Abstract
Alternative splicing and polyadenylation of human cytomegalovirus (HCMV) immediate-early (IE) pre-mRNAs are temporally regulated and rely on cellular RNA-processing factors. This study examined the location and abundance of essential RNA-processing factors, which affect alternative processing of UL37 IE pre-mRNAs, during HCMV infection. Serine/threonine protein kinase 1 (SRPK1) phosphorylates serine/arginine-rich proteins, necessary for pre-spliceosome commitment. It was found that HCMV infection progressively increased the abundance of cytoplasmic SRPK1, which is regulated by subcellular partitioning. The essential polyadenylation factor CstF-64 was similarly increased in abundance, albeit in the nucleus, proximal to and within viral replication compartments (VRCs). In contrast, the location of polypyrimidine tract-binding protein (PTB), known to adversely affect splicing of HCMV major IE RNAs, was temporally regulated during infection. PTB co-localized with CstF-64 in the nucleus at IE times. By early times, PTB was detected in punctate cytoplasmic sites of some infected cells. At late times, PTB relocalized to the nucleus, where it was notably excluded from HCMV VRCs. Moreover, HCMV infection induced the formation of nucleolar stress structures, fibrillarin-containing caps, in close proximity to its VRCs. PTB exclusion from HCMV VRCs required HCMV DNA synthesis and/or late gene expression, whereas the regulation of SRPK1 subcellular distribution did not. Taken together, these results indicated that HCMV increasingly regulates the subcellular distribution and abundance of essential RNA-processing factors, thereby altering their ability to affect the processing of viral pre-mRNAs. These results further suggest that HCMV infection selectively induces sorting of nucleolar and nucleoplasmic components.
INTRODUCTION
The human cytomegalovirus (HCMV) UL36–38 locus produces multiple UL37 transcripts, including an unspliced transcript, UL37 exon 1 (UL37x1), and ten alternatively spliced UL37 transcripts (Adair et al., 2003, 2004, 2006; Goldmacher et al., 1999; Kouzarides et al., 1988; Su et al., 2003a, b; Tenney & Colberg-Poley, 1990, 1991a, b). The UL37x1 anti-apoptotic protein (pUL37x1), also known as viral mitochondrion-localized inhibitor of apoptosis or vMIA, is the product of the predominant UL37x1 RNA (Goldmacher et al., 1999; Kouzarides et al., 1988; Tenney & Colberg-Poley, 1991a). pUL37x1 is essential for HCMV growth in humans (Hayajneh et al., 2001) and for the growth of primary HCMV strains (Jurak & Brune, 2006) and HCMV strain AD169 (Dunn et al., 2003; Reboredo et al., 2004; Sharon-Friling et al., 2006), but not of HCMV strain TownevarATCC (McCormick et al., 2005) in cultured fibroblasts.
Post-transcriptional RNA processing involves 5′ capping, splicing and 3′ polyadenylation. Splicing of pre-mRNA requires the assembly of spliceosomes that consist of five small nuclear ribonucleoprotein particles (snRNPs) and numerous non-snRNPs (Matlin & Moore, 2007; Wahl et al., 2009). Serine/arginine-rich (SR) proteins play roles in pre-spliceosome assembly, branchpoint entry and snRNP interactions (Graveley, 2000; Shen et al., 2004). Their activity is regulated by phosphorylation of serine residues in the arginine/serine-rich domain by protein kinases, including SR protein kinase 1 (SRPK1). SRPK1 activity, in turn, is regulated by subcellular partitioning and anchoring in the cytoplasm (Ding et al., 2006). SRPK1 translocates from the cytoplasm to the nucleus in a cycle-dependent manner before initiation of the M phase (Ding et al., 2006). Cytoplasmic anchoring of SRPK1 results from its interactions with two co-chaperones specific for heat-shock proteins (Zhong et al., 2009). Osmotic stress can also induce nuclear translocation of SRPK1 (Zhong et al., 2009).
Polyadenylation requires recognition of a consensus polyadenylation signal and a downstream cis element, cleavage of the nascent transcript and addition of a poly(A) tail to the 3′ end of cleaved pre-mRNA (Moore & Proudfoot, 2009; Proudfoot, 2004). Cleavage stimulation factor (CstF) recognizes the downstream U- or G/U-rich sequences through its CstF-64 subunit (MacDonald et al., 1994; Takagaki et al., 1992; Wilusz et al., 1990).
UL37 RNA species share a common initiation site, but UL37x1 RNA is unspliced and is polyadenylated at an intronic site (Adair et al., 2003; Goldmacher et al., 1999; Kouzarides et al., 1988; Su et al., 2003b; Tenney & Colberg-Poley, 1991a, b). UL37x1 RNA becomes increasingly predominant over its spliced isoforms during infection (Adair et al., 2004). Alternative processing of UL37 RNAs is dictated, in part, by the competition of cellular RNA factors for its juxtaposed splicing and polyadenylation cis elements (Su et al., 2003a). Similar competition was subsequently found for cis elements in the HCMV major immediate-early (MIE) transcripts, IE1 and IE2 (Sanchez et al., 2004).
The UL37x1 unspliced transcript requires usage of a cleavage site that contains a consensus polypyrimidine tract-binding protein (PTB)-binding site, and its polyadenylation site usage is enhanced by a downstream element, recognized by CstF-64 (Adair et al., 2004; Su et al., 2003a). PTB strongly represses HCMV MIE gene expression and replication (Cosme et al., 2009).
Temporal regulation of alternative UL37 RNA processing during HCMV infection suggests that key post-transcriptional processing factors are manipulated during infection. HCMV infection increases the abundance of critical cellular post-transcriptional processing factors, including the SR protein ASF/SF2 (alternative splicing factor/splicing factor 2), the essential polyadenylation factor CstF-64 and the splicing suppressor PTB (Adair et al., 2003, 2004, 2006; Su et al., 2003a).
Because SRPK1 is regulated primarily by subcellular distribution (Ding et al., 2006) and HCMV immediate-early (IE) RNA processing displays complex regulation, we examined whether HCMV infection manipulates the abundance and location of key cellular RNA-processing factors. We found that the abundance of positive regulators of splicing (SRPK1) and polyadenylation (CstF-64) were increased during infection in the cytoplasm and nucleus, respectively. Surprisingly, CstF-64 was partially included in subnuclear viral replication compartments (VRCs). The negative regulator of splicing, PTB, was partially relocalized at early times of infection from the nucleus to cytoplasmic punctate sites and then at late times to the nucleus, but was selectively excluded from HCMV VRCs. Interestingly, HCMV infection caused selective sorting of a nucleolar component, fibrillarin, into cap structures at late times of infection.
RESULTS
HCMV infection increases the abundance of SRPK1
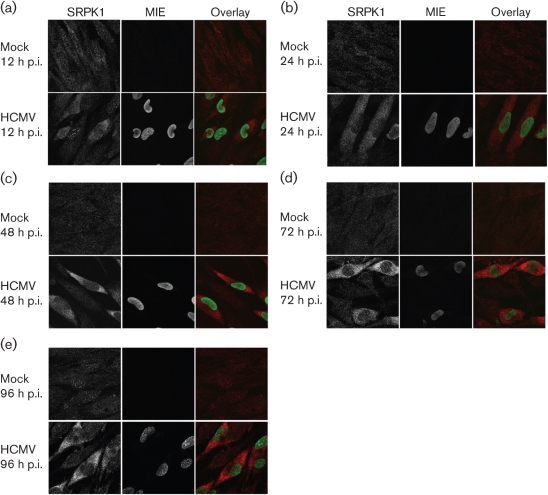
SRPK1, which phosphorylates SR proteins, is regulated by its subcellular distribution rather than by affecting its constitutive enzymic activity (Ding et al., 2006; Zhong et al., 2009). Our previous data demonstrated that the abundance of hypo-phosphorylated ASF/SF2 increases very early during HCMV infection (Adair et al., 2004). We therefore hypothesized that HCMV infection regulates the subcellular distribution of SRPK1. To test this hypothesis, we examined SRPK1 localization in human foreskin fibroblasts (HFFs) in G0 phase (G0-HFFs) using confocal imaging. At 12 h post-infection (p.i.), SRPK1 was localized in the cytoplasm and nucleus of mock-treated and HCMV-infected cells. However, nuclear speckles, some partially co-localized with IE1/2, were more intense in HCMV-infected than in mock-treated cells (Fig. 1a). Within 24 and 48 h p.i., SRPK1 progressively increased in abundance in the cytoplasm of infected cells (Fig. 1b, c). The increase in cytoplasmic SRPK1 continued through late times (Fig. 1d, e). These results suggested that HCMV increases the abundance of SRPK1, primarily in the cytoplasm of infected cells.
Fig. 1.
HCMV infection progressively increases the abundance of SRPK1 in the cytoplasm of infected G0-HFFs. G0-HFFs were HCMV infected (m.o.i.=3) or mock infected. At 12 (a), 24 (b), 48 (c), 72 (d) and 96 (e) h p.i., cells were harvested and sequentially incubated with anti-SRPK1 (red, 1 : 1000) and mAb 810 (specific for MIE, green, 1 : 500), followed by the corresponding secondary antibodies. Probed cells were imaged by confocal microscopy. Panels on the left and in the middle show greyscale imaging, whereas panels on the right show the merged colour images.
SRPK1 abundance is increased in the cytoplasm of HCMV-infected cells by late times of infection
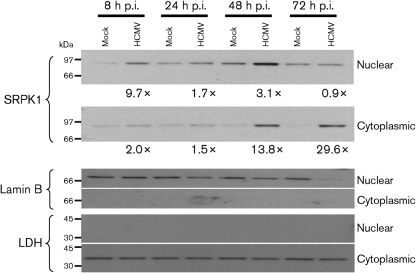
To verify these results, we examined SRPK1 in nuclear and cytoplasmic fractions from mock-infected and HCMV-infected G0-HFFs (Fig. 2). The abundance of SRPK1 increased progressively during HCMV infection. At IE times of infection, the abundance of nuclear SRPK1 was transiently increased (∼9.7-fold) more than cytoplasmic SRPK1 (∼2.0-fold). By 48 and 72 h p.i., the abundances of cytoplasmic SRPK1 (13.8-fold and 29.6-fold, respectively) were progressively increased more than nuclear SRPK1 (3.1-fold and 0.9-fold, respectively) at the same times. The nuclear (lamin B) and cytoplasmic [lactate dehydrogenase (LDH)] markers verified the identity and purity of the fractions. The decrease in lamin B at 72 h p.i. reflects its degradation during HCMV nuclear egress (Marschall et al., 2005; Milbradt et al., 2007, 2009). Taken together with the imaging results above, we concluded that HCMV infection regulates the subcellular distribution of SRPK1, progressively increasing its abundance in the cytoplasm of HCMV-infected cells through to late times of infection.
Fig. 2.
Subcellular distribution of SRPK1 in HCMV-infected G0-HFFs. HFFs (6.0×107 cells) were growth arrested and mock infected or HCMV infected (m.o.i.=3), harvested at the indicated times, and fractionated into nuclear and cytoplasmic fractions. Fractionated proteins (20 μg) were resolved by electrophoresis, transferred to nitrocellulose and probed with anti-SRPK1 (1 : 1000), anti-lamin B (1 : 1000) and anti-LDH (1 : 1000). Probed blots were stripped of bound antibodies after exposure and reprobed with other antibodies. The fold inductions in SRPK1 abundance were determined by comparison of the densities of the bands from HCMV-infected cells divided by the densities of the corresponding mock-infected cells.
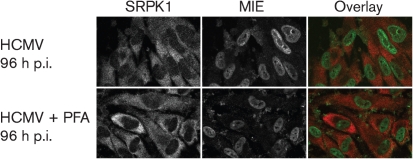
To determine whether HCMV DNA replication or late gene expression is required for the regulation of SRPK1 subcellular distribution, mock-infected or HCMV-infected G0-HFFs were treated with phosphonoformate (PFA), an inhibitor of HCMV DNA replication through oriLyt (a cis-acting sequence that promotes initiation of lytic-phase DNA replication), and examined by confocal microscopy (Fig. 3). PFA treatment did not alter the SRPK1 subcellular distribution or increased abundance in HCMV-infected cells compared with untreated HCMV-infected cells. These results indicated that the regulation of subcellular distribution and abundance of SRPK1 does not require HCMV DNA replication or late gene expression.
Fig. 3.
HCMV induction of cytoplasmic SRPK1 during PFA treatment. G0-HFFs were HCMV infected (m.o.i.=10) and untreated or treated with PFA (400 μg ml−1), following absorption, for 96 h p.i. Cells were harvested, stained for MIE (green) and SRPK1 (red) and visualized as in Fig. 1.
Divergent trafficking of PTB and CstF-64 in HCMV infection
PTB and CstF-64 are known to affect the processing of HCMV IE transcripts (Adair et al., 2004; Cosme et al., 2009; Su et al., 2003a). PTB is a well-characterized repressor of RNA splicing (Castelo-Branco et al., 2004). CstF-64 plays a constitutive role in polyadenylation and its abundance regulates the efficiency of 3′ end processing (Qu et al., 2007; Takagaki & Manley, 1998). Deletion of the UL37x2 downstream element, predicted to bind CstF-64, causes a large decrease in UL37x1 RNA polyadenylation site usage (Adair et al., 2004; Su et al., 2003a).
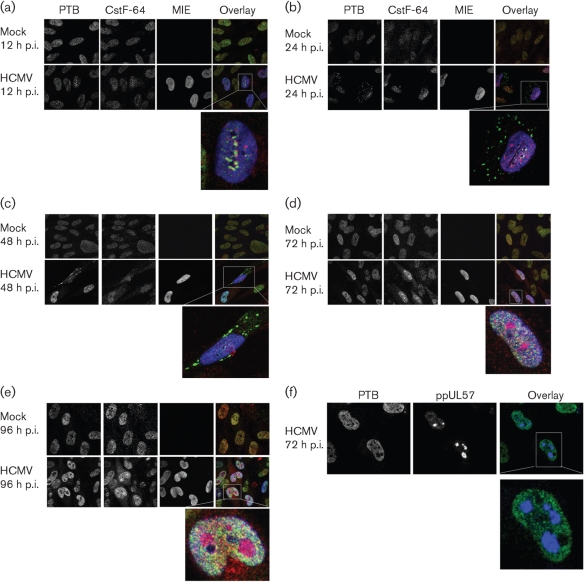
To determine whether their subcellular distributions were temporally and differentially regulated during HCMV infection, we examined the co-localization of PTB and CstF-64 in HCMV-infected G0-HFFs. At 12 h p.i., PTB and CstF-64 were both localized primarily in the nucleus, similarly to mock-treated cells, and their co-localization at subnuclear sites was observed in some HCMV-infected cells (Fig. 4a). By early times of infection (24 and 48 h p.i.), PTB was partially relocalized to punctate bodies in the cytoplasm of some infected cells (Fig. 4b, c). During late times of infection (72 and 96 h p.i.), the cytoplasmic PTB bodies were no longer detected in HCMV-infected cells (Fig. 4d, e). In stark contrast, CstF-64 continuously increased in the nucleus, from IE through to late times of infection. CstF-64 was included in subnuclear bodies that resembled HCMV VRCs, whereas PTB was notably excluded from the same subdomains.
Fig. 4.
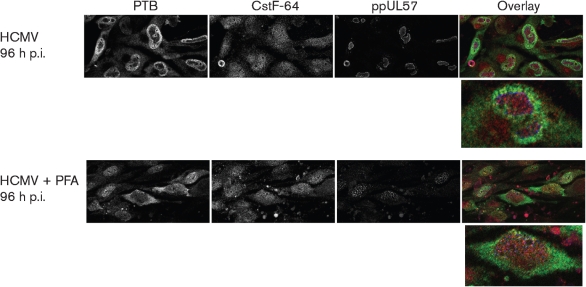
Temporal relocalization of PTB and CstF-64 during HCMV infection. G0-HFFs were treated as in Fig. 1. HCMV-infected and mock-infected cells were methanol fixed at 12 (a), 24 (b), 48 (c), 72 (d) and 96 (e) h p.i., and sequentially incubated with anti-PTB (green, 1 : 250), anti-CstF-64 (red, 1 : 250) and mAb 810 (blue, 1 : 500; MIE) and then with the corresponding secondary antibodies. Cells were imaged by confocal microscopy. The left and middle panels are greyscale, whilst the right panel shows the merged images. (f) PTB is selectively excluded from HCMV VRCs at late times of infection. G0-HFFs were HCMV infected (m.o.i.=3) and fixed at 72 h p.i. as in Fig. 1 and sequentially incubated with anti-PTB (green, 1 : 250) and anti-ppUL57 (blue, 1 : 250). Cells were then probed with secondary antibodies and imaged using a Zeiss Axiovert inverted UV microscope with an ApoTome attachment. The left and middle panels show greyscale images whilst the right panels show an overlay of both channels.
To test whether PTB was excluded from HCMV VRCs, HCMV-infected G0-HFFs were co-stained for PTB and ppUL57 and examined by confocal microscopy (Fig. 4f). ppUL57 is an HCMV-encoded, ssDNA-binding protein and in these experiments served as a marker for VRCs, as previous experiments have demonstrated the presence of HCMV ppUL57 in VRCs at late times of infection (Penfold & Mocarski, 1997). PTB was excluded from large subnuclear compartments at 72 h p.i. and these were found to be HCMV VRCs.
We determined the percentages of infected cells in which PTB and CstF-64 were redistributed (Table 1). PTB did not co-localize with ppUL57 in most of the cells (93.8 %) but partially co-localized with ppUL57 in a low percentage (6.2 %) of the infected cells. Conversely, CstF-64 partially co-localized with ppUL57 in most (97 %) HCMV-infected cells and co-localized with ppUL57 in only a few (3 %) infected cells. These results demonstrated the preponderance of PTB exclusion and CstF-64 inclusion in HCMV VRCs, indicating the probable importance of their subnuclear distribution for HCMV replication.
Table 1.
PTB exclusion and CstF-64 inclusion in HCMV VRCs
| Staining | Subnuclear co-localization* | Dispersed | |||
|---|---|---|---|---|---|
| Complete | Partial | None | Nuclear | Cytoplasmic | |
| PTB and ppUL57 | |||||
| Mock infected | 100 % (264/264) | ||||
| Mock+PFA | 100 % (312/312) | ||||
| HCMV infected | 0 % (0/307) | 6.2 % (19/307) | 93.8 % (288/307) | ||
| HCMV+PFA | 76.4 % (370/484) | 23.6 % (114/484) | |||
| CstF-64 and ppUL57 | |||||
| Mock infected | 100 % (201/201) | ||||
| Mock+PFA | 100 % (312/312) | ||||
| HCMV infected | 0 % (0/298) | 97.0 % (289/298) | 3.0 % (9/298) | ||
| HCMV+PFA | 100 % (351/351) | ||||
*Co-localization of PTB or CstF-64 and ppUL57 (VRC marker) was measured in mock-infected or HCMV-infected G0-HFFs, either untreated or PFA treated at 96 h p.i. as described in Fig. 5.
To determine whether HCMV DNA replication and/or late gene expression is required for PTB exclusion from VRCs, we treated HCMV-infected cells with PFA and examined the localization of PTB and CstF-64 at late times of infection (Fig. 5). In contrast to the SRPK1 results in Fig. 3, PFA blocked the redistribution of PTB in HCMV-infected cells. PTB distribution was dispersed between the nucleus (76.4 %) and cytoplasm (23.6 %) of PFA-treated, HCMV-infected cells at 96 h p.i. (Table 1). These results established that HCMV DNA replication and/or late gene expression is needed for PTB subnuclear relocalization.
Fig. 5.
PFA treatment blocks HCMV-induced redistribution of PTB at late times of infection. G0-HFFs were HCMV infected (m.o.i.=10) and untreated or PFA treated as in Fig. 3. Cells were harvested at 96 h p.i. and stained for PTB (green), CstF-64 (red) and ppUL57 (blue) and imaged by confocal microscopy as in Fig. 4.
Divergent effects of HCMV infection on nuclear and cytoplasmic levels of PTB and CstF-64
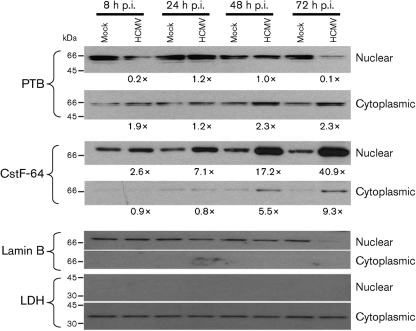
To confirm their subcellular location observed by confocal microscopy, we examined nuclear and cytoplasmic fractions from HCMV-infected G0-HFFs for PTB and CstF-64 abundance (Fig. 6). At late times (48 and 72 h p.i.), full-length cytoplasmic PTB was slightly increased (∼2.3-fold), whilst nuclear PTB was decreased (0.1-fold) at 72 h p.i. In stark contrast, CstF-64 progressively increased in abundance (2.6–40.9-fold), mostly in the nuclear compartment. The nuclear (lamin B) and cytoplasmic (LDH) markers verified the identity and purity of the fractions.
Fig. 6.
Subcellular distribution of PTB and CstF-64 in HCMV-infected G0-HFFs. G0-HFFs were HCMV infected (m.o.i.=3) or mock infected, fractionated and blotted as in Fig. 2. The same membranes were stripped and reprobed with anti-PTB (1 : 250), rabbit anti-CstF-64 (1 : 250), anti-lamin B (1 : 1000) and anti-LDH (1 : 1000). Probed blots were repeatedly stripped of bound primary and secondary antibodies after exposure and reprobed as required. The increases in PTB and CstF-64 abundance were determined using the density of the bands from HCMV-infected cells divided by the density of the corresponding mock-infected band, as in Fig. 2. Lighter exposures of the nuclear CstF-64 blot were used for scanning and quantification.
HCMV infection causes sorting of a nucleolar component, fibrillarin, into nucleolar caps
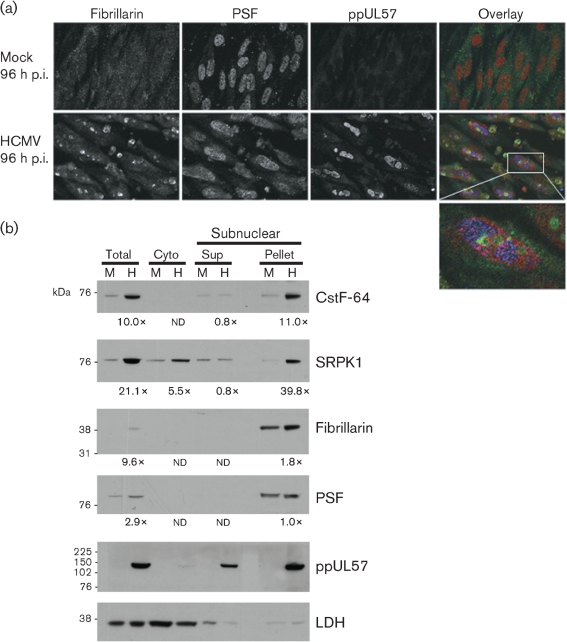
The relocalization of nuclear components resembled features of subnuclear redistribution during stress responses (Shav-Tal et al., 2005). HCMV infection modulates some cellular stress responses by relocalization of cellular proteins (Hakki et al., 2006). Nucleolar cap formation is associated with transcriptional arrest and results from the redistribution and sequestering of nucleolar and nucleoplasmic proteins into distinct bodies. We therefore investigated whether HCMV reorganization of the nucleus caused sorting of nucleolar components. PTB-associated splicing factor (PSF) serves as a marker for dark nucleolar caps (DNCs) in transcriptionally arrested cells (Dye & Patton, 2001; Ghetti et al., 1992). We examined HCMV-infected G0-HFFs at late times of infection (96 h p.i.) for co-localization of PSF with ppUL57 (Fig. 7a). Similar to PTB, PSF was partially excluded to the periphery of HCMV VRCs.
Fig. 7.
(a) HCMV infection induces the formation of fibrillarin-containing caps. G0-HFFs were mock infected or HCMV infected and harvested at 96 h p.i. as in Fig. 3. Cells were stained with anti-fibrillarin (green, 1 : 250), anti-PSF (red, 1 : 250) and anti-ppUL57 (blue, 1 : 250) antibodies and corresponding secondary antibodies. The left and middle panels are greyscale and the rightmost panels show all three channels merged. The bottom panel shows an enlarged HCMV-infected cell. (b) Co-sedimentation of cellular RNA-processing factors and nucleolar components with HCMV ppUL57, a VRC component. G0-HFFs were mock infected (M) or HCMV infected (H, m.o.i.=1) as in Fig. 2 and harvested at 96 h p.i. Cells were fractionated into cytoplasmic fraction, subnuclear pellet and supernatant (Sup) by modification of the nucleolar isolation procedure of Andersen et al. (2002). The cytoplasmic (Cyto) fraction was concentrated 5-fold and proteins (15 μg per well) were resolved by SDS-PAGE and blotted as described in Fig. 2. The blots were reacted with antibodies against CstF-64 (1 : 2000), SRPK1 (1 : 1000), fibrillarin (1 : 1000), PSF (1 : 2000), ppUL57 (1 : 500) or LDH (1 : 1000). Probed blots were stripped of bound primary and secondary antibodies after exposure and reprobed as required. The fold inductions in CstF-64, SRPK1, fibrillarin and PSF abundance were determined by comparison of the densities of the bands from HCMV-infected cells divided by the densities of the corresponding mock-infected cells as in Fig. 2 except where the mock- or HCMV-infected band was not detectable (nd).
Sorting of nucleolar components during RNA transcription can result in the formation of fibrillarin-containing caps that contain high concentrations of the nucleolar protein fibrillarin (Shav-Tal et al., 2005). To determine whether HCMV infection induces the formation of fibrillarin-containing caps, we examined the localization of fibrillarin at late times of HCMV infection. We found that fibrillarin was concentrated in cap structures, similar to light nucleolar caps (LNCs) and closely apposed to nucleoli, in the majority of HCMV-infected cells (78.9 vs 5.2 % of mock-infected HFFs) at late times of infection (96 h p.i.). These studies indicated that HCMV infection causes sorting of nucleolar components into fibrillarin-containing caps in close proximity to VRCs.
To verify the imaging results above, we modified a protocol used for subnuclear fractionation and examined HCMV-infected cells at late times of infection (Fig. 7b). CstF-64, SRPK1, fibrillarin, PSF and ppUL57 were enriched in the subnuclear pelleted fraction at late times of infection, whereas LDH was not. These results together with the confocal microscopy results suggest that key RNA-processing factors and selected nucleolar and nucleoplasmic components are physically associated with subnuclear compartments that contain HCMV VRC components.
DISCUSSION
HCMV infection temporally alters the abundance and subcellular distribution of key post-transcriptional processing factors during infection. At IE times, SRPK1, PTB and CstF-64 co-localized in nuclear punctate sites with the MIE proteins in HCMV-infected cells. IE1 and IE2, visualized as single punctate foci, are enriched in sites called nuclear domain 10 (ND10) at IE times (Ahn & Hayward, 1997). Because IE1 becomes diffused in the nucleus (Korioth et al., 1996) and IE2 remains at punctate sites, the cellular RNA-processing factors appear to contain IE2 at former ND10 sites. ND10s are active sites of HCMV IE gene transcription (Ishov et al., 1997). The combined presence of SRPK1, PTB and CstF-64 will predictably affect alternative splicing and polyadenylation of HCMV IE transcripts. Phosphorylation of SR proteins by SRPK1 targets them to sites of active transcription. PTB is known to negatively regulate MIE pre-mRNA splicing (Cosme et al., 2009), whereas CstF-64 selectively increases HCMV UL37x1 poly(A) site usage (Adair et al., 2004; Su et al., 2003a).
During the cell cycle, SRPK1 is translocated to the nucleus during M phase (Ding et al., 2006). In contrast, during HCMV infection, which causes pseudo-mitosis of the infected cell (Hertel et al., 2007), SRPK1 is increased mainly in the cytoplasm. Regulation of SRPK1 is achieved by partitioning through an anchoring mechanism (Ding et al., 2006) and by its interactions with molecular co-chaperones (Zhong et al., 2009). Heat-shock protein 70, part of the macromolecular complex used to anchor SRPK1 in the cytosol, is induced from IE to late times of HCMV infection (Santomenna & Colberg-Poley, 1990) with kinetics similar to those we observed for the progressively increasing abundance of cytoplasmic SRPK1. HCMV infection is well documented to block cellular stress pathways that would result in apoptosis (Goldmacher et al., 1999; Reeves et al., 2007; Skaletskaya et al., 2001; Terhune et al., 2007). Thus, HCMV infection may effectively block the cellular stress responses that could result in release of SRPK1 from the co-chaperone complex and nuclear translocation. Of particular note, HCMV infection redundantly blocks mitochondrial stress responses (Goldmacher et al., 1999; Reeves et al., 2007), possibly modulating oxidative stress, its consequent release of SRPK1 from its co-chaperone complex and its translocation from the cytoplasm to the nucleus (Zhong et al., 2009).
The SRPK1 cytoplasmic location may allow continued RNA splicing during HCMV infection but inhibit its nuclear functions. HCMV virion egress requires the destabilization of the nuclear lamina through recruitment of the HCMV pUL97 by p32 and the structural modifications of HCMV proteins ppUL50 and ppUL53 through their interaction with protein kinase C (Camozzi et al., 2008; Marschall et al., 2005; Milbradt et al., 2007, 2009). Phosphorylation of lamin B receptor (LBR) by SRPK1 induces its association with lamin B at the inner nuclear membrane (Papoutsopoulou et al., 1999). SRPK1 phosphorylation of LBR also stimulates the binding of the protein to chromatin and the stabilization of the nuclear membrane (Takano et al., 2004). It is therefore possible that continued anchoring of SRPK1 to the cytoplasm sequesters its activity from the nucleus allowing efficient egress of the HCMV nucleocapsids while allowing continued RNA splicing to occur through late times of infection.
As PTB inhibits splicing of the MIE transcripts and has negative effects on HCMV growth (Cosme et al., 2009), HCMV tightly regulates its subcellular distribution during infection. During IE times of infection, PTB, CstF-64 and MIE proteins are enriched in punctate structures, potentially perinucleolar compartments (PNCs). PNCs are subcellular structures, located at the periphery of nucleoli with high concentrations of PTB (Ghetti et al., 1992; Matera et al., 1995). PNCs are thought to be active sites of RNA polymerase II and III transcription. At early times of HCMV infection, PTB was partially translocated to discrete cytoplasmic sites of some infected cells. As PTB was relocalized, the nuclear PNCs in HCMV-infected cells appeared to disintegrate. This dissolution of PNCs following cytoplasmic PTB relocalization is consistent with the requirement for localization of PTB at the periphery of the nucleoli to maintain their integrity (Wang et al., 2003) and may partially underlie the sorting of fibrillarin to nucleolar caps.
By late times of infection, we observed exclusion of PTB from increasingly prominent subnuclear structures and the inclusion of CstF-64 in these. These proved to be VRCs by co-localization with the HCMV DNA-binding protein ppUL57. When HCMV DNA replication was blocked, the formation of the VRCs and the redistribution PTB was reduced, suggesting a role for HCMV DNA replication and/or late gene expression in PTB exclusion.
The presence of CstF-64 and exclusion of PTB suggest that HCMV VRCs are active sites of transcription and post-transcriptional processing, in addition to sites of DNA replication. RNA polymerase II, phosphorylated at Ser-2, was found in HCMV VRCs at 48 h p.i. in HCMV-infected G0-HFFs (Tamrakar et al., 2005). By subnuclear fractionation, we intriguingly found that HCMV ppUL57 mostly co-sedimented with induced nuclear CstF-64 and SRPK1, suggesting that these factors may be functionally associated with HCMV VRCs during infection.
The abundance of CstF-64 is the rate-limiting factor for the assembly of the core polyadenylation machineries, and increases in CstF-64 abundance positively influence 3′-end processing and the use of weak polyadenylation sites. Analysis of the HCMV UL37x1 polyadenylation signal, contained within an intron, showed that this signal is increasingly used during infection and that its usage is enhanced by a downstream element (Adair et al., 2004, 2006; Su et al., 2003a, b). The presence of CstF-64 at sites of HCMV post-transcriptional processing could serve to favour alternative polyadenylation of its transcripts, despite increased levels of viral transcripts at late times of infection.
The HCMV-induced sorting of nuclear components included the redistribution of nucleolar components. Sorting of nucleolar components in response to inhibition of transcription results in the formation of dynamic structure bodies (Shav-Tal et al., 2005). Fibrillarin is normally localized in the dense fibrillar component domain within nucleoli and segregates to LNCs following actinomycin D treatment. We observed similar segregation of fibrillarin to nucleolar caps at late times of HCMV infection and in close proximity to VRCs. During transcription inhibition, DNCs enriched in CstF-64 and PSF are formed (Shav-Tal et al., 2005). Similar to stress-responsive DNCs, HCMV VRCs include CstF-64 and do not include PTB. However, in contrast to DNCs, HCMV VRCs partially exclude PSF.
The question of what promotes nucleolar cap formation in HCMV-infected cells remains unanswered. Several HCMV proteins are known to localize to nucleoli (Salsman et al., 2008). Sorting of nucleolar components into nucleolar caps can result in the disintegration of PNCs (Shav-Tal et al., 2005). PNCs disintegrate at early times of HCMV infection when PTB shuttles to the cytoplasm and nuclear VRCs begin to form.
Nucleolar cap formation has been attributed to transcriptional arrest (Shav-Tal et al., 2005). Transcription of cellular and viral genes continues to occur throughout the HCMV life cycle. However, HCMV infection might differentially inhibit transcription of sensor genes, resulting in the formation of nucleolar caps. Alternatively, the presence of HCMV proteins in nucleoli and Cajal bodies (Salsman et al., 2008) may segregate nucleolar components and cause formation of nucleolar caps. Finally, the addition of pyruvate, whose concentration is markedly increased in HCMV-infected cells (Munger et al., 2008), can lead to the formation of nucleolar caps, as it does in other cell types (Shav-Tal et al., 2005). Taken together, our studies show that HCMV-induced restructuring of subnuclear domains includes relocalization of cellular RNA-processing machineries and nucleolar and nucleoplasmic components during permissive infection.
METHODS
Cell culture.
Primary HFFs were cultured in Dulbecco's modified Eagle's medium (Invitrogen) with 10 % fetal calf serum (FCS; HyClone/Gemini Bio-Products), penicillin/streptomycin (100 U ml−1 and 100 μg ml−1, respectively; Invitrogen) and growth arrested as described previously (Santomenna & Colberg-Poley, 1990). Briefly, HFFs were grown to confluence and fed very-low-serum medium (0.2 % FCS) for 72 h. Conditioned medium was removed from the cells and saved for reapplication after appropriate treatments.
Virus infection.
G0-HFFs were infected with HCMV (strain AD169) grown in low-serum medium (2 % FCS) at the indicated m.o.i. Control cells were treated with mock-infected medium. Following infection, in experiments to examine the requirement for HCMV DNA synthesis and/or late gene expression, cells were treated with PFA (400 μg ml−1; Sigma) for the duration of infection. Control cells were untreated.
Preparation of nuclear and cytoplasmic fractions.
Mock-treated or HCMV-infected HFFs were harvested at various times p.i. by trypsinization and collected by centrifugation. The cell pellets were resuspended in ice-cold PBS, pelleted and immediately used or quickly frozen and stored at −80 °C.
Nuclear and cytoplasmic fractions were prepared using a protocol adapted from Scherl et al. (2002). Briefly, 10 vols hypotonic buffer [10 mM Tris/HCl (pH 7.4), 10 mM NaCl, 1 mM MgCl2, 1 mM EDTA, 1× Protease Inhibitor Complete Cocktail (Roche), 1 mM PMSF, 1 mM sodium orthovanadate and 50 mM NaF] were added to cell pellets and incubated on ice for 1 h with occasional mixing. Swollen cells were lysed by the addition of 0.3 % Triton X-100 (Sigma) and dounce homogenization (25–30 strokes). Nuclei were collected by centrifugation at 1000 g at 4 °C for 10 min. Supernatants were removed from the pelleted nuclei, cleared from cellular debris by centrifugation at 12 879 g at 4 °C for 15 min, and stored for later use as the cytoplasmic fraction.
Pelleted nuclei were resuspended in 0.25 M sucrose buffer containing 10 mM MgCl2 and isolated and pelleted by centrifugation (1200 g for 10 min) through a 0.88 M sucrose cushion containing 0.05 mM MgCl2. Nuclei were resuspended in 0.25 M sucrose buffer and collected by centrifugation (12 879 g for 5 min). Nuclei were resuspended in radioimmunoprecipitation assay (RIPA) buffer I [50 mM Tris/HCl (pH 7.4), 150 mM NaCl, 1 % Triton X-100, 1 % sodium deoxycholate, 0.1 % SDS, 1 mM EDTA, 1× Protease Inhibitor Complete Cocktail, 1 mM PMSF, 1 mM sodium orthovanadate and 50 mM NaF] and incubated on ice for 15 min. To ensure complete lysis, nuclei were sonicated briefly (10 s, power 250–275 W cm−2, continuous pulse) and incubated on ice for an additional 15 min. Supernatants were subsequently cleared by centrifugation (12 879 g for 15 min at 4 °C). Cleared supernatants were used as the nuclear fraction. Both the cytoplasmic and nuclear fractions were stored at −20 °C (short term) or −80 °C (long term) until used.
Subnuclear fractions.
Nuclei from HFFs infected with HCMV (m.o.i.=1) or mock-infected cells were fractionated by a nucleolar isolation procedure (Andersen et al., 2002) with a minor modification of the final recovery step. Briefly, RIPA buffer II was added to the recovered fractions to final concentrations of 50 mM Tris/HCl (pH 7.5), 150 mM NaCl, 1 % NP-40, 0.5 % sodium deoxycholate and 1× Protease Inhibitor Complete Cocktail. The cytoplasmic and nucleoplasmic fractions were then separated by centrifugation at 2800 g for 10 min at 4 °C, and the subnuclear fraction was sonicated again five times for 10 s each. After centrifugation, the supernatant of the cytoplasmic fraction was concentrated ∼5-fold using an Ultra 4 UltraCel 3K centrifugal concentrator (Amicon).
Protein concentration.
Protein concentrations were determined using a BCA Protein Assay kit (Pierce) according to the manufacturer's instructions. Absorbance measurements were quantified using SoftMax Pro software (Molecular Devices).
Western blot analysis.
Proteins (10–20 μg) were separated by electrophoresis in NuPAGE Novex 10 or 12–14 % polyacrylamide Bis/Tris gels (Invitrogen) and transferred to Hybond ECL nitrocellulose (GE Healthcare) membranes at 50 V for 1 h. Western blots were blocked and reacted with the indicated antibodies as described previously (Adair et al., 2004; Mavinakere et al., 2006). Briefly, blotted proteins were incubated with primary antibodies diluted in PBS with 1 % BSA (Sigma) at room temperature for 1 h with gentle shaking. Membranes were washed and probed with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody (diluted in PBS with 1 % BSA) for 1 h at room temperature with gentle shaking. Proteins were visualized by chemiluminescence by briefly incubating the probed membranes in ECL Western Blotting Substrate (Pierce) and exposing the reacted membranes to BioMax MS film (Kodak) or HyBlot CL film (Denville Scientific). Images from exposed film were digitized and formatted using ScanWizard Pro v.7.0 (Microtek), a Bio-Rad GS-800 Calibrated Densitometer, and Quantity One, Adobe Photoshop (v.7.0) and Microsoft PowerPoint 2003. When additional probing was required, membranes were treated with stripping buffer [62.5 mM Tris/HCl (pH 6.4), 100 mM 2-mercaptoethanol, 2 % SDS] for 1 h at 50 °C as described previously (Mavinakere et al., 2006). Fold induction was determined by comparison of the density of bands from infected cells with the corresponding bands from mock-infected cells using Bio-Rad Quantity One basic software.
Antibodies.
The primary antibodies used for these studies at the concentrations indicated in the figure legends were specific for CstF-64 (Bethyl Laboratories), LDH (Millipore), lamin B (Santa Cruz), SRPK1 (BD Transduction Laboratories), PTB C terminus (Zymed Laboratories), fibrillarin (Abcam), PSF (Bethyl Laboratories), HCMV IE1/IE2 (mAb 810; Chemicon) and HCMV ppUL57 (a gift from Dr Lenore Pereira, University of California, CA, USA). Secondary antibodies used in the immunofluorescence assays (IFAs) were Alexa Fluor 488-conjugated goat anti-mouse IgG, Alexa Fluor 488-conjugated goat anti-mouse IgG2a, Alexa Fluor 488-conjugated goat anti-mouse IgG1, Alexa Fluor 568-conjugated goat anti-mouse IgG1, Alexa Fluor 488-conjugated goat anti-mouse IgG2b, Alexa Fluor 647-conjugated goat anti-mouse IgG2a and Alexa Fluor 568-conjugated goat anti-rabbit IgG (Molecular Probes, Invitrogen). All Alexa Fluor secondary antibodies were used at a 1 : 1500 dilution. Secondary antibodies (1 : 2500) used for Western blots were HRP-conjugated goat anti-mouse IgG (Stressgen Bioreagents), HRP-conjugated donkey anti-goat IgG (Santa Cruz Biotechnology) and HRP-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch, Santa Cruz Biotechnology).
IFA.
HFFs were seeded onto 18×18 mm or 18 mm circular glass coverslips (Fisher), allowed to reach confluence and serum starved as described previously (Santomenna & Colberg-Poley, 1990). G0-HFFs were mock infected or HCMV infected at the indicated multiplicities. Mock-infected or HCMV-infected cells were fixed in ice-cold methanol for 10 min. After fixation, cells were air dried and either used immediately or stored at 4 °C until use. At the time of use, cells were washed in PBS+0.05 % Tween 20 (three 5 min washes), followed by incubation with 10 % normal goat serum (Jackson ImmunoResearch) for 30 min at room temperature. Cells were then washed with PBS+0.05 % Tween 20 (three 5 min washes) and incubated with primary antibody for 30 min at 37 °C. Double- and triple-labelled cells were probed sequentially with primary antibodies with washes (three 5 min washes) between incubation with each antibody. Cells were then incubated simultaneously with secondary antibodies, treated to one final wash with PBS alone and permanently mounted onto microscope slides (Fisher) using ProLong Gold antifade reagent (Molecular Probes, Invitrogen) and stored in the dark at 4 °C until use.
Confocal laser-scanning microscopy.
Probed cells were examined by IFA using a Bio-Rad MRC 1024 confocal laser-scanning microscope [Center for Microscopy and Image Analysis, George Washington University (GWU) Washington, DC, USA] as described previously (Bozidis et al., 2008; Colberg-Poley et al., 2000; Mavinakere et al., 2006) or using a Zeiss LSM 510 confocal microscope [Children's Research Institute (CRI), Intellectual and Developmental Disabilities Research Center, Washington, DC, USA]. Images were acquired by sequential excitation at 488, 561 and 633 nm as appropriate using 40× (NA=1.3), 63× (NA=1.4) or 100× [NA=1.35 (GWU) or NA=1.4 (CRI)] oil objectives. Post-acquisition processing and image assembly were performed using ImageJ software (National Institutes of Health, MD, USA), Zeiss ZEN 2008, Adobe Photoshop (v.7.0) and Microsoft PowerPoint 2003.
ApoTome imaging.
Images were acquired with a Zeiss Axiovert inverted UV microscope with an ApoTome attachment following the manufacturer's instructions. Captured images were processed and merged using the Zeiss Axiovision Image Analysis software (v. 4.6).
Acknowledgments
The authors thank Dr Chad D. Williamson for his advice and critical reading of the manuscript and Dr Richard Adair for helpful discussions. Dr Charla Gaddy gratefully acknowledges the Army Medical Service Corps Long-term Health Education and Training Scholarship, United States Army, for supporting her doctoral dissertation work. These studies were funded, in part, by NIAID, NIH R01 AI46459 and R01 AI057906, and by Discovery Funds from the Children's Research Institute and the CNMC Board of Visitors. The confocal microscopy imaging was supported by a core grant (1P30HD40677) to the Children's Mental Retardation and Developmental Disabilities Research Center.
References
- Adair, R., Liebisch, G. W. & Colberg-Poley, A. M. (2003). Complex alternative processing of human cytomegalovirus UL37 pre-mRNA. J Gen Virol 84, 3353–3358. [DOI] [PubMed] [Google Scholar]
- Adair, R., Liebisch, G. W., Su, Y. & Colberg-Poley, A. M. (2004). Alteration of cellular RNA splicing and polyadenylation machineries during productive human cytomegalovirus infection. J Gen Virol 85, 3541–3553. [DOI] [PubMed] [Google Scholar]
- Adair, R., Liebisch, G. W., Lerman, B. J. & Colberg-Poley, A. M. (2006). Human cytomegalovirus temporally regulated gene expression in differentiated, immortalized retinal pigment epithelial cells. J Clin Virol 35, 478–484. [DOI] [PubMed] [Google Scholar]
- Ahn, J. H. & Hayward, G. S. (1997). The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol 71, 4599–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, J. S., Lyon, C. E., Fox, A. H., Leung, A. K., Lam, Y. W., Steen, H., Mann, M. & Lamond, A. I. (2002). Directed proteomic analysis of the human nucleolus. Curr Biol 12, 1–11. [DOI] [PubMed] [Google Scholar]
- Bozidis, P., Williamson, C. D. & Colberg-Poley, A. M. (2008). Mitochondrial and secretory human cytomegalovirus UL37 proteins traffic into mitochondrion-associated membranes of human cells. J Virol 82, 2715–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camozzi, D., Pignatelli, S., Valvo, C., Lattanzi, G., Capanni, C., Dal Monte, P. & Landini, M. P. (2008). Remodelling of the nuclear lamina during human cytomegalovirus infection: role of the viral proteins pUL50 and pUL53. J Gen Virol 89, 731–740. [DOI] [PubMed] [Google Scholar]
- Castelo-Branco, P., Furger, A., Wollerton, M., Smith, C., Moreira, A. & Proudfoot, N. (2004). Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol 24, 4174–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colberg-Poley, A. M., Patel, M. B., Erezo, D. P. & Slater, J. E. (2000). Human cytomegalovirus UL37 immediate-early regulatory proteins traffic through the secretory apparatus and to mitochondria. J Gen Virol 81, 1779–1789. [DOI] [PubMed] [Google Scholar]
- Cosme, R. S., Yamamura, Y. & Tang, Q. (2009). Roles of polypyrimidine tract binding proteins in major immediate-early gene expression and viral replication of human cytomegalovirus. J Virol 83, 2839–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, J. H., Zhong, X. Y., Hagopian, J. C., Cruz, M. M., Ghosh, G., Feramisco, J., Adams, J. A. & Fu, X. D. (2006). Regulated cellular partitioning of SR protein-specific kinases in mammalian cells. Mol Biol Cell 17, 876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, W., Chou, C., Li, H., Hai, R., Patterson, D., Stolc, V., Zhu, H. & Liu, F. (2003). Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100, 14223–14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye, B. T. & Patton, J. G. (2001). An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to subnuclear speckles. Exp Cell Res 263, 131–144. [DOI] [PubMed] [Google Scholar]
- Ghetti, A., Pinol-Roma, S., Michael, W. M., Morandi, C. & Dreyfuss, G. (1992). hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res 20, 3671–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmacher, V. S., Bartle, L. M., Skaletskaya, A., Dionne, C. A., Kedersha, N. L., Vater, C. A., Han, J. W., Lutz, R. J., Watanabe, S. & other authors (1999). A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc Natl Acad Sci U S A 96, 12536–12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B. R. (2000). Sorting out the complexity of SR protein functions. RNA 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakki, M., Marshall, E. E., De Niro, K. L. & Geballe, A. P. (2006). Binding and nuclear relocalization of protein kinase R by human cytomegalovirus TRS1. J Virol 80, 11817–11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayajneh, W. A., Colberg-Poley, A. M., Skaletskaya, A., Bartle, L. M., Lesperance, M. M., Contopoulos-Ioannidis, D. G., Kedersha, N. L. & Goldmacher, V. S. (2001). The sequence and antiapoptotic functional domains of the human cytomegalovirus UL37 exon 1 immediate early protein are conserved in multiple primary strains. Virology 279, 233–240. [DOI] [PubMed] [Google Scholar]
- Hertel, L., Chou, S. & Mocarski, E. S. (2007). Viral and cell cycle-regulated kinases in cytomegalovirus-induced pseudomitosis and replication. PLoS Pathog 3, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov, A. M., Stenberg, R. M. & Maul, G. G. (1997). Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol 138, 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurak, I. & Brune, W. (2006). Induction of apoptosis limits cytomegalovirus cross-species infection. EMBO J 25, 2634–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korioth, F., Maul, G. G., Plachter, B., Stamminger, T. & Frey, J. (1996). The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res 229, 155–158. [DOI] [PubMed] [Google Scholar]
- Kouzarides, T., Bankier, A. T., Satchwell, S. C., Preddy, E. & Barrell, B. G. (1988). An immediate early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology 165, 151–164. [DOI] [PubMed] [Google Scholar]
- MacDonald, C. C., Wilusz, J. & Shenk, T. (1994). The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol 14, 6647–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall, M., Marzi, A., aus dem Siepen, P., Jochmann, R., Kalmer, M., Auerochs, S., Lischka, P., Leis, M. & Stamminger, T. (2005). Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J Biol Chem 280, 33357–33367. [DOI] [PubMed] [Google Scholar]
- Matera, A. G., Frey, M. R., Margelot, K. & Wolin, S. L. (1995). A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol 129, 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin, A. J. & Moore, M. J. (2007). Spliceosome assembly and composition. Adv Exp Med Biol 623, 14–35. [DOI] [PubMed] [Google Scholar]
- Mavinakere, M. S., Williamson, C. D., Goldmacher, V. S. & Colberg-Poley, A. M. (2006). Processing of human cytomegalovirus UL37 mutant glycoproteins in the endoplasmic reticulum lumen prior to mitochondrial importation. J Virol 80, 6771–6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, A. L., Meiering, C. D., Smith, G. B. & Mocarski, E. S. (2005). Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis. J Virol 79, 12205–12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbradt, J., Auerochs, S. & Marschall, M. (2007). Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J Gen Virol 88, 2642–2650. [DOI] [PubMed] [Google Scholar]
- Milbradt, J., Auerochs, S., Sticht, H. & Marschall, M. (2009). Cytomegaloviral proteins that associate with the nuclear lamina: components of a postulated nuclear egress complex. J Gen Virol 90, 579–590. [DOI] [PubMed] [Google Scholar]
- Moore, M. J. & Proudfoot, N. J. (2009). Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700. [DOI] [PubMed] [Google Scholar]
- Munger, J., Bennett, B. D., Parikh, A., Feng, X. J., McArdle, J., Rabitz, H. A., Shenk, T. & Rabinowitz, J. D. (2008). Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol 26, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoutsopoulou, S., Nikolakaki, E. & Giannakouros, T. (1999). SRPK1 and LBR protein kinases show identical substrate specificities. Biochem Biophys Res Commun 255, 602–607. [DOI] [PubMed] [Google Scholar]
- Penfold, M. E. & Mocarski, E. S. (1997). Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239, 46–61. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N. (2004). New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol 16, 272–278. [DOI] [PubMed] [Google Scholar]
- Qu, X., Perez-Canadillas, J. M., Agrawal, S., De Baecke, J., Cheng, H., Varani, G. & Moore, C. (2007). The C-terminal domains of vertebrate CstF-64 and its yeast orthologue Rna15 form a new structure critical for mRNA 3′-end processing. J Biol Chem 282, 2101–2115. [DOI] [PubMed] [Google Scholar]
- Reboredo, M., Greaves, R. F. & Hahn, G. (2004). Human cytomegalovirus proteins encoded by UL37 exon 1 protect infected fibroblasts against virus-induced apoptosis and are required for efficient virus replication. J Gen Virol 85, 3555–3567. [DOI] [PubMed] [Google Scholar]
- Reeves, M. B., Davies, A. A., McSharry, B. P., Wilkinson, G. W. & Sinclair, J. H. (2007). Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 316, 1345–1348. [DOI] [PubMed] [Google Scholar]
- Salsman, J., Zimmerman, N., Chen, T., Domagala, M. & Frappier, L. (2008). Genome-wide screen of three herpesviruses for protein subcellular localization and alteration of PML nuclear bodies. PLoS Pathog 4, e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, V., McElroy, A. K., Yen, J., Tamrakar, S., Clark, C. L., Schwartz, R. A. & Spector, D. H. (2004). Cyclin-dependent kinase activity is required at early times for accurate processing and accumulation of the human cytomegalovirus UL122–123 and UL37 immediate-early transcripts and at later times for virus production. J Virol 78, 11219–11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santomenna, L. D. & Colberg-Poley, A. M. (1990). Induction of cellular hsp70 expression by human cytomegalovirus. J Virol 64, 2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl, A., Couté, Y., Déon, C., Callé, A., Kindbeiter, K., Sanchez, J. C., Greco, A., Hochstrasser, D. & Diaz, J. J. (2002). Functional proteomic analysis of human nucleolus. Mol Biol Cell 13, 4100–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon-Friling, R., Goodhouse, J., Colberg-Poley, A. M. & Shenk, T. (2006). Human cytomegalovirus pUL37x1 induces the release of endoplasmic reticulum calcium stores. Proc Natl Acad Sci U S A 103, 19117–19122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal, Y., Blechman, J., Darzacq, X., Montagna, C., Dye, B. T., Patton, J. G., Singer, R. H. & Zipori, D. (2005). Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell 16, 2395–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H., Kan, J. L. & Green, M. R. (2004). Arginine–serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell 13, 367–376. [DOI] [PubMed] [Google Scholar]
- Skaletskaya, A., Bartle, L. M., Chittenden, T., McCormick, A. L., Mocarski, E. S. & Goldmacher, V. S. (2001). A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc Natl Acad Sci U S A 98, 7829–7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y., Adair, R., Davis, C. N., DiFronzo, N. L. & Colberg-Poley, A. M. (2003a). Convergence of RNA cis elements and cellular polyadenylation factors in the regulation of human cytomegalovirus UL37 exon 1 unspliced RNA production. J Virol 77, 12729–12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y., Testaverde, J. R., Davis, C. N., Hayajneh, W. A., Adair, R. & Colberg-Poley, A. M. (2003b). Human cytomegalovirus UL37 immediate early target minigene RNAs are accurately spliced and polyadenylated. J Gen Virol 84, 29–39. [DOI] [PubMed] [Google Scholar]
- Takagaki, Y. & Manley, J. L. (1998). Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell 2, 761–771. [DOI] [PubMed] [Google Scholar]
- Takagaki, Y., MacDonald, C. C., Shenk, T. & Manley, J. L. (1992). The human 64-kDa polyadenylylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc Natl Acad Sci U S A 89, 1403–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, M., Koyama, Y., Ito, H., Hoshino, S., Onogi, H., Hagiwara, M., Furukawa, K. & Horigome, T. (2004). Regulation of binding of lamin B receptor to chromatin by SR protein kinase and cdc2 kinase in Xenopus egg extracts. J Biol Chem 279, 13265–13271. [DOI] [PubMed] [Google Scholar]
- Tamrakar, S., Kapasi, A. J. & Spector, D. H. (2005). Human cytomegalovirus infection induces specific hyperphosphorylation of the carboxyl-terminal domain of the large subunit of RNA polymerase II that is associated with changes in the abundance, activity, and localization of cdk9 and cdk7. J Virol 79, 15477–15493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney, D. J. & Colberg-Poley, A. M. (1990). RNA analysis and isolation of cDNAs derived from the human cytomegalovirus immediate-early region at 0.24 map units. Intervirology 31, 203–214. [DOI] [PubMed] [Google Scholar]
- Tenney, D. J. & Colberg-Poley, A. M. (1991a). Expression of the human cytomegalovirus UL36–38 immediate early region during permissive infection. Virology 182, 199–210. [DOI] [PubMed] [Google Scholar]
- Tenney, D. J. & Colberg-Poley, A. M. (1991b). Human cytomegalovirus UL36–38 and US3 immediate-early genes: temporally regulated expression of nuclear, cytoplasmic, and polysome-associated transcripts during infection. J Virol 65, 6724–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhune, S., Torigoi, E., Moorman, N., Silva, M., Qian, Z., Shenk, T. & Yu, D. (2007). Human cytomegalovirus UL38 protein blocks apoptosis. J Virol 81, 3109–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl, M. C., Will, C. L. & Luhrmann, R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718. [DOI] [PubMed] [Google Scholar]
- Wang, C., Politz, J. C., Pederson, T. & Huang, S. (2003). RNA polymerase III transcripts and the PTB protein are essential for the integrity of the perinucleolar compartment. Mol Biol Cell 14, 2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz, J., Shenk, T., Takagaki, Y. & Manley, J. L. (1990). A multicomponent complex is required for the AAUAAA-dependent cross-linking of a 64-kilodalton protein to polyadenylation substrates. Mol Cell Biol 10, 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, X. Y., Ding, J. H., Adams, J. A., Ghosh, G. & Fu, X. D. (2009). Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev 23, 482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]