Abstract
Ion channel subunits encoded by KCNQ1 and KCNE1 produce the slowly activating K+ current (IKs) that plays a central role in myocardial repolarization. The KCNQ1α-subunit and the KCNE1 β-subunit assemble with their membrane-spanning segments interacting resulting in transformation of channel activation kinetics. We recently reported a functional interaction involving C-terminal portions of the two subunits with ensuing regulation of channel deactivation. Here we provide evidence characterizing a physical interaction between the C-termini of KCNQ1 and KCNE1. When expressed in cultured cells the C-terminus of KCNE1 (KCNE1-CT) co-localized with KCNQ1, co-immunoprecipitated with KCNQ1 and perturbed deactivation kinetics of the KCNQ1 currents. Purified KCNQ1 C-terminus (KCNQ1-CT) and KCNE1-CT physically interacted in pull-down experiments indicating a direct association. Deletion-analysis of KCNQ1-CT indicated that KCNE1-CT binds to a KCNQ1 region just after the last transmembrane segment but N-terminal to the tetramerization domain. Surface plasmon resonance (SPR) corroborated the pull-down results showing that the most proximal region (KCNQ1 a.a.349–438) contributed most to the bimolecular interaction with dissociation constant of ~4μM. LQT mutants of KCNE1-CT, D76N and W87F retained binding to KCNQ1-CT with comparable affinity, indicating that these disease-causing mutations do not alter channel behavior by disruption of the association. Several LQT mutations involving the C-terminus of KCNQ1 however, showed varying effects on KCNQ1/KCNE1 association. Our results indicate that the C-termini of KCNQ1 and KCNE1 comprise an independent interaction domain that may play a role in IKs channel regulation that is potentially affected in some Long QT Syndrome mutations.
Introduction
KCNQ1 (Kv7.1) is a voltage-gated K+ channel that performs diverse physiological functions in a variety of cell types [1]. KCNE1, a single transmembrane domain peptide, assembles with the KCNQ1 to form the slowly activating cardiac delayed rectifier current (IKs), which is essential for normal myocardial repolarization (Figure 1A&B) [2, 3]. Mutations in KCNQ1 and KCNE1 genes that result in a loss of function of the channel by various means are a major cause of the inherited Long QT Syndrome (LQTS)1 characterized by ventricular arrhythmia, syncope, and sudden death (often triggered by exercise) [4, 5].
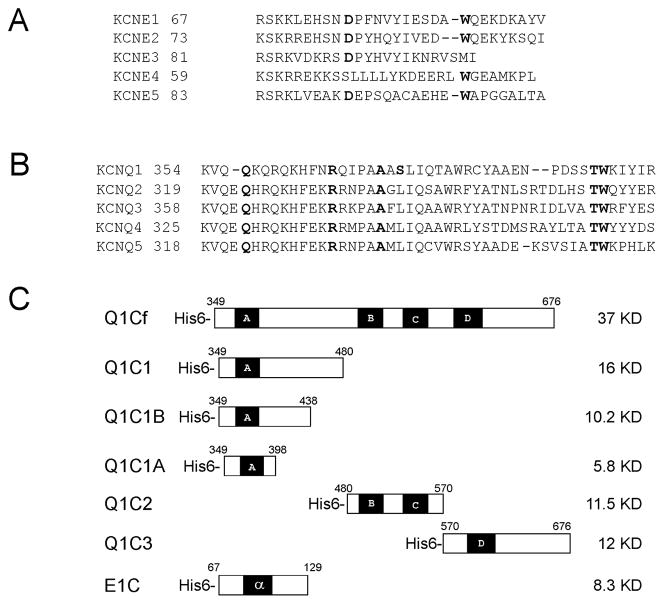
Figure 1. KCNQ and KCNE family K channel subunits.
(A) Sequence alignment of KCNE1 cytoplasmic domains with C-termini of the other KCNE family members. (B) Alignments of the partial amino acid sequences of KCNQ1 C-terminal region containing helix A compared to other KCNQ family members. (C) Schematic representation of recombinant constructs use in this study. Q1Cf refers to the full-length KCNQ1-C-terminus and Q1C1, Q1C2, Q1C3, Q1C1A, Q1C1B each refer to truncated Versions as illustrated. E1C refers to the KCNE1-C-terminus. Boldface residues are those subjected to mutagenesis. All recombinant proteins were expressed in E. coli BL21 (DE3) pLsS strains, and purified with a Ni-affinity column followed by Superdex 200 gel filtration chromatography. (Numbers indicate amino acid position; black and shaded boxes represent α-helical segments)
KCNQ1 is a member of the Kv7 channel subfamily that shares a common topology consisting of six transmembrane domains with intracellular N- and C-termini (Figure 1A). Individual channels are comprised of tetramers of KCNQ subunits with or without closely associated KCNE subunits for regulation. The C-termini of KCNQ channels (Figure 1B) are thought to posses multifunctional domains and play an essential role in K+ channel biological processes including channel folding, assembly, trafficking, and gating [6–10]. Many LQTS mutations in the C-terminus of KCNQ1 result in hereditary cardiac arrhythmia syndromes including the autosomal dominant Romano-Ward Syndrome and the autosomal recessive Jervell and Lange-Nielsen. Primary sequence analysis predicts that the C-terminus of KCNQ family contains four α-helices (Figure 2A). Helices A and B harbor binding sites for calmodulin (CaM) that is required for the proper folding of the channel C-terminus [8, 11, 12]. Confocal immunocytofluorescence analysis suggests that helix C may be involved in channel trafficking [10]. The tetramerization domain of the KCNQ channels is located within helix D of the C-terminus [9, 10].
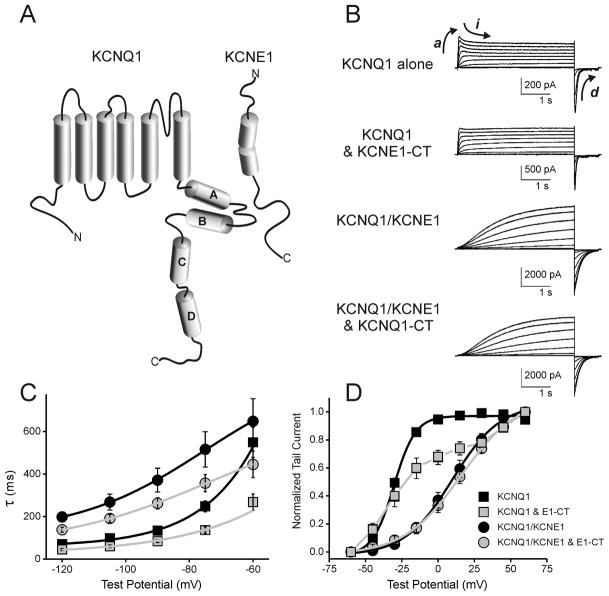
Figure 2. Functional interaction of free-KCNE1-CT with KCNQ1 channels.
(A) Schematic Diagram of KCNQ1 and KCNE1 channel subunits with cylinders representing predicted α-helices. (B) Voltage clamp current traces (elicited by depolarizing voltage steps) of KCNQ1 alone, KCNQ1/KCNE1-CT, KCNQ1/KCNE1 and KCNQ1/KCNE1/KCNE1-CT channels expressed in CHO cells. (C) Deactivation rates plotted against voltage from CHO cells expressing KCNQ1 either without KCNE1, or with KCNE1, KCNE1-CT, or with both KCNE1 and KCNE1-CT. (D) Normalized voltage-dependence of activation curves from same cells as in panel C.
As a potassium ion channel ancillary subunit, KCNE1 modulates gating of KCNQ1 and other K+ channels. Previous studies have shown that the transmembrane region of KCNE1 adopts an α-helical structure [13], and interacts with the KCNQ1 pore-lining S6 domain to modulate channel activity [14]. Additional studies have identified the specific amino acids within the transmembrane region that impact KCNQ1 activation gating and differentiate between KCNE1- and KCNE3-mediated effects [14–17]. A functional interaction between the C-termini of KCNQ1 and KCNE1 was first suggested by chimera studies we conducted between KCNQ1 and KNCQ4 in which full recapitulation of KCNE1-dependent regulation required both the KCNQ1 pore structures and C-terminus [14]. We also reported results supporting a functional interaction between KCNQ1 and KCNE1 C-termini that regulates channel deactivation [18]. In that study we also showed that recombinant KCNE1-C-terminus (KCNE1-CT) co-precipitated with full-length KCNQ1. Recently, Haitin and colleagues showed that the C-terminus of KCNQ1 (KCNQ1-CT) co-immunoprecipitated with C-terminal KCNE1 in vitro and provided FRET evidence for a dynamic interaction during channel gating [19].
The KCNE family sequence homology is primarily limited to a few conserved or identical residues in the transmembrane segment and the proximal intracellular segment (Figure 1A). Portions more C-terminal in the cytoplasmic tail (after KCNE F78) diverge within the KCNE family. LQTS mutations within the C-terminus of KCNE1, (S74L, D76N, Y81C, and W87F) reduce IKs current [5, 20], suggesting that this region of the protein is involved in subunit interaction. Chimeric and deletion analysis furthermore supports the importance of KCNE1 carboxyl terminus for KCNQ1 gating [21]. Compared to the considerable knowledge concerning precise physical and functional interactions between transmembrane segments of KCNQ1 and KCNE1, less is understood with respect to C-terminal interaction between these proteins; and lack of three-dimensional structure of the KCNQ1/KCNE1 channel complex limits full understanding of the molecular basis of subunit interaction, and mechanism of gating modulation. In the present study, we used biophysical and biochemical methods to characterize the physical interaction between the C-terminal cytoplasmic domains of KCNQ1 and KCNE1.
Materials and Methods
Cloning and Expression of KCNQ1 and KCNE1 C-termini in E. coli
The DNA fragments encoding KCNQ1 C-terminus (Q1Cf, Q1C1, Q1C1A, Q1C1B, Q1C2, and Q1C3; graphically illustrated in Figure 1C) and KCNE1 C-terminus were obtained by PCR amplification of the human KCNQ1 and KCNE1 genes using Pfu DNA polymerase and the primers containing Nde I and Hind III restriction sites. The PCR products were cloned into an Nde I- and Hind III - digested pET23a(+) plasmid (Novagen) such that the HIS6-tag was followed by a linker (ELAA) and the KCNQ1 fragment. The amino acid boundaries for each fragment are as follows: Q1Cf 349–676; Q1C1 349–480; Q1C1A 349–398; Q1C1B 349–438; Q1C2 480–570; and Q1C3 570–676. The maltose binding protein (MBP) fusion proteins of KCNQ1 (MBP-KCNQ1) MBP-Q1Cf, MBP-Q1C1, MBP-Q1C2 and MBP-Q1C3, were constructed by cloning the DNA fragments of Q1Cf, Q1C1, Q1C2, and Q1C3 into EcoR I- and Hind III-digested pMAL-2C vector. These recombinant plasmids were expressed in the E. coli strain BL21 (DE3) pLysS grown at 37°C to an A600 of 0.5 in LB medium containing 50 μg/mL carbenicillin, and 34 μg/mL chloramphenicol. Cultures were induced with 0.5 mM IPTG and growth was continued for an additional 6–8 hours at 25°C.
Human KCNE1 gene was obtained by PCR amplification of the human heart cDNA using Pfu DNA polymerase and the primers containing Nde I and Hind III restriction sites, respectively and a His6 affinity tag. The PCR products were cloned into an Nde I- and Hind III - digested pET23a(+) plasmid (Novagen). The recombinant plasmid was expressed using Expressway Cell-Free E. coli Expression System (Invitrogen) according to the manufacturer’s instructions. The protein synthesis reaction mixture was centrifuged at 4°C at 20,000 g for 15 minutes. The pellet was suspended in 20 mM Tris, 150 mM NaCl, pH 7.2, and centrifuged three times at 4°C for 15 minutes. The pellet was suspended in binding buffer (20 mM Tris, 150 mM NaCl, pH 7.2, 8 M Urea, 0.1% (w/v) SDS) and centrifuged at 25 °C at 20,000 g for 15 minutes to remove insoluble debris. The supernatant containing solubilized KCNE1 product was incubated with Ni(II)-NTA resin, which was shacked at 25 °C for 1 ~ 2 hours. The resin was then packed into a gravity-flow column and washed with 10 fold bed volumes of binding buffer, followed by washing with 5 fold bed volumes of wash buffer (20 mM Tris-HCl, 150 mM NaCl, pH 7.2, 0.5% DDM, 5 mM β-mercaptoethanol). KCNE1 product was then eluted using a wash buffer containing 200 mM imidazole, pH 6.2.
Mutagenesis
The LQT mutants of KCNE1 C-terminus (D76N and W87F), and that of KCNQ1 (Q357R, R366W, A371T, S373P, T391I and W392R) were generated using the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions and pET23a(+):E1C as the template. The mutants were sequenced in their entirety to ensure that no undesired mutations occurred. The mutants were expressed in the E. coli strain BL21 (DE3) pLysS cells as above.
Expression of KCNQ1, C-termini of KCNQ1 and KCNE1 in HEK and CHO cells
Myc-tagged human KCNQ1 expression vector and HEK 293 cells stably expressing KCNQ1 (HQ5) were generated as previously described [18]. The DNA fragments encoding KCNE1 C-terminus and KCNQ1 C-terminus were amplified by PCR with Taq DNA polymerase, and cloned into p3XFLAG-CMV-10 vector (Sigma) to generate N-terminal Flag-tagged KCNE1 C-terminus and N-terminal Flag-tagged KCNQ1 C-terminus expression plasmids. For electrophysiological experiments CHO cells were chosen for their low electrical background.
HEK 293 cells were grown in 5% CO2 humidified atmosphere at 37°C in RPMI 1640 medium (Mediatech) containing 10% fetal calf serum (HyClone) and 100 units/ml penicillin and 100 μg/ml streptomycin (Mediatech). CHO cells were grown in Ham’s F-12 (Mediatech Inc.) supplemented with 10% FBS (Hyclone) and Penicillin/Streptomycin (Mediatech Inc.). HQ5 cells, a HEK 293 clonal cell line stably expressing KCNQ1, were transfected with Flag-tagged KCNE1 C-terminus expression plasmids using Lipofectamine 2000 (Invitrogen). HEK 293 and CHO cells were co-transfected with Flag-tagged KCNQ1 C-terminus and Flag-tagged KCNE1 C-terminus using Lipofectamine 2000 (Invitrogen).
Electrophysiology
Transfected CHO cells were grown on sterile glass coverslips and placed in an acrylic/polystyrene perfusion chamber (Warner Instruments) mounted in an inverted microscope outfitted with fluorescence optics and patch pipette micromanipulators. The external bath consisted of NaCl 150 mM, CaCl2 1.8 mM, KCl 4 mM, MgCl2 1 mM, glucose 5 mM, and HEPES buffer 10 mM (pH 7.4) at room temperature (20–22°C). The internal pipette solution contained KCl 126 mM, K-ATP 4 mM, MgSO4 1 mM, EGTA 5 mM, CaCl2 0.5 mM, and HEPES buffer 25 mM (pH 7.2) at room temperature. The whole-cell configuration of the patch clamp technique was used to measure potassium currents using a MultiClamp 700B patch-clamp amplifier controlled via PC running pCLAMP10 acquisition and analysis software (Axon Instruments). Pipette tips were heat-polished to obtain a resistance of 2–3 mΩ in the test solutions. The pipette offset potential in these solutions was zeroed just prior to seal formation. The junction potential for these solutions was calculated between 3–4mV (by pClamp analysis software) and was not corrected for analyses. Whole cell capacitance (generally 10–25pF) was compensated electronically through the amplifier. Whole cell series resistance of 6–12MΩ was compensated to 75–90% by amplifier circuitry such that the voltage errors for currents of 2nA were always less than 6 mV. Data was filtered using an 8-pole Bessel filter at 1 kHz. Activation kinetics were quantified as a rise time to half max following an 60 mV depolarization from −80 mV. Deactivation kinetics were fit to a single exponential. For the inactivation in KCNQ1 homotetrameric channels we compared the maximal outward current amplitude just after depolarization (between 0–200msec) to the current amplitude between 400–800msec. Inactivation of KCNQ1 currents was quantified as the percentage of maximal current after depolarizing step to 60mV subtracted by the current remaining after 250msec of sustained depolarization.
Purification and Analysis of the proteins
Cells (~10 grams) were suspended in 40mL of 50mM Tris-HCl, pH 7.8, 150mM NaCl, 1mM β-mercaptoethanol containing protease inhibitors (Boehringer Mannheim) and 100μg/mL of DNase I. After sonication, cells debris was removed by centrifugation for 45 min at 15 000 rpm at 4°C. The supernatant was dialyzed against 50mM Tris-HCl containing 150mM NaCl, 1mM β-mercaptoethanol, pH 7.8, for 2 hours at 4°C. After centrifugation, the supernatant was applied to a 4mL Ni2+-NTA His-Bind affinity column (Qiagen), and proteins were eluted with a linear gradient (25 to 250mM) of imidazole at 1mL/min. Correct size fractions were pooled, concentrated to 4mL, and loaded onto a 1.6 × 70 cm Superdex 200 gel filtration column. The proteins were eluted with 50mM Tris-HCl containing 150mM NaCl, 1mM β-mercaptoethanol, pH 7.8, at 0.5mL/min and the correct size fractions were pooled and concentrated.
MBP-KCNQ1 C-terminal fragments were purified by amylose resin column. Briefly, cells (~10 grams) were suspended in MBP buffer, 20mM Tris-HCl, pH 7.4, 200mM NaCl, 1mM EDTA, and 1mM β-mercaptoethernol containing protease inhibitors (Boehringer Mannheim), 100μg/mL of DNase I. Cells were lysed by sonication, and debris was removed by centrifugation. The supernatant was loaded onto 3mL amylose column (New England BioLabs), and the proteins were eluted with 3 column volumes of MBP buffer containing 10mM maltose. After concentration to 5mL, the proteins were applied to a 1.6 × 70 cm Superdex 200 gel filtration column. The proteins were eluted with 20mM Hepes containing 150mM NaCl, 5mM KPO4, 1mM β-mercaptoethanol, pH 7.8, at 0.5mL/min and the correct size fractions were pooled and concentrated. Protein concentration was measured using a Bio-Rad protein assay kit with bovine serum albumin as a standard. The purity of proteins was determined by SDS-polyacrylamide gel electrophoresis according to the method of Laemmli. Immunoblot assay was performed to confirm the specificity of each peptide. The native molecular weight was estimated by using a Superdex-200 gel filtration column calibrated using Bio-Rad molecular weight markers. Electrospray ionization/mass spectrometry was performed on the purified protein to determine the subunit molecular mass of KCNE1 C-terminus and KCNQ1 C-terminal fragments.
Immunoprecipitation
5 μg of KCNQ1 C-terminus was incubated with 5 μg KCNE1 C-terminus in 50mM Tris-HCl, pH 7.5, 130mM KCl, 1mM β-mercaptoethanol for 20 minutes, and goat anti-KCNQ1 C-terminal specific antibody was added in a volume of 500 μl and gently agitated for 4 hours at 4°C. The immunocomplexes were incubated with protein A-agarose beads for 2 hours and washed three times with the suspension buffer. The protein complexes were eluted in 2x sample loading buffer and resolved in 4–20% SDS-PAGE followed immunoblot analysis using anti KCNE1 antibody.
MBP Pull-down Assay
10 μg MBP fusion proteins, MBP-Q1Cf, -Q1C1, -Q1C2 and -Q1C3, were incubated with 10 μg KCNE1 C-terminus in 20mM Tris-HCl, pH 7.5, 150mM KCl, 1mM EDTA and 1mM β-mercaptoethanol for 10 minutes, respectively. 50 μl of amylose beads was added to each reaction and incubated for 2 hours at 4°C with gentle agitation. After 5 washes with MBP buffer, the bound proteins were eluted in MBP buffer containing 10mM maltose and examined by SDS-PAGE followed by immunoblot analysis using KCNE1 C-terminal specific antibody.
Immunoprecipitation and Western blotting
72-hours post-transfection, cells were lysed with ice-cold NDET buffer (1% NP-40, 0.4% Deoxycholic acid, 5 mM EDTA, 25 mM Tris,150 mM NaCl, pH 7.5) with complete protease inhibitor cocktail (Roche) for 15 minutes. Cell lysates were centrifuged at 13,000 rpm at 4°C for 10 minutes, and the supernatants were incubated with a 1:100 dilution of the anti-KCNQ1 antibody (Santa Cruz Biotechnology) for 2 hours at room temperature. Protein-G-agarose (Pierce) was used to capture antibody–protein complexes, and bound proteins were eluted with SDS-PAGE loadingbuffer. Proteins were separated by 4–20% SDS-PAGE and transferred to nitrocellulose membranes.
The nitrocellulose membranes were blocked with Tris-buffered saline (TBS) containing 10%dry milk for 30 minutes, and then incubated with a 1:5000 dilution of the anti-FLAG antibody (Sigma) for 2 hours at room temperature. The membrane was washed with TBS containing 0.5% Tween-20 (TBS-T) and then incubated with horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Pierce) for 1-hour. Immunoreactivity was detected by ECL kit (Amersham).
Immunofluorescence and Confocal Microscopy
HEK 293 cells were transfected as above and grown on glass-bottomed 35-mm dishes. 48-hours after transfection cells were fixed with 4% paraformaldehyde for 20-minutes, permeabilized with 0.3% Triton X-100 in PBS for 5 minutes and blocked with PBS containing5% bovine serum albumin for 30 minutes at room temperature. Fixed and blocked specimens were then incubated with rabbit anti-Flag antibody (Sigma. dilution 1:1000), goat anti-Myc antibody (Santa Cruz. dilution 1:100) and mouse>cadherin antibody (Abcam. Dilution 1:100) in PBS containing 5% BSA and 0.1% Triton X-100 at room temperature for 2-hours. After 3-washes with PBS, the cells were incubated with Alexa 488 donkey anti-rabbit, Alexa 568 donkey anti-goat, and Alexa 647 donkey anti-mouse secondary antibodies (Molecular Probes. dilution 1:8000) in PBS buffer for 1 hour. Immunofluorescence was visualized using the Leica (Deerfield, IL) AOBS confocal microscope.
Surface Plasmon Resonance Analysis
Quantitative interaction studies were carried out on a Biacore 3000 instrument (Biacore, Inc., Piscataway, NJ) using running buffer (20mM KPO4, 130mM KCl, 3.4mM EDTA and 0.005% Tween 20). KCNE1 C terminus or E1C mutants D76N and W87F (ligand) were immobilized on biosensor chip CM5, and KCNQ1 C terminal fragments, Q1Cf, Q1C1, Q1C1A, Q1C1B, Q1C2, Q1C3, or Q1C1 mutants (analytes) were injected over this interaction surface, respectively. An equivalent volume of each protein sample (analyte) was injected over a chip surface with no protein immobilized to serve as a blank phase for the background subtraction. MBP was injected over the surface with immobilized KCNE1 C terminus as negative controls. For kinetic analysis, different concentrations of analytes (0.1 – 12.5 μM) were tested. Data were analyzed using BIAevaluation software (GE Healthcare). The binding curves of various Q1C concentrations were fitted to the two-state binding model described by the equation 1 [22].
| (1) |
where A and B represent the analyte (Q1C domain) and the ligand (E1C domain), AB* and AB represent an initial complex (transition state) and final docked complex (conformational change), respectively. The equilibrium constants of each binding steps are:
| (2) |
And the overall equilibrium binding constant (KA) is calculated as
| (3) |
The binding response (R) of the sensorgram for each reaction was subsequently plotted as a function of analyte (Q1C) concentration (A) to determine the overall binding affinity. The data were fitted to equation 4 [23]:
| (4) |
where Rmax is the maximum binding response value and Kd is the dissociation constant.
Results
Functional evidence of isolated KCNE1 C-terminal interaction with KCNQ1 channels
To determine if the C-terminus of KCNE1 could interact with and affect KCNQ1 channels, in situ, we co-transfected full length KCNQ1 with KCNE1 C-terminus lacking the transmembrane segment into CHO cells. Subsequent voltage clamp analysis showed that the C-terminus of KCNE1 had functional impact on the KCNQ1 K+ currents (Figure 2). The addition of free KCNE1 peptide accelerated the voltage-dependent deactivation of both KCNQ1 alone and KCNQ1/KCNE1 channels (Figure 2B,C. p<0.01 between KCNQ1 and KCNQ1/E1-CT at all voltages; p<0.05 between KCNQ1/KCNE1 and KCNQ1/KCNE1/E1-CT at −120mV). Additional changes included reduction in voltage-dependent-inactivation of KCNQ1 channels, measured as the time-dependant relaxation from peak outward (illustrated by arrow marked i in Figure 2B) current during a sustained depolarizing step to 60mV (20±0.03% current inactivation for KCNQ1 versus 5±0.03%, p<0.01, n=6). Effects of free E1-CT expressed with KCNQ1 upon the voltage-dependence of activation were more complex and could not be adequately fit to a Boltzman function (Figure 2D). Although it is an indirect measure, such an activation curve could be possible if there were various stoichiometris or saturation of possible interaction sites on the channel. We have previously noted similar results with KCNE1 truncation mutants yielding a mix of currents depending on the saturation of the KCNQ1 with KCNE1 [18]. KCNQ1/KCNE1 channels were not appreciably perturbed by the soluble KCNE1 fragment. Activation kinetics of KCNQ1 or KCNQ1/KCNE1 channels were not significantly altered by the presence of free KCNE1-CT.
Co-immunoprecipitation of C-termini of KCNQ1 and KCNE1, and co-localization of KCNQ1 with the C-terminus of KCNE1 in HEK cells
We have previously determined that the C-terminus of KCNE1 could physically interact with full-length KCNQ1 in transfected cells [18]. To investigate the possible interaction between the C-terminus of KCNE1 and the C-terminus of KCNQ1, we conducted co-immunoprecipitation experiments. KCNE1 C-terminus was co-expressed with KCNQ1 C-terminus in HEK 293 cells. 72-hours after transfection, the cells were lysed and immunoprecipitated with an antibody against KCNQ1. As shown in Figure 3A, KCNE1 C-terminus interacted with KCNQ1 C-terminus, and was immunoprecipitated with the KCNQ1 antibody.
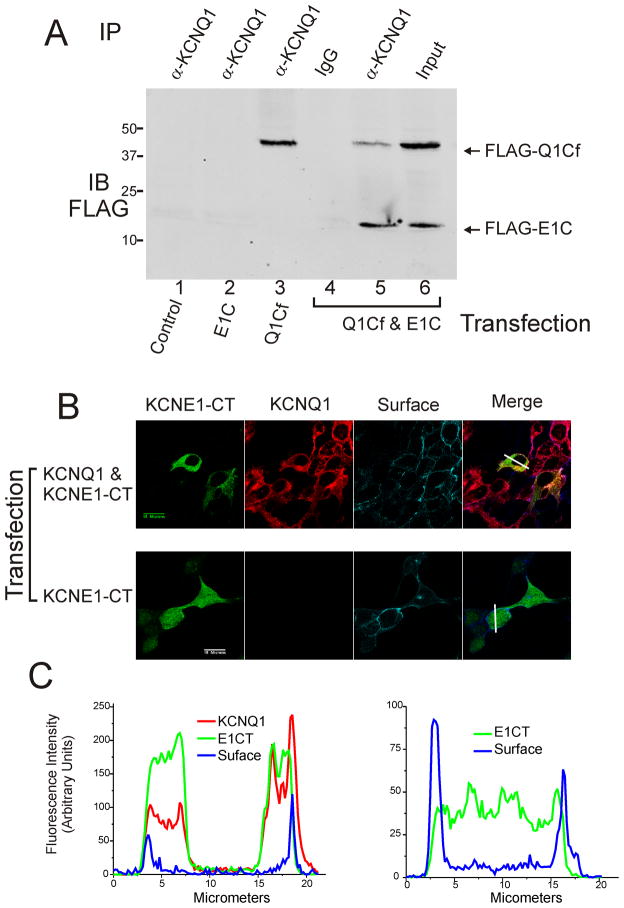
Figure 3.
Co-immunoprecipitation of the C-terminal cytoplasmic regions of KCNQ1 and KCNE1 and co-localization of KCNE1 C-terminus and KCNQ1 in the surface of live HEK cells. (A) Immunoblot analysis from HEK 293 cells co-transfected with E1C (lane 2), Q1Cf (lane 3), or both (lanes 4–6). E1C was co-immunoprecipitated with Q1Cf by anti-KCNQ1 antibody (Santa Cruz) (lane 5). Lane 4 shows results from nonspecific antibody IP and lane 6 shows input lysate prior to IP demonstrating expression levels. (B) HEK 293 cell line stably expressing full-length KCNQ1 channels (top row of micrographs) or HEK 293 cells (bottom row of micrographs) were transfected with Flag-tagged KCNE1 C-terminus expression plasmids, and the subcellular distribution of the KCNE1 C-terminus was detected by immunofluorescence using confocal microscopy. KCNE1-CT staining is shown in green, KCNQ1 in red and cadherin (surface) in blue. (C) Graphical analysis of signal strength across co-transfected cells (indicated by solid white line in right panels of 3B) for KCNQ1 plus KCNE1-CT (on left) and KCNe1-CT without KCNQ1 (on right).
To determine whether KCNE1 C-terminus and KCNQ1 can co-localize in HEK 293 cells, confocal microscopy was used to analyze the cellular distribution of the KCNE1 C-terminus. HQ5 cells (HEK 293 cell line stably expressing KCNQ1) transfected with the FLAG-tagged KCNE1 C-terminus were stained with antibodies to FLAG, KCNQ1 and cadherin (for surface marking)(Figure 3B). KCNE1-C-terminus immunofluorescence staining and that of KCNQ1 were largely overlapping with exclusion of the nucleus (Left panels, Figure 3B,C). When KCNE1-C-terminus was transfected in the parent HEK cell line without KCNQ1 the immunofluorescent distribution was uniform throughout the cell (Right panels of Figures 3B,C). These results were consistent and uniform between two separate transfection and immunofluorescence experiments. No visible immunofluorescence staining was observed on untransfectedHQ5 or HEK cells.
Purification and properties of the recombinant KCNQ1 C-termini and KCNE1 C-terminus
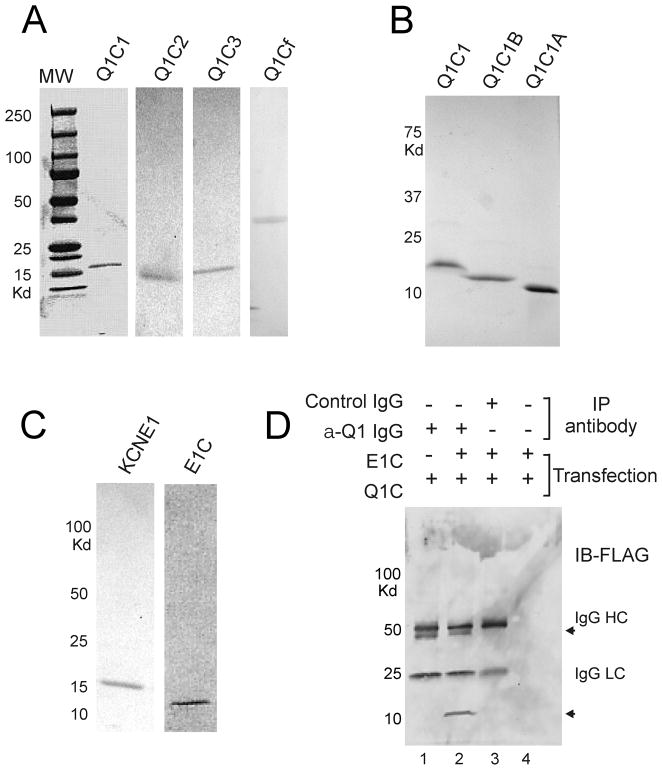
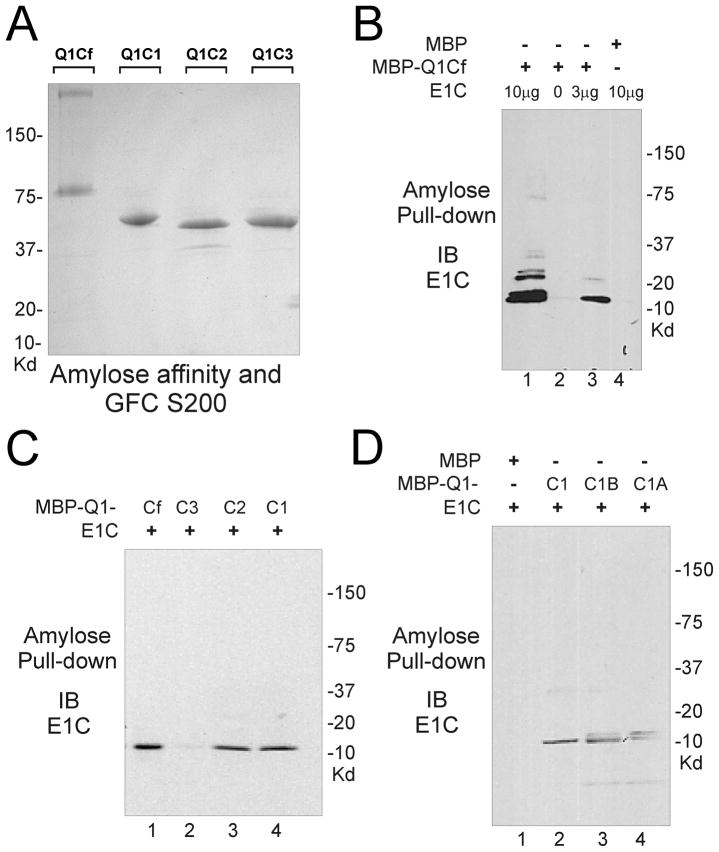
His-tagged human KCNQ1 C-terminal domains and KCNE1 C-terminus were purified from E. coli cells by using Ni2+-NTA His-Bind affinity and Superdex 200 gel filtration column chromatography. Recombinant KCNQ1 fragments were designed to incorporate either the full-length C-terminus (Q1Cf) or smaller domains based on predicted α-helices (Q1C1, Q1C1B, Q1C1A, Q1C2 and Q1C3) as schematically demonstrated in Figure 1C. Subunit molecular mass of His-tagged Q1Cf, -Q1C1, -Q1C1A,-Q1C1B,-Q1C2, -Q1C3, and -E1C recombinant proteins determined by SDS-polyacrylamide gel electrophoresis is 37, 16, 5.8, 10, 11.5, 12 and 8.3 kDa, respectively (Figure 4A–C), consistent with the predicted molecular weight calculated from the amino acid sequences (Figure 1C). A Superose-6 gel filtration column calibrated using Bio-Rad molecular weight markers was used to estimate the molecular weights of native proteins, indicating that Q1Cf, Q1C1, Q1C2 and Q1C3 purify as higher-order oligomers. Gel filtration chromatography on a Superdex-200 column was used to estimate the molecular weight of the native protein as 17 KDa, suggesting the KCNE1 C-terminus purifies as a dimmer.
Figure 4.
Association of purified, recombinant C-terminal cytoplasmic regions of KCNQ1-KCNE1. (A) Coomassie-stained SDS-PAGE of purified recombinant KCNQ1-C-terminal protein fragments, Q1C1, Q1C2, Q1C3, and Q1Cf (1–4). (B) Coomassie-stained SDS-PAGE of purified recombinant proteins, Q1C1, Q1C1B and Q1C1A (1–3). (C) Coomassie-stained SDS-PAGE of purified recombinant protein, KCNE1 (1st lane) and KCNE1 C-terminus (2nd lane). (D) Immunoblot of Co-IP analysis of the interaction between the full length C-termini of KCNQ1 and KCNE1. Reactions included KCNQ1-Cf (all lanes) and KCNE1-CT (lanes 2–4). Anti-KCNQ1 antibody was used for IP in lanes 1 & 2, control antibody in lane 3 and protein-G beads alone in lane 4 for controls. Immunoblots for FLAG-tagged KCNE1-CT and KCNQ1-Cf show that only IP of KCNQ1-Cf co-precipitated KCNE1-CT (arrows indicate KCNQ1-Cf and KCNE1-CT).
MBP-fusion KCNQ1 C-terminal domains were purified from E. coli cells by using amylose affinity column followed by Superdex-200 gel filtration column chromatography. Subunit molecular weight of MBP-fusion proteins was estimated by SDS-PAGE electrophoresis (Figure 5).
Figure 5.
Interaction of KCNQ1 C-terminal cytoplasmic domains with KCNE1 C-terminus. (A) Coomassie-stained SDS-PAGE of purified MBP-fusion proteins, MBP- Q1Cf, MBP-Q1C1, MBP-Q1C2, and MBP-Q1C3. (B) Immunoblot analysis of association of KCNE1 C-terminus with KCNQ1-Cf. Pulldown of MBP-Q1Cf with immobilized amylose co-precipitated KCNE1-CT (lanes 1 & 3) however MBP alone did not pull down KCNE1-CT (lane 4). (C) Immunoblot analysis of MBP-pulldown reaction demonstrates KCNQ1-C-terminal fragments Q1Cf, Q1C2 and Q1C1 each co-precipitate KCNE1-CT but not KCNQ1-C3. (D) Immunoblot analysis of MBP-pulldown reaction demonstrates KCNQ1-C-terminal fragments Q1C1, Q1C1B and Q1C1A each co-precipitate KCNE1-CT.
Interaction between cytoplasmic domains of KCNQ1 and KCNE1 in vitro
To further examine possible physical interactions between KCNE1 and KCNQ1 C-termini we performed immunoprecipitation experiments using purified recombinant proteins after mixing in vitro. Physical binding of KCNQ1 C-terminus with KCNE1 C-terminal region was readily observed using His-tagged fusion proteins (Figure 4D). To further characterize potential binding interactions of KCNQ1 C-terminus with the intracellular sequence of KCNE1, a series of MBP fusion proteins were prepared that contained the subdivided fragments of the KCNQ1 C-terminus (Figure 5A). MBP pull-down assay using immobilized amylose showed that His-tagged KCNE1 C-terminus directly associated with MBP-Q1C1 and MBP-Q1C2 regions but not with MBP-Q1C3 (Figure 5B,C).
To further delineate the binding regions of KCNQ1 C1 for KCNE1 C-terminus, we generated Q1C1A and Q1C1B MBP fusion proteins for MBP binding test (Figures 1C, 4B). Pull-down assay shows that both Q1C1A and Q1C1B bind to KCNE1 cytoplasmic domain (Figure 5D), indicating KCNQ1 C-terminal region (349–398) was sufficient for interaction with His-tagged KCNE1 C-terminus.
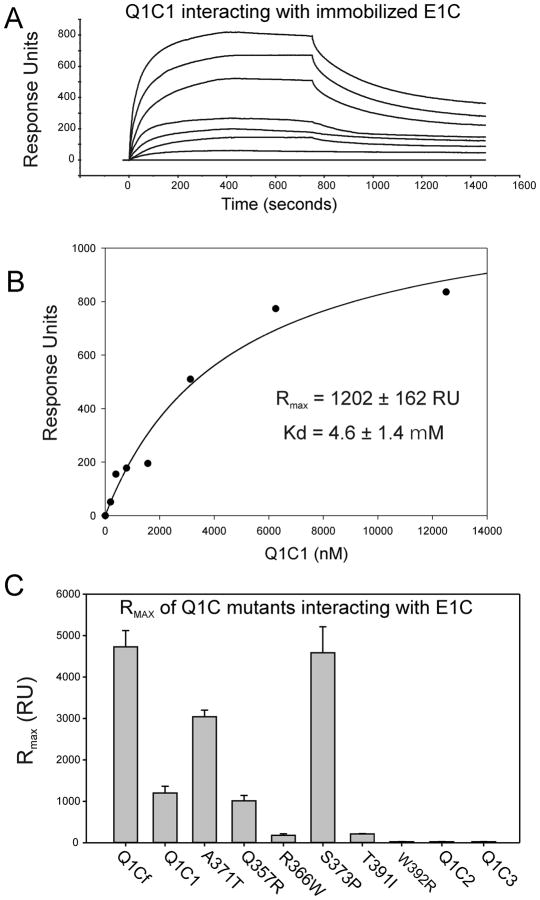
To probe the specificity of the interaction between C-terminal cytoplasmic domains of KCNQ1 and KCNE1 proteins in a more quantitative manner, surface plasmon resonance analysis was performed. KCNE1 C-terminus was immobilized on biosensor chip equilibrated with running buffer, and increasing concentrations of a series of KCNQ1 C-terminal fragments were then run over the chips and binding of the proteins was monitored (Figure 6A,B). The kinetics of binding could not be globally fitted perfectly to a one-to-one or one-to-two binding model. This may reflect a higher oligomeric state or mixed stoichiometry of the purified KCNQ1-CT that could present more than one binding site per with variable affinity dependent on degree of oligomer saturation. Thus, the kinetics of binding determined on the BIAcore may be a summation of interactions between KCNE1 C-terminus on chip surface and oligomeric form of KCNQ1 C-terminal domains in solution. The sensorgrams (as shown in the example in Figure 6A,B) could be fitted to a two-state binding model (as outlined in Materials and Methods) indicating that KCNQ1-Q1C1 associated with KCNE1 involving a conformational change. A steady-state binding response value (R) for each concentration of Q1C1 was subsequently plotted as a function of the concentration of Q1C1 (example in Figure 6B). The data were fitted to equation (4) to derive the dissociation constant (Kd) (Table 1). The kinetic parameters in table 1 shows that the full length KCNQ1 C-terminus (Q1Cf) associated with KCNE1 cytoplasmic domain with estimated Kd of 3.2 ± 0.7 μM. A similar binding reaction was observed on Q1C1 with estimated Kd of 4.6 ± 1.4± μM. Kd for KCNE1 binding to Q1C2 and Q1C3 could not be obtained given the lack of measurable binding as indicated by the negligible RU. These results indicate that Q1C1 region predominantly contributes to the bimolecular interaction. This result is mainly consistent with MBP-pull-down studies that KCNE1-CT can interact with Q1C1 but not Q1C3. The association of KCNE1 C-terminus with Q1C2 observed in MBP-pull-down assay but not in BIAcore analysis may be due to an improper orientation of a binding motif caused by E1C immobilization on a BIAcore sensor chip. To date, we have not been able to successfully immobilize KCNQ1 C-terminal fragments onto the SPR chips due to either precipitation or degradation during the linking reaction. We also tested for KCNE1-C-terminal dimerization by applying solutions of E1C to chips with E1C immobilized and no interaction was observed (data not shown). To test whether the disease-causing LQT mutants of KCNE1 C-terminus impair the binding ability, we performed the binding assay for E1C D76N and W87F to Q1C1, respectively. For these experiments separate SPR chips were constructed with each KCNE1 mutant. Both E1C mutant proteins were still able to bind to Q1C1 with the comparable affinity (Table 2).
Figure 6.
Surface plasmon resonance analysis of the interactions of KCNQ1 C-terminal cytoplasmic domains with KCNE1 C-Terminus. SPR analysis was carried out as described in Materials and Methods. (A) Illustrates a typical sensorgram for the interaction of various concentrations of the Q1C1 fragment of KCNQ1 with E1C immobilized on a CI sensor chip. Each trace represents the response to increasing concentrations of Q1C1. Introduction of each concentration of Q1C1 is started at time=0 and washout at 750 seconds. (The concentration ranges used were 0.5–10 μM for KCNQ1 C-terminal fragments.) (B) A typical plot of sensorgram data is show for a Q1C1-E1C interaction experiment. The data were fitted separately for each experiment to the two-state binding model where A + B ↔ AB* ↔ AB, as described in the Methods section. (C) The graph shows maximum response values (RUmax) for the highest concentration (10μM) of various KCNQ1-C-terminal fragments and LQT1 mutants. These data were used to derive Kd calculations shown in Table 1.
Table 1.
SPR results for KCNQ1 C-terminal domains and mutants binding to Immobilized KCNE1 C-terminus
| Q1CT | Kd (μM) | Rmax (RU) |
|---|---|---|
| Q1Cf | 3.2 ± 0.7 | 4729 ± 391 |
| Q1C1 | 4.6 ± 1.4 | 1202 ± 162 |
| Q1C1 A371T | 6.5 ± 0.7 | 3039 ± 161 |
| Q1C1 Q357R | 2.4 ± 0.9 | 1011 ± 129 |
| Q1C1 R366W | Nd* | 178 ± 39 |
| Q1C1 S373P | 7.0 ± 1.9 | 4586 ± 627 |
| Q1C1 T391I | * ND | 213 ± 10 |
| Q1C1 W392R | * ND | 25 ± 4 |
| Q1C1A | * ND | 56 ± 9.1 |
| Q1C1B | 2.4 ± 0.6 | 936 ± 75 |
| Q1C2 | * ND | <25 |
| Q1C3 | * ND | <25 |
ND Not determined due to low interaction signal.
Table 2.
SPR results for KCNQ1 C-terminal domain binding to immobilized KCNE1 C-terminus and LQT5 mutants of KCNE1.
| Immobilized E1C | Kd | Rmax |
|---|---|---|
| WT | 4.5 ± 1.1 | 102 ± 11 |
| D76N | 3.1 ± 1.5 | 166 ± 32 |
| W87F | 4.1 ± 1.8 | 130 ± 25 |
To more precisely locate the binding site(s) within Q1C1 terminus, we performed SPR analysis of KCNE1 binding to Q1C1, Q1C1A, and Q1C1B. Interactions were detected in Q1C1, Q1C1A and Q1C1B. The Q1C1B fragment showed comparably strong binding as did Q1C1 while Q1C1A bound to KCNE1-CT with greatly reduced Response Units making accurate determination of binding affinity unreliable (Table 1). This result suggests to us that the KCNQ1 C-terminal region encompassing the initial 349–398 amino acids is sufficient for physical interaction with KCNE1 C-terminus. That Q1Cf showed higher RU and affinity than Q1C1 and that Q1C1B showed higher affinity than Q1C1A suggests that additional regions contributed to enhanced or accelerated conformational changes as might occur in a 2-state binding reaction (Equation 1). These contributory regions may include the liner between Helices A and B or Helix B itself. These results are also consistent with MBP-pull down data investigating the same regions (Figure 5C,D).
SPR analysis also showed that KCNQ1 LQT mutants, Q357R, R366W, A371T, S373P, T391I and W392R each exhibited varying association with KCNE1 C-terminus in terms of RU and Kd suggesting that these disease-causing mutations may affect channel function via perturbed interactions with KCNE1 C-terminus (Table 1). Of these, R366W, T391I, W392R exhibited a significant decrease or undetectable RU while Q357R, A371T registered comparable RU values, and S373P showed an increase in RU compared to wild-type Q1C1. The fitting of sensorgrams in the cases of R366W, T391I, W392R was quite poor, resulting in uncertainty for their dissociation constants values (Table 1). The difficulty in fitting these three mutants to two-state model is likely due to low affinity of the interaction resulting from local structural change at the binding interface.
Discussion
Assembly of the KCNE1 β subunit with KCNQ1 α subunits transforms the channel current into IKs and also affects trafficking of the KCNQ1 to the cell surface [2, 3, 24]. This assembly is mediated by the interactions between specific regions of the α and β subunits. The precise region(s) that account for all subunit association and gating regulation are still a matter of intense investigation.
The data we present here characterize a physical and functional interaction between the two K+ channel subunits, KCNQ1 and KCNE1 residing in their intracellular C-termini. The interaction is direct, not requiring other adaptor proteins or the transmembrane segments of the channel subunits. Heterologous expression of soluble KCNE1 C-terminus physically and functionally interacts with KCNQ1 channels, in situ, and perturbs pre-formed KCNQ1/KCNE1 C-terminal interaction as assayed by voltage clamp analysis. Deletion analysis of KCNQ1 indicates that the KCNE1 C-terminus binds robustly to a limited region of ~50 amino acids (349–398) encompassing Helix-A of the C-terminus as measured by both MBP-pulldown studies and SPR analyses. Our results suggest that more C-terminal regions (Helices-B-C or inter-helix linkers) play a role in the interaction with either direct binding (pull-down assays, Figure 5D) or participation in conformational changes in the binding site that allow more rapid association (SPR data, Figure 6, Table 1). Our results are largely in agreement with those of Haitin et al, [19] in terms of C-terminal interaction between KCNQ1 and KCNE1. The main exception being that GST-pulldown experiments in their paper showd that KCNQ1-C-terminal truncations after the proximal C-Helix diminished KCNE1 binding leading to the conclusion the C-Helix was the biding site. Our results suggest that portions A-Helix and its linker to the transmembrane S6 bind KCNE1using both pulldown and SPR assays while the fragment encompassing isolated B- and C-helices binds in solution but not with immobilized KCNE1. Several differences in the experimental approach and analyses may account for apparent discrepancies between the two studies. The KCNQ1-C-terminal fragments were not entirely the same for the two studies and multiple portions of the protein may contribute indirectly to binding efficiency. There may be differences in the solution behavior of recombinant GST- and MBP-fusion proteins compared to more immobilized forms as used for SPR. We also used purification tags (HIS6 or MBP) on the N-termini of the KCNQ1 fragments. The use of SPR (with one ligand immobilized) may allow recognition of qualitative and quantitative differences in protein interactions. Moreover, the Haitin group’s in situ FRET data with full-length C-termini suggest that the interaction may not be static but rather a dynamic process that could be affected by environmental forces (voltage, immobilization, pH and ionic concentrations). At present, further experimentation using additional approaches will be needed to resolve the remaining discrepancies.
Many LQTS mutations have been described within the interaction regions for KCNE1 and KCNQ1 C-termini, suggesting that this interaction may be perturbed by these disease-causing variants. The nature of the perturbation may not be the same for each mutation. This is supported by our finding that the each KCNQ1 mutant exhibited a different SPR binding profile while the KCNE1 mutants had no appreciable effect on binding. These findings may be due to differing sites within the binding interface, or alternatively allosteric effects from sites at a distance from the interaction site. Further structural studies are needed to more fully understand these results. That the KCNE1 mutations (at the conserved D76 and W87 sites) did not perturb SPR profiles suggest that they may qualitatively affect the functional result of the interaction without preventing overall binding to KCNQ1. The FRET data from the Haitan study [19] would suggest that it is the dynamic changes in the interaction that are altered by the D76N mutation rather than binding, per se.
Previous studies have examined, in depth, KCNQ1/KCNE1 interaction sites in the transmembrane segments and extracellular portions [14–16, 21, 24, 25]. The physical interaction interface comprised of the membrane-spanning segments appears to be extensive and the same for KCNE1 and KCNE3 in the KCNQ1 channel. Many other voltage-gated K+ channels associate with KCNE family members. The binding site for KCNE1 within the transmembrane region of various K+ channels however, is not a conserved sequence motif, but probably a common structural conformation encompassing the pore-forming helices. The initial mapping of the functional interaction sites in the membrane-spanning segments of KCNE1 and KCNQ1 showed that activation kinetics were controlled by specific interacting residues [15]. Chimera studies of KCNE1/3 showed that control of deactivation did not co-segregate with regions controlling activation. Functional analysis of the KCNE1 C-terminus subsequently placed deactivation kinetics regulation of KCNQ1 in the KCNE1 C-terminus [18]. Our results here provide additional support for this structure-function analysis. Another functional parameter controlled by the interaction of KCNE1 with KCNQ1 is the voltage-dependence of activation (VDA). VDA appears to be affected by perturbations within both the transmembrane interaction site and the C-termini of KCNQ1/KCNE1. Whether this is due to specific interaction sites versus more general changes in subunit anchoring and allosteric effects cannot be resolved with available data.
An anchoring effect of C-terminal interactions between KCNQ1 and KCNE1 is supported by mutant expression studies of KCNE1 C-terminal truncation where activation kinetics were preserved but VDA and deactivation kinetics were altered [18]. This is seemingly at odds with a previous report [21] where investigators provided evidence that KCNE1 C-terminus was necessary for activation kinetics when expressed in Xenopus oocytes. Our recent finding that KCNE1 C-terminal truncation reduces the apparent affinity of KCNE1 for KCNQ1 but can be surmounted by relative over-expression of the KCNE1 mutant expressed in CHO cells (which lack endogenous KCNE subunits that are present in Xenopus oocytes [26]). We suggest that the anchoring effects of the C-terminus may be responsible for this apparent discrepancy and probably reflects relative amounts of subunit binding and overall structure stability of the channel complex.
Recent structural data from NMR studies of KCNE1 purified in liposomes suggests that the C-terminus is comprised, in part, of an interrupted α-helix, which is consistent with a scanning mutagenesis report [27–29]. The remainder of the KCNE1 C-terminus did not appear to exhibit structure in solution. Molecular modeling for the location of the KCNE1 C-terminus suggested that it folded back towards the membrane leading the authors to predict that it interacted with internal pore structures such as the S4–S5 linker [28]. While this model is not consistent with data presented here and that from the Haitin study [19] however, it could be explained by the fact that the NMR structure did not resolve the non-helical portions of the C-terminus and its structure was determined in the absence of KCNQ1. An alternative possibility is that KCNE1 C-terminus has dynamic interactions with KCNQ1 that could involve alternative interaction with differing portions of the channel during gating or a more complex interaction such as bridging between KCNQ1 C-terminus and the S4–S5 linker. The peptides representing the distal C-terminus of KCNQ1 (Helix-D, ~40amino acids) has crystal structure solved showing a coiled-coil tetrameric structure [9, 10]. In our analysis all the binding of KCNE1 C-terminus reside proximal to this region of KCNQ1, where structure is unknown.
Our results also raise several new questions. The precise functional and physical sites of C-terminal interaction are yet to be resolved. Complex dynamics of the interaction during gating is a possibility that could resolve some of the experimental differences between investigators. Solving the structure of the binding interaction may resolve some of these issues. Functional issues that must be kept in mind include not only how the interaction regulates channel deactivation rates but how it may be involved with the newer results indicating KCNE1 C-terminal effects on channel trafficking and recycling [30, 31], response to cAMP/PKA signaling [32] and phenotypes of LQT1 and LQT5 mutations.
Acknowledgments
We thank Michael Brenowitz of the Albert Einstein College of Medicine Department of Biochemistry for advice and support with the SPR analysis. This work was supported by grants to TVM from the NIH [HL075615, HL093440].
Footnotes
Abbreviations: LQTS, long QT syndrome; SPR, surface plasmon resonance; CaM, calmodulin; IKs, slowly activation delayed rectifier potassium current; RU, Response Units; VDA, voltage dependence of activation
AUTHOR CONTRIBUTION: Renjian Zheng participated and helped design all experiments and wrote the initial manuscript draft. Keith Thompson, Edmond Obeng-Gyimah, and Dana Alessi each participated in construction of recombinant plasmids, protein purification and in vitro binding experiments. Jerri Chen performed all electrophysiology experiments. Huiyong Cheng supervised and assisted in all surface plasmon resonance experiments. Thomas V. McDonald was responsible for overall project design, data analysis and final manuscript preparation.
References
- 1.Robbins J. KCNQ potassium channels: physiology, pathophysiology, and pharmacology. Pharmacol Ther. 2001;90:1–19. doi: 10.1016/s0163-7258(01)00116-4. [DOI] [PubMed] [Google Scholar]
- 2.Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. KVLQT1 and IsK (minK) proteins associate to form IKs cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 3.Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Keating MT. Coassembly of KVLQT1 and minK (IsK) proteins to form cardiac IKs potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 4.Keating MT, Sanguinetti MC. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 2001;104:569–580. doi: 10.1016/s0092-8674(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 5.Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nat Genet. 1997;17:338–340. doi: 10.1038/ng1197-338. [DOI] [PubMed] [Google Scholar]
- 6.Schwake M, Athanasiadu D, Beimgraben C, Blanz J, Beck C, Jentsch TJ, Saftig P, Friedrich T. Structural determinants of M-type KCNQ (Kv7) K+ channel assembly. J Neurosci. 2006;26:3757–3766. doi: 10.1523/JNEUROSCI.5017-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shamgar L, Ma L, Schmitt N, Haitin Y, Peretz A, Wiener R, Hirsch J, Pongs O, Attali B. Calmodulin is essential for cardiac IKS channel gating and assembly: impaired function in long-QT mutations. Circ Res. 2006;98:1055–1063. doi: 10.1161/01.RES.0000218979.40770.69. [DOI] [PubMed] [Google Scholar]
- 8.Ghosh S, Nunziato DA, Pitt GS. KCNQ1 assembly and function is blocked by long-QT syndrome mutations that disrupt interaction with calmodulin. Circ Res. 2006;98:1048–1054. doi: 10.1161/01.RES.0000218863.44140.f2. [DOI] [PubMed] [Google Scholar]
- 9.Howard RJ, Clark KA, Holton JM, Minor DL., Jr Structural insight into KCNQ (Kv7) channel assembly and channelopathy. Neuron. 2007;53:663–675. doi: 10.1016/j.neuron.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiener R, Haitin Y, Shamgar L, Fernandez-Alonso MC, Martos A, Chomsky-Hecht O, Rivas G, Attali B, Hirsch JA. The KCNQ1 (Kv7.1) COOH terminus, a multitiered scaffold for subunit assembly and protein interaction. J Biol Chem. 2008;283:5815–5830. doi: 10.1074/jbc.M707541200. [DOI] [PubMed] [Google Scholar]
- 11.Wen H, Levitan IB. Calmodulin is an auxiliary subunit of KCNQ2/3 potassium channels. J Neurosci. 2002;22:7991–8001. doi: 10.1523/JNEUROSCI.22-18-07991.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yus-Najera E, Santana-Castro I, Villarroel A. The identification and characterization of a noncontinuous calmodulin-binding site in noninactivating voltage-dependent KCNQ potassium channels. J Biol Chem. 2002;277:28545–28553. doi: 10.1074/jbc.M204130200. [DOI] [PubMed] [Google Scholar]
- 13.Aggeli A, Bannister ML, Bell M, Boden N, Findlay JB, Hunter M, Knowles PF, Yang JC. Conformation and ion-channeling activity of a 27-residue peptide modeled on the single-transmembrane segment of the IsK (minK) protein. Biochemistry. 1998;37:8121–8131. doi: 10.1021/bi972112h. [DOI] [PubMed] [Google Scholar]
- 14.Melman YF, Um SY, Krumerman A, Kagan A, McDonald TV. KCNE1 binds to the KCNQ1 pore to regulate potassium channel activity. Neuron. 2004;42:927–937. doi: 10.1016/j.neuron.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Melman YF, Domenech A, de la Luna S, McDonald TV. Structural determinants of KvLQT1 control by the KCNE family of proteins. J Biol Chem. 2001;276:6439–6444. doi: 10.1074/jbc.M010713200. [DOI] [PubMed] [Google Scholar]
- 16.Melman YF, Krumerman A, McDonald TV. A Single Transmembrane Site in the KCNE-encoded Proteins Controls the Specificity of KvLQT1 Channel Gating. J Biol Chem. 2002;277:25187–25194. doi: 10.1074/jbc.M200564200. [DOI] [PubMed] [Google Scholar]
- 17.Panaghie G, Abbott GW. The role of S4 charges in voltage-dependent and voltage-independent KCNQ1 potassium channel complexes. J Gen Physiol. 2007;129:121–133. doi: 10.1085/jgp.200609612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Zheng R, Melman YF, McDonald TV. Functional interactions between KCNE1 C-terminus and the KCNQ1 channel. PLoS ONE. 2009;4:e5143. doi: 10.1371/journal.pone.0005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haitin Y, Wiener R, Shaham D, Peretz A, Cohen EB, Shamgar L, Pongs O, Hirsch JA, Attali B. Intracellular domains interactions and gated motions of I(KS) potassium channel subunits. Embo J. 2009 doi: 10.1038/emboj.2009.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bianchi L, Shen Z, Dennis AT, Priori SG, Napolitano C, Ronchetti E, Bryskin R, Schwartz PJ, Brown AM. Cellular dysfunction of LQT5-minK mutants: abnormalities of IKs, IKr and trafficking in long QT syndrome. Hum Mol Genet. 1999;8:1499–1507. doi: 10.1093/hmg/8.8.1499. [DOI] [PubMed] [Google Scholar]
- 21.Tapper AR, George AL., Jr MinK subdomains that mediate modulation of and association with KvLQT1 [In Process Citation] J Gen Physiol. 2000;116:379–390. doi: 10.1085/jgp.116.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipschultz CA, Li Y, Smith-Gill S. Experimental design for analysis of complex kinetics using surface plasmon resonance. Methods. 2000;20:310–318. doi: 10.1006/meth.1999.0924. [DOI] [PubMed] [Google Scholar]
- 23.Schuck P, Boyd L, Andersen P. Measuring Protein Interactions by Optical Biosensors. In: Colgin JE, Dunn BM, Ploegh HL, Speicher DW, Wingfield PT, editors. Current Protocols in Protein Science. John Wiley & Sons; 1999. pp. 20.22.21–20.22.22. [Google Scholar]
- 24.Xu X, Jiang M, Hsu KL, Zhang M, Tseng GN. KCNQ1 and KCNE1 in the IKs channel complex make state-dependent contacts in their extracellular domains. J Gen Physiol. 2008;131:589–603. doi: 10.1085/jgp.200809976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung DY, Chan PJ, Bankston JR, Yang L, Liu G, Marx SO, Karlin A, Kass RS. Location of KCNE1 relative to KCNQ1 in the I(KS) potassium channel by disulfide cross-linking of substituted cysteines. Proc Natl Acad Sci U S A. 2009;106:743–748. doi: 10.1073/pnas.0811897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anantharam A, Lewis A, Panaghie G, Gordon E, McCrossan ZA, Lerner DJ, Abbott GW. RNAi reveals endogenous Xenopus MiRPs govern mammalian K+ channel function in oocyte expression studies. J Biol Chem. 2003;15:15. doi: 10.1074/jbc.M212751200. [DOI] [PubMed] [Google Scholar]
- 27.Tian C, Vanoye CG, Kang C, Welch RC, Kim HJ, George AL, Jr, Sanders CR. Preparation, functional characterization, and NMR studies of human KCNE1, a voltage-gated potassium channel accessory subunit associated with deafness and long QT syndrome. Biochemistry. 2007;46:11459–11472. doi: 10.1021/bi700705j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang C, Tian C, Sonnichsen FD, Smith JA, Meiler J, George AL, Jr, Vanoye CG, Kim HJ, Sanders CR. Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel. Biochemistry. 2008;47:7999–8006. doi: 10.1021/bi800875q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rocheleau JM, Gage SD, Kobertz WR. Secondary structure of a KCNE cytoplasmic domain. J Gen Physiol. 2006;128:721–729. doi: 10.1085/jgp.200609657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seebohm G, Strutz-Seebohm N, Birkin R, Dell G, Bucci C, Spinosa MR, Baltaev R, Mack AF, Korniychuk G, Choudhury A, Marks D, Pagano RE, Attali B, Pfeufer A, Kass RS, Sanguinetti MC, Tavare JM, Lang F. Regulation of endocytic recycling of KCNQ1/KCNE1 potassium channels. Circ Res. 2007;100:686–692. doi: 10.1161/01.RES.0000260250.83824.8f. [DOI] [PubMed] [Google Scholar]
- 31.Xu X, Kanda VA, Choi E, Panaghie G, Roepke TK, Gaeta SA, Christini DJ, Lerner DJ, Abbott GW. MinK-dependent internalization of the IKs potassium channel. Cardiovasc Res. 2009 doi: 10.1093/cvr/cvp047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurokawa J, Bankston JR, Kaihara A, Chen L, Furukawa T, Kass RS. KCNE variants reveal a critical role of the beta subunit carboxyl terminus in PKA-dependent regulation of the I(Ks) potassium channel. Channels (Austin) 2009:3. doi: 10.4161/chan.3.1.7387. [DOI] [PMC free article] [PubMed] [Google Scholar]