (B) or PCAF (C) WT and mutant constructs and the corresponding null cells. *p < 0.05. Expression of the transfected co-activator constructs is shown in Supplemental Figure 2.
KLF8 physically interacts with p300, PCAF and CBP in a Q118 and Q248 dependent manner
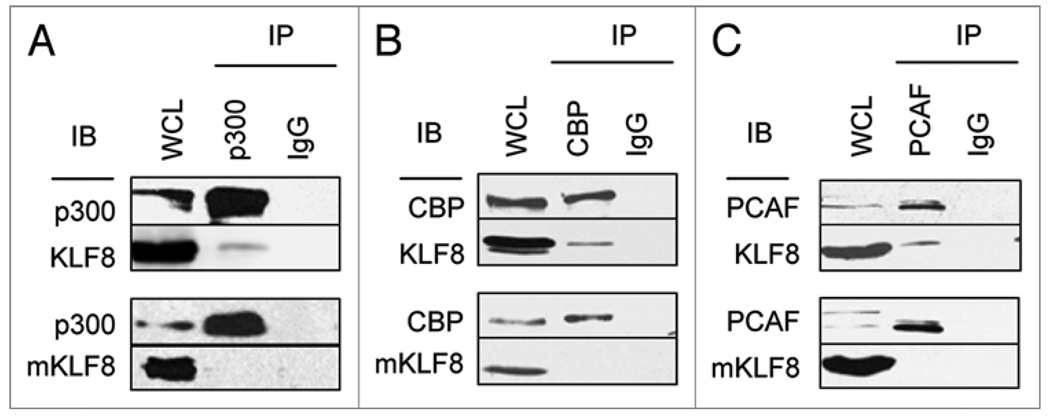
Several other KLF family members have been shown to interact with the p300, CBP or PCAF co-activators to mediate activation of target gene promoters.20–25 To test whether KLF8 physically interacts with these co-activators in vivo, HA-KLF8 and p300, CBP or PCAF were co-transfected into HEK293 cells for co-immunoprecipitation analyses. We found that KLF8 interacted with all three proteins but mKLF8 did not (Fig. 5). These results suggest that the Q118 and Q248 residues of KLF8 are critical for recruiting these co-activators to target gene promoters.
Figure 5.
KLF8 physically interacts with p300, CBP and PCAF in a Q118 and Q248 dependent manner. (A) p300 was co-transfected into HEK293 cells with either HA-tagged KLF8 (KLF8) or its Q118N-Q248N mutant (mKLF8). Whole cell extracts were prepared and subjected to immunoprecipitation with a p300 antibody or control IgG followed by anti-p300 or anti-HA blotting. (B and C) Similar experiments were performed using CBP (B) or FLAG-PCAF (C) and corresponding antibodies.
KLF8 promotes histone acetylation at the cyclin D1 gene promoter in a Q118 and Q248 and co-activator dependent manner
When recruited to gene promoters p300, CBP and/or PCAF can acetylate histone tails, allowing for the unraveling of DNA and the activation of transcription. To test whether KLF8 promotes histone acetylation at the cyclin D1 gene promoter and whether it requires the KLF8 activation domain, we transfected KLF8 or mKLF8 into NIH3T3 cells for ChIP experiments. We found that when KLF8 bound to the promoter, histone acetylation was greatly increased (Fig. 6A and compare the middle with top panel for AcH3/4). In contrast, when the mutant KLF8 was present at the promoter, histone acetylation is significantly decreased (Fig. 6A and compare bottom with middle panel for AcH3/4, and Fig. 6B). To test whether p300, CBP and/or PCAF are required for the KLF8-mediated histone acetylation, we transfected KLF8 into the p300−/−, CBP−/− or PCAF−/− cells and performed similar histone ChIP experiments. We found that histone acetylation at the promoter was significantly decreased in all the null cells compared to WT cells (Fig. 6C and compare columns 3, 5 or 7 with 1). By contrast, the mKLF8 was unable to promote histone acetylation to equivalent levels in either the WT or null cells (Fig. 6C and compare column 2 with 1, 4 with 3, 6 with 5 and 8 with 7). These results suggest that a functional activation domain of KLF8 is required to recruit the co-activators to the cyclin D1 promoter to acetylate histones.
Figure 6.
KLF8 requires interaction with p300, CBP and PCAF for histone acetylation at the KLF8 binding region of cyclin D1 gene promoter. (A) NIH3T3 cells were transfected with either HA-KLF8 (KLF8) or its Q118N-Q248N mutant (mKLF8) for ChIP experiments. The KLF8 binding promoter region was probed for associated KLF8 (HA) or acetylated histones 3 and 4 (AcH3/4) using anti-HA or anti-AcH3/4 IP followed by PCR. Empty vector and control IgG IP were included as negative controls. (B) AcH3/4 and KLF8 levels were normalized to Pre-IP. Plotted is the ratio of AcH3/4 to KLF8 relative to that in the KLF8-transfected cells *p < 0.05. (C) The WT, p300−/−, CBP−/− or PCAF−/− MEF cells were transfected with HA-KLF8 (lanes 1, 3, 5 and 7) or HA-mKLF8 (lanes 2, 4, 6 and 8) and ChIP experiments were performed as in (A). (D) AcH3/4 and KLF8 levels were normalized to Pre-IP. Plotted is the ratio of AcH3/4 to KLF8 relative to WT KLF8 in WT MEF cells. *p < 0.05 comparing columns 3, 5 or 7 to 1; **p < 0.05 comparing columns 2 with 1, 4 with 3, 6 with 5, or 8 with 7.
The Q118 and Q248 residues are required for KLF8 to promote cyclin D1 protein expression and cell cycle progression
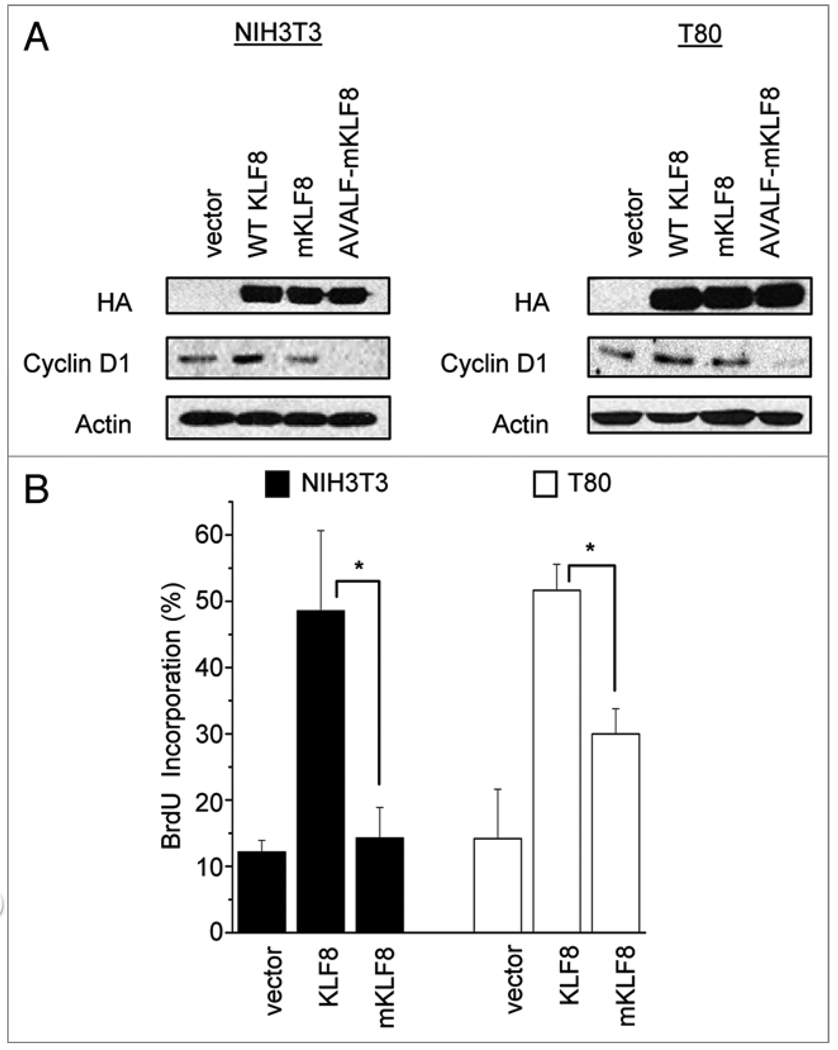
We have previously shown that KLF8 overexpression can promote cyclin D1 protein expression and cell cycle progression.1,3 Here we show that overexpression of KLF8 induced endogenous cyclin D1 protein expression in both NIH3T3 and T80 cells, whereas mKLF8 did not (Fig. 7A). Additionally, we show that the AVALF-Q118N-Q248N mutant caused a decrease in endogenous cyclin D1 expression compared to vector control, again confirming our notion that the repression motif of KLF8 is also required to regulate the cyclin D1 expression. To test whether the KLF8 activation domain is essential for promoting cell cycle progression we transfected KLF8, or mKLF8 into NIH3T3 or T80 cells for BrdU incorporation assays. We found that in both cell types, the KLF8 mutant was unable to promote DNA synthesis at levels comparable to wild type KLF8 (Fig. 7B), suggesting that the activation domain is required for KLF8 mediated cell cycle progression from G1 to S phase.
Figure 7.
The Q118 and Q248 residues are required for KLF8 to promote cyclin D1 protein expression and cell cycle progression. (A) NIH3T3 cells or T80 cells were transfected with empty vector, KLF8, the Q118N-Q248N mutant (mKLF8), or the AVALF-Q118N-Q248N mutant (AVALF-mKLF8). After 16 h lysates were collected and subjected to western blot analysis using anti-HA, anti-cyclin D1 and anti-actin antibodies. (B) NIH3T3 or T80 cells were transfected with empty vector, KLF8, or mKLF8. The G1 cell cycle progression of the cells was determined by measuring BrdU incorporation as described in Matherials and Methods. *p < 0.05.
Discussion
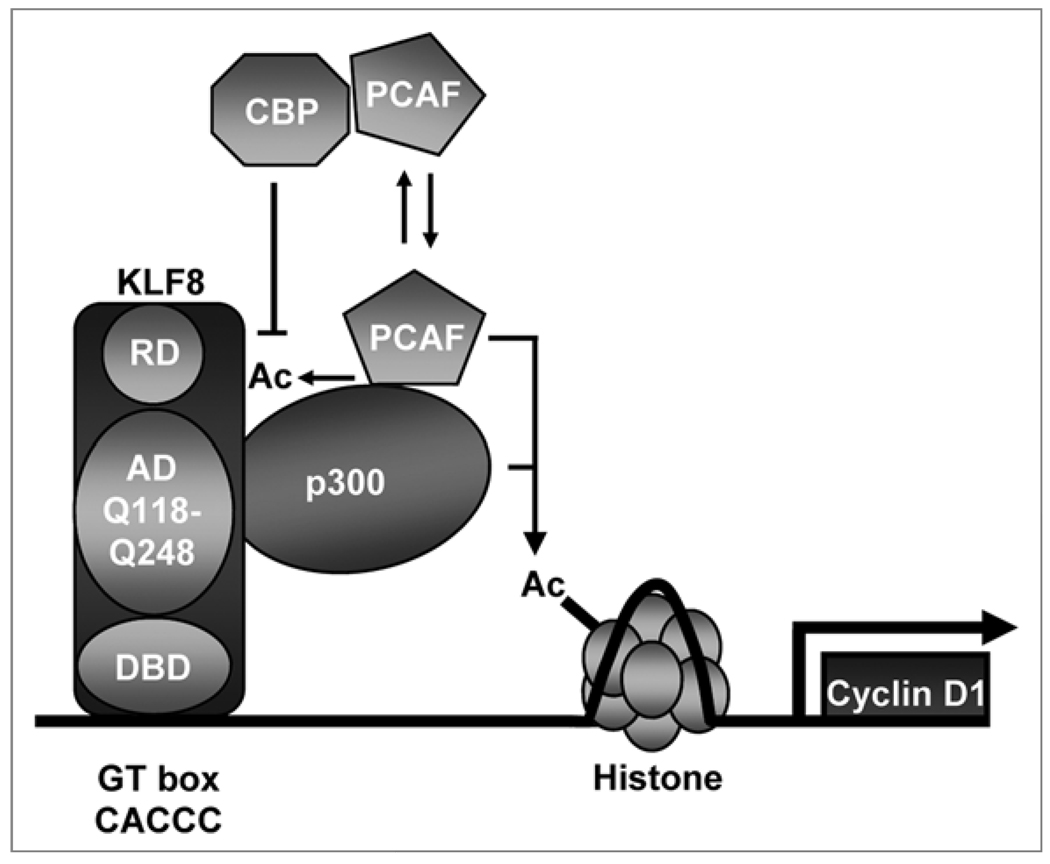
Like several other KLFs,26 KLF8 can both repress3,6,10–12 and activate1,3,4 the transcription of target genes to regulate important cellular processes. While the KLF8 repression motif and an associated co-repressor have been previously identified,10 the molecular mechanisms by which KLF8 activates the transcription of target genes has not been investigated. In this study, we mapped the KLF8 activation domain within the 101–260 aa region and identified Q118 and Q248 as essential residues for recruiting the p300 and PCAF co-activators to acetylate histones and subsequently activate the cyclin D1 gene promoter. We also demonstrated that this KLF8 activation domain is required for its promotion of cyclin D1 protein expression and cell cycle progression through the G1 phase. The similar findings in both mouse fibroblasts and human epithelial cells highlight a potentially high conservation of the KLF8-mediated transactivation mechanisms across species and tissue types. Based on these novel observations, we propose a model for KLF8 transactivation of cyclin D1 gene promoter (Fig. 8) which could generally apply to KLF8 transactivation of targets other than cyclin D1.
Figure 8.
A hypothesized model for KLF8 mediated transcription. KLF8 binds to a GT box within the promoter of its cyclin D1 target gene through its DNA binding domain (DBD). Its activation domain (AD) is located between the DBD and its repression domain (RD). The AD possesses two glutamine residues (Q118 and Q248) that are essential for recruitment of the p300, CBP and PCAF co-activators to acetylate the histones for promoter activation. We hypothesize that p300 and/or PCAF, but not CBP, may acetylate KLF8. We propose that when CBP binds to KLF8, the p300/PCAF interaction with KLF8 and subsequent acetylation on KLF8 may be blocked.
As illustrated in the model, KLF8 binds target promoters via its C-terminal zinc finger domain at the GT box in the promoter. The activation domain of KLF8 then primarily recruits the p300 and PCAF co-activators to the promoter. The KLF8-associated co-activators then acetylate the histones, and perhaps other proteins at the promoter including the basic transcription machinery proteins and KLF8 itself, to activate the promoter. Although CBP interaction with KLF8 appears to play a similar role for histone acetylation, this interaction could interfere with p300/PCAF mediated acetylation of the other proteins due to the competition for KLF8 binding between p300 and CBP, resulting in a reduced activation of the promoter.
Indeed, recent studies using a variety of experimental systems have suggested that, despite the high homology between p300 and CBP, these two co-activators are not completely redundant in function as previous studies have suggested. In fact, they play differential or even opposing roles depending upon the contexts of cellular processes.27,28 For example, Kawasaki et al. showed that p300 was required for p21cip1, whereas CBP was required for p27kip1 upregulation to mediate retinoic-induced differentiation of F9 embryonal carcinoma cells.29 Another study demonstrated that CBP, but not p300, is required for TCF/β-catenin mediated aberrant transcription of target genes including cyclin D1.30,31
Differential function of p300 versus CBP has not been shown in the context of other KLFs. However, while studies have suggested that PCAF usually interacts and cooperates with p300 or CBP, the studies of other KLF family members have shown that this is not always the case. For instance, a recent study by Song et al.24 showed that CBP-mediated acetylation of KLF13 resulted in disruption of KLF13 binding to DNA, which can be blocked by PCAF in a HAT-independent manner. Intriguingly, the HAT activity of PCAF but not p300 or CBP was demonstrated by the same group to be required for KLF13’s ability to bind DNA and activate transcription.25 Although not discussed by the authors, it is possible that competition between p300 and CBP for interacting with KLF13 and/or PCAF may play a dynamic role similarly as in the case of KLF8. In contrast, p300 and CBP can acetylate and activate KLF1 to increase its activity, but PCAF can not acetylate KLF1 but actually inhibits KLF1 activity.20 These results appear to further complicate the inter-relationship between p300, CBP and PCAF. Nevertheless, it will be interesting to test whether KLF8 requires p300, CBP and PCAF differentially to regulate its own post-translational modification, its interactions with other proteins at the target gene promoter, and subsequent activation of the target transcription.
KLF family proteins share a highly conserved C-terminal DNA binding zinc finger domain, and therefore they use their N-terminal difference as one of the mechanisms to distinguish their specificities in the regulation of target gene transcription.32 We have found that glutamines 118 and 248 are essential for KLF8 mediated transcriptional activation. The glutamine-rich motifs are believed to play a crucial role in mediating transcriptional activation for many transcription activators although the underlying mechanisms remain largely unknown. For example, mutation of the glutamine rich motif in Sp1 transcription factor only slightly alters its activation domain sequence but greatly reduces its transactivity,33 suggesting that the glutamines may be critical in conferring secondary or tertiary active conformation of transcription activators.17 It will be interesting to determine, using X-ray crystallographic studies for example, whether the Q118 and Q248 of KLF8 play a major role in maintaining the active protein conformation and whether these glutamines are precise contact sites for the p300 and PCAF co-activators.
The majority of the KLF family proteins have a transcriptional activating function and more than half of them function as both an activator and repressor depending upon target promoter or cell contexts.26 Several KLFs, including KLF1, KLF4, KLF5, KLF6 and KLF13, have been demonstrated to interact with p300, CBP and/or PCAF.20–25,34 The activation domains have been mapped for KLF1 and KLF13 also within the N-termini35,36 although core residues within these domains remain unclear. Interestingly, these activation domains are not necessarily required for interaction with p300 and/or CBP.25,34 Our identification of the KLF8 activation domain and the unique conservation of the Q118 and Q248 core residues among species but not across the KLF family proteins (Suppl. Fig. 4A and B) highlight an important role of these residues for transcriptional activation of KLF8-specific target genes.
It is not clear at present why the P110/3A mutation causes a significant decrease in cyclin D1 gene promoter activity in the full length but not ΔN100 context (see Fig. 1D and E). Our previous studies1,3 suggest that the KLF8 repression domain-mediated inhibition of KLF4 promoter and subsequent de-repression of the cyclin D1 promoter contribute partially to the activation of cyclin D1 transcription by KLF8. We have also demonstrated that sumoylation of the Lys67 inhibits KLF8 transcriptional activity.3 Therefore it appears that the P110-P113 residues play some role to influence the aa 1–100 region associated KLF8 functions such as CtBP interaction and sumoylation. This will be another interesting area for future studies.
In addition to targeting the histones, the p300, CBP and/or PCAF co-activators also acetylate transcription factors to regulate transcriptional activation. Recent studies have demonstrated that several KLFs, including KLF1,20,21 KLF4,22 KLF6,23 and KLF13,24 can be acetylated by these co-activators. Interestingly, the acetylation sites on the KLFs are either located within their zinc finger domains (KLF1, KLF6 and KLF13)21,23,24,35,36 or repression domains (KLF4),22 but not activation domains. Experiments are in progress to determine the potential KLF8 acetylation by the co-activator proteins and the molecular mechanisms by which this modification modulates KLF8 transcriptional activity and cellular functions.
In summary, we have identified the KLF8 activation domain, its essential core residues and associated co-activators. This work significantly advances our understanding of the mechanisms by which KLF8 activates its target genes. This study also provides information that will be useful in the future for designing strategies to study or target KLF8-activated genes-specific cellular functions in both physiological and pathological conditions.
Materials and Methods
Plasmid construction
The mammalian expression plasmids pKH3, pKH3-KLF8, pHAN and pHAN-KLF8 were previously described.1,3 All the KLF8 mutants were generated by site-directed mutagenesis PCR or overlapping PCR using pKH3-KLF8 or pHAN-KLF8 as the template and mutation-specific primers (Suppl. Table 1) paired with one of the master primers (forward: 5'-CCC AAG CTT CTG CAG GTC G-3' and reverse: 5'-GGA CAA ACC ACA ACT AGA ATG CAG-3'). Amplified DNA fragments were gel-purified using the Qiaquick Gel Extraction Kit (QIAGEN), digested with SmaI and EcoRI and inserted into the pKH3 or pHAN vector at the same sites. Correct mutations were confirmed by DNA sequencing. Protein expression was verified by western blotting (Suppl. Fig. 1), and nuclear localization was verified by immunofluorescent staining (Data not shown and our previously published studies).4 The pCX/Flag-PCAF, pCAFΔHATA (Δ579–608), pCAFΔHATB (Δ609–624), pCMVβ-HA-p300, pCL/p300 (ΔH), pRc/RSV/CBP and pRc/RSV/CBP (HAT-) (F1541A mutation) were previously described.20,21,37–41
Cell culture and transfection
NIH3T3 mouse fibroblasts, T80 human ovarian epithelial,2,42 HEK293, or mouse embryonic fibroblast (MEF) cells were maintained in DMEM supplemented with either 10% calf serum (NIH3T3), or 10% fetal bovine serum (T80, HEK293 and MEF). The wild type, p300−/−, CBP−/− or PCAF−/− MEF cells were previously described.43 Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Promoter luciferase reporter assays
Experiments were performed similarly as previously described.1,3 Briefly, cells were grown to ~75% confluency in a 12-well dish. The cyclin D1 promoter luciferase reporter vector1,3 (0.2 µg) was co-transfected into NIH3T3 or T80 cells with 0.2 µg of the expression vectors encoding KLF8, KLF8 mutants, or empty vector alone along with the pRlSV40 Renilla luciferase reporter internal control plasmid (4 ng). After 16 hours, cell lysates were prepared and luciferase activity was analyzed using the Dual Luciferase Reporter Assay System (Promega) and the 20/20n Luminometer (Turner Biosystems). For experiments using the MEF cells, the weight ratio of the cyclin D1 promoter:KLF8:co-activator:the internal control vector was 1:1:5:0.02 and appropriate amounts of the empty vector were included in each transfection to ensure an equal DNA content in total (1.404 µg). Re-expression of p300, CBP or PCAF was verified in null cells by western blotting (Suppl. Fig. 2).
Co-immunoprecipitation and western blotting
HEK293 cells were co-transfected with HA-KLF8 or its Q118N–Q248N mutant plus p300, CBP or FLAG-PCAF. Cells were lysed in NP40 lysis buffer with 1 mM Na3VO4, 1 mM PMSF, 20 µg/ml leupeptin and 0.06 TIU/ml aprotinin. The lysates (500 µg) were pre-cleared with 5 µl of Protein A/G agarose beads (Santa Cruz Biotechnology) for 30 min at 4°C with rotation. The supernatants were incubated at 4°C for 2 h with 1.6 µg of p300, CBP, FLAG antibody (p300 (N-15), CBP (A-22) and FLAG (OctA-Probe D-8), Santa Cruz Biotechnology) or control rabbit IgG, and followed by an overnight rotation at 4°C with 25 µl of Protein A/G beads. After washing, the precipitates were boiled in 1x SDS-sample buffer and resolved on a SDS-PAGE gel followed by western blotting using an antibody specific for HA (1:2,000 dilution, HA-F-7, Santa Cruz Biotechnology), p300, CBP or FLAG (1:500 dilution). Detection of endogenous cyclin D1 or actin was determined using an antibody specific for cyclin D1 (M-20, Santa Cruz Biotechnology) or actin (C4, Santa Cruz Biotechnology).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed essentially as we previously described2,3,6 using the Upstate Acetyl-Histone H3 and H4 Immunoprecipitation (ChIP) Assay Kit according to their protocol with minor modifications. pKH3, pKH3-KLF8, or its Q118N–Q248N mutant was transiently transfected into NIH3T3 or the MEF cells. After 16 hours, the cells were treated with 5 µM TSA and 5 mM sodium butyrate for 8 hours before being lysed. Sonicated lysates prepared from 2 × 106 cells were subjected to immunoprecipitation overnight at 4°C the anti-HA antibody, a mixture of Anti-acetyl-Histone 4 and Anti-acetyl-Histone 3 antibodies (Upstate cat # 06–866 and 06–599 respectively) or control mouse IgG, followed by a rotation for 1 hour at 4°C with 60 µl of Protein A/G agarose beads. The precipitated DNA fragments were then purified using Mini Elute PCR Purification Kit (QIAGEN) and eluted into a final volume of 50 µl for each sample. Three µl of the DNA was used as template for PCR using primers that are specific for the mouse cyclin D1 promoter (forward: 5'-ATT GAA AGA CAG GGA CGC TGG-3' and reverse: 5'-AAG CCA AGG AAG AAT GTA TGG AAG G-3') to amplify the KLF8 binding region.
BrdU incorporation assays
NIH3T3 or T80 cells were transiently transfected with either pKH3, pKH3-KLF8, or its Q118N–Q248N mutant and BrdU incorporation rates were analyzed as previously described.2,3,44
Supplementary Material
Acknowledgements
We thank Drs. James Bieker of Mount Sinai School of Medicine, Jonathan Harton of Albany Medical College and James DiCaprio of Dana-Farber Cancer Institute for kindly providing plasmid constructs or cell lines. We also thank our colleagues Drs. Jonathan Harton and Chunhong Yan, and Ms. Diane Colello for critical reading of and helpful comments on this manuscript. This research was supported by grants from the American Cancer Society (#RSG CCG-111381), NIH-NCI (CA132977) and Susan G. Koman for the Cure (KG090444) to J.Z.
Abbreviations
- KLF8
krüppel-like factor 8
- CtBP
C-terminal binding protein
- CBP
creb-binding protein
- PCAF
p300/CBP associated factor
- HAT
histone acetyltransferase
- MEF
mouse embryonic fibroblast
- NLS
nuclear localization signal
- RD
repression domain
- AD
activation domain
- mKLF8
the Q118N–Q248N KLF8 mutant
- ChIP
chromatin immunoprecipitation
- H3/4
histone 3 and histone 4
- aa
amino acid
Footnotes
Supplementary materials can be found at: www.landesbioscience.com/supplement/UrvalekCC9-3-Sup.pdf
References
- 1.Zhao J, Bian ZC, Yee K, Chen BP, Chien S, Guan JL. Identification of transcription factor KLF8 as a downstream target of focal adhesion kinase in its regulation of cyclin D1 and cell cycle progression. Mol Cell. 2003;11:1503–1515. doi: 10.1016/s1097-2765(03)00179-5. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Urvalek AM, Liu J, Zhao J. Activation of KLF8 transcription by focal adhesion kinase in human ovarian epithelial and cancer cells. J Biol Chem. 2008;283:13934–13942. doi: 10.1074/jbc.M709300200. [DOI] [PubMed] [Google Scholar]
- 3.Wei H, Wang X, Gan B, Urvalek AM, Melkoumian ZK, Guan JL, Zhao J. Sumoylation delimits KLF8 transcriptional activity associated with the cell cycle regulation. J Biol Chem. 2006;281:16664–16671. doi: 10.1074/jbc.M513135200. [DOI] [PubMed] [Google Scholar]
- 4.Mehta TS, Lu H, Wang X, Urvalek AM, Nguyen KH, Monzur F, et al. A unique sequence in the N-terminal regulatory region controls the nuclear localization of KLF8 by cooperating with the C-terminal zinc-fingers. Cell Res. 2009 doi: 10.1038/cr.2009.64. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Zhao J. KLF8 transcription factor participates in oncogenic transformation. Oncogene. 2007;26:456–461. doi: 10.1038/sj.onc.1209796. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Zheng M, Liu G, Xia W, McKeown-Longo PJ, Hung MC, et al. Kruppel-like factor 8 induces epithelial to mesenchymal transition and epithelial cell invasion. Cancer Res. 2007;67:7184–7193. doi: 10.1158/0008-5472.CAN-06-4729. [DOI] [PubMed] [Google Scholar]
- 7.Cox BD, Natarajan M, Stettner MR, Gladson CL. New concepts regarding focal adhesion kinase promotion of cell migration and proliferation. J Cell Biochem. 2006;99:35–52. doi: 10.1002/jcb.20956. [DOI] [PubMed] [Google Scholar]
- 8.Ding Q, Grammer JR, Nelson MA, Guan JL, Stewart JEJr, Gladson CL. p27Kip1 and cyclin D1 are necessary for focal adhesion kinase regulation of cell cycle progression in glioblastoma cells propagated in vitro and in vivo in the scid mouse brain. J Biol Chem. 2005;280:6802–6815. doi: 10.1074/jbc.M409180200. [DOI] [PubMed] [Google Scholar]
- 9.Eaton SA, Funnell AP, Sue N, Nicholas H, Pearson RC, Crossley M. A network of Kruppel-like Factors (Klfs). Klf8 is repressed by Klf3 and activated by Klf1 in vivo. J Biol Chem. 2008;283:26937–26947. doi: 10.1074/jbc.M804831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Vliet J, Turner J, Crossley M. Human Kruppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription. Nucleic Acids Res. 2000;28:1955–1962. doi: 10.1093/nar/28.9.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu JH, Navas P, Cao H, Stamatoyannopoulos G, Song CZ. Systematic RNAi studies on the role of Sp/KLF factors in globin gene expression and erythroid differentiation. J Mol Biol. 2007;366:1064–1073. doi: 10.1016/j.jmb.2006.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang P, Basu P, Redmond LC, Morris PE, Rupon JW, Ginder GD, et al. A functional screen for Kruppel-like factors that regulate the human gamma-globin gene through the CACCC promoter element. Blood Cells Mol Dis. 2005;35:227–235. doi: 10.1016/j.bcmd.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 15.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 16.Mermod N, O’Neill EA, Kelly TJ, Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989;58:741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- 17.Courey AJ, Holtzman DA, Jackson SP, Tjian R. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell. 1989;59:827–836. doi: 10.1016/0092-8674(89)90606-5. [DOI] [PubMed] [Google Scholar]
- 18.Sadoul K, Boyault C, Pabion M, Khochbin S. Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie. 2008;90:306–312. doi: 10.1016/j.biochi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Imhof A, Yang XJ, Ogryzko VV, Nakatani Y, Wolffe AP, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Bieker JJ. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, Kadam S, Emerson BM, Bieker JJ. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol Cell Biol. 2001;21:2413–2422. doi: 10.1128/MCB.21.7.2413-2422.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. 2007;282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- 23.Li D, Yea S, Dolios G, Martignetti JA, Narla G, Wang R, et al. Regulation of Kruppel-like factor 6 tumor suppressor activity by acetylation. Cancer Res. 2005;65:9216–9225. doi: 10.1158/0008-5472.CAN-05-1040. [DOI] [PubMed] [Google Scholar]
- 24.Song CZ, Keller K, Chen Y, Stamatoyannopoulos G. Functional interplay between CBP and PCAF in acetylation and regulation of transcription factor KLF13 activity. J Mol Biol. 2003;329:207–215. doi: 10.1016/s0022-2836(03)00429-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song CZ, Keller K, Murata K, Asano H, Stamatoyannopoulos G. Functional interaction between coactivators CBP/p300, PCAF and transcription factor FKLF2. J Biol Chem. 2002;277:7029–7036. doi: 10.1074/jbc.M108826200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 27.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka Y, Naruse I, Hongo T, Xu M, Nakahata T, Maekawa T, Ishii S. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech Dev. 2000;95:133–145. doi: 10.1016/s0925-4773(00)00360-9. [DOI] [PubMed] [Google Scholar]
- 29.Kawasaki H, Eckner R, Yao TP, Taira K, Chiu R, Livingston DM, Yokoyama KK. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature. 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 30.Teo JL, Ma H, Nguyen C, Lam C, Kahn M. Specific inhibition of CBP/beta-catenin interaction rescues defects in neuronal differentiation caused by a presenilin-1 mutation. Proc Natl Acad Sci USA. 2005;102:12171–12176. doi: 10.1073/pnas.0504600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma H, Nguyen C, Lee KS, Kahn M. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene. 2005;24:3619–3631. doi: 10.1038/sj.onc.1208433. [DOI] [PubMed] [Google Scholar]
- 32.Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 33.Courey AJ, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 34.Matsumura T, Suzuki T, Aizawa K, Munemasa Y, Muto S, Horikoshi M, Nagai R. The deacetylase HDAC1 negatively regulates the cardiovascular transcription factor Kruppel-like factor 5 through direct interaction. J Biol Chem. 2005;280:12123–12129. doi: 10.1074/jbc.M410578200. [DOI] [PubMed] [Google Scholar]
- 35.Pandya K, Donze D, Townes TM. Novel transactivation domain in erythroid Kruppel-like factor (EKLF) J Biol Chem. 2001;276:8239–8243. doi: 10.1074/jbc.M008457200. [DOI] [PubMed] [Google Scholar]
- 36.Yet SF, McA’Nulty MM, Folta SC, Yen HW, Yoshizumi M, Hsieh CM, et al. Human EZF, a Kruppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J Biol Chem. 1998;273:1026–1031. doi: 10.1074/jbc.273.2.1026. [DOI] [PubMed] [Google Scholar]
- 37.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani YA. p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 38.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, et al. Molecular cloning and functional analysis of the adenovirus E1A–associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 39.Kwok RP, Laurance ME, Lundblad JR, Goldman PS, Shih H, Connor LM, et al. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 40.Harton JA, Zika E, Ting JP. The histone acetyltransferase domains of CREB-binding protein (CBP) and p300/CBP-associated factor are not necessary for cooperativity with the class II transactivator. J Biol Chem. 2001;276:38715–38720. doi: 10.1074/jbc.M106652200. [DOI] [PubMed] [Google Scholar]
- 41.Jiang H, Lu H, Schiltz RL, Pise-Masison CA, Ogryzko VV, Nakatani Y, et al. PCAF interacts with tax and stimulates tax transactivation in a histone acetyltransferase-independent manner. Mol Cell Biol. 1999;19:8136–8145. doi: 10.1128/mcb.19.12.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Yang G, Thompson-Lanza JA, Glassman A, Hayes K, Patterson A, et al. A genetically defined model for human ovarian cancer. Cancer Res. 2004;64:1655–1663. doi: 10.1158/0008-5472.can-03-3380. [DOI] [PubMed] [Google Scholar]
- 43.Poulin DL, Kung AL, DeCaprio JA. p53 targets simian virus 40 large T antigen for acetylation by CBP. J Virol. 2004;78:8245–8253. doi: 10.1128/JVI.78.15.8245-8253.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao JH, Reiske H, Guan JL. Regulation of the cell cycle by focal adhesion kinase. J Cell Biol. 1998;143:1997–2008. doi: 10.1083/jcb.143.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.