Abstract
The haemagglutinin (HA) glycoprotein of influenza A virus is a major antigen that initiates humoral immunity against infection; however, the cellular immune response against HA is poorly understood. Furthermore, HA-derived cytotoxic T-lymphocyte (CTL) epitopes are relatively rare in comparison to other internal gene products. Here, CTL epitopes of the HA serotype H5 protein were screened. By using in silico prediction, in vitro refolding and a T2 cell-binding assay, followed by immunization of HLA-A2.1/Kb transgenic mice, an HLA-A*0201-restricted decameric epitope, RI-10 (H5 HA205–214, RLYQNPTTYI), was shown to elicit a robust CTL epitope-specific response. In addition, RI-10 and its variant, KI-10 (KLYQNPTTYI), were also demonstrated to be able to induce a higher CTL epitope-specific response than the influenza A virus dominant CTL epitope GL-9 (GILGFVFTL) in peripheral blood mononuclear cells of HLA-A*0201-positive patients who had recovered from H5N1 virus infection. Furthermore, the crystal structures of RI-10–HLA-A*0201 and KI-10–HLA-A*0201 complexes were determined at 2.3 and 2.2 Å resolution, respectively, showing typical HLA-A*0201-restricted epitopes. The conformations of RI-10 and KI-10 in the antigen-presenting grooves in crystal structures of the two complexes show significant differences, despite their nearly identical sequences. These results provide implications for the discovery of diagnostic markers and the design of novel influenza vaccines.
INTRODUCTION
Influenza A virus has always been a major cause of morbidity and mortality throughout history. Both the ability of the virus genome to mutate at a very high rate and its special gene segment-reassortment mechanism often lead to the emergence of highly virulent strains that may potentially become the cause of a pandemic, or at least some panic worldwide, for example the recent outbreak of swine-origin influenza A (H1N1).
The highly pathogenic avian influenza virus (HPAIV) H5N1 has aroused global attention since 1997, when it was transmitted directly from chicken to humans in Hong Kong and resulted in a severe respiratory disease in 18 humans, of whom six died (Claas et al., 1998). From late 2003, H5N1 re-emerged and reached epizootic levels in domestic fowl and even spread to wild birds. In April 2005, a large-scale outbreak of H5N1 infection occurred in migratory waterfowl in Qinghai Lake Nature Reserve of western China, killing more than 6000 wild birds (Chen et al., 2005, 2006; Liu et al., 2005; Wang et al., 2008; Zhou et al., 2006a). Since then, migratory birds have been recognized as a significant source of international spread of HPAIV (Kilpatrick et al., 2006; Normile, 2006; Olsen et al., 2006; Wang et al., 2008). Between December 2003 and 23 April 2009, 421 human H5N1 infection cases and 257 deaths were detected in 15 countries (WHO, 2009), although so far no sustained human-to-human transmission has been found. However, the possibility of a human influenza pandemic resulting from either mutations or avian-origin virus reassortments with human strains caused great international concern and pandemic preparedness has been accelerated (Salomon & Webster, 2009).
In addition to the antibody-induced humoral immune response, cellular immunity plays a variety of roles in defending against influenza virus infection and these have been increasingly documented (Bender & Small, 1992; Rimmelzwaan et al., 2007; Thomas et al., 2006; Yap et al., 1978). In many observations, influenza virus-specific cytotoxic T lymphocytes (CTLs) target mainly viral internal proteins (Gotch et al., 1987; Kees & Krammer, 1984; Townsend & Skehel, 1984; Wahl et al., 2009). As a result, many studies have focused on the cross-reactive CTL response against internal proteins conserved between the H5N1 influenza subtype and seasonal influenza viruses, assuming that a cross-reactive CTL response against the conserved regions of the viral proteome may provide partial protection against H5 influenza virus and dampen the impact of the next pandemic (Ahmed et al., 2007; Doherty & Kelso, 2008; Heiny et al., 2007; Kreijtz et al., 2008; Lee et al., 2008).
Although there are lines of evidence that the viral surface haemagglutinin (HA) elicits a CTL response (Bennink et al., 1984; Townsend et al., 1986; Wabuke-Bunoti et al., 1984), HA-specific CTL epitopes are rare compared with those specific for the viral internal proteins. HA glycoproteins are responsible for virus binding to host receptors, enabling entry into the host cell through endocytosis and subsequent membrane fusion (Wilson & Cox, 1990). HA is recognized as the primary target of neutralizing antibodies and is an important target for both drug and vaccine development (Stevens et al., 2006). However, the role of HA in CTL-based cellular immunity and the major histocompatibility complex (MHC or HLA in humans)-restricted HA epitopes are poorly understood. In the present study, we have successfully identified CTL epitopes in H5 HA restricted by HLA-A*0201 and have shown an immune response in patients who have recovered from H5N1 infection. HLA-A*0201 is an allele expressed by nearly half of the world's population and the peptide–HLA (pHLA) complexes defined here are clearly seen by X-ray crystallography, confirming the authenticity of the epitope presentation.
RESULTS
Selection of potential HLA-A*0201-binding peptides within the H5 protein
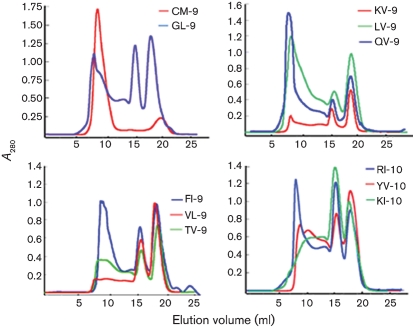
Based on bimas software analysis, 15 candidate nonameric and decameric peptides with the highest estimated half-life of dissociation were synthesized (Table 1). To evaluate the binding ability of these peptides to the HLA-A*0201 molecule, we employed a peptide-induced stabilization assay (complex refolding) of HLA-A*0201 heavy chain and β2 microglobulin (β2m) in vitro. In this experiment, the correctly refolded complex is detected if the right peptide is presented by the HLA heavy chain and β2m. Among the 15 synthesized peptides, nine could be refolded with HLA-A*0201 heavy chain and β2m molecules (Fig. 1). To further test the peptide presentation of HLA-A*0201, a T2 cell-binding assay was employed, which measures the increase of HLA-A*0201 molecules induced on the surface of T2 cells following exposure to an exogenous HLA-A*0201-binding peptide. As they are a TAP-defective HLA-A2-expressing cell line, T2 cells cannot present endogenous peptides. However, when T2 cells are exposed to an exogenous HLA-A2-binding peptide, the expression of HLA-A2 molecules on the cell surface will increase, because peptide binding stabilizes the HLA-A2 molecules on the cell surface. Among nine candidate peptides that can refold with HLA-A*0201, seven could be presented on the cell surface (Table 1).
Table 1.
Predicted HLA-A*0201-restricted peptides for H5 HA epitopes and presentation by and binding to HLA-A*0201
The prediction is based on the HA sequence of AIV strain A/bar-headed goose/Qinghai/1/05 (H5N1).
| No. | Start position* | Name† | Sequence | Score‡ | Refolding§ | T2 binding|| |
|---|---|---|---|---|---|---|
| P1 | 6 | LV-9 | LLLAIVSLV | 1006.209 | + | – |
| P2 | 86 | FI-10 | FLNVPEWSYI | 448.715 | + | + |
| P3 | 5 | VLL-9 | VLLLAIVSL | 309.050 | – | – |
| P4 | 463 | NL-10 | NLYDKVRLQL | 280.275 | – | – |
| P5 | 314 | TV-9 | TIGECPKYV | 215.655 | + | + |
| P6 | 205 | RI-10 | RLYQNPTTYI | 183.617 | + | + |
| P7 | 404 | KV-9 | KMNTQFEAV | 163.681 | + | + |
| P8 | 34 | TV-10 | TIMEKNVTV | 145.077 | – | – |
| P9 | 446 | VL-9 | VLMENERTL | 110.183 | + | + |
| P10 | 429 | KMV-9 | KMEDGFLDV | 87.653 | – | – |
| P11 | 531 | QV-9 | QILSIYSTV | 53.077 | + | + |
| P12 | 207 | YV-10 | YQNPTTYISV | 48.657 | + | + |
| P13 | 304 | SL-10 | SMPFHNIHPL | 35.485 | – | – |
| P14 | 547 | IM-10 | IMVAGLSLWM | 33.548 | – | – |
| P15¶ | 205 | KI-10 | KLYQNPTTYI | 642.660 | + | + |
| P16# | 58 | GL-9 | GILGFVFTL | 550.927 | + | + |
| P17** | 181 | CM-9 | CTPYDINQM | 0.159 | – | – |
*The position of the first amino acid of the signal peptide in the sequence is defined as 1.
†The peptides are denominated with their first and last residue code and the number of their residues. If two peptides have the same name, then one of them is denominated by the first two residue codes, the last residue code and the number of their residues (e.g. VLL-9).
‡Estimated half-time of dissociation (T1/2) of HLA-A*0201 peptide complexes, calculated by using bimas (http://bimas.dcrt.nih.gov/molbio/hla_bind/index.html).
§In the refolding assay, if an elution peak representing the complex of the peptide and HLA-A*0201 appears in gel filtration, the peptide is thought to be able to bind to HLA-A*0201 and is defined as +; otherwise it is defined as –.
||In the T2-cell binding assay, the affinity of the peptide to HLA-A*0201 is measured by the fluorescence index (FI; see Methods). If the FI of a given peptide is ≥1, it is defined as +; otherwise it is defined as –.
¶P15 (KI-10) is a variant of P6 (RI-10), found in the HA sequence of some human H5N1 virus isolates.
#P16 (GL-9), the HLA-A*0201-restricted influenza virus M1 (58–66) peptide, was used as a positive control for HLA-A*0201-binding ability.
**P17 (CM-9), the Mamu-A*01 restricted (SIV)-derived peptide gag (aa 181–189), was used as negative control.
Fig. 1.
Peptide-induced stabilization assay of HLA-A*0201 molecules by refolding. In the refolding assay, the refolded complexes, eluted with the expected molecular mass of 45 kDa, were analysed by FPLC Superdex G200 gel-filtration chromatography. The positive control GL-9 peptide is HLA-A*0201-restricted influenza virus M1 aa 58–66 (GILGFVFTL) and the negative control CM-9 peptide is a non-HLA-A*0201-restricted unrelated peptide derived from SIV. Peptides KV-9, LV-9, QV-9, VL-9, TV-9, FI-10, RI-10, YV-10 and KI-10, which could refold with HLA-A*0201 heavy chain and β2m, showed peaks with an elution volume of 15 ml in comparison with other peptides.
CTL epitope-induced gamma interferon (IFN-γ) production in HLA-A2.1/Kb transgenic (Tg) mice by vaccinia–H5 or pcDNA3.0–H5 immunization
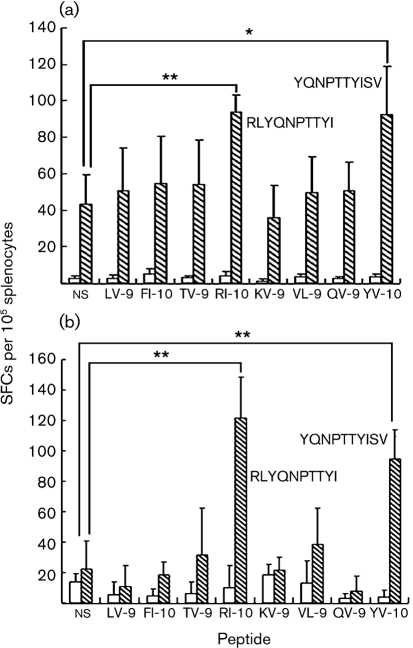
In order to examine whether the peptides that can bind to the HLA-A*0201 molecule are able to induce a CTL epitope-specific response, we immunized HLA-A2.1/Kb Tg mice using vaccinia virus expressing the H5 HA of H5N1 virus strain A/bar-headed goose/Qinghai/1/05 (vaccinia–H5) three times at an interval of 4 weeks. The splenocytes were isolated 10 days after the last immunization and a peptide-specific putative CTL response was detected by ELISPOT assay. (As cells were stimulated with CTL epitopes, we presumed that the IFN-γ was produced by CD8+ cells and referred to it as a putative CTL response, although we did not do a CTL killing assay. IFN-γ-producing CD4+ or γδ T cells cannot be ruled out completely.) A significantly higher number of spot-forming cells (SFCs) were produced in the splenocytes from vaccinia-immunized HLA-A2.1/Kb Tg mice compared with those from PBS-immunized HLA-A2.1/Kb Tg mice (P<0.05, t test), suggesting that vaccinia–H5 immunization induced a high IFN-γ-producing response. Noticeably, in the splenocytes of mice immunized with vaccinia–H5, the numbers of SFCs increased significantly in the presence of two overlapping decameric peptides, RI-10 (RLYQNPTTYI) and YV-10 (YQNPTTYISV), compared with that in the absence of peptide stimulation (ns) (P<0.01 and P<0.05, respectively; Student's t test). In contrast, with the stimulation of other candidate peptides, the numbers of SFCs did not increase significantly (Fig. 2a).
Fig. 2.
Detection of CTL-epitope-specific IFN-γ-producing cells in vaccinia–H5- (a) or pcDNA3.0–H5- (b) immunized HLA-A2.1/Kb Tg mice (hatched bars) by ELISPOT assay. Empty bars represent negative controls [PBS (a); pcDNA3.0 (b)]. Female HLA-A2.1/Kb Tg mice, 6–8 weeks old, were immunized with 3–106 p.f.u. vaccinia–H5 in 20 μl PBS or PBS alone at weeks 0, 4 and 8. Ten days after the final boosting, splenocytes were isolated and CTL epitope-specific IFN-γ-producing cells were evaluated by ELISPOT assay. In the ELISPOT assay, splenocytes were stimulated in vitro in the presence of 20 U interleukin-2 (IL-2) ml−1 and 20 μM of the designated peptide for 24 h. ns represents cells that were cultured in the absence of any peptide. * (P<0.05) and ** (P<0.01) indicate significant differences between peptides and ns, using Student's t test.
Because there were many IFN-γ-producing cells in the absence of peptide stimulation in the splenocytes of vaccinia–H5-immunized Tg mice, the SFCs increased by only about two times with the stimulation of RI-10 and YV-10, although the statistical difference is significant. However, when we stimulated the splenocytes of the Tg mice immunized with pcDNA3.0–H5 with the eight peptides refolded with HLA-A*0201 molecules, RI-10 and YV-10 elicited a vigorous response (P<0.01 compared with ns; Student's t test). These results suggested that, among the eight peptides that could refold with HLA-A*0201, only RI-10 and YV-10 were naturally processed CTL epitopes with potent antigenicity (Fig. 2b).
Induction of CTL epitope-specific IFN-γ-producing T cells in HLA-A2.1/Kb Tg mice by peptide immunization
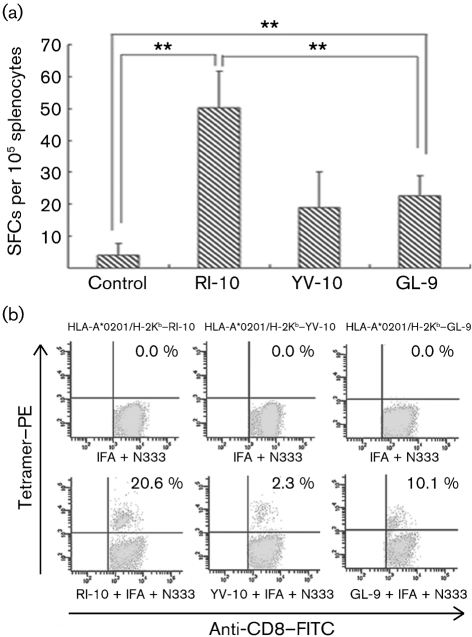
In order to further examine the antigenicity of RI-10 and YV-10, we immunized HLA-A2.1/Kb Tg mice with RI-10, YV-10 or GL-9 peptide. This was done following our previous regime by using incomplete Freund's adjuvant and murine GP96 N-terminal fragment N333 as adjuvant, which has been confirmed to be very effective (Li et al., 2005). The peptide-specific IFN-γ-producing T-cell-response was detected by ELISPOT assay and tetramer staining. After immunization with the peptides three times at intervals of 2 weeks in vivo and stimulation with the peptides for 2 weeks in vitro, peptide-specific T cells were detected. The ELISPOT assay showed that RI-10 induced a significant IFN-γ response that was even higher than that with the influenza dominant epitope GL-9 (P<0.01, Student's t test). YV-10 also induced a certain degree of IFN-γ response, but it was much weaker than that induced by RI-10 (P<0.01, Student's t test) and was not significantly different from the control group (P>0.05, Student's t test; Fig. 3a). Similarly, tetramer staining also showed that RI-10 elicited a robust CTL epitope-specific CD8+ T-cell response and that RI-10-specific CD8+ cells accounted for 20.6 % of CD8+ T cells, whereas the proportion of GL-9-specific CD8+ cells was 10.1 % (Fig. 3b). However, YV-10 induced a much weaker specific CD8+ T-cell response and the proportion of YV-10-specific CD8+ T cells was only 2.3 %. These results suggest that RI-10 is a CTL epitope with potent antigenicity, but that the antigenicity of YV-10 is much weaker in this scenario.
Fig. 3.
Detection of peptide-specific IFN-γ-producing cells in peptide-immunized HLA-A2.1/Kb Tg mice by ELISPOT assay and tetramer staining. Female Tg mice, 6–8 weeks old, were immunized with a mixture of peptides, incomplete Freund's adjuvant (IFA) and the N-terminal fragment N333 (aa 22–355) of murine gp96 three times with intervals of 2 weeks. Control mice were immunized with only IFA and N333. Splenocytes were harvested 10 days after the last immunization and stimulated in the presence of 20 U IL-2 ml−1 and 10 μM peptide for 2 weeks in vitro. Then, the cells were harvested for ELISPOT assay (a) and tetramer staining (b) as described in Methods. ** indicates a significant difference (P<0.01, Student's t test).
The reason that YV-10 elicited a comparable response with RI-10 in vaccinia–H5-immunized HLA-A*0201/Kb Tg mice, but induced a much weaker CD8+ T-cell response in peptide-immunized Tg mice, is unknown. However, YV-10 may not bind well to HLA-A2 because it does not harbour a typical anchor motif for HLA-A2. Indeed, it showed a much lower binding affinity for the HLA-A*0201 molecule on the cell surface in the T2 cell-binding assay compared with RI-10 (data not shown), in spite of its ability to bind to HLA-A2 in the refolding assay.
We did not test KI-10 in the Tg mice, although it showed obvious binding to the HLA-A*0201 molecule in the refolding/T2 assays, because it is different from RI-10 only in the first residue. The first residue of both peptides is a basic amino acid. We speculated that KI-10 would share similar antigenicity with RI-10. This was confirmed by detection of a specific CD8+ T-cell response in the peripheral blood mononuclear cells (PBMCs) of patients who had recovered from H5N1 infection in the following experiments.
Detection of CTL epitope-specific CD8+ T cells in PBMCs of patients who had recovered from avian influenza virus (AIV) infection
In order to confirm whether the results obtained in HLA-A2 Tg mice could also be applied to humans, we examined whether RI-10 and its variant KI-10 could induce a CTL epitope-specific CD8+ T-cell response in PBMCs from human samples. We do not exclude the possibility that other candidate peptides that can bind to the HLA-A2 molecule, but do not induce a CTL response in HLA-A*0201/Kb Tg mice, may induce a CTL epitope-specific response in humans. All HLA-A2-binding peptides will be tested in human samples when more blood samples of HLA-A2-positive patients who have recovered from H5N1 virus infection are available to us. Here, we focus on the most promising candidates, RI-10 and its variant KI-10, due to sample limitation.
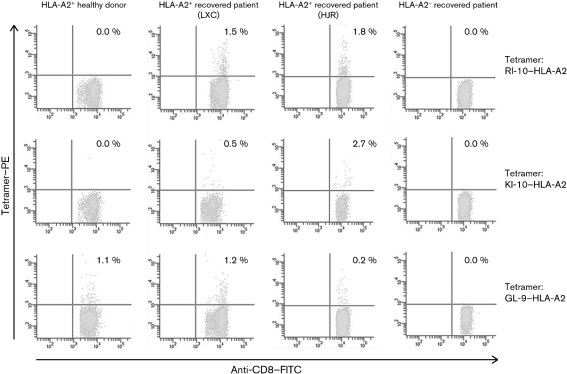
PBMC samples were collected from an HLA-A2-positive healthy donor and three survivors of AIV H5N1 infection. Two of the survivors are HLA-A*0201-positive (named LXC and HJR) and one is HLA-A2-negative. The samples of the three patients were collected about 15–18 months after the onset of disease. The PBMCs were stimulated for 7 days in the presence of 10 μM RI-10, KI-10 or GL-9 peptide and 20 U recombinant human IL-2 (rhIL-2) ml−1. They were then stained with RI-10–HLA-A*0201, KI-10–HLA-A*0201 or GL-9–HLA-A*0201 tetramer. The proportion of peptide-specific and cross-reactive CD8+ T cells was detected by flow cytometry. As shown in Fig. 4, when stained with RI-10 or KI-10-tetramer, CTL epitope-specific CD8+ T cells were detected in the PBMCs of patient LXC at 1.5 and 0.5 % in CD8+ T cells, respectively. The proportions of CTL epitope-specific CD8+ T cells for RI-10 and KI-10 in the PBMCs of patient HJR were 1.8 and 2.7 %, respectively. When staining the PBMCs with GL-9 (a common HLA-A*0201-restricted CTL epitope) tetramer, specific CTLs were detected at 1.2 and 0.2 % in CD8+ T cells in the two PBMC samples, which are both slightly lower than the values of the specific RI-10 or KI-10 tetramer staining. In contrast, the CTL epitope-specific CD8+ T cells could not be detected with RI-10, KI-10 or GL-9 tetramer staining in PMBCs of the HLA-A2-negative AIV-recovered patient. Specific CTLs were not detectable with RI-10 or KI-10 tetramer staining in the PBMCs of an HLA-A2-positive healthy control, but the proportion of GL-9-specific CTLs was 1.1 % in his CD8+ T cells.
Fig. 4.
Measurement of RI-10-, KI-10- and GL-9-specific CD8+ T cells in the PBMCs of AIV-infection-recovered donors and a healthy donor by tetramer staining. PBMCs of an HLA-A2+ healthy donor and three patients recovered from AIV infection (two HLA-A2+ AIV patients, named LXC and HJR, and an HLA-A2− AIV patient) were stimulated with 10 μM RI-10, KI-10 or GL-9 in the presence of 20 U IL-2 ml−1 for 1 week and the cells were harvested and stained using phycoerythrin (PE)-conjugated HLA-A*0201–RI-10, HLA-A*0201–KI-10 or HLA-A*0201–GL-9 tetramer, along with FITC-conjugated anti-CD8 monoclonal antibodies (mAbs) for cell staining. The numbers shown represent the percentage of tetramer+CD8+ cells within CD8+ T lymphocytes. The results are the representative of three independent experiments.
Crystal structures of the HLA-A*0201 complexes with RI-10 or KI-10 peptide
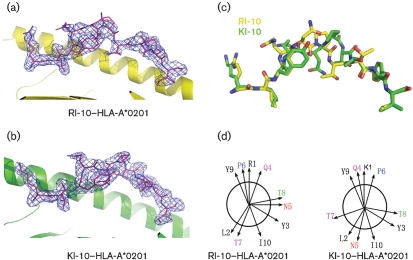
The RI-10–HLA-A*0201 and KI-10–HLA-A*0201 complexes were both crystallized in the P1 space group. The crystals diffracted X-rays to 2.3 and 2.2 Å resolution for the RI-10 and KI-10 complexes, respectively (Table 2). The structures were determined by molecular replacement, using a previously determined HLA-A2 structure as the search probe (PDB accession no. 1JF1). The RI-10–HLA-A2 complex structure has been refined to Rwork and Rfree of 20.2 and 24.7 %, respectively. The KI-10–HLA-A2 complex structure has been refined to Rwork and Rfree of 19.2 and 23.7 %, respectively. The crystallographic asymmetrical unit contains four complex molecules with the peptides adopting the same conformation and there were no direct lattice contacts involving the peptides. The electron densities of RI-10 and KI-10 peptides unambiguously identify their anchor residues Leu2 and Ile10, which point downwards and tether the peptides in the antigen-binding groove (Fig. 5a, b).
Table 2.
Data collection and refinement statistics
| RI-10 | KI-10 | |
|---|---|---|
| Non-hydrogen atoms | ||
| Protein | 13679 | 12680 |
| Water | 975 | 1087 |
| Space group | P1 | P1 |
| Unit cell dimension | a=63.21, b=79.32, c=87.15 ; α=90.00, β=90.00, γ=90.02 | a=63.172, b=79.304, c=86.739; α=90.02, β=89.99, γ=89.96 |
| Molecules in AU | 4 | 4 |
| Resolution (Å) | 33.6−2.30 | 25.5–2.20 |
| Measured reflections | 282436 | 165362 |
| Unique reflections | 75342 | 76982 |
| Data completeness (%) | 96.2 | 95.8 |
| I/σ (I) | 15.1 | 23.0 |
| Rwork | 0.2022 | 0.1920 |
| Rfree | 0.2470 | 0.2367 |
| RMS* deviations from ideality | ||
| Bond lengths (°) | 0.002 | 0.004 |
| Bond angles (°) | 0.575 | 0.817 |
| Dihedrals (°) | 15.309 | 16.316 |
| Ramachandran plot | ||
| Most favoured | 91.8 | 91.5 |
| Allowed region | 7.9 | 8.2 |
| Generously allowed region | 0.3 | 0.3 |
| B factors (Å2) | ||
| Average main chain | 32.09 | 31.34 |
| Average side chain | 33.50 | 32.12 |
| Average water molecule | 36.73 | 37.14 |
*RMS, Root mean square.
Fig. 5.
Crystal structures of RI-10/KI-10 in complex with HLA-A*0201. (a, b) 2Fo−Fc electron density maps contoured at 1.0 σ for RI-10 (a) and KI-10 (b). (c) Superimposition of RI-10 and KI-10. (d) Orientation of the peptide side chains in the HLA-A*0201-bound RI-10 and KI-10, viewing along the peptide from the N-terminal to the C-terminal end. Magenta (Gln4), red (Asn5), blue (Pro6), purple (Thr7) and green (Thr8) lettering indicates the central residues that show significant positional variation between RI-10 and KI-10.
Except for the side chain of the first amino acid (Arg for RI-10 and Lys for KI-10), the three N-terminal and the two C-terminal residues of RI-10 and KI-10 superimpose well. However, the conformations of the main chains and the side chains of the central residues of RI-10 and KI-10, from Gln4 to Thr8, show significant differences, in spite of their similar sequences (Fig. 5c, d). The root mean square (RMS) deviation is 1.367–1.606 between any RI-10 and KI-10 peptide chain of the four complex molecules in their respective asymmetrical unit. The peptides are in the same conformation in the four molecules in one asymmetrical unit (RMS deviation ≤0.280 between any two peptide chains of the four complex molecules in the same asymmetrical unit). The conformational difference between RI-10 and KI-10 is not caused by lattice contact. A previous study suggested that a single amino acid substitution could alter the entire conformation of the bound peptide (Madden et al., 1993). Subsequent studies have demonstrated that even subtle changes in anchor residues can change the peptide–MHC (pMHC) surface. For example, as in the HLA-B8 human immunodeficiency virus 1 (HIV-1) gag p17 structure, substitution of arginine for lysine at the anchor of position 5 (Reid et al., 1996). More importantly, evidence has shown that N-terminal residues are energetically important and that chemical modification or removal of the N-terminal residue of the bound peptide lowers the stability of complexes of HLA-A2 and peptide. In these studies, chemical modification or removal of the N-terminal residue does not alter the conformation of the peptides significantly unless they are involved in direct lattice contact (Bouvier et al., 1998; Khan et al., 2000). Therefore, it is a novel finding that a subtle change of the N-terminal residue of the bound peptide may profoundly impact the whole conformation of the central residues.
For both RI-10 and KI-10, the main chains of the centrally located residues formed arches centred at Pro6 and tended to protrude out of the antigen-binding groove at this site. Gln4, Pro6 and Tyr9, whose side chains point upwards, are the most exposed residues as measured by the solvent-accessible surface and these are the most probable recognizing sites for the T-cell receptors (TCRs).
DISCUSSION
Immune responses against influenza A virus have been characterized intensively for several decades and over 100 CTL epitopes within numerous subtypes and strains have been identified. Nevertheless, little work has been done on the highly pathogenic H5N1 subtype (Bui et al., 2007). In this study, we successfully identified a strongly immunogenic CTL epitope in the H5 HA protein by using a strategy that was applied in screening and identification of CTL epitopes of severe acute respiratory syndrome coronavirus in our laboratory, i.e. starting from a computer motif prediction, followed by in vitro complex refolding and then coming back to a standard T2-binding assay (Zhou et al., 2006b). We found that nine of 15 H5 HA-derived peptides could bind to HLA*0201 in the refolding assay and seven of them could bind to HLA*0201 on the cell surface in the T2-binding assay. Among the nine peptides that could refold, two overlapping peptides, RI-10 (H5 HA205-214, RLYQNPTTYI) and YV-10 (H5 HA207-216, YQNPTTYISV), induced a vigorous CTL epitope-specific response in HLA-A*0201/Kb Tg mice immunized with vaccinia–H5 or pcDNA3.0–H5. An intriguing coincidence is that two overlapping Kd-restricted CTL epitopes, H2 HA204–212 (LYQNVGTYV) and H2 HA210–219 (TYVSVGTSTL), have been defined in the same region of the HA molecule of the influenza virus A/JAP/305/57 (H2N2) (Braciale et al., 1989; Cao et al., 1996). The former epitope H2 HA204–212 is immunodominant, eliciting vigorous CTL responses in BALB/c mice immunized with A/Japan/57 virus, whereas H2 HA210–219 is sub-immunodominant (Braciale et al., 1989; Cao et al., 1996). The RI-10 and KI-10 epitopes identified in this study share certain sequence similarity with H2 HA204–212. This may imply that this region is a ‘hot spot' for a putative CTL response against influenza A virus. Although YV-10 elicits a comparable CTL epitope-specific response with RI-10 in Tg mice immunized with vaccinia–H5 or pcDNA3.0–H5, it induces a much weaker CTL epitope-specific response in the peptide-immunized Tg mice. Therefore, we focused on RI-10 and its variant KI-10 in this study.
Here, we show that RI-10/KI-10 induced a significant CTL epitope-specific response in patients who had recovered from H5N1 infection and carried the HLA-A2+ haplotype. It is noteworthy that virus sequence from patient HJR has a KI-10 epitope, but that patient LXC's virus sequence is not available for this region. Our results are somewhat unexpected, because RI-10 and KI-10 induced a higher CTL epitope-specific response than the GL-9 epitope. The GL-9 epitope is suggested as a dominant CTL epitope in influenza A virus infection (Stewart-Jones et al., 2003). As the HA of influenza A virus does not have more copies and is not expressed earlier than the matrix protein, the dominance of RI-10 and KI-10 observed here is worthy of future investigation. Furthermore, the sequences of RI-10 and KI-10 not only harbour typical anchor residues for HLA-A2, but also match the requirement of TAP transporter binding perfectly. Namely, they each possess a basic amino acid residue (Arg or Lys) stabilized at position 1, and contain favoured hydrophobic and aromatic residues at positions 2 and 3 (Uebel & Tampe, 1999; Uebel et al., 1997). Our results suggest that the hierarchy of CTL response against highly pathogenic H5N1 virus may be different from those of other human influenza A virus subtypes, such as H1N1 and H3N2. It is necessary in the future study to re-evaluate the relative roles played by these newly identified CTL epitopes in comparison with those in other influenza A virus subtypes.
We present the crystal structures of RI-10 and KI-10 in a complex with HLA-A*0201 and β2m. To the best of our knowledge, these are the second crystal structures to be released of influenza virus-derived CTL epitopes restricted by HLA-A*0201 in addition to the dominant epitope GL-9. The most interesting finding is that the two peptides, RI-10 and KI-10, adopt different conformations in their central residues, from Gln4 to Thr8, despite their sequence similarity. It is unknown whether RI-10 and KI-10 can be recognized by a single TCR or whether their responding TCR repertoires are actually not cross-reactive. Despite the conformational difference between the two peptides, they may still be recognized by a single TCR, because conformational adjustment can be adopted at the binding interface when pMHC engages TCR. For example, Lee et al. (2004) demonstrated that the immunodominant HLA-A2-restricted HIV gag epitope (SLFNTVATL) and its variant (SLYNTVATL) show marked differences in structure when bound to HLA-A2. However, their on-rate kinetics of TCR binding were identical, implying that conformational changes at the TCR–peptide–MHC binding interface occur after an initial permissive antigen contact (Lee et al., 2004). In our observations, RI-10 and KI-10 are cross-reactive in tetramer staining in the PBMCs of patients who have recovered from H5N1 virus infection. The characterization of their responding TCR repertoires should be demonstrated in the near future.
HA sequence alignment shows that the sequences of RI-10/KI-10 are unique compared with relevant 10-mer peptide sequences from seasonal 'flu (H1 or H3) and other subtypes of influenza virus. However, the sequences relevant to RI-10/KI-10 in the HA of serotypes H1 and H3 (and most other serotypes) also have a leucine as the second residue and most of these sequences have a valine or isoleucine as the last residue (Fig. 6). In other words, most of these sequences have typical anchor residues for HLA-A2. Therefore, it is highly possible that these relevant peptides can also bind to HLA-A2. However, whether they can also elicit a significant CD8+ T-cell response should be demonstrated in a future study. Even if they can induce a certain CD8+ T-cell response, it is less likely that the response is cross-reactive to RI-10/KI-10, because the central part of RI-10/KI-10 is largely different from that of the HA proteins of other serotypes (Fig. 6). At least in our experiment, we did not observe any cross-reactivity between RI-10/KI-10 and seasonal 'flu (H1 or H3). As shown in Fig. 4, GL-9-specific CD8+ T cells could be detected in the PBMCs of the HLA-A2+ healthy donor, suggesting that the donor used to be infected by seasonal 'flu. However, no RI-10/KI-10-specific CD8+ T cells were detected in the healthy donor's PBMCs, so our data do not support a cross-reaction between RI-10/KI-10 and their relevant putative epitopes in seasonal 'flu.
Fig. 6.
HA sequence alignment of H5 and seasonal 'flu (H1 and H3) at the position of RI-10/KI-10 CTL epitope. The sequence of RI-10/KI-10 and their relevant sequence in H1 and H3 are shaded grey. Circles show that all of these sequences have a conserved leucine at position 2 and a valine or isoleucine at the C terminals, suggesting that they all have a typical HLA-A2-binding motif. However, central parts of RI-10/KI-10 in H5 and the relevant positions in seasonal 'flu (H3 and H1) are largely different.
The strong immunogenicity of RI-10 and KI-10 that we observed suggests that they are potential targets for the design of new vaccines. However, whether they can induce any protection against H5N1 virus or whether they actually may have a pathogenic effect during H5N1 infection remains to be elucidated. Nevertheless, RI-10 and KI-10 might be useful diagnostic markers for the surveillance of a specific CTL response associated with H5N1 virus infection.
METHODS
Prediction of epitopes and synthesis of peptides.
Sequences of potential HLA-A*0201-binding peptides within the HA protein were based on a Qinghai strain of H5N1 isolated from Qinghai Lake in May 2005 (A/bar-headed goose/Qinghai/1/05) (Liu et al., 2005). A computer-based program was applied with access through the BioInformatics and Molecular Analysis Section (bimas) HLA Peptide Binding Predictions website (http://bimas.dcrt.nih.gov/molbio/hla_bind/index.html; Bertoletti et al., 1994). Nonamer and 10-mer peptides with a high estimated half-life of dissociation (T1/2) were synthesized and the purity was determined to be approximately 90 % by analytical HPLC profile and mass spectrometry. As a positive control for HLA-A*0201-binding ability, the HLA-A*0201-restricted influenza virus matrix 1 (M1) protein-derived peptide GL-9 (GILGFVFTL; synthesized) was used. The Mamu-A*01-restricted simian immunodeficiency virus type 1 (SIV-1)-derived peptide CM-9 (CTPYDINQM; synthesized) (Chu et al., 2007) was used as a negative control. Lyophilized peptides were stored in aliquots at −80 °C.
Detection of the binding of potential T-cell epitopic peptides to HLA-A*0201 molecules on T2 cells.
The T2 cell line was a generous gift from the late Professor Weifeng Chen (Peking University Health Science Center, PR China). A peptide-induced stabilization assay of the HLA-A*0201 molecules expressed by T2 cells was performed by using a previously described method (Gricks et al., 2001; Kuzushima et al., 2001; Passoni et al., 2002). Briefly, T2 cells were incubated with 50 μM candidate peptides and 1 μM human β2m (Sigma) in serum-free RPMI 1640 medium (Life Technologies) for 18 h at 37 °C in a 5 % CO2 incubator. Expression of HLA-A*0201 on the T2 cells was then determined by staining with fluorescein isothiocyanate (FITC)-labelled anti-HLA-A2 mAb BB7.2 (Serotech) and detected by flow cytometry using FACScan (BD Biosciences). Data analysis was performed using CellQuest software (BD Biosciences). The fluorescence index (FI) was calculated as follows: FI=(mean FITC fluorescence with the given peptide−mean FITC fluorescence with the negative control peptide)/(mean FITC fluorescence with the negative peptide). Peptides with FI≥1 were arbitrarily regarded as high-affinity candidate epitopes (Zhou et al., 2006b).
Refolding of computer-predicted peptides with HLA-A*0201 heavy chain and β2m.
Refolding was performed as described previously with minor modifications (Garboczi et al., 1992). Briefly, HLA-A*0201 heavy chain (extracellular domain aa 1–275) and β2m were expressed in Escherichia coli with the pET prokaryotic expression system (Novagen Inc.) as inclusion bodies. The inclusion bodies were separately dissolved in a solution of 10 mM Tris/HCl (pH 8.0) and 8 M urea. The synthetically prepared peptide was dissolved in DMSO (Sigma). HLA-A*0201 heavy chain, β2m and peptide were subsequently injected at a molecular ratio of 1 : 1 : 3 into a diluted solution (100 mM Tris/HCl, 400 mM l-arginine-HCl, 2 mM sodium EDTA, 0.5 mM oxidized glutathione, 5 mM reduced glutathione; Sigma). After stirring at 4 °C for 24–48 h, the soluble refolded portion was concentrated and then purified by chromatography on a Superdex G-200 size exclusion column (Amersham Pharmacia Biotech) to monitor the correctly refolded pHLA complex.
Immunization of mice with vaccinia virus or pcDNA3.0 vector expressing H5 HA.
Vaccinia virus (Tiantan strain) with an insert of the Qinghai Lake H5N1 virus HA-protein-encoding gene (vaccinia–H5) was a gift from Professor Zhiwei Chen (AIDS Institute, University of Hong Kong, PR China). The pcDNA3.0 vector inserted with the Qinghai Lake H5N1 virus HA-protein-encoding gene (pcDNA3.0–H5) was constructed by cloning the Qinghai Lake H5N1 virus HA-protein-encoding gene into the pcDNA3.0 vector using BamHI and XhoI restriction sites. HLA-A2.1/Kb Tg mice (Vitiello et al., 1991), in which a chimeric form of human HLA-A*0201 heavy-chain α1 and α2 domains and mouse H2-2Kb α3, transmembrane and cytoplasmic domains were produced, were a generous gift from Professor Xuetao Cao (Institute of Immunology, Second Military Medical University, PR China). Tg mice were bred in a pathogen-free facility. Cell-surface HLA-A2.1 expression was assessed by flow cytometry using FITC-labelled anti-HLA-A2 mAb BB7.2. Female HLA-A2.1/Kb Tg mice, 6–8 weeks old, were immunized by intramuscular injection with vaccinia–H5 (3.0×106 p.f.u. in 20 μl PBS) at weeks 0, 4 and 8. Spleens were recovered 10 days after the last immunization, dispersed with a syringe plunger and passed through cell strainers. Erythrocytes were lysed with 0.83 % ammonium chloride lysis solution (0.83 % NH4Cl, 0.26 % Tris–HCl, pH 7.2). Splenocytes were then washed and resuspended. Specific IFN-γ-producing cells were detected by an ELISPOT assay (Lagging et al., 1995; Wang et al., 2008). For pcDNA3.0–H5 immunization, the Tg mice were injected intramuscularly with 60 μg pcDNA3.0–H5 plasmid at weeks 0, 4 and 8. The splenocytes were prepared and the IFN-γ-producing cells were measured as described above. All studies and procedures involving Tg mice were approved by the Animal Care and Use Committee of the Institute of Microbiology, CAS.
Immunization of mice with peptide using gp96 N-terminal fragment as an adjuvant.
The peptide-immunization experiments were carried out as described previously using the N-terminal fragment of the murine glycoprotein 96 (gp96) as an adjuvant (Li et al., 2005). Briefly, female HLA-A2.1/Kb Tg mice (Vitiello et al., 1991), 6–8 weeks old, were immunized subcutaneously at multiple sites with a mixture of peptides, incomplete Freund's adjuvant (Difco) and the N-terminal fragment N333 (aa 22–355) of murine gp96, three times with intervals of 2 weeks. The injection volume was adjusted to 200 μl for each animal. Specific IFN-γ-producing cells were detected by ELISPOT assay and tetramer staining 10 days after the last immunization.
Detection of peptide–CTL epitope-specific CD8+ T cells in PBMCs of AIV-recovered donors.
PBMCs were isolated from the whole blood of donors who had recovered from AIV infection in addition to blood from healthy controls. The AIV-infection-recovered donors were sampled between 15 and 18 months after onset of disease. The expression of HLA-A2 was determined by FITC-conjugated anti-HLA-A2 mAb BB7.2 (Serotech) staining and cytometry. PBMCs were cultured with the peptides at a concentration of 10 μM in RPMI 1640 medium containing 10 % fetal calf serum (Hydone) and 20 U rhIL-2 ml−1 (R&D Systems) in a 24-well culture plate. Half of the media were changed every 3 days supplemented with rhIL-2 at 20 U ml−1. After 1 week, cells were harvested and tested for the presence of peptide-specific CD8+ T cells by tetramer staining.
ELISPOT assay.
The ELISPOT assay was performed by using a commercially available kit (U-Cytech). A 96-well PVDF-backed plate was pre-coated with an anti-IFN-γ mAb overnight at 4 °C. The plate was blocked with blocking buffer for 1 h at 37 °C. Murine splenocytes or human PBMCs were dispensed at a predetermined density of 106 ml−1 in triplicate wells. To stimulate the effector cells, 20 μM peptide was added. The plate was incubated at 37 °C for 24 h. Subsequently, cells were removed and the plate was processed according to the manufacturer's instructions. The coloured spots, representing epitope-specific IFN-γ-producing T cells, were counted by using an automatic ELISPOT reader. Only brown-coloured spots with fuzzy borders were scored as SFCs.
Tetramer preparation and staining.
Tetrameric HLA-A*0201–peptide complexes (tetramers) containing RI-10 (RLYQNPTTYI), KI-10 (KLYQNPTTYI) or GL-9 (GILGFVFTL) were prepared by using a previously described method (Sourdive et al., 1998). Briefly, a DNA sequence containing the BirA enzymic biotinylation site was added to the C terminus of HLA-A*0201 heavy chain or HLA-A*0201/H-2Kb chimeric heavy chain, which was created by fusing human α1 and α2 domains to H-2Kb mouse α3 domain. The entire construct was expressed in E. coli. Purified recombinant HLA-A*0201 or HLA-A*0201/H2 Kb chimeric heavy chain containing the BirA site and human β2m were refolded with peptide RI-10, KI-10 or GL-9. The complexes were purified by chromatography using a Superdex G-200 size-exclusion column (Pharmacia) followed by Mono Q (Pharmacia) anion-exchange chromatography, and biotinylated using BirA enzyme (Avidity) as described previously (Altman et al., 1996). The biotinylated complexes were assembled into tetramers by mixing biotinylated HLA-A*0201–peptide or HLA-A*0201/H2 Kb–peptide complexes and phycoerythrin (PE)-labelled streptavidin (Biosource) at a molar ratio of 4 : 1. Murine splenocytes or human PBMCs were incubated at room temperature for 20 min in staining buffer [PBS with 0.1 % bovine serum albumin (Sigma) and 0.1 % sodium azide (Amresco)] containing PE-labelled tetrameric complex. Cells were washed twice with staining buffer and then incubated at 4 °C in staining buffer containing saturating amounts of FITC-labelled anti-CD8 mAb (Becton Dickinson). Samples were detected by flow cytometry as described above. Tetramer+ cells gated from CD8+ T lymphocytes were counted as epitope-specific CTLs.
X-ray crystallography.
Proteins of RI-10 or KI-10 in complex with HLA-A*0201/β2m were prepared by refolding as described above and purified by gel filtration with Superdex 75 and then Superdex 200 (Amersham Pharmacia Biotech) chromatography. The initial crystals were obtained from vapour-diffusion hanging drops containing equal volumes of protein [10 mg ml−1; 25 mM 2-(N-morpholino)ethanesulphonic acid (MES), pH 6.5] and 16 % polyethylene glycol 6000 in 25 mM MES (pH 6.5). The diffractable crystals were obtained by subsequent microseeding. For data collection, the crystals were soaked in reservoir solution supplemented with 20 % glycerol, flash-cooled and maintained at 100 K in a cryostream. The data were collected on a Rigaku R-AXIS IV++ image plate with a Rigaku MM007 rotation Cu Kα rotating anode in the house X-ray generator at 40 kV and 20 mA (λ=1.5418 Å). The data were scaled using the HKL2000 suite of programs (HKL Research Inc.). The structures were solved by the molecular-replacement method (PDB accession no. 1JF1) using the Phenix program suite (Zwart et al., 2008).
Acknowledgments
We thank Dr Fuliang Chu, Dr Jinghua Yan, Dr Yiwei Liu, Dr Jianxuan Qi, Ms Zhenying Liu and Dr Christopher Vavricka for assistance and discussion. This study is supported by the National Natural Science Foundation of China (NSFC, 30599434), the Ministry of Science and Technology of China (MOST, Project 973, 2005CB523001) and the National Institutes of Health, USA (NIH, 3 U19 AI051915-05S1). This work is also partly supported by China National Grand S&T Special Project no. 2009ZX10004-305. The China–Japan Joint Laboratory of Molecular Immunology and Molecular Microbiology is, in part, supported by Japan MEXT (Ministry of Education, Culture, Sports, Science and Technology). G. F. G. is a Distinguished Young Investigator of the NSFC (30525010).
References
- Ahmed, R., Oldstone, M. B. & Palese, P. (2007). Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat Immunol 8, 1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman, J. D., Moss, P. A., Goulder, P. J., Barouch, D. H., McHeyzer-Williams, M. G., Bell, J. I., McMichael, A. J. & Davis, M. M. (1996). Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96. [DOI] [PubMed] [Google Scholar]
- Bender, B. S. & Small, P. A., Jr (1992). Influenza: pathogenesis and host defense. Semin Respir Infect 7, 38–45. [PubMed] [Google Scholar]
- Bennink, J. R., Yewdell, J. W., Smith, G. L., Moller, C. & Moss, B. (1984). Recombinant vaccinia virus primes and stimulates influenza haemagglutinin-specific cytotoxic T cells. Nature 311, 578–579. [DOI] [PubMed] [Google Scholar]
- Bertoletti, A., Costanzo, A., Chisari, F. V., Levrero, M., Artini, M., Sette, A., Penna, A., Giuberti, T., Fiaccadori, F. & other authors (1994). Cytotoxic T lymphocyte response to a wild type hepatitis B virus epitope in patients chronically infected by variant viruses carrying substitutions within the epitope. J Exp Med 180, 933–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier, M., Guo, H. C., Smith, K. J. & Wiley, D. C. (1998). Crystal structures of HLA-A*0201 complexed with antigenic peptides with either the amino- or carboxyl-terminal group substituted by a methyl group. Proteins 33, 97–106. [DOI] [PubMed] [Google Scholar]
- Braciale, T. J., Sweetser, M. T., Morrison, L. A., Kittlesen, D. J. & Braciale, V. L. (1989). Class I major histocompatibility complex-restricted cytolytic T lymphocytes recognize a limited number of sites on the influenza hemagglutinin. Proc Natl Acad Sci U S A 86, 277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui, H. H., Peters, B., Assarsson, E., Mbawuike, I. & Sette, A. (2007). Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proc Natl Acad Sci U S A 104, 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, W., Myers-Powell, B. A. & Braciale, T. J. (1996). The weak CD8+ CTL response to an influenza hemagglutinin epitope reflects limited T cell availability. J Immunol 157, 505–511. [PubMed] [Google Scholar]
- Chen, H., Smith, G. J., Zhang, S. Y., Qin, K., Wang, J., Li, K. S., Webster, R. G., Peiris, J. S. & Guan, Y. (2005). Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 436, 191–192. [DOI] [PubMed] [Google Scholar]
- Chen, H., Li, Y., Li, Z., Shi, J., Shinya, K., Deng, G., Qi, Q., Tian, G., Fan, S. & other authors (2006). Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. J Virol 80, 5976–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, F., Lou, Z., Chen, Y. W., Liu, Y., Gao, B., Zong, L., Khan, A. H., Bell, J. I., Rao, Z. & other authors (2007). First glimpse of the peptide presentation by rhesus macaque MHC class I: crystal structures of Mamu-A*01 complexed with two immunogenic SIV epitopes and insights into CTL escape. J Immunol 178, 944–952. [DOI] [PubMed] [Google Scholar]
- Claas, E. C., Osterhaus, A. D., van Beek, R., De Jong, J. C., Rimmelzwaan, G. F., Senne, D. A., Krauss, S., Shortridge, K. F. & Webster, R. G. (1998). Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351, 472–477. [DOI] [PubMed] [Google Scholar]
- Doherty, P. C. & Kelso, A. (2008). Toward a broadly protective influenza vaccine. J Clin Invest 118, 3273–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garboczi, D. N., Hung, D. T. & Wiley, D. C. (1992). HLA-A2–peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc Natl Acad Sci U S A 89, 3429–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotch, F., McMichael, A., Smith, G. & Moss, B. (1987). Identification of viral molecules recognized by influenza-specific human cytotoxic T lymphocytes. J Exp Med 165, 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gricks, C. S., Rawlings, E., Foroni, L., Madrigal, J. A. & Amlot, P. L. (2001). Somatically mutated regions of immunoglobulin on human B-cell lymphomas code for peptides that bind to autologous major histocompatibility complex class I, providing a potential target for cytotoxic T cells. Cancer Res 61, 5145–5152. [PubMed] [Google Scholar]
- Heiny, A. T., Miotto, O., Srinivasan, K. N., Khan, A. M., Zhang, G. L., Brusic, V., Tan, T. W. & August, J. T. (2007). Evolutionarily conserved protein sequences of influenza A viruses, avian and human, as vaccine targets. PLoS One 2, e1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kees, U. & Krammer, P. H. (1984). Most influenza A virus-specific memory cytotoxic T lymphocytes react with antigenic epitopes associated with internal virus determinants. J Exp Med 159, 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A. R., Baker, B. M., Ghosh, P., Biddison, W. E. & Wiley, D. C. (2000). The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J Immunol 164, 6398–6405. [DOI] [PubMed] [Google Scholar]
- Kilpatrick, A. M., Chmura, A. A., Gibbons, D. W., Fleischer, R. C., Marra, P. P. & Daszak, P. (2006). Predicting the global spread of H5N1 avian influenza. Proc Natl Acad Sci U S A 103, 19368–19373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreijtz, J. H., de Mutsert, G., van Baalen, C. A., Fouchier, R. A., Osterhaus, A. D. & Rimmelzwaan, G. F. (2008). Cross-recognition of avian H5N1 influenza virus by human cytotoxic T-lymphocyte populations directed to human influenza A virus. J Virol 82, 5161–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzushima, K., Hayashi, N., Kimura, H. & Tsurumi, T. (2001). Efficient identification of HLA-A*2402-restricted cytomegalovirus-specific CD8 (+) T-cell epitopes by a computer algorithm and an enzyme-linked immunospot assay. Blood 98, 1872–1881. [DOI] [PubMed] [Google Scholar]
- Lagging, L. M., Meyer, K., Hoft, D., Houghton, M., Belshe, R. B. & Ray, R. (1995). Immune responses to plasmid DNA encoding the hepatitis C virus core protein. J Virol 69, 5859–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. K., Stewart-Jones, G., Dong, T., Harlos, K., Di Gleria, K., Dorrell, L., Douek, D. C., van der Merwe, P. A., Jones, E. Y. & other authors (2004). T cell cross-reactivity and conformational changes during TCR engagement. J Exp Med 200, 1455–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, L. Y., Ha Do, L. A., Simmons, C., de Jong, M. D., Chau, N. V., Schumacher, R., Peng, Y. C., McMichael, A. J., Farrar, J. J. & other authors (2008). Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest 118, 3478–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Zhou, M., Han, J., Zhu, X., Dong, T., Gao, G. F. & Tien, P. (2005). Generation of murine CTL by a hepatitis B virus-specific peptide and evaluation of the adjuvant effect of heat shock protein glycoprotein 96 and its terminal fragments. J Immunol 174, 195–204. [DOI] [PubMed] [Google Scholar]
- Liu, J., Xiao, H., Lei, F., Zhu, Q., Qin, K., Zhang, X. W., Zhang, X. L., Zhao, D., Wang, G. & other authors (2005). Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 309, 1206. [DOI] [PubMed] [Google Scholar]
- Madden, D. R., Garboczi, D. N. & Wiley, D. C. (1993). The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell 75, 693–708. [DOI] [PubMed] [Google Scholar]
- Normile, D. (2006). Avian influenza. Evidence points to migratory birds in H5N1 spread. Science 311, 1225. [DOI] [PubMed] [Google Scholar]
- Olsen, B., Munster, V. J., Wallensten, A., Waldenstrom, J., Osterhaus, A. D. & Fouchier, R. A. (2006). Global patterns of influenza A virus in wild birds. Science 312, 384–388. [DOI] [PubMed] [Google Scholar]
- Passoni, L., Scardino, A., Bertazzoli, C., Gallo, B., Coluccia, A. M., Lemonnier, F. A., Kosmatopoulos, K. & Gambacorti-Passerini, C. (2002). ALK as a novel lymphoma-associated tumor antigen: identification of 2 HLA-A2.1-restricted CD8+ T-cell epitopes. Blood 99, 2100–2106. [DOI] [PubMed] [Google Scholar]
- Reid, S. W., McAdam, S., Smith, K. J., Klenerman, P., O'Callaghan, C. A., Harlos, K., Jakobsen, B. K., McMichael, A. J., Bell, J. I. & other authors (1996). Antagonist HIV-1 Gag peptides induce structural changes in HLA B8. J Exp Med 184, 2279–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmelzwaan, G. F., Fouchier, R. A. & Osterhaus, A. D. (2007). Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development. Curr Opin Biotechnol 18, 529–536. [DOI] [PubMed] [Google Scholar]
- Salomon, R. & Webster, R. G. (2009). The influenza virus enigma. Cell 136, 402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourdive, D. J., Murali-Krishna, K., Altman, J. D., Zajac, A. J., Whitmire, J. K., Pannetier, C., Kourilsky, P., Evavold, B., Sette, A. & other authors (1998). Conserved T cell receptor repertoire in primary and memory CD8 T cell responses to an acute viral infection. J Exp Med 188, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, J., Blixt, O., Tumpey, T. M., Taubenberger, J. K., Paulson, J. C. & Wilson, I. A. (2006). Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312, 404–410. [DOI] [PubMed] [Google Scholar]
- Stewart-Jones, G. B., McMichael, A. J., Bell, J. I., Stuart, D. I. & Jones, E. Y. (2003). A structural basis for immunodominant human T cell receptor recognition. Nat Immunol 4, 657–663. [DOI] [PubMed] [Google Scholar]
- Thomas, P. G., Keating, R., Hulse-Post, D. J. & Doherty, P. C. (2006). Cell-mediated protection in influenza infection. Emerg Infect Dis 12, 48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, A. R. & Skehel, J. J. (1984). The influenza A virus nucleoprotein gene controls the induction of both subtype specific and cross-reactive cytotoxic T cells. J Exp Med 160, 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, A. R., Bastin, J., Gould, K. & Brownlee, G. G. (1986). Cytotoxic T lymphocytes recognize influenza haemagglutinin that lacks a signal sequence. Nature 324, 575–577. [DOI] [PubMed] [Google Scholar]
- Uebel, S. & Tampe, R. (1999). Specificity of the proteasome and the TAP transporter. Curr Opin Immunol 11, 203–208. [DOI] [PubMed] [Google Scholar]
- Uebel, S., Kraas, W., Kienle, S., Wiesmuller, K. H., Jung, G. & Tampe, R. (1997). Recognition principle of the TAP transporter disclosed by combinatorial peptide libraries. Proc Natl Acad Sci U S A 94, 8976–8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitiello, A., Marchesini, D., Furze, J., Sherman, L. A. & Chesnut, R. W. (1991). Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human-mouse class I major histocompatibility complex. J Exp Med 173, 1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wabuke-Bunoti, M. A., Taku, A., Fan, D. P., Kent, S. & Webster, R. G. (1984). Cytolytic T lymphocyte and antibody responses to synthetic peptides of influenza virus hemagglutinin. J Immunol 133, 2194–2201. [PubMed] [Google Scholar]
- Wahl, A., Schafer, F., Bardet, W., Buchli, R., Air, G. M. & Hildebrand, W. H. (2009). HLA class I molecules consistently present internal influenza epitopes. Proc Natl Acad Sci U S A 106, 540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., Zhan, D., Li, L., Lei, F., Liu, B., Liu, D., Xiao, H., Feng, Y., Li, J. & other authors (2008). H5N1 avian influenza re-emergence of Lake Qinghai: phylogenetic and antigenic analyses of the newly isolated viruses and roles of migratory birds in virus circulation. J Gen Virol 89, 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2009). Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to WHO. http://www.who.int/csr/disease/avian_influenza/country/cases_table_2009_04_23/en/index.html
- Wilson, I. A. & Cox, N. J. (1990). Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol 8, 737–771. [DOI] [PubMed] [Google Scholar]
- Yap, K. L., Ada, G. L. & McKenzie, I. F. (1978). Transfer of specific cytotoxic T lymphocytes protects mice inoculated with influenza virus. Nature 273, 238–239. [DOI] [PubMed] [Google Scholar]
- Zhou, J. Y., Shen, H. G., Chen, H. X., Tong, G. Z., Liao, M., Yang, H. C. & Liu, J. X. (2006a). Characterization of a highly pathogenic H5N1 influenza virus derived from bar-headed geese in China. J Gen Virol 87, 1823–1833. [DOI] [PubMed] [Google Scholar]
- Zhou, M., Xu, D., Li, X., Li, H., Shan, M., Tang, J., Wang, M., Wang, F. S., Zhu, X. & other authors (2006b). Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J Immunol 177, 2138–2145. [DOI] [PubMed] [Google Scholar]
- Zwart, P. H., Afonine, P. V., Grosse-Kunstleve, R. W., Hung, L. W., Ioerger, T. R., McCoy, A. J., McKee, E., Moriarty, N. W., Read, R. J. & other authors (2008). Automated structure solution with the PHENIX suite. Methods Mol Biol 426, 419–435. [DOI] [PubMed] [Google Scholar]