Abstract
Merino Walk virus (MWV), a proposed novel tentative species of the family Arenaviridae, was isolated from a rodent, Myotomys unisulcatus, collected at Merino Walk, Eastern Cape, South Africa, in 1985. Full-length genomic sequence confirmed MWV as an arenavirus related distantly to Mobala, Mopeia and Ippy viruses, all members of the Old World arenavirus complex. We propose MWV as a tentative novel species in the Lassa–lymphocytic choriomeningitis virus complex, based on its isolation from a novel rodent species and its genetic and serological characteristics.
INTRODUCTION
The family Arenaviridae currently includes 23 antigenically related viruses classified into two groups: Old World (OW; Lassa–lymphocytic choriomeningitis complex) and New World (NW; Tacaribe complex) (http://www.ictvonline.org/virusTaxonomy.asp?), based on geographical, genetic, antigenic and host relationships (Bowen et al., 1997; Moncayo et al., 2001). The arenavirus genome is bisegmented, comprising a larger (L) and a smaller (S) segment, each encoding two open reading frames (ORFs) using an ambisense coding strategy (Auperin et al., 1984; Salvato & Shimomaye, 1989). Nonetheless, members of the Arenaviridae, together with the families Orthomyxoviridae and Bunyaviridae, are classified as segmented, single-stranded, negative-sense RNA viruses. Some members of the family Arenaviridae, such as the South American haemorrhagic fever viruses [Junín (JUNV; Parodi et al., 1958; Pirosky et al., 1959), Machupo (MACV; Johnson et al., 1965), Guanarito (GTOV; Salas et al., 1991) and Sabiá (SABV; Lisieux et al., 1994) viruses] and the African Lassa virus (LASV) are listed as potential biowarfare or bioterrorism agents (http://www.cdc.gov/od/sap) because of their human pathogenicity and aerosol-transmission potential. These latter five viruses are classified as biosafety level 4 agents.
Arenavirus species are usually maintained in and transmitted by a single rodent species (Bowen et al., 1997), the only exceptions thus far being Tacaribe virus (TCRV; Downs et al., 1963), which was isolated from fruit bats (Artibeus sp.), and Amapari virus (AMAV), which has been isolated from two rodent species, Oryzomys gaeldi and Neacomys guianae (Pinheiro & Woodall, 1969). The NW arenaviruses are associated with rodents in the subfamily Sigmodontinae of the family Cricetidae, whereas the OW arenaviruses are associated with rodents in the subfamily Murinae of the family Muridae. LASV and Mopeia virus (MOPV; Wulff et al., 1977) are hosted by members of the genus Mastomys, Mobala virus (MOBV; Gonzalez et al., 1983) by Praomys spp., Ippy virus (IPPYV; Meunier et al., 1985; Swanepoel et al., 1985) by Arvicanthis spp., and lymphocytic choriomeningitis virus (LCMV; Peters, 2006) by the ubiquitous Mus musculus.
Of the OW arenavirus species, only LCMV and LASV were known to cause severe disease in humans (Buckley & Casals, 1970) until September 2008, when a cluster of undiagnosed fatal haemorrhagic fever cases in South Africa and Zambia led to the identification of a novel OW arenavirus, designated Lujo virus (LUJV; Briese et al., 2009; Paweska et al., 2009). The rodent host of this species remains unknown, but efforts to survey wildlife are under way to identify its origin. This sudden outbreak of viral haemorrhagic fever highlights the importance of surveying wildlife, in this case the rodent population, to describe the virome diversity and its ecology. As an example of such surveys, here we describe rapid full-genome sequencing for genetic characterization of another novel OW arenavirus, tentatively termed Merino Walk virus (MWV).
RESULTS AND DISCUSSION
Pathogenicity
Newborn mice inoculated intracerebrally with MWV were moribund and died 10–11 days after infection. Adult ICR mice inoculated intraperitoneally with a 10 % suspension of the infected newborn mouse brains did not become ill, but subsequently developed antibodies to the virus.
In comparison, IPPYV kills intracerebrally inoculated suckling mice between 5 and 8 days after inoculation, with a mortality of 60 % (MOBV and MOPV show also a similar mortality) (Swanepoel et al., 1985). In general, LCMV and LASV are pathogenic for weaned mice, but not suckling mice (Salvato et al., 2005). We do not have any data on MWV serology in mice or humans. Given that MWV was isolated in 1985 and that other OW arenaviruses are associated with outbreaks of viral haemorrhagic fever in the same geographical region, serology should be pursued to determine whether MWV infects humans.
Serology
Results of complement fixation (CF) tests comparing MWV with four other African arenaviruses (MOBV, MOPV, IPPYV and LCMV) are shown in Table 1. By this method, MWV was shown to be related antigenically to MOBV, MOPV and IPPYV, but unrelated to LCMV. However, the cross-CF tests indicate that MWV is antigenically distinct from these other arenaviruses.
Table 1.
Results of CF tests comparing MWV with other African arenaviruses
Values are shown as reciprocal of highest antiserum dilution/reciprocal of highest antigen dilution. Homologous titres are shown in bold.
| Antigen | Antibody | ||||
|---|---|---|---|---|---|
| MWV | MOBV | MOPV | IPPYV | LCMV* | |
| MWV | 256/32 | 16/8 | 64/32 | 8/8 | 0† |
| MOBV | 16/32 | 512/128 | ≥1024/128 | 32/32 | – |
| MOPV | 16/32 | 128/128 | ≥1024/128 | 32/32 | – |
| IPPYV | 16/32 | 128/128 | 512/128 | 512/512 | – |
| Control | 0 | 0 | 0 | 0 | 0 |
*LCMV homologous titre=128/256.
†0=<8/<8.
Transmission electron microscopy
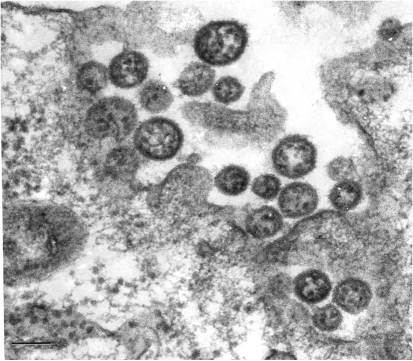
In ultrathin sections of infected Vero cells, virions with typical arenavirus morphology were observed at the cell surface and in the process of budding from it (Fig. 1). They had diameters of 80–190 nm.
Fig. 1.
MWV as observed at the plasma membrane of an infected Vero cell in an ultrathin section. Bar, 100 nm.
Sequence acquisition and analysis
Consistent with the ambisense genome organization characteristic of members of the family Arenaviridae (Auperin et al., 1984; Charrel et al., 2008; Salvato & Shimomaye, 1989), the genome of MWV comprises two RNA segments: an L segment that encodes a large (L) polymerase-related ORF in the negative-sense orientation and a small RING-finger protein Z in the positive-sense orientation (GenBank accession no. GU078661), and an S segment that encodes a nucleocapsid protein (NP) in the negative-sense orientation and a glycoprotein precursor protein (GPC) in the positive-sense orientation (GenBank accession no. GU078660).
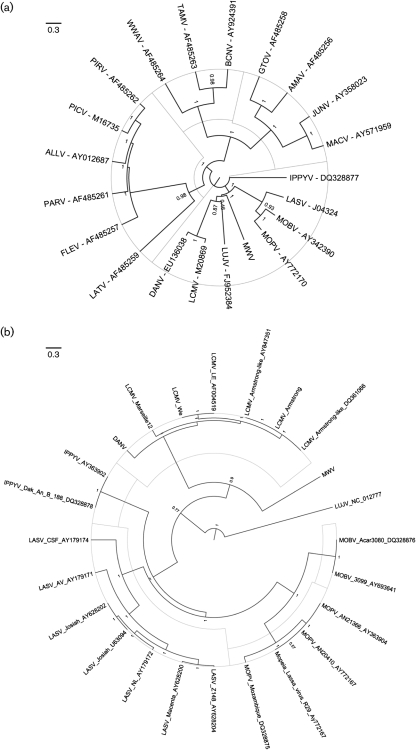
The result of the phylogenetic analysis of the GPC and L (Fig. 2a, b, respectively) and NP (Supplementary Fig. S1, available in JGV Online) ORFs indicated that, although distant from any other known OW arenavirus, MWV is consistently associated with MOBV, MOPV and IPPYV. No evidence of RNA segment reassortment or intersegmental recombination was discovered. The NP gene sequence of MWV differs from those of other arenaviruses by between 34.5 % (MOPV) and 49.5 % (Tamiami virus, TAMV) on the nucleotide level, and between 31.4 % (MOPV) and 55.5 % (TAMV) on the amino acid level (Table 2). Historically, phylogenetic classification of arenaviruses was based on analysis of partial areas of the NP gene. The degree of divergence is significantly higher than proposed cut-off values within (<10–12 %) or between (>21.5 %) OW arenavirus species (Bowen et al., 2000; Emonet et al., 2006).
Fig. 2.
Phylogenetic analyses of MWV were inferred based on primary nucleotide alignments of (a) GPC and (b) L sequences. Maximum clade credibility trees generated from analysis of arenavirus GPC or L sequences are shown. Posterior probabilities are listed below the branches for supported nodes.
Table 2.
Percentage differences in NP sequence between arenavirus isolates
Nucleotide sequence differences (%) are shown above the diagonal; amino acid sequence differences (%) are shown below the diagonal.
| Virus | LUJV | DANV | LCMV | MOPV | MOBV | IPPYV | LASV | MWV | GTOV | AMAV | MACV | CPXV | SABV | TCRV | JUNV | CHPV | OLVV | LATV | FLEV | CATV | PIRV | ALLV | BCNV | SKTV | NAAV | TAMV | PARV | WWAV | PICV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LUJV | 38.7 | 38.9 | 39.7 | 42.0 | 40.4 | 41.6 | 40.7 | 46.9 | 47.6 | 45.0 | 46.6 | 46.5 | 46.0 | 46.7 | 46.7 | 46.9 | 48.0 | 45.5 | 45.6 | 44.0 | 45.1 | 46.2 | 47.1 | 46.4 | 49.4 | 46.2 | 47.2 | 45.8 | |
| DANV | 41.3 | 19.5 | 39.0 | 38.4 | 38.4 | 39.7 | 37.3 | 45.0 | 46.7 | 45.2 | 45.3 | 45.3 | 46.4 | 44.8 | 45.3 | 46.0 | 44.4 | 44.0 | 45.2 | 43.2 | 45.1 | 44.8 | 45.3 | 46.2 | 49.2 | 45.3 | 45.2 | 45.3 | |
| LCMV | 40.8 | 5.8 | 38.1 | 38.3 | 38.4 | 39.6 | 37.9 | 45.8 | 46.6 | 44.3 | 45.4 | 44.4 | 45.7 | 44.8 | 45.8 | 46.2 | 44.1 | 44.0 | 45.3 | 43.9 | 44.9 | 44.2 | 46.1 | 46.1 | 48.6 | 44.2 | 45.7 | 47.3 | |
| MOPV | 43.7 | 34.9 | 36.3 | 29.8 | 33.7 | 31.9 | 34.5 | 46.9 | 46.3 | 44.7 | 43.6 | 45.8 | 46.2 | 46.0 | 46.3 | 45.3 | 44.8 | 45.3 | 44.7 | 44.6 | 44.5 | 45.1 | 44.8 | 46.0 | 49.3 | 44.6 | 44.6 | 44.2 | |
| MOBV | 42.4 | 37.0 | 37.5 | 21.6 | 36.1 | 32.0 | 35.3 | 46.3 | 46.7 | 45.2 | 43.4 | 44.6 | 45.7 | 47.3 | 45.5 | 45.7 | 46.1 | 45.5 | 45.1 | 45.6 | 44.6 | 46.3 | 46.6 | 46.9 | 48.3 | 44.3 | 45.8 | 46.1 | |
| IPPYV | 44.4 | 37.1 | 37.2 | 29.7 | 29.3 | 35.0 | 35.2 | 46.6 | 46.9 | 45.2 | 45.3 | 46.9 | 46.9 | 45.4 | 46.3 | 46.7 | 47.9 | 47.8 | 45.5 | 44.7 | 46.7 | 46.3 | 45.5 | 47.1 | 49.0 | 45.7 | 45.8 | 44.8 | |
| LASV | 42.3 | 37.4 | 38.4 | 26.2 | 27.2 | 31.0 | 35.2 | 45.1 | 47.2 | 46.6 | 45.2 | 45.6 | 45.1 | 45.9 | 47.1 | 46.0 | 45.4 | 45.7 | 45.8 | 47.2 | 44.7 | 45.3 | 46.2 | 46.1 | 47.9 | 45.6 | 46.7 | 44.4 | |
| MWV | 43.2 | 34.2 | 34.5 | 31.4 | 33.3 | 31.6 | 32.3 | 45.8 | 46.1 | 44.7 | 46.6 | 45.2 | 46.5 | 45.2 | 45.7 | 47.3 | 46.7 | 45.6 | 44.8 | 44.5 | 44.9 | 45.0 | 44.9 | 46.6 | 49.5 | 44.7 | 46.2 | 44.0 | |
| GTOV | 52.6 | 49.6 | 48.5 | 48.7 | 49.9 | 49.6 | 49.4 | 47.7 | 24.5 | 30.7 | 26.6 | 34.0 | 32.3 | 30.4 | 32.4 | 39.2 | 37.6 | 41.2 | 42.2 | 41.3 | 42.8 | 42.8 | 42.3 | 41.8 | 45.2 | 41.4 | 42.4 | 41.3 | |
| AMAV | 52.6 | 49.3 | 48.3 | 49.6 | 49.7 | 49.3 | 50.1 | 49.0 | 14.5 | 32.7 | 26.6 | 33.5 | 32.4 | 31.9 | 31.9 | 36.3 | 37.8 | 42.2 | 43.2 | 42.4 | 42.1 | 43.2 | 43.0 | 43.5 | 47.0 | 41.3 | 43.7 | 41.3 | |
| MACV | 52.1 | 49.8 | 49.0 | 50.0 | 49.9 | 49.8 | 49.6 | 49.5 | 23.7 | 26.3 | 31.3 | 32.9 | 28.7 | 23.7 | 34.3 | 36.9 | 37.8 | 40.7 | 42.0 | 40.0 | 41.6 | 41.0 | 42.4 | 42.8 | 44.8 | 41.2 | 42.3 | 42.0 | |
| CPXV | 51.6 | 47.7 | 46.7 | 47.9 | 48.2 | 49.4 | 48.4 | 50.0 | 17.9 | 17.4 | 25.9 | 34.2 | 32.6 | 32.0 | 33.0 | 37.3 | 37.2 | 42.4 | 42.5 | 42.5 | 41.4 | 43.9 | 43.9 | 45.1 | 47.0 | 41.9 | 43.2 | 42.7 | |
| SABV | 51.7 | 48.5 | 47.6 | 49.4 | 48.7 | 49.9 | 49.3 | 48.6 | 29.6 | 28.2 | 28.3 | 29.1 | 35.3 | 33.3 | 25.2 | 40.1 | 38.8 | 43.1 | 42.7 | 43.0 | 41.3 | 41.7 | 43.9 | 44.5 | 44.9 | 42.2 | 42.4 | 42.3 | |
| TCRV | 52.2 | 48.8 | 49.1 | 49.6 | 50.4 | 49.4 | 49.5 | 49.8 | 28.6 | 30.0 | 20.7 | 30.2 | 33.5 | 27.8 | 33.6 | 39.0 | 37.9 | 42.2 | 43.5 | 41.6 | 41.0 | 43.0 | 43.5 | 44.0 | 44.7 | 41.4 | 43.7 | 41.9 | |
| JUNV | 53.0 | 49.5 | 49.2 | 50.2 | 51.4 | 50.9 | 50.4 | 50.1 | 25.4 | 26.6 | 12.2 | 27.3 | 29.8 | 21.5 | 32.8 | 37.4 | 37.7 | 41.3 | 42.7 | 40.9 | 41.3 | 43.7 | 42.7 | 44.5 | 45.4 | 42.3 | 43.7 | 42.3 | |
| CHPV | 52.6 | 47.5 | 47.6 | 49.0 | 49.3 | 49.7 | 49.5 | 48.0 | 28.2 | 27.7 | 29.1 | 28.2 | 16.2 | 31.7 | 30.1 | 39.2 | 38.6 | 42.0 | 41.7 | 41.5 | 42.0 | 42.0 | 42.8 | 43.0 | 46.1 | 41.4 | 43.5 | 42.6 | |
| OLVV | 53.5 | 49.6 | 49.9 | 50.4 | 50.5 | 51.4 | 49.7 | 50.6 | 38.0 | 37.6 | 37.8 | 36.1 | 38.2 | 40.0 | 39.1 | 38.0 | 27.7 | 39.7 | 41.8 | 39.7 | 39.1 | 41.6 | 42.8 | 42.6 | 44.5 | 40.3 | 42.5 | 39.7 | |
| LATV | 53.3 | 47.7 | 48.1 | 49.0 | 50.4 | 51.3 | 50.7 | 48.6 | 35.9 | 37.3 | 37.2 | 35.6 | 37.9 | 39.7 | 38.8 | 38.1 | 19.2 | 41.0 | 41.2 | 38.7 | 39.6 | 41.8 | 42.4 | 40.9 | 43.0 | 38.4 | 42.0 | 41.5 | |
| FLEV | 53.8 | 48.9 | 48.8 | 50.4 | 51.5 | 51.3 | 50.3 | 49.4 | 41.7 | 42.9 | 43.1 | 44.4 | 43.7 | 44.2 | 42.0 | 41.2 | 41.6 | 42.8 | 36.8 | 36.0 | 31.8 | 37.5 | 37.3 | 38.7 | 41.1 | 29.8 | 38.0 | 35.3 | |
| CATV | 53.7 | 50.1 | 49.3 | 48.6 | 50.5 | 52.5 | 50.1 | 50.6 | 45.0 | 46.4 | 45.5 | 46.9 | 45.4 | 46.8 | 45.9 | 43.8 | 44.2 | 43.1 | 35.8 | 36.7 | 36.5 | 28.4 | 22.7 | 23.0 | 29.8 | 37.0 | 23.0 | 37.1 | |
| PIRV | 49.8 | 45.1 | 44.8 | 48.0 | 50.6 | 48.0 | 50.6 | 47.5 | 41.7 | 43.3 | 43.6 | 44.1 | 43.5 | 43.1 | 43.3 | 41.6 | 40.4 | 40.3 | 32.4 | 37.5 | 34.8 | 36.4 | 36.8 | 38.4 | 39.8 | 35.8 | 38.0 | 34.6 | |
| ALLV | 51.9 | 47.9 | 48.5 | 48.6 | 49.5 | 50.0 | 47.2 | 47.9 | 43.5 | 43.0 | 42.3 | 42.9 | 43.6 | 41.5 | 42.4 | 41.8 | 41.0 | 40.5 | 25.7 | 38.5 | 29.6 | 37.0 | 36.2 | 37.6 | 39.1 | 32.6 | 36.6 | 30.0 | |
| BCNV | 52.5 | 48.8 | 47.8 | 48.9 | 51.4 | 50.7 | 50.6 | 49.4 | 45.2 | 45.9 | 45.9 | 47.0 | 45.1 | 46.1 | 45.7 | 43.6 | 43.2 | 41.8 | 34.9 | 17.8 | 36.4 | 35.4 | 26.6 | 28.6 | 30.8 | 36.3 | 27.9 | 37.3 | |
| SKTV | 53.6 | 49.7 | 49.5 | 48.7 | 50.5 | 51.6 | 50.1 | 49.9 | 45.3 | 47.1 | 47.0 | 47.4 | 45.6 | 46.8 | 46.8 | 44.7 | 44.4 | 42.0 | 37.1 | 11.4 | 37.6 | 37.2 | 16.9 | 24.2 | 29.6 | 38.0 | 22.9 | 38.3 | |
| NAAV | 52.2 | 49.1 | 48.6 | 48.5 | 51.1 | 52.4 | 50.4 | 50.4 | 44.2 | 45.1 | 46.1 | 46.2 | 45.3 | 45.8 | 44.5 | 43.2 | 44.7 | 41.7 | 36.3 | 14.6 | 38.1 | 36.7 | 20.5 | 15.4 | 30.2 | 37.4 | 16.0 | 38.1 | |
| TAMV | 57.2 | 54.5 | 53.6 | 53.4 | 55.1 | 56.8 | 55.3 | 55.5 | 49.1 | 50.4 | 49.1 | 49.7 | 47.8 | 49.6 | 48.0 | 47.6 | 45.7 | 46.1 | 41.8 | 22.5 | 40.9 | 42.9 | 25.0 | 22.3 | 23.9 | 39.2 | 31.3 | 40.6 | |
| PARV | 51.9 | 48.4 | 47.5 | 46.6 | 50.3 | 50.0 | 49.4 | 47.5 | 42.1 | 43.2 | 43.5 | 44.2 | 43.7 | 43.4 | 42.5 | 41.6 | 41.9 | 41.4 | 21.9 | 38.5 | 32.3 | 28.1 | 34.6 | 38.7 | 38.2 | 41.8 | 37.1 | 34.8 | |
| WWAV | 52.8 | 49.9 | 49.3 | 47.8 | 50.5 | 50.9 | 50.3 | 50.3 | 44.8 | 45.3 | 44.4 | 46.3 | 44.9 | 45.5 | 45.0 | 43.6 | 44.1 | 42.2 | 37.8 | 13.5 | 37.1 | 36.9 | 18.4 | 14.3 | 9.3 | 23.7 | 39.2 | 36.7 | |
| PICV | 52.2 | 50.2 | 50.5 | 48.0 | 51.0 | 49.8 | 50.1 | 47.7 | 43.1 | 43.3 | 44.2 | 45.7 | 43.0 | 44.0 | 44.0 | 42.8 | 42.9 | 42.6 | 30.8 | 37.0 | 29.6 | 23.8 | 36.1 | 37.0 | 36.9 | 40.4 | 32.9 | 35.8 |
ORFs
Z protein.
A conserved RING motif and shorter C-terminal domain resembling those present in NW arenaviruses is found in the 89 aa Z protein of MWV (10 kDa, pI=7.6). MWV includes the conserved N-terminal myristoylation site G2, implicated in membrane association during budding and interaction of Z with the viral glycoprotein complex (Capul et al., 2007; Perez et al., 2004; Strecker et al., 2006). The tryptophan residue conserved in the RING motif of other arenavirus Z proteins and in cellular ubiquitin ligases is found in the RING motif of MWV (Joazeiro et al., 1999) (W44; Supplementary Fig. S2, available in JGV Online).
The Z protein of arenaviruses is critical in the budding of viral progeny, similar to matrix proteins of other negative-sense RNA viruses; mutation analysis of the late (L) domains of the LASV and LCMV Z proteins demonstrated that their abrogation impairs budding completely (Perez et al., 2003; Strecker et al., 2003). Two L-domain motifs have been identified in the C terminus of the LASV Z protein. The first motif (PTAP) mediates recognition of the tumour-susceptibility gene 101 (Tsg101; Perez et al., 2003), whilst the second motif (PPPY) acts as a Nedd4-like ubiquitin ligase-recognition motif (Staub et al., 1996). Those motifs are also conserved in the OW arenaviruses MOBV, MOPV and IPPYV. In contrast, only one motif has been recognized in the LCMV Z protein (P85PPY) (Perez et al., 2003). It is not possible to recognize any known L domains in the MWV Z ORF. MWV is not unique in this feature; TCRV also does not show canonical L domains. Instead, TCRV Z contains an ASAP sequence that partially mimics the L domain, PTAP. Similarly, MWV contains a PTCP domain that might also mimic the PTAP domain, although more distantly due to its polarity properties. Recently, similar L-domain motifs have been described in other viruses, including YP(x)nL in the p9 region of equine infectious anemia virus Gag protein (Puffer et al., 1997) and the YRKL sequence in the matrix protein of influenza virus (Hui et al., 2003).
Large RNA-dependent RNA polymerase protein (RdRp; L).
The less divergent areas of the 2001 aa MWV RdRp (230 kDa, pI=6.8) overlap conserved areas among all other arenaviruses, confirming their association with function: region I (position 1–250), region II (position 500–900) (Supplementary Fig. S3a, available in JGV Online), region III (position 1000–1650; RNA replicase domain) and region IV (position 1750–1900). Region III is characterized by the motifs in the catalytic domain termed pre-A, A, B, C, D and E (Delarue et al., 1990; Müller et al., 1994; Poch et al., 1989; Vieth et al., 2004) (Supplementary Fig. S3b).
NP.
The 561 aa NP of MWV (62.2 kDa, pI=9.0) shows between 45 and 53 % conservation at the amino acid level (51–56 % at the nucleotide level) to those of NW arenaviruses, and between 57 and 72 % (59–66 % at the nucleotide level) to those of OW arenaviruses (Table 2; Supplementary Fig. S4, available in JGV Online). The following amino acid motifs previously described in the family are all well-conserved (Gonzalez et al., 1996): (a) mixed-charge segment at the N-terminal region (Parisi et al., 1996); (b) the atypical ribonucleoprotein consensus sequence motif (RNP-1) (Parisi et al., 1996); (c) zinc-finger motif in the C-terminal region (Parisi et al., 1996); and (d) the cytotoxic T-lymphocyte (CTL) epitope G123VYMGNL described in LCMV (Whitton et al., 1989). Another potential CTL epitope, Y315IACRTSIV as found in LCMV, is not well-conserved, similar to LUJV. A potential antigenic site in the mixed-charge area of the LASV NP (RKSKRND; Gonzalez et al., 1995) has the sequence R55KEKRDD in MWV.
GPC.
The 497 aa MWV GPC preprotein precursor (56.3 kDa, pI=8.5) is cleaved cotranslationally into a stable signal peptide and an immature precursor, which is subsequently processed by the host signal signalase (S1P) into the G1 and G2 fragments (Beyer et al., 2003; Burns & Buchmeier, 1991, 1993; Lenz et al., 2001; Neuman et al., 2005; Rojek et al., 2008b). The G1 segment is the area of lowest conservation of the whole GPC (Supplementary Fig. S5a, available in JGV Online).
Using the SignalP algorithm, S1P is predicted to cleave the signal peptide of MWV between A and T60. The two hydrophobic domains found in arenaviruses are in the 59 aa signal peptide of MWV (6.4 kDa, pI=4.9) (Eichler et al., 2004). The sequence also displays a conserved G2 that has been implicated in myristoylation of JUNV (York et al., 2004).
By comparison with other arenaviruses, S1P cleavage of RRLK265 may lead to separation of G1 (261 aa, 29.2 kDa, pI=6.8) from G2 (232 aa, 26.6 kDa, pI=8.5) (Beyer et al., 2003; Lenz et al., 2000; Rojek et al., 2008a). Although the G2 fragment contains unique amino acid residues in several positions, it is generally well-conserved with respect to other arenaviruses (Supplementary Fig. S5b).
Two receptors are reported to interact with the G1 glycoprotein moiety of arenaviruses. Alpha-dystroglycan (α-DG) (Cao et al., 1998) binds OW arenaviruses LASV and LCMV and the non-pathogenic clade C NW arenaviruses Oliveros virus (OLVV) and Latino virus (LATV) (Spiropoulou et al., 2002). Transferring receptor 1 (TfR1) binds the pathogenic NW arenaviruses JUNV, MACV, GTOV and SABV (Radoshitzky et al., 2007) (Fig. 3). Structural alignment of the MWV G1 with those of representative members of the OW and NW clade C arenaviruses might correlate with its receptor usage.
Fig. 3.
Phylogenetic analysis of MWV based on secondary structure alignments of G1 sequences. Receptors reported to interact with the G1 glycoprotein moiety are shown, where • represents NW arenaviruses using TfR1, ▪ represents viruses using host TfR1, but not human receptors, and ○ indicates OW and clade C NW arenaviruses that use α-DG.
Non-translated sequence analysis
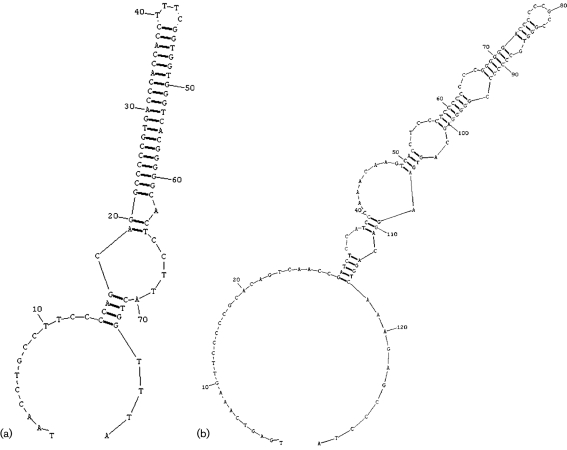
The 5′ and 3′ non-coding regions (NCRs) of the S segment are 52 and 60 nt long, respectively. The S segment intergenic region (IR) of MWV comprises 72 nt and is predicted to form a single, highly stable stem–loop structure (Fig. 4a). The S segment IR of SABV has been reported to form three such structures (Gonzalez et al., 1996), whereas those of JUNV, TCRV, OLVV, Flexal virus (FLEV), GTOV, LATV, MACV, AMAV, Cupixi virus (CPXV) (all NW) and of the OW virus MOPV can fold into two (Archer & Rico-Hesse, 2002; Bowen et al., 1996; Ghiringhelli et al., 1991; Iapalucci et al., 1991; Wilson & Clegg, 1991). Those of the NW viruses Pichinde virus (PICV), Paraná virus (PARV), Pirital virus (PIRV), Allpahuayo virus (ALLV; Moncayo et al., 2001) and Whitewater Arroyo virus (WWAV; Fulhorst et al., 1996), and of the OW viruses LASV and LCMV, appear to form only a single stem–loop structure (Archer & Rico-Hesse, 2002; Auperin et al., 1984, 1986; Charrel et al., 2001; Moncayo et al., 2001; Romanowski & Bishop, 1985). The L segment has a 5′ NCR of 99 nt, a 3′ NCR of 23 nt and an IR of 122 nt that can be assumed to form a complex folding structure, as observed in other arenavirus L segment IRs (Fig. 4b) (Archer & Rico-Hesse, 2002).
Fig. 4.
Predicted secondary structures of the (a) S and (b) L segments. The S segment (a) is predicted to form a single, highly stable stem–loop structure, whereas the L segment (b) is assumed to form a complex folding structure.
Conclusion
Our analysis of the genomic and biological characteristics of MWV indicates that it is a novel virus related only distantly to other known arenaviruses. MWV fulfils the majority of the parameters established by the International Committee on Taxonomy of Viruses for species demarcation within the genus Arenavirus: (i) association with a specific host species. Myotomys unisulcatus is a member of the rodent subfamily Muridae; this is the first arenavirus detected in rodents of the genus Myotomys. (ii) Substantive differences in antigenic cross-reactivity. MWV has antigenic cross-reactivity with MOBV, MOPV and IPPYV, but no reactivity with LCMV. (iii) Substantive sequence difference from other known species in the genus. The observed lowest nucleotide sequence divergence with the closest arenavirus species, MOPV, was 34.5 %. The lowest amino acid sequence divergence with the same species was 31.4 %. Two parameters remain undefined: (iv) the distribution of this virus within a defined geographical area (southern Africa) and (v) pathogenicity (or not) for humans. We have tentatively named this species Merino Walk virus (MWV), following the standard nomenclature of naming arenaviruses by their geographical origin, and we propose SPU 353-85 as its reference genome (prototype).
METHODS
Source of virus.
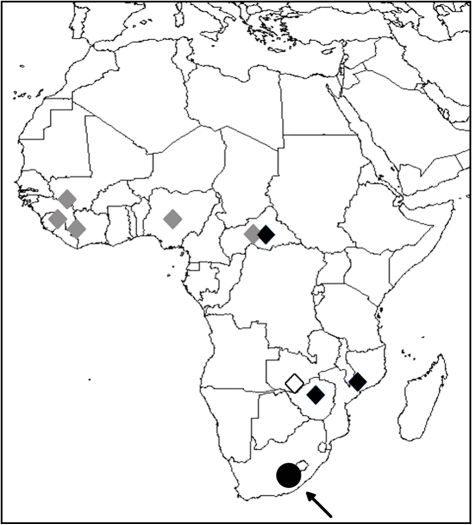
The prototype strain of MWV, SPU 353-85, was isolated in 1985 from a bush Karoo rat, Myotomys unisulcatus, captured at Merino Walk, Eastern Cape, South Africa (Fig. 5). Myotomys unisulcatus is endemic to southern Africa (Skinner & Smithers, 1990), confined primarily to the semi-arid Karoo region of the South-West Arid Zone (Davis, 1962; Skinner & Smithers, 1990), but occasionally found in the South-West Cape Zone (Davis, 1962). Myotomys unisulcatus (previously known as Otomys unisulcatus) is a member of the family Muridae (Musser & Carleton, 2005).
Fig. 5.
Known geographical distribution of arenaviruses in Africa based on isolation. MOBV, MOPV and IPPYV, which have not been associated with disease in humans, are shown by ⧫. LASV (⧫) and LUJV (◊) are associated with haemorrhagic fever. The arrow indicates the approximate location where MWV was recovered (•).
Virus isolation and immunological characterization.
Tissue samples were homogenized mechanically in 650 μl virus isolation medium [1× minimum essential medium (MEM), 2.2 g NaHCO3 l−1, 10 % fetal bovine serum (FBS) and 2× antibiotic/antimycotic solution (400 U penicillin ml−1, 400 μg streptomycin ml−1, 1 μg amphotericin B ml−1)]. Homogenized tissues were centrifuged (6700 g for 10 min) to pellet debris, and an aliquot (100 μl) of the clarified supernatant was used to inoculate Vero cells. Cells were maintained in MEM supplemented with 5 % FBS and 2× antibiotic/antimycotic solution, and incubated at 37 °C in a humidified atmosphere containing 5 % CO2.
Antigens and immunological reagents.
In addition to MWV, the following viruses were used to prepare the antigens and immune reagents: IPPYV (strain Dak An B 188d), MOPV (strain AN 20410) and MOBV (strain ACAR 3080-MRC 5P2). Antigens for use in CF tests were prepared from infected newborn mouse brain by the sucrose–acetone extraction method (Beaty et al., 1995). Immunizing antigens were 10 % crude brain suspensions of infected mice in PBS.
Specific hyperimmune mouse ascitic fluids (MAF) were prepared against the arenavirus prototype strains listed above. The immunization schedule consisted of four weekly intraperitoneal injections of mouse brain antigen mixed with Freund's adjuvant, as described previously (Fulhorst et al., 1997). Following the fourth injection, sarcoma 180 cells were also injected intraperitoneally to induce ascites formation. The lymphocytic choriomeningitis MAF was produced by the same method, but was obtained from the NIAID Reference Reagents Program (V-580-701-562). All animal work was carried out under an animal protocol approved by the University of Texas Medical Branch.
Serology.
CF tests were performed, using a microtechnique (Beaty et al., 1995), with two full units of guinea pig complement. Titres were recorded as the highest dilutions giving 3+ or 4+ fixation of complement on a scale of 0 (complete haemolysis) to 4+ (no haemolysis).
Transmission electron microscopy.
For ultrastructural analysis, infected cells were fixed in 2 % paraformaldehyde/2.5 % glutaraldehyde (Polysciences Inc.) in 100 mM phosphate buffer, pH 7.2, for 1 h at room temperature. Samples were washed in phosphate buffer and post-fixed in 1 % osmium tetroxide (Polysciences Inc.) for 1 h. Samples were then rinsed extensively in distilled H2O (dH2O) prior to en bloc staining with 1 % aqueous uranyl acetate (Ted Pella Inc.) for 1 h. Following several rinses in dH2O, samples were dehydrated in a graded series of ethanol and embedded in Eponate 12 resin (Ted Pella Inc.). Sections of 95 nm were cut with uranyl acetate and lead citrate, and viewed on a JEOL 1200 EX transmission electron microscope (JEOL USA Inc.).
Genomic sequencing.
The protocol for preparation of genomic material for pyrosequencing has been published in detail elsewhere (Palacios et al., 2008). MWV was extracted by using TRIzol LS (Invitrogen). Total RNA extracts were treated with DNase I (DNA-free; Ambion) and cDNA was subjected to a modified DOP-PCR procedure (Palacios et al., 2007). Products >70 bp were selected by column purification (MinElute; Qiagen) and ligated to specific adapters for sequencing on a 454 Genome Sequencer FLX (454 Life Sciences) without fragmentation of the cDNA (Cox-Foster et al., 2007; Margulies et al., 2005; Palacios et al., 2008). Software programs accessible through the analysis applications at the GreenePortal website (http://tako.cpmc.columbia.edu/Tools/) were used for removal of primer sequences, redundancy filtering and sequence assembly. With primers designed by using pyrosequence data, gaps between the aligned fragments were rapidly filled by specific PCR amplification. Conventional PCRs were performed with HotStar polymerase (Qiagen) on PTC-200 thermocyclers (Bio-Rad): an enzyme-activation step of 5 min at 95 °C was followed by 45 cycles of denaturation at 95 °C for 1 min, annealing at 55 °C for 1 min and extension at 72 °C for 1–3 min depending on the expected amplicon size. Specific primer sequences are available upon request. Amplification products were run on 1 % agarose gels, purified (MinElute; Qiagen) and sequenced directly in both directions with ABI PRISM Big Dye Terminator 1.1 Cycle Sequencing kits on ABI PRISM 3700 DNA Analyzers (Perkin-Elmer Applied Biosystems). Terminal sequences were generated by using a universal arenavirus primer targeting the conserved viral termini (5′-CGCACMGDGGATCCTAGGC), combined with four specific primers positioned near the ends of each segment. Sequence was verified by classical dideoxy sequencing using primers designed along the draft sequence. The assembled data revealed a classical arenavirus genome structure (GenBank accession nos GU078660 and GU078661).
Phylogenetic analysis.
A set of OW and NW arenavirus sequences (50 for the L segment; 95 for the GPC gene; 116 for the NP gene) comprising all arenavirus sequences available from GenBank (June 2009) were used to assess the phylogenetic history of MWV. All arenavirus sequences were aligned by using the clustal algorithm [as implemented in the mega package (version 3); Kumar et al., 2004] at the amino acid level, with additional manual editing to ensure the highest possible quality of the alignment. Bayesian phylogenetic analyses of the sequence differences among the S and L segments of arenaviruses were conducted by using the beast, beauti and Tracer analysis software packages (Drummond & Rambaut, 2007). Preliminary analyses were run for 10 000 000 generations with the HKY+G nucleotide-substitution model to select the clock and demographic models most appropriate for the GPC, NP and L datasets. An analysis of the marginal likelihoods indicated that the relaxed lognormal molecular clock and constant population size model was decisively chosen (log10 Bayes factors=78.416 for GPC ORF, 11.127 for NP and 25.097 for L). Final data analyses included Markov chain Monte Carlo chain lengths of 10 000 000 generations, with sampling every 1000 states.
Reference arenavirus G1 sequences were aligned by using promals 3D software and 3D T-Coffee to obtain a secondary-structure alignment. Resulting alignments were analysed with the same parameters as described above.
Sequence analysis.
Programs of the Geneious package (Biomatters, NZ) were used for sequence assembly and analysis; percentage sequence identities were calculated by using mega. Topology and targeting predictions were generated by employing SignalP, NetNGlyc, tmhmm (http://www.cbs.dtu.dk/services), the web-based version of TopPred2 (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html) and Phobius (http://phobius.sbc.su.se/) (Bendtsen et al., 2004; Claros & von Heijne, 1994; Käll et al., 2004; Krogh et al., 2001).
Supplementary Material
Acknowledgments
We are grateful to Robert Swanepoel for providing isolate SPU 353-85. This work was supported by National Institutes of Health awards AI051292, U54 AI57158 (North-east Biodefense Center – W. I. L.) and NO1 AI30027, and by Google.org and the US Department of Defense.
Footnotes
References
- Archer, A. M. & Rico-Hesse, R. (2002). High genetic divergence and recombination in arenaviruses from the Americas. Virology 304, 274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auperin, D. D., Romanowski, V., Galinski, M. & Bishop, D. H. (1984). Sequencing studies of pichinde arenavirus S RNA indicate a novel coding strategy, an ambisense viral S RNA. J Virol 52, 897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auperin, D. D., Sasso, D. R. & McCormick, J. B. (1986). Nucleotide sequence of the glycoprotein gene and intergenic region of the Lassa virus S genome RNA. Virology 154, 155–167. [DOI] [PubMed] [Google Scholar]
- Beaty, B., Calisher, C. & Shope, R. (1995). Arboviruses. In Diagnostic Procedures for Viral, Rickettsial and Chlamydial Infections, 7th edn, pp. 189–212. Edited by E. H. Lennette, D. A. Lennette & E. T. Lennette. Washington, DC: American Public Health Association.
- Bendtsen, J. D., Nielsen, H., von Heijne, G. & Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Beyer, W. R., Popplau, D., Garten, W., von Laer, D. & Lenz, O. (2003). Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J Virol 77, 2866–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen, M. D., Peters, C. J. & Nichol, S. T. (1996). The phylogeny of New World (Tacaribe complex) arenaviruses. Virology 219, 285–290. [DOI] [PubMed] [Google Scholar]
- Bowen, M. D., Peters, C. J. & Nichol, S. T. (1997). Phylogenetic analysis of the Arenaviridae: patterns of virus evolution and evidence for cospeciation between arenaviruses and their rodent hosts. Mol Phylogenet Evol 8, 301–316. [DOI] [PubMed] [Google Scholar]
- Bowen, M. D., Rollin, P. E., Ksiazek, T. G., Hustad, H. L., Bausch, D. G., Demby, A. H., Bajani, M. D., Peters, C. J. & Nichol, S. T. (2000). Genetic diversity among Lassa virus strains. J Virol 74, 6992–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese, T., Paweska, J. T., McMullan, L. K., Hutchison, S. K., Street, C., Palacios, G., Khristova, M. L., Weyer, J., Swanepoel, R. & other authors (2009). Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog 5, e1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley, S. M. & Casals, J. (1970). Lassa fever, a new virus disease of man from West Africa. 3. Isolation and characterization of the virus. Am J Trop Med Hyg 19, 680–691. [DOI] [PubMed] [Google Scholar]
- Burns, J. W. & Buchmeier, M. J. (1991). Protein–protein interactions in lymphocytic choriomeningitis virus. Virology 183, 620–629. [DOI] [PubMed] [Google Scholar]
- Burns, J. W. & Buchmeier, M. J. (1993). Glycoproteins of the arenaviruses. In The Arenaviridae, pp. 17–35. Edited by M. S. Salvato. New York: Plenum Press.
- Cao, W., Henry, M. D., Borrow, P., Yamada, H., Elder, J. H., Ravkov, E. V., Nichol, S. T., Compans, R. W., Campbell, K. P. & Oldstone, M. B. (1998). Identification of α-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282, 2079–2081. [DOI] [PubMed] [Google Scholar]
- Capul, A. A., Perez, M., Burke, E., Kunz, S., Buchmeier, M. J. & de la Torre, J. C. (2007). Arenavirus Z–glycoprotein association requires Z myristoylation but not functional RING or late domains. J Virol 81, 9451–9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel, R. N., de Lamballerie, X. & Fulhorst, C. F. (2001). The Whitewater Arroyo virus: natural evidence for genetic recombination among Tacaribe serocomplex viruses (family Arenaviridae). Virology 283, 161–166. [DOI] [PubMed] [Google Scholar]
- Charrel, R. N., de Lamballerie, X. & Emonet, S. (2008). Phylogeny of the genus Arenavirus. Curr Opin Microbiol 11, 362–368. [DOI] [PubMed] [Google Scholar]
- Claros, M. G. & von Heijne, G. (1994). TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci 10, 685–686. [DOI] [PubMed] [Google Scholar]
- Cox-Foster, D. L., Conlan, S., Holmes, E. C., Palacios, G., Evans, J. D., Moran, N. A., Quan, P. L., Briese, T., Hornig, M. & other authors (2007). A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318, 283–287. [DOI] [PubMed] [Google Scholar]
- Davis, D. (1962). Distribution patterns of southern African Muridae, with some of their fossil antecedents. Ann Cape Prov Mus 2, 56–76. [Google Scholar]
- Delarue, M., Poch, O., Tordo, N., Moras, D. & Argos, P. (1990). An attempt to unify the structure of polymerases. Protein Eng 3, 461–467. [DOI] [PubMed] [Google Scholar]
- Downs, W. G., Anderson, C. R., Spence, L., Aitken, T. H. G. & Greenhall, A. H. (1963). Tacaribe virus, a new agent isolated from Artibeus bats and mosquitoes in Trinidad, West Indies. Am J Trop Med Hyg 12, 640–646. [DOI] [PubMed] [Google Scholar]
- Drummond, A. J. & Rambaut, A. (2007). beast: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler, R., Lenz, O., Strecker, T., Eickmann, M., Klenk, H. D. & Garten, W. (2004). Lassa virus glycoprotein signal peptide displays a novel topology with an extended endoplasmic reticulum luminal region. J Biol Chem 279, 12293–12299. [DOI] [PubMed] [Google Scholar]
- Emonet, S., Lemasson, J. J., Gonzalez, J. P., de Lamballerie, X. & Charrel, R. N. (2006). Phylogeny and evolution of Old World arenaviruses. Virology 350, 251–257. [DOI] [PubMed] [Google Scholar]
- Fulhorst, C. F., Bowen, M. D., Ksiazek, T. G., Rollin, P. E., Nichol, S. T., Kosoy, M. Y. & Peters, C. J. (1996). Isolation and characterization of Whitewater Arroyo virus, a novel North American arenavirus. Virology 224, 114–120. [DOI] [PubMed] [Google Scholar]
- Fulhorst, C. E., Bowen, M. D., Salas, R. A., de Manzione, N. M., Duno, G., Utrera, A., Ksiazek, T. G., Peters, C. J., Nichol, S. T. & other authors (1997). Isolation and characterization of pirital virus, a newly discovered South American arenavirus. Am J Trop Med Hyg 56, 548–553. [DOI] [PubMed] [Google Scholar]
- Ghiringhelli, P. D., Rivera-Pomar, R. V., Lozano, M. E., Grau, O. & Romanowski, V. (1991). Molecular organization of Junin virus S RNA: complete nucleotide sequence, relationship with other members of the Arenaviridae and unusual secondary structures. J Gen Virol 72, 2129–2141. [DOI] [PubMed] [Google Scholar]
- Gonzalez, J. P., McCormick, J. B., Saluzzo, J. F., Herve, J. P., Georges, A. J. & Johnson, K. M. (1983). An arenavirus isolated from wild-caught rodents (Pramys species) in the Central African Republic. Intervirology 19, 105–112. [DOI] [PubMed] [Google Scholar]
- Gonzalez, J. P., Sanchez, A. & Rico-Hesse, R. (1995). Molecular phylogeny of Guanarito virus, an emerging arenavirus affecting humans. Am J Trop Med Hyg 53, 1–6. [PubMed] [Google Scholar]
- Gonzalez, J. P., Bowen, M. D., Nichol, S. T. & Rico-Hesse, R. (1996). Genetic characterization and phylogeny of Sabiá virus, an emergent pathogen in Brazil. Virology 221, 318–324. [DOI] [PubMed] [Google Scholar]
- Hui, E. K., Barman, S., Yang, T. Y. & Nayak, D. P. (2003). Basic residues of the helix six domain of influenza virus M1 involved in nuclear translocation of M1 can be replaced by PTAP and YPDL late assembly domain motifs. J Virol 77, 7078–7092. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Iapalucci, S., Lopez, N. & Franze-Fernandez, M. T. (1991). The 3′ end termini of the Tacaribe arenavirus subgenomic RNAs. Virology 182, 269–278. [DOI] [PubMed] [Google Scholar]
- Joazeiro, C. A., Wing, S. S., Huang, H., Leverson, J. D., Hunter, T. & Liu, Y. C. (1999). The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286, 309–312. [DOI] [PubMed] [Google Scholar]
- Johnson, K. M., Wiebenga, N. H., Mackenzie, R. B., Kuns, M. L., Tauraso, N. M., Shelokov, A., Webb, P. A., Justines, G. & Beye, H. K. (1965). Virus isolations from human cases of hemorrhagic fever in Bolivia. Proc Soc Exp Biol Med 118, 113–118. [DOI] [PubMed] [Google Scholar]
- Käll, L., Krogh, A. & Sonnhammer, E. L. (2004). A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338, 1027–1036. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K. & Nei, M. (2004). mega3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5, 150–163. [DOI] [PubMed] [Google Scholar]
- Lenz, O., ter Meulen, J., Feldmann, H., Klenk, H. D. & Garten, W. (2000). Identification of a novel consensus sequence at the cleavage site of the Lassa virus glycoprotein. J Virol 74, 11418–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz, O., ter Meulen, J., Klenk, H. D., Seidah, N. G. & Garten, W. (2001). The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc Natl Acad Sci U S A 98, 12701–12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisieux, T., Coimbra, M., Nassar, E. S., Burattini, M. N., de Souza, L. T. M., Ferreira, I. B., Rocco, I. M., da Rosa, A. P. A. T., Vasconcelos, P. F. C., Pinheiro, F. P. & other authors (1994). New arenavirus isolated in Brazil. Lancet 343, 391–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies, M., Egholm, M., Altman, W. E., Attiya, S., Bader, J. S., Bemben, L. A., Berka, J., Braverman, M. S., Chen, Y. J. & other authors (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier, D. Y., McCormick, J. B., Georges, A. J., Georges, M. C. & Gonzalez, J. P. (1985). Comparison of Lassa, Mobala, and Ippy virus reactions by immunofluorescence test. Lancet 1, 873–874. [DOI] [PubMed] [Google Scholar]
- Moncayo, A. C., Hice, C. L., Watts, D. M., Travassos de Rosa, A. P., Guzman, H., Russell, K. L., Calampa, C., Gozalo, A., Popov, V. L. & other authors (2001). Allpahuayo virus: a newly recognized arenavirus (Arenaviridae) from arboreal rice rats (Oecomys bicolor and Oecomys paricola) in northeastern Peru. Virology 284, 277–286. [DOI] [PubMed] [Google Scholar]
- Müller, R., Poch, O., Delarue, M., Bishop, D. H. & Bouloy, M. (1994). Rift Valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J Gen Virol 75, 1345–1352. [DOI] [PubMed] [Google Scholar]
- Musser, G. & Carleton, M. (2005). Superfamily Muroidea. In Mammal Species of the World: a Taxonomic and Geographic Reference, pp. 894–1531. Edited by D. Wilson & D. Reeder. Washington, DC: Smithsonian Institution Press.
- Neuman, B. W., Adair, B. D., Burns, J. W., Milligan, R. A., Buchmeier, M. J. & Yeager, M. (2005). Complementarity in the supramolecular design of arenaviruses and retroviruses revealed by electron cryomicroscopy and image analysis. J Virol 79, 3822–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios, G., Quan, P. L., Jabado, O. J., Conlan, S., Hirschberg, D. L., Liu, Y., Zhai, J., Renwick, N., Hui, J. & other authors (2007). Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis 13, 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios, G., Druce, J., Du, L., Tran, T., Birch, C., Briese, T., Conlan, S., Quan, P. L., Hui, J. & other authors (2008). A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med 358, 991–998. [DOI] [PubMed] [Google Scholar]
- Parisi, G., Echave, J., Ghiringhelli, D. & Romanowski, V. (1996). Computational characterisation of potential RNA-binding sites in arenavirus nucleocapsid proteins. Virus Genes 13, 247–254. [DOI] [PubMed] [Google Scholar]
- Parodi, A. S., Greenway, D. J., Rugiero, H. R., Frigerio, M., De La Barrera, J. M., Mettler, N., Garzon, F., Boxaca, M., Guerrero, L. & Nota, N. (1958). Concerning the epidemic outbreak in Junin. Dia Med 30, 2300–2301 (in Spanish). [PubMed] [Google Scholar]
- Paweska, J., Sewlall, N., Ksiazek, T., Blumberg, L., Hale, M., Lipkin, W. I., Weyer, J., Nichol, S. T., Rollin, P. E. & other authors (2009). Nosocomial outbreak of novel arenavirus infection, Southern Africa. Emerg Infect Dis 15 [DOI] [PMC free article] [PubMed]
- Perez, M., Craven, R. C. & de la Torre, J. C. (2003). The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci U S A 100, 12978–12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, M., Greenwald, D. L. & de la Torre, J. C. (2004). Myristoylation of the RING finger Z protein is essential for arenavirus budding. J Virol 78, 11443–11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, C. J. (2006). Lymphocytic choriomeningitis virus – an old enemy up to new tricks. N Engl J Med 354, 2208–2211. [DOI] [PubMed] [Google Scholar]
- Pinheiro, F. P. & Woodall, J. P. (1969). Ecological Studies on Amapari Virus, p. 18. Rio de Janeiro: Fundacao Servico Especial de Saude Publica Rio de Janeiro.
- Pirosky, I., Zuccarini, J., Molinelli, E. A. & Di Pietro, A. (1959). Virosis hemorragica del noroeste bonaerense. II. Recuperacion del virus causal a partir de acaros (Mesostigmata) capturados (1958) en la zona epidemica. Orientacion Medica 8, 156(in Spanish) [Google Scholar]
- Poch, O., Sauvaget, I., Delarue, M. & Tordo, N. (1989). Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J 8, 3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puffer, B. A., Parent, L. J., Wills, J. W. & Montelaro, R. C. (1997). Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol 71, 6541–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoshitzky, S. R., Abraham, J., Spiropoulou, C. F., Kuhn, J. H., Nguyen, D., Li, W., Nagel, J., Schmidt, P. J., Nunberg, J. H. & other authors (2007). Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek, J. M., Lee, A. M., Nguyen, N., Spiropoulou, C. F. & Kunz, S. (2008a). Site 1 protease is required for proteolytic processing of the glycoproteins of the South American hemorrhagic fever viruses Junin, Machupo, and Guanarito. J Virol 82, 6045–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek, J. M., Perez, M. & Kunz, S. (2008b). Cellular entry of lymphocytic choriomeningitis virus. J Virol 82, 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski, V. & Bishop, D. H. (1985). Conserved sequences and coding of two strains of lymphocytic choriomeningitis virus (WE and ARM) and Pichinde arenavirus. Virus Res 2, 35–51. [DOI] [PubMed] [Google Scholar]
- Salas, R., de Manzione, N., Tesh, R. B., Rico-Hesse, R., Shope, R. E., Betancourt, A., Godoy, O., Bruzual, R., Pacheco, M. E. & other authors (1991). Venezuelan haemorrhagic fever. Lancet 338, 1033–1036. [DOI] [PubMed] [Google Scholar]
- Salvato, M. S. & Shimomaye, E. M. (1989). The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology 173, 1–10. [DOI] [PubMed] [Google Scholar]
- Salvato, M., Clegg, J., Buchmeier, M., Charrel, R., Gonzalez, J., Lukashevich, I., Peters, C., Rico-Hesse, R. & Romanowski, V. (2005). Family Arenaviridae. In Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses, pp. 725–733. Edited by C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger & L. A. Ball. London: Elsevier Academic Press.
- Skinner, J. & Smithers, R. (1990). The Mammals of the Southern African Subregion. Pretoria: University of Pretoria.
- Spiropoulou, C. F., Kunz, S., Rollin, P. E., Campbell, K. P. & Oldstone, M. B. (2002). New World arenavirus clade C, but not clade A and B viruses, utilizes α-dystroglycan as its major receptor. J Virol 76, 5140–5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub, O., Dho, S., Henry, P., Correa, J., Ishikawa, T., McGlade, J. & Rotin, D. (1996). WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J 15, 2371–2380. [PMC free article] [PubMed] [Google Scholar]
- Strecker, T., Eichler, R., Meulen, J., Weissenhorn, W., Dieter Klenk, H., Garten, W. & Lenz, O. (2003). Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles. J Virol 77, 10700–10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker, T., Maisa, A., Daffis, S., Eichler, R., Lenz, O. & Garten, W. (2006). The role of myristoylation in the membrane association of the Lassa virus matrix protein Z. Virol J 3, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanepoel, R., Leman, P. A., Shepherd, A. J., Shepherd, S. P., Kiley, M. P. & McCormick, J. B. (1985). Identification of Ippy as a Lassa-fever-related virus. Lancet 1, 639. [DOI] [PubMed] [Google Scholar]
- Vieth, S., Torda, A. E., Asper, M., Schmitz, H. & Gunther, S. (2004). Sequence analysis of L RNA of Lassa virus. Virology 318, 153–168. [DOI] [PubMed] [Google Scholar]
- Whitton, J. L., Tishon, A., Lewicki, H., Gebhard, J., Cook, T., Salvato, M., Joly, E. & Oldstone, M. B. (1989). Molecular analyses of a five-amino-acid cytotoxic T-lymphocyte (CTL) epitope: an immunodominant region which induces nonreciprocal CTL cross-reactivity. J Virol 63, 4303–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, S. M. & Clegg, J. C. (1991). Sequence analysis of the S RNA of the African arenavirus Mopeia: an unusual secondary structure feature in the intergenic region. Virology 180, 543–552. [DOI] [PubMed] [Google Scholar]
- Wulff, H., McIntosh, B. M., Hamner, D. B. & Johnson, K. M. (1977). Isolation of an arenavirus closely related to Lassa virus from Mastomys natalensis in south-east Africa. Bull World Health Organ 55, 441–444. [PMC free article] [PubMed] [Google Scholar]
- York, J., Romanowski, V., Lu, M. & Nunberg, J. H. (2004). The signal peptide of the Junín arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1–G2 complex. J Virol 78, 10783–10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.