Summary
Animal models suggest a role for osteonectin/SPARC in determination of bone mass. We found haplotypes consisting of three single nucleotide polymorphisms (SNPs) in the 3′ untranslated region (UTR) of the osteonectin gene are associated with bone density in Caucasian men with idiopathic osteoporosis.
Introduction
Osteonectin is a matricellular protein regulating matrix assembly, osteoblast differentiation, and survival. Animal studies indicate that osteonectin is essential for normal bone mass. The 3′ UTR is a regulatory region controlling mRNA stability, trafficking, and translation, and we determined whether osteonectin 3′ UTR haplotypes could be associated with bone mass and/or idiopathic osteoporosis.
Methods
Single strand conformation polymorphism and allele-specific PCR analysis were used to assess alleles at osteonectin cDNA bases 1046, 1599, and 1970, using genomic DNA from middle-aged Caucasian men with idiopathic, low turnover osteoporosis (n=56) and matched controls (n=59). Bone density was measured by DXA at spine, hip and radius. Allele and haplotype frequencies were analyzed by Chi square analysis and Fisher's exact test.
Results
Five common osteonectin 3′ UTR haplotypes were identified. The frequency of one haplotype (1046C-1599C-1970T) was higher in controls compared with patients, and this haplotype was also associated with higher bone densities at multiple sites in patients. In contrast, a second haplotype (1046C-1599G-1970T) was associated with lower bone densities in patients at multiple sites.
Conclusions
Osteonectin regulates skeletal remodeling and bone mass in animals, and haplotypes in the 3′ UTR of this gene are associated with bone density in Caucasian men with idiopathic osteoporosis.
Keywords: 3′ Untranslated region, Bone mineral density, Idiopathic osteoporosis, Non-collagen matrix protein, Osteonectin/SPARC, SNP/polymorphism
Introduction
Heritability estimates suggest that genetic factors account for at least 50% of the observed variation in bone mineral density (BMD) [1]. Thus, there is potential clinical value in identifying the genetic components of osteoporosis and in characterizing genotypes associated with decreased bone density. Men with idiopathic osteoporosis (IOM) represent a well-defined cohort of individuals who have undergone extensive clinical evaluation for osteoporosis, but for whom no etiology has been found [2, 3]. While there is recent evidence that low body mass index (BMI) and perturbed sex steroid status may be contributory to the pathophysiology of this disorder, there is a growing body of data suggesting that the IOM phenotype may be attributed to genetic determinants [4-7]. First, there is a much higher prevalence of osteoporosis in first-degree male relatives of these men than might be expected in the general population [5, 7]. In addition, there is evidence suggesting that men with idiopathic osteoporosis exhibit a defect in bone acquisition, and acquisition of bone mass is largely genetically determined [1, 5, 8]. Because it is postulated that genetic polymorphisms may result in small functional or regulatory differences that can affect BMD over the lifetime of an individual, studying single nucleotide polymorphisms (SNPs) may yield valuable information about the genetic etiologies of osteoporosis, and men with IOM appear to be an optimal cohort in which to do so [8].
One candidate gene that may impact bone mass is osteonectin (SPARC or secreted protein acidic and rich in cysteine; BM-40), a 43 kiloDalton intracellular and extracellular matrix glycoprotein [9-11]. Abundant in bone, secreted osteonectin binds both collagen and hydroxyapatite and regulates collagen fibril assembly. Osteonectin modulates cell behavior, at least in part by modifying signaling initiated at selected transmembrane receptors [10, 12]. Osteoblasts from patients with osteogenesis imperfecta synthesize less osteonectin than age-matched controls, and animal models of osteopenia and osteogenesis imperfecta suggest that osteonectin may be important in the maintenance of bone mass [13-16]. Osteonectin-null mice develop low turnover osteopenia characterized by decreased mineral apposition rate and decreased osteoblast numbers. Osteonectin-haploinsufficient mice also display osteopenia, particularly at the lumbar spine [11, 17]. In vitro studies indicate that osteonectin-null osteoblasts are more sensitive to apoptotic stress and develop a less differentiated phenotype. These studies highlight the importance of osteonectin in normal bone remodeling and in maintenance of skeletal mass [18].
The 3′ untranslated region (UTR) of a gene is an important regulatory region that controls mRNA stability, trafficking, and translational efficiency through its interaction with trans-acting factors [19]. Thus, polymorphisms in 3′ UTRs have the potential to modulate gene expression levels, and SNPs in the 3′ UTR of the osteonectin gene have been associated with systemic sclerosis, a fibrotic disease in which osteonectin expression is increased in multiple organ systems [20]. Because of the important role osteonectin plays in the mammalian skeleton, we sought to explore the relationship between osteonectin 3′ UTR haplotypes and bone mass [11, 17, 18]. To this end, we studied SNPs at bases 1046, 1599 and 1970 in the osteonectin 3′ UTR, in a well-characterized cohort of men with idiopathic osteoporosis and in normal, matched controls [2, 3].
Methods
Study population
A total of 56 unrelated Caucasian men with idiopathic low turnover osteoporosis were enrolled at Columbia University Medical Center (CUMC). Inclusion criteria for the study were Z-score less than −2.0 at the lumbar spine, and patients with any known secondary causes of osteoporosis were excluded. This patient group has been previously described [2, 3]. Clinical characteristics include low bone turnover as reflected by reduced indices of bone formation on histomorphometry, and serum and urinary markers of bone turnover in the lower normal range [2]. A control group consisting of 59 healthy Caucasian men with normal BMD (Z-score > −1.0 at all sites), matched for age and body mass index was also enrolled at CUMC. The characteristics of each group are shown in Table 1. The patient and control groups were not significantly different with regard to alcohol use, smoking or level of physical activity, although the patient group reported an average ~two fold higher intake of calcium and vitamin D compared with the controls (data not shown). Our analysis was limited to Caucasians, because they comprised the largest ethnic group in the cohort of patients recruited and we wished to minimize genetic variation that might result from population admixture. This study was conducted with the approval of the institutional review boards of CUMC and University of Connecticut Health Center, and all subjects gave written informed consent.
Table 1.
Clinical characteristics for Caucasian male idiopathic osteoporosis patients and matched controls
| Characteristic | Patients, N=56 mean ± SEM (range) |
Controls, N=59 mean ± SEM (range) |
|---|---|---|
| Age (yrs) | 47.8±1.4 (30 to 64) | 45.6±1.5 (28 to 64) |
| Body mass index (kg/m2) | 24.40±0.50* (16.0 to 30.6) | 26.24±0.40 (20.4 to 36.5) |
| Z-score lumbar spine | −2.93 ±0.23 (−4.70 to −2.0) | 0.37±0.13 (−0.98 to +4.62) |
| Z-score femoral neck | −1.50±0.17 (−2.94 to +2.3) | 0.39±0.08 (−0.70 to +2.43) |
| Z-score total hip | −1.19±0.23 (−2.93 to +2.10) | 0.49±0.08 (−0.80 to +2.19) |
| Z-score distal radius | −0.09±0.57 (−3.88 to +2.16) | 0.72±0.15 (−1.53 to +3.52) |
| Prevalence of fragility fracture | 23%* | 0% |
significantly different from control, p<0.05 (ANOVA). Differences in bone density not applicable because of study selection criteria
Bone mineral density (BMD)
Bone density of the lumbar spine, right hip, and nondominant 1/3 radius was measured by dual energy x-ray absorptiometry using either the QDR 4500 Delphi C, the QDR 4500 A, or the Hologic 1000 W bone densitometer (Hologic, Inc., Bedford, MA). All controls were scanned on the 4500 C on-site; 28 patients were scanned on the 4500 C, seven patients were scanned on a Hologic 4500 C off-site, six patients were scanned on a Hologic 4500 A, and 14 patients were scanned on the Hologic1000 W. One patient was scanned on a GE Lunar densitometer off-site. To maintain consistency between machines, bone density is expressed as the Z-score, which compares, by SD, individual bone measurements to a database of age- and sex-matched normal population.
Single nucleotide polymorphism (SNP) analysis
All subjects had 20 cc of whole blood drawn for DNA extraction. After preparation of Buffy coat, DNA was extracted using the Puregene Kit (Gentra Systems, Minneapolis, MN) according to manufacturer's instructions. Osteonectin SNPs are listed according to their position in the cDNA, as obtained from the dbsnp database (http://www.ncbi.nlm.nih.gov/projects/SNP). This numbering is slightly different from that reported by Zhou et al.: SNP 1046 corresponds to 998 of Zhou et al., 1599 corresponds to 1551 of Zhou et al., and 1970 corresponds to 1922 of Zhou et al. [20]). The primer sets used for each analysis are detailed in Table 2. SNPs 1046 and 1970 were characterized by allele specific PCR, and each reaction contained a positive control for amplification. For SNP 1046, the allele specific PCR primers and PCR reaction conditions corresponded to those described by Lagan et al. [21]. PCR reactions consisted of genomic DNA, 0.2 mM dNTP mix, 1.5 mM MgCl2, 0.5 U Taq polymerase (Life Technologies), 20 mM Tris-Cl, pH 8.4, 50 mM KCI, 1.5 μM allele specific forward primer, 0.125 μM control forward primer, and 1.5 μM reverse primer. The cycling parameters used were: 2 min 96°C followed by ten cycles of 15 s at 96°C, 60 s at 65°C, then 20 cycles of 10 s 96°C, 50 s 61°C, and 30 s 72°C. PCR products were resolved on 2% agarose gels and visualized by ethidium bromide staining. For SNP 1970, the allele specific PCR primers and PCR reaction conditions corresponded to those described by Lagan et al. [21]. PCR reactions consisted of genomic DNA, 0.2 mM dNTP mix, 1.5 mM MgCl2, 0.5 U Taq polymerase, 20 mM Tris-Cl, pH 8.4, 50 mM KCI, 1.5 μM allele specific forward primer, 1.5 μM reverse primer, 0.125 μM control forward primer and 0.125 μM control reverse primer. The cycling parameters used were: 2 min 96°C followed by 30 cycles of 30 s at 96°C, 45 s at 56°C, and 45 s at 72°C. PCR products were resolved on 1.5% agarose gels and visualized by ethidium bromide staining. SNP 1599 was characterized by single strand conformation polymorphism analysis with primer pairs designed using online software (Massachusetts Institute of Technology; web.mit.edu/cgi-bin/primer/primer3) [22]. PCR reactions consisted of genomic DNA, 0.2 mM dNTP mix, 1.5 mM MgCl2, 0.5 U Taq polymerase, 0.2 μM each primer, 20 mM Tris-Cl, pH 8.4, and 50 mM KCl. PCR was performed using 34 cycles of 94°C for 30 sec, 58°C for 30 sec, 72°C for 30 sec, and a final extension at 72°C for 5 min. Single strand conformation polymorphism (SSCP) analysis was performed by mixing an aliquot of PCR product with an equal volume of stop solution containing 95% formamide, 20 mM EDTA, 10 mM NaOH, 0.05% bromophenol blue and 0.05% xylene cyanol. Samples were denatured for 3 minutes at 95°C and subjected to electrophoresis on 8.5% polyacrylamide (acrylamide:bis ratio 39:1), 10% glycerol, 0.5X Tris-borate EDTA gels at 5 W for 15 hours, with cooling (Life Technologies), and visualized by silver staining [23, 24]. Patterns were scored by two blinded observers. Genotyping accuracy was confirmed by sequencing of PCR products (4 for each SNP) using an automated fluorescence sequencer according to manufacturer's instructions (Big-Dye chemistry, Applied Biosystems). Haplotypes were constructed using SNPHap software (http://www-gene.cimr.cam.ac.uk/clayton/software/).
Table 2.
Primer pairs for analysis of osteonectin 3′ UTR SNPs
| SNP | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| 1046 C | TCC ACA GTA CCG GAT TCT C | CTG CAA TGT GTG TTT AAG GC |
| 1046 G | TCC ACA GTA CCG GAT TCT G | |
| 1599 | TGG GAC TAG AGG CTC AGT GG | GCT CCC AAA AGT TTG AAC CA |
| 1970 T | TTC TTC ACC CTC CCC TTT T | CTG ATA ATG GTG GTA GTG ATC |
| 1970 G | TCT TCA CCC TCC CCT TTG | |
| Control | TCC TTG GGA AGT GCG TTA TC | TGT CTC CAG GCA GAA CAA CA |
Statistical analysis
To illustrate the heterogeneity in BMD within the patient and control groups, BMI adjusted Z-scores for lumbar spine, total hip, femoral neck and radius were subjected to tertile analysis. Within each group and for each site, BMI adjusted Z-scores were ranked from low to high, then divided into three equal groups (17–18 individuals/tertile for patients; 19–20 individuals/tertile for controls). Allele and haplotype frequencies in the idiopathic osteoporosis patients were compared to the matched healthy controls using Chi square analysis and Fisher's Exact test. ANOVA was used to determine whether the patient and control groups had significant differences in potentially confounding variables, such as anthropometrics. All analyses were conducted with SAS/STAT v.9.1 software (SAS Institute, Inc., Cary, NC).
Results
Table 1 summarizes the group characteristics of the osteoporosis patients and matched controls. All the men with idiopathic osteoporosis had Z-scores ≤ −2.0 at the lumbar spine, with 23% having Z-scores ≤ −2.0 at both the femoral neck and total hip, and 20% having Z-scores ≤ −2.0 at the cortical radius. Healthy controls, by definition, had Z-scores of > −1.0 at all sites as per inclusion criteria (Table 1). Further, 23% of the osteoporotic patients had experienced fragility fractures, predominantly in the axial skeleton, while there was no history of fragility fracture in the control group. Despite efforts to match patients to controls for age, height and weight, there was a 9% difference in BMI between groups (p<0.05). BMI is an important factor contributing to variance in BMD in men with idiopathic osteoporosis [5-7]. Therefore, BMD Z-scores were adjusted for BMI in all analyses.
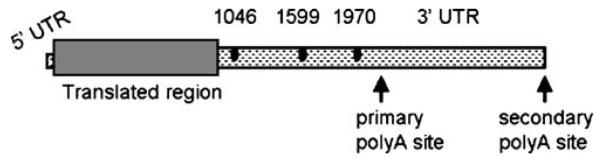
A schematic diagram of the human osteonectin cDNA is presented in Fig. 1. SNPs at bases 1046, 1599 and 1970 in the 3′ UTR of the osteonectin cDNA were characterized in osteoporosis patients and controls using allele-specific PCR and SSCP. Genotype frequencies for each SNP were calculated as shown in Table 3, and frequencies conformed to Hardy–Weinberg equilibrium. For each SNP, genotype frequencies were not significantly different between osteoporosis patients and controls. The dbsnp database at the National Center for Biotechnology Information (NCBI) was interrogated for the distribution of the three osteonectin SNPs of interest in populations of European Americans, African Americans, and Asian Americans. When genotype frequencies for the osteonectin 3′ UTR SNPs were calculated for individuals of European, African or Asian descent, there was no significant difference in genotype frequencies based on ethnic group (Table 3).
Fig. 1.
Schematic diagram of the human osteonectin cDNA. The 5′ UTR is from 1–105; protein coding region/translated region is from 106 to 1017; the 3′ UTR is from 1018 to 3178. The primary polyadenylation signal is at 2126–2131; the secondary polyadenylation signal is at 3018 to 3023. The characterized SNPs are at 1046, 1599, and 1970, denoted by the dark ovals
Table 3.
Osteonectin 3′ UTR genotype frequencies, by SNP, in osteoporosis patients, controls, and as obtained from the dbsnp database for European-, African- and Asian Americans (AFD panel)
| SNP | Patients | Controls | dbsnp European | Dbsnp African | Dbsnp Asian |
|---|---|---|---|---|---|
| 1046 C/C | 41 (73.2) | 44 (73.3) | 15 (62.5) | 18 (78.3) | 10 (41.7) |
| C/G | 12 (21.4) | 15 (25.0) | 9 (37.5) | 4 (17.2) | 11 (45.8) |
| G/G | 3 (5.4) | 1 (1.7) | 0 (0) | 1 (4.3) | 3 (12.5) |
| 1599 G/G | 18 (32.1) | 12 (20.0) | 24 (28.6) | 48 (57.8) | 17 (19.1) |
| G/C | 26 (46.4) | 33 (55.0) | 39 (46.4) | 29 (34.9) | 56 (49.6) |
| C/C | 12 (21.4) | 15 (25.0) | 21 (25.0) | 6 (7.2) | 29 (32.6) |
| 1970 T/T | 40 (71.4) | 49 (81.7) | 62 (73.8) | 74 (90.2) | 27 (39.7) |
| T/G | 13 (23.2) | 9 (15.0) | 20 (23.8) | 8 (9.8) | 31 (45.6) |
| G/G | 3 (5.4) | 2 (3.3) | 2 (2.4) | 0 (0) | 10 (14.7) |
Number of individuals (percent of population)
In our cohort of 115 participants, we observed ten different genotype patterns across the three SNPs studied (Table 4). The SNPHap software assigned five haplotypes to 228/230 strands of DNA that we termed A-E (Tables 4 and 5). Two men from the control group had an additional haplotype not predicted by SNPHap, “G_C_T” and we termed this haplotype “X”. Haplotypes were assigned to the genotype patterns, and Table 4 shows the groups arranged in order from the most common to the least common haplotype pairs. The three most common haplotypes found in our study (A, B, and C) are also the most common haplotypes constructed from the dbsnp data (Table 4 and 5).
Table 4.
Osteonectin 3′ UTR genotype frequencies in osteoporosis patients and control subjects, compared with genotypes of European-, African- and Asian Americans (AFD panel, dbsnp database)
| 1046_1599_1970 genotype |
Haplotype | Genotype as haplotype pairs |
Patients number (%) |
Controls number (%) |
dbsnp European number (%) |
dbsnp African number (%) |
dbsnp Asian number (%) |
|---|---|---|---|---|---|---|---|
| C/C_G/C_T/T | C_G_T | A | 16 (28.6) | 24 (40.7) | 9 (37.5) | 5 (22.7) | 2 (8.3) |
| C_C_T | B | ||||||
| C/C_G/G_T/T | C_G_T | A | 16 (28.6) | 11 (18.6) | 5 (20.8) | 13 (59.1) | 8 (33.3) |
| C_G_T | A | ||||||
| C/G_G/C_T/G | C_G_T | A | 7 (12.5) | 5 (8.5) | 4 (16.7) | 3 (13.6) | 11 (45.8) |
| G_C_G | C | ||||||
| C/C_C/C_T/T | C_C_T | B | 6 (10.7) | 8 (13.6) | 1 (4.2) | 0 (0) | 0 (0) |
| C_C_T | B | ||||||
| C/G_C/C_T/G | C_C_T | B | 3 (5.4) | 3 (5.1) | 5 (20.8) | 0 (0) | 0 (0) |
| G_C_G | C | ||||||
| G/G_C/C_G/G | G_C_G | C | 3 (5.4) | 1 (1.7) | 0 (0) | 0 (0) | 3 (12.5) |
| G_C_G | C | ||||||
| C/C_G/C_T/G | C_G_T | A | 3 (5.4) | 1 (1.7) | 0 (0) | 0 (0) | 0 (0) |
| C_C_G | D | ||||||
| C/G_G/G_T/T | C_G_T | A | 2 (3.6) | 1 (1.7) | 0 (0) | 0 (0) | 0 (0) |
| G_G_T | E | ||||||
| C/G_C/G_T/T | C_C_T | B | 0 (0) | 3 (5.1) | 0 (0) | 1 (4.5) | 0 (0) |
| G_G_T | E | ||||||
| C/G_C/C_T/T | C_C_T | B | 0 (0) | 2 (3.4) | 0 (0) | 0 (0) | 0 (0) |
| G_C_T | X | ||||||
| Total | 56 | 59 | 24 | 22 | 24 |
One genotype, which contained only one representative, was not included in the analysis due to statistical considerations Number of individuals (percent of population)
Table 5.
Osteonectin 3′ UTR haplotype frequency distribution in osteoporosis patients and controls, and in European, African and Asian Americans (AFD panel, dbsnp database)
| Haplotype 1046_1599_1970 | OP patients | Healthy controls | Dbsnp Euro | Dbsnp African | Dbsnp Asian |
|---|---|---|---|---|---|
| “A” (C_G_T) | 60 (53.8) | 53 (44.9) | 23 (47.9) | 34 (77.3) | 29 (60.4) |
| “B” (C_C_T) | 31 (27.7) * | 48 (40.7) | 16 (33.3) | 6 (13.6) ** | 2 (4.2) **, # |
| “C” (G_C_G) | 16 (14.3) | 10 (8.5) | 9 (18.8) | 3 (6.8) ** | 17 (35.4) **# |
| “D” (C_C_G) | 3 (2.7) | 1 (0.8) | 0 (0) | 0 (0) | 0 (0) |
| “E” (G_G_T) | 2 (1.8) | 4 (3.4) | 0 (0) | 1 (2.3) | 0 (0) |
| “X” (G_C_T) | 0 (0) | 2 (1.7) | 0 (0) | 0 (0) | 0 (0) |
Number of individuals (percent of population)
Percent of each population with a given haplotype may not total to 100% due to rounding
=significantly different between patients and controls p<0.01;
= significantly different from European Americans p<0.02;
=significantly different from African Americans p<0.04
When osteonectin 3′ UTR haplotype frequency distribution was calculated, we found a significant decrease in the frequency of the B haplotype in osteoporosis patients compared with controls, 28% vs. 41%, respectively (p<0.01; Table 5). The distribution of the other haplotypes was not significantly different between the patient and control groups.
Differences were also noted in osteonectin 3′ UTR haplotype frequency between ethnic groups. The frequency of the B haplotype was significantly lower in African Americans and Asian Americans compared with European Americans (Table 5). Similarly, the frequency of the C haplotype was significantly lower in African Americans than European Americans, but was highest in Asian Americans (Table 5). There were no significant differences among ethnic groups noted in distribution of the most common A haplotype or the rare D and E haplotypes.
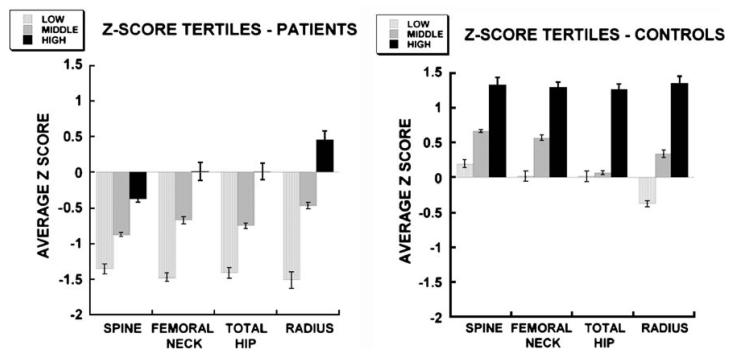
Although the osteoporosis patients as a group were defined as having lumbar spine Z-scores ≤ −2.0, some patients were, in addition, osteoporotic at the hip and radius, while others were unaffected at these sites. Further, although men in the control group were defined as having Z-scores > −1.0 at all sites, Z-scores exhibited a broad range (i.e., from −0.98 to +4.62 for lumbar spine) (Table 1). When Z-scores were corrected for BMI, a fairly broad range remained in both patient and control groups. Recognizing this diversity, these groups were evenly subdivided into tertiles according to BMI-adjusted Z-score at each skeletal site examined (spine, femoral neck, total hip and radius) (Fig. 2). The tertile analysis illustrates the degree of heterogeneity within the patient and control groups, suggesting sub-classifications of bone mass within each group: low, middle, and high.
Fig. 2.
BMI-adjusted Z-scores (mean ± SEM) in osteoporosis patients and controls divided into tertile groups (low, middle and high) according to Z-score. Tertile groups are significantly different from each other for each site (spine, femur, hip, radius) (p<0.001)
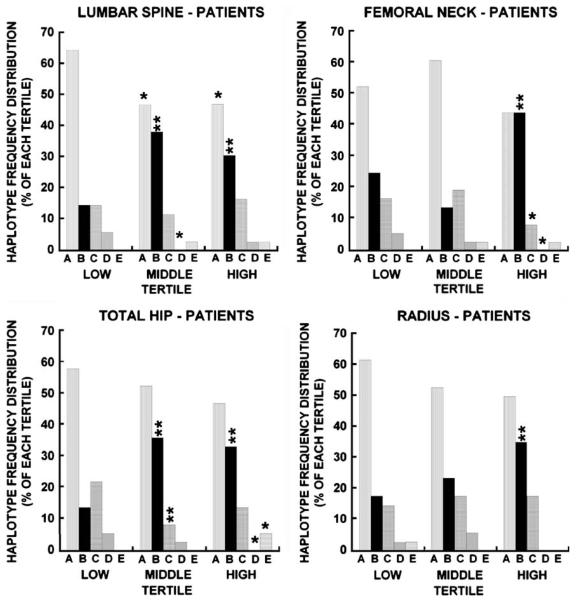
To test whether diversity in bone mass would correlate with genetic diversity, osteonectin 3′ UTR haplotype frequency distribution was determined for each tertile in the patient group (Fig. 3), analyzing lumbar spine, femoral neck, total hip and cortical radius separately. There were significant differences in osteonectin 3′ UTR haplotype frequency across all tertiles and at all sites (Fig. 3). The A haplotype was most common in the most severely affected patients, with significantly increased frequency of “A” at the lowest tertile of lumbar spine compared with mid and high tertiles (p<0.05), and a trend toward this finding at the other three sites examined (p=NS; Fig. 3). To a lesser extent, the C and D haplotypes were also associated with lower bone density, with increased frequency of these haplotypes in the lowest tertile compared with high and middle tertiles at the femoral neck and total hip for “C” and at the lumbar spine, femoral neck, and total hip for the D haplotype, respectively (Fig. 3). In contrast, the B haplotype was associated with higher bone density. At the lumbar spine and total hip, there was a significantly increased frequency of haplotype B in the middle and high Z-score tertiles, while at the femoral neck and radius this was only appreciated when the highest tertile was compared with the lowest (p<0.01; Fig. 3). Finally, there was a significantly higher frequency of the rare E haplotype in the highest Z-score tertile at the total hip, with similar but not significant trends observed for the spine and femoral neck.
Fig. 3.
Significant differences in osteonectin 3′ UTR haplotype frequency distribution in low, middle and high Z-score tertiles for osteoporosis patients (p<0.001 for total hip and femoral neck; p<0.01 for lumbar spine; p<0.03 for cortical radius). **= haplotype frequency significantly different from low tertile, p<0.01; *= haplotype frequency significantly different from low tertile p<0.05
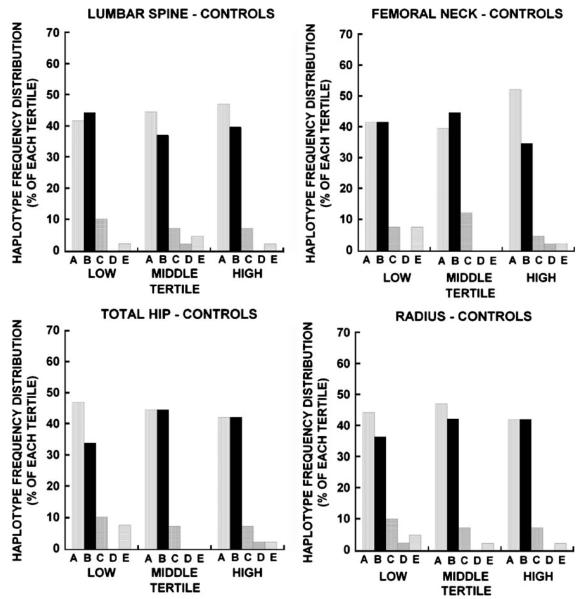
When osteonectin 3′ UTR haplotype frequency distribution was determined for each tertile in the control group (Fig. 4), consistent significant differences in bone density by haplotype were not found. In fact, there appeared to be a fairly similar frequency distribution of haplotypes A and B across all bone mass tertiles, at all sites, in the controls. This observation contrasted with the apparent skewed frequency in patients, in which the A haplotype was over-represented in the lowest tertiles and the B haplotype was over-represented at the higher patient BMD tertiles (Figs. 3 and 4).
Fig. 4.
Osteonectin 3′ UTR haplotype frequency distribution in low, middle and high Z-score tertiles for controls
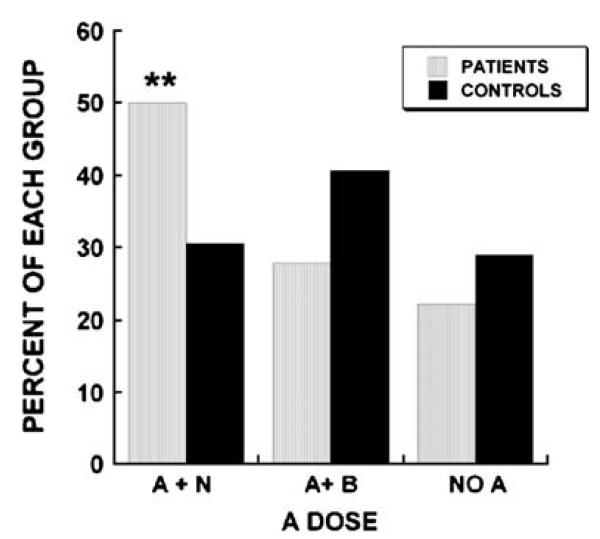
Since haplotype A is associated with low bone density and haplotype B is associated with higher bone density, we determined the percentage of patients and controls with haplotype pairs lacking A (no A), an A haplotype combined with a B (A + B), or an A haplotype combined with any haplotype other than B (A + N). We found that 50% of patients had at least one A haplotype in the absence of B, compared with only 30% of controls (Fig. 5). Patients were 2.33 times more likely than controls to have an A haplotype unopposed by a B haplotype (p-value for odds ratio n=0.0549). There was no significant difference between groups in the frequency of the “A+B” haplotype pairing or the frequency of haplotype pairs lacking A. These data suggest that in Caucasian men, osteonectin 3′ UTR haplotype combinations can contribute to bone mass expression.
Fig. 5.
Increased percentage of individuals with at least one osteonectin 3′ UTR haplotype A (A + N) in osteoporosis patients (**= p≤0.006). The percentage of patients and controls with one A and one B (A + B) or without any A (No A) was not significantly different
Discussion
This study provides three novel observations with regard to the candidate gene osteonectin and its relationship to bone mass in men with idiopathic osteoporosis. First, SNP haplotypes in the osteonectin 3′ UTR are associated with idiopathic osteoporosis in Caucasian men. This conclusion is supported by the significant difference in the frequency of the B haplotype between patient and control groups, and the finding of a significant difference in the way haplotypes are paired together in each group. Second, when tertile analysis was performed, osteonectin 3′ UTR haplotype frequency distribution was significantly different between the highest and lowest Z-score tertiles for patients at multiple skeletal sites, indicating haplotypes associated with higher (B) or lower (A, C, D) bone density. Finally, this study identifies osteonectin as a putative candidate gene that affects cortical as well as trabecular bone.
The ethnic variations observed between our Caucasian cohort and the data available from the dbsnp database are noteworthy. Differences in haplotype distribution with ethnicity (European vs. African vs. Asian) are readily apparent (Table 5), underscoring the necessity of limiting our analysis to a single ethnic group. In addition, the ethnic variation suggests that the osteonectin 3′ UTR haplotype likely operates in a multigenic fashion. We make this inference because African Americans have the highest BMD among ethnic groups [25]. Yet among African Americans, the B haplotype, associated with the control group and with higher bone density in the osteoporotic Caucasians, is significantly less common compared with the sampling of European Americans. The fact that African Americans do not have the osteonectin 3′ UTR profile we are ascribing to Caucasians would suggest that interaction between the osteonectin gene and other genes or factors is likely in the association with bone density.
The osteonectin gene contains ten exons and gives rise to mRNAs containing a short 105 base 5′ untranslated region (UTR), a 912 base protein coding region, and a 1114 or 2121 base 3′ UTR, due to differential usage of polyadenylation signals [26] (Fig. 1). The shorter mRNA, containing the 1114 base 3′ UTR, is the predominant transcript in humans and the only osteonectin transcript found in mice, rats, cows and other species [26-29]. Overall, the amino acid sequence of osteonectin, its intron/exon structure, and portions of the osteonectin 3′ UTR are highly conserved across species. Further, the three SNPs we characterized are found within a relatively small region, ~1000 bases in length. We hypothesize that regulatory motifs are contained within this region and these polymorphisms may affect UTR function. Further, regulatory motifs may interact with each other in this limited span of nucleotides, underscoring the importance of considering 3′ UTR haplotypes, in addition to individual SNPs. It is noteworthy that the osteonectin gene, located at chromosome 5q31.3–32, is in a region that has shown evidence of linkage to bone mass in several large genome scans [30]. The osteonectin 3′ UTR SNPs of interest may be contributing to that linkage signal.
Three previous studies have attempted to find an association between osteonectin gene polymorphisms and connective tissue/musculoskeletal disorders. A study by Zhou et al. suggested an association of osteonectin 3′ UTR SNPs with the clinical presentation of systemic sclerosis [20]. However, subsequent study of a different cohort of systemic sclerosis patients failed to document an association between osteonectin SNPs and fibrotic disease [21]. In this second systemic sclerosis study, eight SNPs were examined: five in the 3′ UTR, two in the putative promoter region, and one in exon 3, which does not affect amino acid coding. The only previous study assessing osteonectin gene polymorphisms and bone mass involved analysis of a polymorphic dinucleotide (CA) repeat in an intragenic region of osteonectin in pre- and perimenopausal women participating in the Michigan Bone Health Study. Analysis of this intragenic region failed to reveal an association between osteonectin polymorphisms and bone mass [31].
In our study of the osteonectin 3′ UTR, substitution of a single base pair at SNP 1599 (G→ C) accounts for the difference between haplotypes A and B (i.e., 1046_1599_1970: C_G_T vs. C_C_T), that we have associated with lower or higher Z-score tertiles within the patient group, respectively (Table 5 and Fig. 3). Further, the presence of at least one B, even in the presence of an A, seems to confer a bone density advantage, and that haplotype pairs consisting of an A without a B are significantly more common in the patients than the controls. Although the 3′ UTR can regulate mRNA stability and translation, we have not yet confirmed whether the SNPs themselves regulate osteonectin expression or contribute to the observed differences in BMD. Alternatively, it is possible that the 3′ UTR SNPs could be in linkage disequilibrium with an unidentified key regulatory element regulating gene expression.
Despite the importance of osteonectin in the skeleton and in extraskeletal tissues such as skin, eye, and blood vessels, data on mechanisms regulating its expression are limited [9]. Promoter elements necessary for basal level and cell-type specific expression of osteonectin have been mapped for the human, bovine and murine genes [32-35]. Osteonectin gene transcription can be stimulated by transforming growth factor β in fibroblasts, and induction of osteonectin promoter activity by dexamethasone or retinoic acid has been reported [36, 37]. However, there is little information available on the post-transcriptional regulation of osteonectin. Although fibroblast growth factor-2 has been shown to decrease osteonectin mRNA stability in osteoblasts, the trans-acting factors and 3′ UTR elements necessary for post-transcriptional regulation of osteonectin have not been defined [38].
The present study has several caveats. First, the sample size is not as large as is optimal for identifying additional positive findings, particularly with regard to the more rare alleles (i.e., SNP 1046_G and SNP 1970_G) and 3′ UTR haplotypes (i.e., D and E). We also note that our results only apply to Caucasian men. We do not know if our observations would hold in women or in other ethnic groups. These limitations notwithstanding, the results of this candidate gene study are compelling and warrant further investigation. Our findings highlight osteonectin as a plausible candidate gene contributing to bone mass in Caucasian men. By expanding our cohort size in future studies, by studying women as well as men, by exploring the way this gene segregates with BMD in family members of osteoporotic men, and ultimately, by studying the regulatory function of the osteonectin 3′ UTR in osteoblasts, future work will further define the role of the osteonectin gene in modulating bone density in both disease and health.
Acknowledgements
The authors gratefully acknowledge the expert technical assistance of Jonathan Shubert-Coleman, Ritu Bahl and Catherine Kessler. We thank Dr. Steve Mackey for his careful review of this manuscript and for helpful suggestions. We thank Dr. Mansoor Sarfarazi for his help in initiating the SNP analysis.
Funding This work was supported by NIH grants AR44877 (AMD), DK31775 (DAG), NS27941 (DAG), AG20725 (ESK), as well as MO1RR06192 (University of Connecticut Health Center) and RR00645 (Columbia University Medical Center), and by the Donaghue Medical Research Foundation (AMD) and the National Osteoporosis Foundation/Mazess Research Award (ESK).
Footnotes
Conflicts of interest None.
References
- 1.Stewart TL, Ralston SH. Role of genetic factors in the pathogenesis of osteoporosis. J Endocrinol. 2000;166:235–245. doi: 10.1677/joe.0.1660235. [DOI] [PubMed] [Google Scholar]
- 2.Kurland ES, Rosen CJ, Cosman F, McMahon D, Chan F, Shane E, Lindsay R, Dempster D, Bilezikian JP. Insulin-like growth factor-I in men with idiopathic osteoporosis. J Clin Endocrinol Metab. 1997;82:2799–2805. doi: 10.1210/jcem.82.9.4253. [DOI] [PubMed] [Google Scholar]
- 3.Kurland ES, Chan FK, Rosen CJ, Bilezikian JP. Normal growth hormone secretory reserve in men with idiopathic osteoporosis and reduced circulating levels of insulin-like growth factor-I. J Clin Endocrinol Metab. 1998;83:2576–2579. doi: 10.1210/jcem.83.7.4971. [DOI] [PubMed] [Google Scholar]
- 4.Cardon LR, Garner C, Bennett ST, Mackay IJ, Edwards SM, Cornish J, Hegde M, Murray MAF, Reid IR, Cundy T. Evidence for a major gene for bone mineral density in idiopathic osteoporotic families. J Bone Miner Res. 2000;15:1132–1137. doi: 10.1359/jbmr.2000.15.6.1132. [DOI] [PubMed] [Google Scholar]
- 5.Van Pottelbergh I, Goemaere S, Zmierczak H, Kaufman JM. Perturbed sex steroid status in men with idiopathic osteoporosis and their sons. J Clin Endocrinol Metab. 2004;89:4949–4953. doi: 10.1210/jc.2003-032081. [DOI] [PubMed] [Google Scholar]
- 6.Evans SF, Davie MWJ. Low body size and elevated sex-hormone binding globulin distinguish men with idiopathic vertebral fracture. Calcif Tissue Int. 2002;70:9–15. doi: 10.1007/s00223-001-2018-6. [DOI] [PubMed] [Google Scholar]
- 7.Kurland ES, Khosla S, Colvin TL, Heller SL, Powell J, Seltzer B, McMahon D, Bilezikian JP. The relative role of the sex steroids in idiopathic osteoporosis in men. J Bone Miner Res. 2003;18(Suppl):SA331. abstract. [Google Scholar]
- 8.Giguere Y, Rousseau F. The genetics of osteoporosis: “complexities and difficulties”. Clin Genet. 2000;57:161–169. doi: 10.1034/j.1399-0004.2000.570301.x. [DOI] [PubMed] [Google Scholar]
- 9.Bradshaw AD, Sage EH. SPARC, a matricellular protein that functions in cellular differentiation and tissue response to injury. J Clin Invest. 2001;107:1049–1054. doi: 10.1172/JCI12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barker TH, Baneyx G, Cardo-Vila m, Workman GA, Weaver M, Menon PM, Dedhar S, Rempel SA, Arap W, Pasqualini R, Vogel V, Sage EH. SPARC regulates extracellular matrix organization through its modulation of integrin-linked kinase. J Biol Chem. 2005;280:36483–36493. doi: 10.1074/jbc.M504663200. [DOI] [PubMed] [Google Scholar]
- 11.Delany AM, Amling M, Priemel M, Howe C, Baron R, Canalis E. Osteopenia and decreased bone formation in osteonectin-null mice. J Clin Invest. 2000;105:915–923. doi: 10.1172/JCI7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kessler C, Delany AM. Increased notch 1 expression and attenuated stimulatory G protein coupling to adenylyl cyclase in osteonectin-null osteoblasts. Endocrinology. 2007;148:1666–1674. doi: 10.1210/en.2006-0443. [DOI] [PubMed] [Google Scholar]
- 13.Fedarko NS, Moerike M, Brenner R, Gehron Robey P, Vetter U. Extracellular matrix formation by osteoblasts from patients with osteogenesis imperfecta. J Bone Miner Res. 1992;7:921–930. doi: 10.1002/jbmr.5650070809. [DOI] [PubMed] [Google Scholar]
- 14.Fedarko NS, Gehron Robey P, Vetter UK. Extracellular matrix stoichiometry in osteoblasts from patients with osteogenesis imperfecta. J Bone Miner Res. 1995;10:1122–1129. doi: 10.1002/jbmr.5650100718. [DOI] [PubMed] [Google Scholar]
- 15.Muriel MP, Bonaventure J, Stanescu R, Maroteaux P, Guenet JL, Stanescu V. Morphological and biochemical studies of a mouse mutant (fro/fro) with bone fragility. Bone. 1991;12:241–248. doi: 10.1016/8756-3282(91)90070-y. [DOI] [PubMed] [Google Scholar]
- 16.Fisher LW, Drum MA, Gehron Robey P, Conn KM, Termine JD. Osteonectin content in human osteogenesis imperfecta bone shows a range similar to that of two bovine models of OI. Calcif Tissue Int. 1987;40:260–264. doi: 10.1007/BF02555258. [DOI] [PubMed] [Google Scholar]
- 17.Kessler C, Delany AM. Osteonectin/SPARC is critical for anabolic response to PTH in the skeleton. J Bone Miner Res. 2006;21S1:S134. [Google Scholar]
- 18.Delany AM, Kalajzic I, Bradshaw AD, Sage EH, Canalis E. Osteonectin-null mutation compromises osteoblast formation, maturation and survival. Endocrinology. 2003;144:2588–2596. doi: 10.1210/en.2002-221044. [DOI] [PubMed] [Google Scholar]
- 19.Mendell JT, Dietz HC. When the message goes awry: disease-producing mutations that influence mRNA content and performance. Cell. 2001;107:411–414. doi: 10.1016/s0092-8674(01)00583-9. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, Tan FK, Reveille JD, Wallis D, Milewicz DM, Ahn C, Wang A, Arnett FC. Association of novel polymorphisms with expression of SPARC in normal fibroblasts and with susceptibility to scleroderma. Arthritis Rheum. 2002;46:2900–2999. doi: 10.1002/art.10601. [DOI] [PubMed] [Google Scholar]
- 21.Lagan AL, Pantelidis P, Renzoni EA, Fonseca C, Beirne P, Taegtmeyer AB, Denton CP, Black CM, Wells AU, duBios RM, Welsh KI. Single-nucleotide polymorphisms in the SPARC gene are not associated with susceptibility to scleroderma. Rheumatology. 2005;44:197–201. doi: 10.1093/rheumatology/keh460. [DOI] [PubMed] [Google Scholar]
- 22.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformantics methods and protocols: methods in molecular biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 23.Evans AL, Brice G, Sotirova V, Mortimer P, Beninon J, Burnand K, Rosbotham J, Chile A, Sarfarazi M. Mapping of primary congenital lymphedema to the 5q35.3 region. Am J Hum Genet. 1999;64:547–555. doi: 10.1086/302248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassam BJ, Caitano-Annoles G, Gresshoff PM. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- 25.Barrett-Connor E, Siris ES, Wehren LE, Miller PD, Abbott TA, Berger ML, Santora AC, Sherwood LM. Osteoporosis and fracture risk in women of different ethnic groups. J Bone Miner Res. 2005;20:185–194. doi: 10.1359/JBMR.041007. [DOI] [PubMed] [Google Scholar]
- 26.Swaroop A, Hogan BL, Franke U. Molecular analysis of the cDNA for human SPARC/osteonectin/BM-40: sequence, expression and localization of the gene to chromosome 5q31–33. Genomics. 1988;2:37–47. doi: 10.1016/0888-7543(88)90107-3. [DOI] [PubMed] [Google Scholar]
- 27.Villarreal XC, Mann KG, Long GL. Structure of human osteonectin based upon analysis of cDNA and genomic sequences. Biochemistry. 1989;28:6483–6491. doi: 10.1021/bi00441a049. [DOI] [PubMed] [Google Scholar]
- 28.Bolander ME, Young MF, Fisher LW, Yamada Y, Termine JD. Osteonectin cDNA sequence reveals potential binding regions for calcium and hydroxyapatite and shows homologies with both a basement membrane protein (SPARC) and a serine proteinase inhibitor. Proc Natl Acad Sci USA. 1988;85:2919–2923. doi: 10.1073/pnas.85.9.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McVey JH, Nomura S, Kelly P, Mason IJ, Hogan BL. Characterization of the mouse SPARC/osteonectin gene. J Biol Chem. 1988;263:11111–11116. [PubMed] [Google Scholar]
- 30.Liu Y-J, Shen H, Xiao P, Xiong D-H, Li L-H, Recker RR, Deng H-W. Molecular genetic studies of gene identification for osteoporosis. A 2004 update. J Bone Miner Res. 2006;21:1511–1535. doi: 10.1359/JBMR.051002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willing M, Sowers M, Aron D, Clark MK, Burns T, Bunten C, Crutchfield M, D'Agostino D, Jannausch M. Bone mineral density and its change in white women: estrogen and vitamin D receptor genotypes and their interaction. J Bone Miner Res. 1998;13:695–705. doi: 10.1359/jbmr.1998.13.4.695. [DOI] [PubMed] [Google Scholar]
- 32.Ibaraki K, Gehron Robey P, Young MF. Partial characterization of a novel “GGA” factor which binds to the osteonectin promoter in bovine bone cells. Gene. 1993;130:225–232. doi: 10.1016/0378-1119(93)90423-z. [DOI] [PubMed] [Google Scholar]
- 33.Hafner M, Zimmermann K, Pottgiesser J, Kreig T, Nischt R. A purine-rich sequence in the human BM-40 gene promoter region is a prerequisite for maximum transcription. Matrix Biol. 1995;14:733–741. doi: 10.1016/s0945-053x(05)80016-2. [DOI] [PubMed] [Google Scholar]
- 34.Dominguez P, Ibaraki K, Gehron Robey P, Hefferan TE, Termine JD, Young MF. Expression of the osteonectin gene potentially controlled by multiple cis- and trans-acting factors in cultured bone cells. J Bone Miner Res. 1991;6:1127–1136. doi: 10.1002/jbmr.5650061015. [DOI] [PubMed] [Google Scholar]
- 35.Nomura S, Hashmi S, McVey JH, Ham J, Parker M, Hogan BLM. Evidence for positive and negative regulatory elements in the 5′-flanking sequence of the mouse SPARC (osteonectin) gene. J Biol Chem. 1989;264:12201–12207. [PubMed] [Google Scholar]
- 36.Wrana JL, Overall CM, Sodek J. Regulation of the expression of a secreted acidic protein rich in cysteine (SPARC) in human fibroblasts by transforming growth factor β. Eur J Biochem. 1991;197:519–528. doi: 10.1111/j.1432-1033.1991.tb15940.x. [DOI] [PubMed] [Google Scholar]
- 37.Ng KW, Manji SS, Young MF, Findlay DM. Opposing influences of glucocorticoid and retinoic acid on transcriptional control in preosteoblasts. Mol Endocrinol. 1989;3:2079–2085. doi: 10.1210/mend-3-12-2079. [DOI] [PubMed] [Google Scholar]
- 38.Delany AM, Canalis E. Basic fibroblast growth factor destabilizes osteonectin mRNA in osteoblasts. Am J Physiol. 1998;274:C734–C740. doi: 10.1152/ajpcell.1998.274.3.C734. Cell Physiol. 43. [DOI] [PubMed] [Google Scholar]