Abstract
We propose a novel Bayesian registration formulation in which image location is represented as a latent random variable. Location is marginalized to determine the maximum a priori (MAP) transform between images, which results in registration that is more robust than the alternatives of omitting locality (i.e. global registration) or jointly maximizing locality and transform (i.e. iconic registration). A mathematical link is established between the Bayesian registration formulation and the mutual information (MI) similarity measure. This leads to a novel technique for selecting informative image regions for registration, based on the MI of image intensity and spatial location. Experimental results demonstrate the effectiveness of the marginalization formulation and the MI-based region selection technique for ultrasound (US) to magnetic resonance (MR) registration in an image-guided neurosurgical application.
1 Introduction
The task of image registration is central to many medical image analysis applications, and involves determining a geometrical mapping or transform relating different images. Registration is often formulated as determining a transform in the form of a smooth one-to-one mapping between images [1]. However, in a number of medical imaging contexts, a smooth one-to-one mapping may not exist or may be difficult to identify. Consider the context of image-guided surgery, for instance, in which intra-operative ultrasound (US) imagery is to be registered to pre-operative magnetic resonance (MR) imagery for the purpose of surgical guidance. In the case of resection, anatomical tissues present prior to surgery may be severely deformed or removed over the course of the procedure, and a valid one-to-one mapping between pre- and intra-operative images will not exist [2]. Furthermore, the intensity characteristic of US is spatially-varying [3], and certain tissues may be absent or incorrectly represented in the image due to tissue echogenicity or to artifacts relating to sensor gain, acoustic shadowing, reverberation, etc., further compounding the difficulty in estimating a transform.
A body of research focuses on registration based on local image regions [4, 2, 5, 7–9] in order to address the situation where a smooth one-to-one mapping is difficult to estimate. Working with local regions improves the ability to identify and cope with missing or altered tissue in images, and simplifies the modeling of geometrical and intensity relationships between images, which can be approximated as locally smooth and stationary. This paper makes three contributions to this line of research. First, a Bayesian registration formulation is proposed, in which image location is incorporated as a latent random variable. The key insight is that, while location is important in describing image content and the registration process, it is ultimately incidental, and should thus be marginalized in order to obtain the maximum a posteriori (MAP) transform relating images. Marginalization provides a principled means of leveraging local regional information for registration, in contrast to omitting location information or maximizing a joint distribution over locations and the transform. Second, a novel mathematical link is established between Bayesian registration and the mutual information (MI) similarity measure, motivating the use of MI in a Bayesian setting. Third, a new strategy is proposed for selecting informative local image regions for registration, based on the MI of image intensity and location within image regions. This strategy generalizes previous work advocating the selection of high-entropy image regions [10], by imposing the additional constraint that the intensity be informative with respect to image location.
The remainder of this paper is organized as follows. Section 2 outlines related work in the literature. Section 3 presents our Bayesian framework for location marginalization and informative region selection. Section 4 presents experiments involving registration of intra-operative US slices to pre-operative MR imagery in an image-guided neurosurgical (IGNS) application. In this challenging registration scenario, location marginalization is shown to outperform standard registration. Furthermore, mutual information-based selection of image regions is shown to outperform both uniform sampling and entropy-based region selection. A discussion follows in Section 5.
2 Previous Work
2.1 Incorporating Local Regions
The task of image registration, or determining a spatial mapping T between images I and J, is a cornerstone of medical image analysis. Cachier et al. propose two classes of techniques by which local image regions X are incorporated into image registration formulations [4]: geometrical feature-based and iconic feature-based. Geometrical feature-based techniques involve the segmentation of localized geometrical structures from image data, e.g. points or shapes, which are then aligned between different images to estimate T. In contrast, iconic feature-based techniques generally determine correspondences between images based on local image patches or blocks, after which these correspondences are used to improve estimation of T. Our work is most closely related to iconic feature-based techniques, which do not impose hard geometrical feature segmentations in both images to be registered.
A number of authors propose registration based on local regions which capture locally stable relationships. Hellier et al. present a technique which combines local constraints in the form of segmented anatomical structures into a deformable registration framework [11]. Clatz et al. describe a robust hierarchical block matching technique [2], where a fraction of blocks are iteratively rejected based on their displacement error with respect to the current transform. Toews et al. [5] and Loeckx et al. [8] propose computing the MI similarity measure locally to overcome spatially-varying multi-modal intensity relationships. Zhang et al. refine cardiological registration by considering locally affine deformations [7]. Wein et al. [9] propose a similarity measure based on a linear relationship between imaging parameters extracted in local windows.
Most work to date has advocated optimizing a function E(X, T) over the joint space of the transform T and local regions X. In contrast, we note that while X may be important in modeling the image and the registration process, it is ultimately a nuisance parameter that should not be optimized but rather marginalized in order to obtain the optimal transform T.
2.2 Local Region Selection
Using local image information requires a sampling strategy in order to define the specific local regions to be used (e.g. their location, size, distribution). A simple and commonly used strategy is to consider a uniform sampling of regions over the image [4, 2, 7, 9]. In general, however, certain regions are more informative than others, in terms of the image patterns they contain, and registration can be improved by considering a subset of highly informative regions. Domain-specific regions or structures can be defined by experts, e.g. sulci for registering cortical imagery [11]. Fully automatic region selection methods can be used when the structures most useful for registration are not known a priori. In the medical imaging literature, Rohr et al. investigate automatic selection of landmarks for registration based on derivative operators [12]. A popular body of computer vision literature has focused on automatically identifying salient or informative regions in general imagery, typically via a search for image regions which maximize a particular saliency criterion. Examples of saliency criteria include the magnitude of image derivatives in scale-spaces [13, 14] and image entropy [10].
2.3 Multi-modal US Registration
Although the formulation we propose in this paper is generally applicable in many different contexts, experiments focus on the registration of intra-operative US imagery to pre-operative modalities. US is an attractive imaging modality due primarily to the low cost and portability of US sensors. The major drawback of US is the poor image quality, due to the shift-variant nature of the sensor [3] and numerous modality related artifacts. The US image formation process is the result of acoustic backscattering or echoing off of boundaries between tissues of differing acoustic impedance. Specular echoing occurs in the presence of smooth tissue boundaries which are significantly larger than the US wavelength, and results in hyperintense image measurements. Scattered echoing occurs in the presence of poorly reflective or irregularly-shaped microstructures of size equal to or smaller than the US wavelength, and results in the formation of US speckle [15]. While speckle can be treated as noise to be removed [16], speckle brightness patterns do reflect the echogenic properties of the underlying tissues, and have been shown to be useful for segmentation, classification and registration of anatomical tissues [15, 17, 18, 9].
Modeling the image intensity relationship is a key focus of inter-modality US registration, and the wide variety of techniques in the literature speak to the difficulty of modeling. Roche et al. [19] propose an information-based similarity measure combining MR intensity and gradient information. Wein et al. [9] simulate US imagery from CT, then compute similarity using a specialized correlation measure accounting for both specular and scattered US echoing. Arbel et al. [20] register the US image to a pseudo US image, generated from gradients in segmented MR imagery. Toews et al. [5] register MR and US imagery using MI coupled with Bayesian estimation of image intensity distributions. Brooks et al. compute MI based on image gradients alone [6]. Reinertsen et al. [21] propose MR-US registration of brain images based on vasculature trees extracted in both modalities, using doppler US imagery to highlight blood vessels. Zhang et al. register MR-US cardiological imagery using the MI of local phase information [7]. Penney et al. propose estimating vasculature images independently from both US and MR modalities, which are then registered [22]. Letteboer et al. report registration success using standard MI between MR and US images, however suggest that incorporation of gradient information may be beneficial [23].
3 Registration via Location Marginalization
The Bayesian registration formulation presented in this section is based on the observation that although local image regions X are helpful in describing image content and the registration process, identifying meaningful regions or their correspondences between images is incidental to computing T. In probabilistic terms, local regions can thus be considered as nuisance variables. While the majority of techniques in the literature advocate maximizing a joint function of X and T, this approach does not generally result in the most probable T given the data. Intuitively, the most probable pair X, T may be determined by noisy, spurious instantiations of X which coincide with potentially incorrect or suboptimal values of T. Bayesian theory suggests X should instead be marginalized, in order to determine the most probable T given the images to be registered [24].
3.1 Bayesian Formulation
Image registration is the task of determining a geometrical mapping T between images I and J. Bayesian techniques typically pose registration as the posterior probability of T conditional on the image data to be registered [25]. Registration can then be computed by searching for the MAP transform TMAP maximizing the posterior:
| (1) |
While the Bayesian posterior provides a principled probabilistic basis for registration, there are other variables which, although not specifically required for registration, may offer important sources of information. Local anatomical structures produce distinct, spatially localized intensity patterns which drive registration, e.g. sulci in brain imagery [11], and information regarding image location can be incorporated in order to improve registration. Here, we propose a random variable of spatial location X, defined over a set of discrete spatial regions within one of the images to be registered I. The posterior in Equation (1) can then be expressed as the marginalization of the joint posterior distribution of X and T given images I and J:
| (2) |
By marginalizing over X as in Equation (2), TMAP can be obtained while leveraging important information regarding spatial location. The joint posterior in Equation (2) can be expressed as:
| (3) |
using Bayes rule, and by the fact that p(I, J) is constant given fixed images I, J. Here, the likelihood factor p(I, J|X, T) contains all expressions relating to image data. p(X) is a prior probability over image regions, which can generally be defined according to relative sizes or overlap of regions X. p(T|X), the conditional probability of T given X, can be defined to reflect the nature of the relationship between the specific transform and set of regions used.
3.2 Informative Region Selection
Utilizing local image regions requires a sampling scheme to determine X, and an ideal region sampling would represent a set of informative image patterns which can be localized with certainty in new images. Here, we develop a criterion to quantify both the informativeness and the localizability of image regions within a single image prior to registration. A mathematical link between Bayesian registration and the MI similarity measure motivates our MI-based selection criterion.
Bayesian Registration and Mutual Information
Although Bayesian techniques and the MI similarity measure [26, 27] are both widely used in medical image registration, their relationship is not immediately obvious. Maximum likelihood has been linked to joint entropy minimization [28] and MI maximization [29], here we provide a novel connection between Bayesian registration and MI maximization. To begin, recall the data likelihood p(I, J|X, T) in Equation (3), which can be represented as a discrete distribution over joint pixel intensity events. Let {mi,j} represent a set of observed counts corresponding to possible joint intensity outcomes (I, J), obtained from an experiment resulting in M independent joint intensity samples. Furthermore, let {mi} and {mj} represent counts of marginal pixel intensity events in histograms I and J, respectively, obtained in the same experiment. The number of unique experiments which could have generated a histogram {mi} is given by the multinomial coefficient . The number of experiments which could have resulted in the same set of joint/marginal histograms can thus be described by the following ratio of counts:
| (4) |
where 1 ≤ P ≤ ∞. Note here that P−1 is the probability mass function of the hypergeometric distribution, which quantifies the probability that a particular joint histogram was generated from independent marginal histograms. Registration can thus be achieved by maximizing P or a suitable monotonically increasing function thereof. In particular, the Stirling's approximation log x! ≈ x log x − x of M−1 log P converges to the mutual information of the empirical event count distributions as M → ∞ [30]:
| (5) |
Note that Equation (5) implies that maximizing the probability that data (I, J) did not arise from independent I and J is equivalent to minimizing the conditional conditional mutual information of I and J given region X and transform T:
| (6) |
Mutual Information-based Region Selection
The previous section showed how the data term of a Bayesian registration formulation is tied to the mutual informativeness of regions to be matched. As the MI of images I and J is upper-bounded by the marginal entropy of individual images,
| (7) |
intuition would suggest that selecting regions with high image entropy H(I) would be beneficial for registration. This technique is proposed by Kadir et al. [10], who use the Shannon entropy of the intensity distribution within an image region as a measure of informativeness. While H(I) measures the informativenss of the intensity distribution within an image region, it does not necessarily reflect the ability to register or localize the region in new images, as illustrated in Figure 1.
Fig. 1.
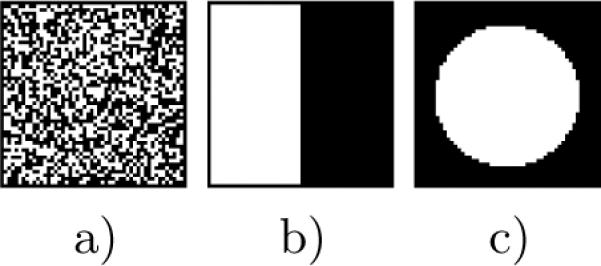
Images a), b) and c) represent three different binary images for which H(I) is maximal. Image a) represents a region of homogenous image texture which may be unlocalizable. Image b) represents a boundary between two tissues, which may only be localizable in the direction normal to the boundary. Image c) represents a pattern whose localization can be constrained in all directions.
In particular, image intensity must be mutually informative with respect to image location, in order to correctly localize homologous regions in different images. We propose instead to quantify region informativeness in terms of the MI between image intensity I and spatial location S:
| (8) |
where H(I|S) represents the conditional entropy of I given spatial location:
| (9) |
In Equations (8) and (9), S represents a random variable of spatial location at sub-region image scale, defined over a set of n discrete spatial labels. As conditioning reduces entropy, 0 ≤ H(I|S) ≤ H(I) and thus 0 ≤ MI(I, S) ≤ H(I). Note that like the H(I) criterion, the MI(I, S) criterion in Equation (8) favors high entropy intensity distributions, however it imposes the additional constraint that the entropy of intensity conditioned on spatial location be low. The two criteria are thus equivalent when H(I|S) = 0, i.e. when the intensity distribution is perfectly predicted by spatial location.
An important implementation detail is to choose a suitable sub-region sampling scheme S. A generally useful sampling scheme would favor patterns which can be localized in space in an unbiased manner. We experiment with a two-class binary sampling scheme, where p(S1) and p(S2) are defined by Gaussian densities of differing variance G(0, σ1) and G(0, σ2). As Gaussian densities are rotationally symmetric, they are consistent with an uncommitted visual front-end feature extractor exhibiting no specific bias as to the orientation of image patterns [31]. Figure 2 illustrates the result of applying both criteria on a US image.
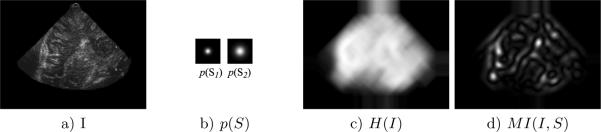
Fig. 2.
Image a) represents a transdural B-mode US slice of the brain. b) represents the distributions associated with the binary spatial variable S defined within a local region, where light regions indicate high p(S). Images c) and d) represent the entropy H(I) and MI(I, S) criteria calculated at square regions of size 39 pixels in the US image. Note that most regions within the US fan in b) are of high H(I), while a smaller number of US regions corresponding to distinctive image patterns in a) result in high M(I, S).
4 Experiments
The context for experiments is an IGNS application, in which pre-operative anatomical MR imagery has been acquired and used as the basis for surgical planning. The brain may shift and deform over the course of surgery, invalidating the pre-operative MR imagery as a basis for guidance. The goal is thus to register intra-operative US imagery to the pre-operative MR, and thereby update the pre-operative MR imagery to correctly reflect changes due to brain shift. Brain shift correction and US-MR registration remain significant research challenges, due to the impoverished nature of the US modality.
The goals of the experiments are two-fold. First, location marginalization of X is compared with the alternative of registration in the absence of local regions. Second, region selection based on the MI(I, S) criterion is compared with the alternatives of uniform sampling and selection based on the H(I) criterion. Experiments involve registration trials aligning B-mode US slices acquired during surgery with a pre-operative MR image. The 3D position and orientation of US the probe are tracked externally and calibrated geometrically with respect to the pre-operative MRI within a neuronavigation system. The approximate geometry of the US slice relative to the MR volume can thus be established and used to evaluate registration accuracy with respect to ground truth. Experiments use a sequence of ten (320×240)-pixel US images acquired after skull removal but prior to opening the dura, in which brain deformation is relatively minor and ground truth can be considered reliable. Registration remains challenging, however, due the difficulty in modeling the US/MR intensity relationship, see Figure 3. As deformation is minimal, registration aims to recover T in the form of a global translation between the US image and the MR slice corresponding to the tracked position of the US probe. The data likelihood p(I, J|X, T) is represented by the MI measure as in Equation (5). Factor p(X) reflects the prior probability of image regions, which can generally be defined according to the relative sizes or overlap of regions. A set of regions of equal size is used here and p(X) is thus taken to be uniform. Finally, as T does not depend X in an obvious manner and we have no particular prior assumptions regarding T, and so p(T|X) = p(T) is taken to be a uniform distribution.
Fig. 3.
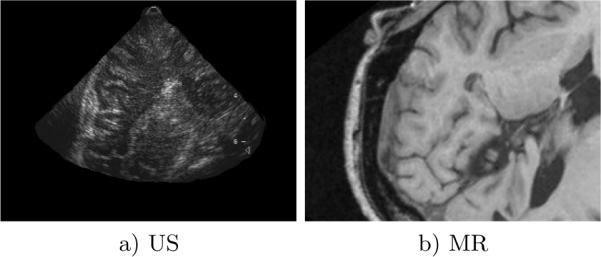
Images a) and b) illustrate an example of an US-MR image pair used in registration trials. Note the generally poor image quality of the US imagery.
To cope with the issue of data sparsity when calculating MI at a local scale, image intensities are first transformed into a mixture model over a small set of tissue classes as in [32]. Briefly, let C = {C1,…, CK} represent a discrete random variable over K tissue classes. Mixture modeling and expectation-maximization [33] can be used to estimate a set of class labels and associated parameters that maximize the likelihood of the image data I:
| (10) |
Once the parameters of mixture components in (10) are estimated, a conditional probability distribution over class labels can be associated with each image intensity I:
| (11) |
Here, the K-means algorithm [33] is used to estimate Gaussian class-conditional densities p(I|Ci). K = 3 tissue classes are used in experiments, and thus the size of the joint intensity histogram is 3 × 3 = 9. Varying K between 2–5 labels does not significantly affect registration accuracy, larger numbers of labels result in higher computational complexity however.
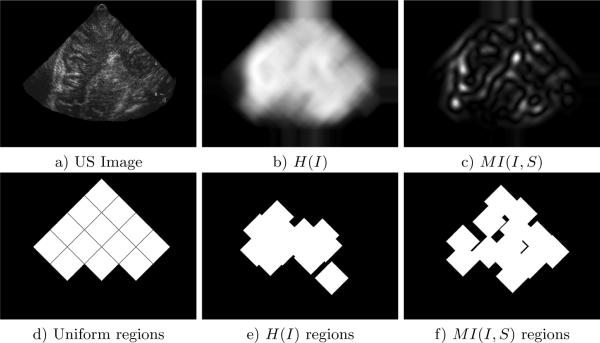
For each trial, local regions X are first defined in the US image as shown in Figure 4. The posterior is then calculated at all discrete pixel displacements T between the US image and its corresponding slice in the 3D MR volume, as determined by the geometry of the tracked US probe. In the case of the marginalized posterior, Equation (2) is evaluated by integrating over X for each displacement T. The results of registration based on various registration formulations and selection strategies are listed in Table 1. Note that MI-based region selection and localization marginalization results in both the highest number of successes (9 out of 10) and the lowest mean error. Incorporating no local regions resulted the fewest successes and high mean error. On this data, entropy-based region selection results in fewer successes and higher mean error than uniform sampling.
Fig. 4.
a) is a US image, images b) and c) are entropy and MI region selection criteria evaluated at all pixel locations in image a). Images d)–f) illustrate the three region sampling/selection strategies applied to the US image. For each strategy, 13 diamond-shaped regions of size 39 × 39 are chosen in the US image. Region size is determined empirically, smaller regions tend to result in spurious matching and larger regions tend to be poorly localized. 13 regions are used to uniformly sample the US image in c), and automatic selection identifies the 13 image regions with the highest value of the particular criterion in d) and e). A minimum distance of 20 pixels is enforced between automatically selected regions to ensure even distribution throughout the image.
Table 1.
Results for registration trials. A trial is considered successful if TMAP is a global maximum, i.e. corresponds to the peak of the posterior closest to the known ground truth displacement. The mean and standard deviation are calculated for successfully identified TMAP solutions.
| Selection Technique | Registration Formulation | Successes (out of 10) | Error Mean (pix) | Error Stdev (pix) |
|---|---|---|---|---|
| MI(I|S) | 9 | 2.5 | 1.7 | |
| Uniform | 9 | 3.3 | 1.9 | |
| H(I) | 8 | 4.1 | 2.6 | |
| - | 6 | 3.9 | 2.9 |
5 Discussion
This paper investigates a framework for the use of local image regions in registration, based on probability and information theory. Local regions are incorporated into a Bayesian registration framework in order to improve the ability to cope with missing tissue and non-stationary image intensity characteristics. Local region information is represented as a nuisance random variable, which is marginalized in order to obtain the MAP transform relating images. A novel link between a Bayesian registration formulation and the MI similarity measure is outlined, showing how MI results from the Bayesian data factor when the distribution of intensity events is modeled by a maximum entropy prior. This in turn motivates a new criterion for identifying a set of informative and localizable image regions for registration, the MI of intensity and location, which generalizes the entropy-based criterion of Kadir et al. [10].
A challenge of marginalizing location is computing the integral over local regions. Experiments here recover global translation via efficient numerical integration, where computation time is linear in the image size and in the number of regions used. Efficient marginalization with more complicated transforms could be addressed via coarse-to-fine methods or Monte Carlo integration, depending on the class of transforms used. The MI-based feature selection criterion is currently evaluated at a fixed image window size, but could be used to automatically determine more elaborate descriptions of local region geometry, including region size or shape, in order identify a wider variety of local image structures.
6 Acknowledgements
This work was funded in part by NIH grants U41 RR019703 and P41 RR13218, and by an NSERC postdoctoral fellowship. Image data was provided by D. Louis Collins of the Montreal Neurological Institute. Catherine Laporte was involved in helpful discussions regarding ultrasound.
References
- 1.Hajnal, et al., editors. Medical Image Registration. CRC Press; 2003. [Google Scholar]
- 2.Clatz O, et al. Robust nonrigid registration to capture brain shift from intraoperative mri. IEEE TMI. 2005;24(11) doi: 10.1109/TMI.2005.856734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng J, et al. Modelling ultrasound imaging as a linear, shift-variant system. IEEE Trans. on Ultrasonics, Ferroelectrics and Frequency Control. 2006;53(3):549–563. doi: 10.1109/tuffc.2006.1610563. [DOI] [PubMed] [Google Scholar]
- 4.Cachier P, et al. Iconic feature based nonrigid registration: the PASHA algorithm. CVIU. 2003;89(2–3):272–298. [Google Scholar]
- 5.Toews M, et al. Maximum a posteriori local histogram estimation for image registration. MICCAI. 2005:163–170. doi: 10.1007/11566489_21. [DOI] [PubMed] [Google Scholar]
- 6.Brooks R, et al. Deformable Ultrasound Registration without Reconstruction. MICCAI. 2008:1023–1031. doi: 10.1007/978-3-540-85990-1_123. [DOI] [PubMed] [Google Scholar]
- 7.Zhang W, et al. Adaptive non-rigid registration of real time 3d ultrasound to cardiovascular mr images. IPMI. 2007:50–61. doi: 10.1007/978-3-540-73273-0_5. [DOI] [PubMed] [Google Scholar]
- 8.Loeckx D, et al. Nonrigid image registration using conditional mutual information. IPMI. 2007:725–737. doi: 10.1007/978-3-540-73273-0_60. [DOI] [PubMed] [Google Scholar]
- 9.Wein W, et al. Automatic ct-ultrasound registration for diagnostic imaging and image-guided intervention. Medical Image Analysis. 2008;12:577–585. doi: 10.1016/j.media.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Kadir T, et al. Saliency, scale and image description. IJCV. 2001;45(2):83–105. [Google Scholar]
- 11.Hellier P, et al. Coupling dense and landmark-based approaches for non rigid registration. IEEE TMI. 2003;22(2):217–227. doi: 10.1109/TMI.2002.808365. [DOI] [PubMed] [Google Scholar]
- 12.Rohr K, et al. Landmark-based elastic registration using approximating thin-plate splines. IEEE TMI. 2001;20(6):526–534. doi: 10.1109/42.929618. [DOI] [PubMed] [Google Scholar]
- 13.Lowe DG. Distinctive image features from scale-invariant keypoints. IJCV. 2004;60(2):91–110. [Google Scholar]
- 14.Mikolajczyk K, et al. Scale and affine invariant interest point detectors. IJCV. 2004;60(1):63–86. [Google Scholar]
- 15.Wagner RF, et al. Statistics of speckle in ultrasound B-scans. IEEE Transactions on Sonics and Ultrasonics. 1983;30(3):156–163. [Google Scholar]
- 16.Coupe P, et al. Bayesian non local means-based speckle filtering. ISBI. 2008:1291–1294. [Google Scholar]
- 17.Thijssen JM. Ultrasonic speckle formation, analysis and processing applied to tissue characterization. Pattern Recognition Letters. 2003;24(4–5):659–675. [Google Scholar]
- 18.Milko S, et al. Segmentation of the liver in ultrasound: a dynamic texture approach. Int. J. of Computer Assisted Radiology and Surgery. 2008;3(1–2):143–150. [Google Scholar]
- 19.Roche A, et al. Rigid registration of 3D ultrasound with MR images: a new approach combining intensity and gradient information. IEEE TMI. 2001;20(10):1038–1049. doi: 10.1109/42.959301. [DOI] [PubMed] [Google Scholar]
- 20.Arbel T, et al. Automatic non-linear mri-ultrasound registration for the correction of intra-operative brain deformations. Comput Aided Surg. 2004;9(4):123–136. doi: 10.3109/10929080500079248. [DOI] [PubMed] [Google Scholar]
- 21.Reinertsen I, et al. Validation of vessel-based registration for correction of brain shift. Medical Image Analysis. 2007;11(4):374–388. doi: 10.1016/j.media.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Penney G, et al. Registration of freehand 3d ultrasound and magnetic resonance liver images. Medical Image Analysis. 2004;(8):81–91. doi: 10.1016/j.media.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Letteboer M, et al. Brain shift estimation in image-guided neurosurgery using 3-d ultrasound. IEEE Transactions on Biomedical Engineering. 2005;52(2):268–276. doi: 10.1109/TBME.2004.840186. [DOI] [PubMed] [Google Scholar]
- 24.Gelman A, et al. Bayesian Data Analysis. Chapman & Hall/CRC; 2000. [Google Scholar]
- 25.Gee J, et al. Probabilistic matching of brain images. IPMI. 1995 [Google Scholar]
- 26.Wells W, et al. Multi-modal volume registration by maximization of mutual information. MIA. 1996;1:35–52. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- 27.Maes F, et al. Multimodality image registration by maximization of mutual information. IEEE TMI. 1997;16(2):187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 28.Zollei L, et al. A marginalized map approach and em optimization for pair-wise registration. IPMI. 2007:662–674. doi: 10.1007/978-3-540-73273-0_55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roche A, et al. Unifying maximum likelihood approaches in medical image registration. IJIST. 2000;11:71–80. [Google Scholar]
- 30.Jaynes E. Prior probabilities. IEEE Transactions on systems, science, and cybernetics. 1968;SSC-4(3):227–241. [Google Scholar]
- 31.Romeny B.t.H. Front-End Vision and Multi-Scale Image Analysis. Kluwer Academic Publisher; 2003. [Google Scholar]
- 32.D'Agostino E, et al. An information theoretic approach for non-rigid image registration using voxel class probabilities. MICCAI. 2003:812–820. doi: 10.1016/j.media.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Duda RO, et al. Pattern classification. 2nd edn. Wiley; 2001. [Google Scholar]