Abstract
The degradation of complex substrates, like salivary mucins, requires an arsenal of glycosidases and proteases to sequentially degrade the oligosaccharides and polypeptide backbone. The mucin MUC5B is a complex oligomeric glycoprotein, heterogeneous in molecular mass (14–40×106 Da), with a diverse repertoire of oligosaccharides, differing in composition and charge. The aim of this study was to investigate whether proteolytic degradation of the mucin polypeptide backbone could be identified and if cooperation of dental biofilm bacteria was required. Cooperative bacteria-mediated proteolysis of MUC5B was determined by comparing individual species and mixed consortia of strains isolated from supragingival plaque, and freshly harvested supragingival plaque. Proteolytic activity was analysed using fluorescent labelled substrate and by visualizing mucin degradation by SDS-PAGE. Dental plaque degraded the polypeptide backbone of the salivary MUC5B mucin. The mucin was also degraded by a specific consortium of isolated species from supragingival plaque, although individual species and other consortia did not. Certain bacteria in supragingival dental plaque therefore cooperate as a consortium to proteolyse human salivary MUC5B and hydrolyse glycosides.
INTRODUCTION
Oral microbial biofilms act in support of host defences to prevent colonization by exogenous pathogenic microbes. The integrity of mucosal surfaces relies, in part, on the ability of the host-derived mucus layer to support colonization of commensal bacterial biofilms. In the oral microbial biofilm known as supragingival dental plaque, salivary proteins serve as a major ecological determinant of the composition of the microbial community, support adhesion and growth, and are the major continuously available source of nutrients (Marsh, 2003). To yield nutrients, the degradation of complex substrates, including salivary mucins, requires an arsenal of glycosidases and proteases. Several bacterial species, which possess complementary patterns of glycosidases and proteases, have been suggested to catabolize glycoproteins through a concerted action (e.g. Bradshaw et al., 1994). When bovine serum albumin was incubated with mutans streptococci, degradation depended on the presence of Streptococcus oralis (Homer & Beighton, 1992). As no single species of commensal micro-organism apparently expresses all of the requisite enzymes to digest available complex substrates, inter-species cooperation is needed.
Oral streptococci express a wide range of glycosidases, including sialidases, N-acetyl-β-d-glucosaminidase, β-d-galactosidase and α-l-fucosidase (Beighton & Whiley, 1990; Bradshaw et al., 1994; Byers et al., 2000). Oral streptococci have been shown to be able to degrade both the carbohydrate moieties and the protein backbone of human immunoglobulin A1 (e.g. Reinholdt et al., 1990), and in that study Streptococcus mitis biovar 2 was shown to remove sialic acid, galactose and N-acetylglucosamine from the molecule. S. oralis expresses the needed range of glycosidases to digest the N-linked glycans of plasma human α1-acid glycoprotein (Byers et al., 1999). Furthermore, S. oralis releases and metabolizes carbohydrate moieties from porcine gastric mucins (Homer et al., 1996). Availability of additional nutrients by catabolic cooperation in the bacterial biofilm might contribute to the diversity of the resident microbial communities (Bradshaw et al., 1994).
The need for cooperation in the oral biofilm to produce a wide range of enzyme specificities reflects the complexity of the substrate. Within the group of oral streptococci, interspecies cooperation to provide proteolytic and coordinated glycosidase activities to digest naturally occurring substrates is not well studied. Pure cultures of oral streptococci do express endopeptidase activity (e.g. Homer et al., 1990) and several streptococci species, e.g. S. oralis and Streptococcus mitis biovar 1, have been found to produce human immunoglobulin A1 protease (Reinholdt et al., 1990). Streptococcus gordonii degrades proline-rich proteins in saliva, leading to the release of a potentially immunoreactive peptide (Li et al., 2000), and also expresses a gelatinase, which hydrolyses type IV collagen (Juarez & Stinson, 1999). Streptococcus mutans and Streptococcus sanguinis also contain complex proteolytic pathways necessary for growth (e.g. Cowman & Baron, 1997). In contrast, supragingival plaque from macaque monkeys expresses both exoglycosidases and a wide range of proteolytic activities (Smith & Beighton, 1986, 1987).
The salivary MUC5B mucin is a complex oligomeric glycoprotein, heterogeneous in molecular mass (14–40×106 Da) (Thornton et al., 1999), depending on the number of disulfide-linked subunits and the composition and charge of the repertoire of oligosaccharides (Levine et al., 1987; Veerman et al., 1992; Wickström et al., 1998; Thomsson et al., 2002). In addition, the processed final structure of MUC5B and possibly its gel-forming properties depend on putative post-translational modifications at the C- and N-termini, resulting in co-secreted protein fragments (Wickström et al., 2000; Wickström & Carlstedt, 2001). Hence, MUC5B exhibits a very diverse and complex macromolecular organization. These post-translational modifications and tertiary structure increase resistance to proteolysis, which must be overcome if MUC5B is to serve as a nutrient source for the normal flora. Indeed, one of two subpopulations of salivary MUC5B appears to serve as a nutrient source, while the other provides a substrate for interfacial adhesion with oral bacteria (Wickström et al., 2009), reflecting the seemingly paradoxical functions of MUC5B in the oral ecosystem.
The aims of this study were to elucidate whether isolated species from dental supragingival plaque and a representative consortium degrade human salivary mucin MUC5B to enable growth, simulating dental plaque. The study also addressed whether degradation of MUC5B requires cooperation between species. In this report we present evidence that human salivary MUC5B is proteolytically degraded by supragingival dental plaque. While individual species could not replicate the spectrum of necessary enzymes, a four-species consortium showed specialized enzymic activities to collectively simulate dental plaque in the digestion of MUC5B.
METHODS
Purification of human salivary MUC5B.
Human salivary mucin MUC5B was isolated as previously described (Wickström & Svensäter, 2008) using a modified version of the method described by Raynal et al. (2003). In short, whole saliva was mixed with an equal volume of 0.2 M NaCl followed by incubation overnight with stirring at 4 °C. After gentle centrifugation (4400 g for 30 min at 4 °C), the supernatant was subjected to density-gradient centrifugation in CsCl/0.1 M NaCl (Beckman Optima LE-80K, rotor 50.2Ti, 36 000 r.p.m., 96 h, 15 °C, start density 1.45 g ml−1) and fractions were collected from the bottom of the tube. Fractions were analysed for density by weighing and measuring absorbance at 280 nm. To detect the core protein rather than the N- and C-terminal proteolyic fragments that might be cleaved by endogenous proteases, MUC5B-containing fractions were identified using an antiserum, LUM5B-2, which recognizes the central domain of the MUC5B polypeptide backbone (Wickström et al., 1998). The MUC5B-containing fractions were pooled and dialysed against PBS (0.15 M NaCl, 5 mM NaH2PO4, pH 7) and then stored at −20 °C until used. MUC5B concentration was determined by extensive dialysis against water, freeze-drying and weighing, and estimated to be 0.3 mg ml−1.
Bacterial strains.
The isolates employed in this study, previously described by Wickström & Svensäter (2008), were Streptocccus cristatus, S. mitis biovar 2 (previously identified as S. oralis using phenotypic tests; (Wickström & Svensäter, 2008) and S. gordonii, identified on the basis of 16S rRNA sequencing, and Actinomyces naeslundii, identified on the basis of metG and atpA sequencing (Henssge et al., 2009). S. mitis biovar 2 was positive for N-acetylgalactosaminidase, N-acetylglucosaminidase, β-galactosidase and sialidase. S. gordonii was positive for N-acetylglucosaminidase, N-acetylgalactosaminidase, α-fucosidase and β-galactosidase. S. cristatus was positive for α-fucosidase and N-acetylgalactosaminidase. A. naeslundii was positive for sialidase.
Growth and induction of enzymic activity.
The individual strains were inoculated onto pre-reduced blood agar for 1 day, resuspended in PBS and left for 72 h at 37 °C. Purified MUC5B was added in 1.5 ml tubes to an equal volume of the four individual strains or a consortium containing all four strains, each at a starting optical density (OD600) of 0.6. The proportion of the four species in the consortium was set to the ratio of the individual strains found in the dental plaque sample from which they were isolated: S. mitis biovar 2 (10), S. gordonii (1), S. cristatus (1) and A. naeslundii (2) (Wickström & Svensäter, 2008). OD600 was measured after 3, 6, 24, 48 and 72 h. The experiment was repeated three times and the mean±se is reported.
At 0, 3, 6 and 24 h, sialidase and protease activities were measured. Sialidase activity was assayed using a fluorogenic substrate, 2′-(4-methylumbelliferyl) α-d-N-acetylneuraminic acid (Sigma-Aldrich) in 96-well microtitre plates (Nunc Maxisorb). Aliquots (50 μl) of the samples were mixed with the substrate (100 μg ml−1 in 50 mM TES buffer, pH 7.5). The plates were incubated for 1 h at 37 °C, analysed in a fluorimeter (FLUOstar OPTIMA, Labvision) fitted with a 96-well plate reader, and fluorescence was recorded at excitation and emission wavelengths of 355 and 460 nm, respectively. The experiment was repeated three times and the mean±se is reported. Statistically significant differences were calculated using the Mann–Whitney U-test.
Proteolytic activity was assessed using FITC-labelled casein (QuantiCleave Fluorescent Protease Assay kit, Pierce) as a substrate. Samples (50 μl aliquots) were mixed with 50 μl substrate (10 μg ml−1 in 25 mM Tris, 150 mM NaCl, pH 7.2), incubated for 1 h at 37 °C, and fluorescence was recorded as above at excitation and emission wavelengths of 488 and 538 nm, respectively. The experiment was repeated three times and the mean±se is reported. Differences were calculated and statistical significance was estimated using the Mann–Whitney U-test.
Proteolytic degradation of MUC5B.
Pure cultures of S. mitis biovar 2 and S. gordonii, a combination of S. cristatus and A. naeslundii and a four-species consortium, approximately 10 μg of each sample, measured using a 10 μl ‘plaque-spoon’ designed by Jordan et al. (1968), were added to 1 ml of a MUC5B solution (0.3 mg ml−1) in 1.5 ml tubes and incubated at 37 °C for 0, 8 and 16 h, respectively. Following incubation, bacteria were removed from the MUC5B solution by gentle centrifugation (12 000 g for 2 min) and the supernatants were collected for analyses. In a separate experiment, 24 h supragingival dental plaque was collected as described previously (Wickström & Svensäter, 2008). Plaque was sampled from the buccal and lingual surfaces of premolar and molar regions from one 38-year-old healthy male before breakfast, and pooled. The individual refrained from normal dental hygiene procedures the night before and on the morning before collection. The dental plaque was mixed by vortexing for 10 s and approximately 10 μg plaque (measured as described above) was added to 1 ml of a MUC5B solution and treated as described above.
SDS-PAGE and Western blotting.
The supernatants recovered from the incubation of MUC5B and the different bacterial combinations were analysed by SDS-PAGE followed by Western blotting. Samples (15 μl) were mixed with 5 μl NuPAGE LDS sample buffer (Invitrogen) and run on a NuPAGE gel (4–12 % Bistris) (Invitrogen) at 180 V for 1 h. Protein bands were electroblotted onto PVDF membranes (Millipore, Immobilon-P, 0.4 μm) overnight using a Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad). After blotting, membranes were blocked with 5 % (w/v) dry milk in TBST (20 mM Tris, 137 mM NaCl, 0.05 % Tween 20) for 1 h, and incubated (2 h) with antiserum LUM5B-2 (1 : 5000) diluted in 5 % BSA/TBST. The membranes were then incubated (1 h) with a horseradish peroxidase-conjugated goat anti-rabbit antibody (Dako, P0448) diluted (1 : 2500) in 5 % dry milk/TBST, and bands were visualized using the ECL Western detection kit (Pierce ECL Western blotting substrate, product no. 32106)
Distribution of proteolytic activity in dental plaque.
The distribution of proteolytic activities in supragingival dental plaque was visualized with an Eclipse TE2000 inverted confocal scanning laser microscope (CSLM) (Nikon) using the FITC-labelled casein substrate described above. Equal volumes of plaque samples and substrate were mixed and incubated for 1 h at 37 °C. Confocal illumination was provided by an Ar laser (488 nm laser excitation) using a ×60 magnification oil immersion objective.
Rate-zonal centrifugation.
To show that the degradation visualized by SDS-PAGE/Western blotting was due to proteolysis, rate-zonal centrifugation was performed on supernatants recovered from dental plaque and consortia at 0 and 8 h. Samples were reduced using 6 M guanidinium chloride, 0.1 M Tris/HCl buffer, pH 8.0, containing 5 mM sodium EDTA and 10 mM DTT, for 5 h at 37 °C, and alkylated by the addition of iodoacetamide (2.5 molar excess over DTT) for 15 h in the dark. Samples were dialysed against 5 M guanidinium chloride and subjected to rate-zonal centrifugation, using guanidinium chloride gradients (6–8 M) formed in 14×89 mm centrifuge tubes with a Hoefer gradient maker connected to an LKB 2232 Microperpex pump at a flow rate of 50 ml h−1. Samples were layered onto the top of the gradients, and tubes were spun in a Beckman L-70 Optima centrifuge (Beckman SW41 Ti rotor, 287 000 g, 20 °C for 2 h 45 min). Depending on the sedimentation coefficient (s value) of the molecule, which is in turn determined by the molecular mass, density, size and shape of the molecule, larger molecules travel further into the gradient. Fractions (400 μl) were collected from the top of the tubes and analysed for reactivity with the LUM5B-2 antiserum.
RESULTS
Growth of single strains and consortium
The four speciated isolates from freshly harvested plaque and these four species as a consortium were monitored for growth for 72 h, in the presence of MUC5B as sole substrate (Fig. 1a). At 3 and 6 h, no growth was seen in any of the samples. After 24 h, however, growth was seen in the consortium and to a limited extent in the four single-strain cultures. At 48 h, the consortium showed linear growth, whereas the single strains did not appear to increase in density. Between 48 and 72 h, a plateau in growth was reached for the consortium. These results suggest that utilization of the MUC5B for growth required cooperation between species.
Fig. 1.
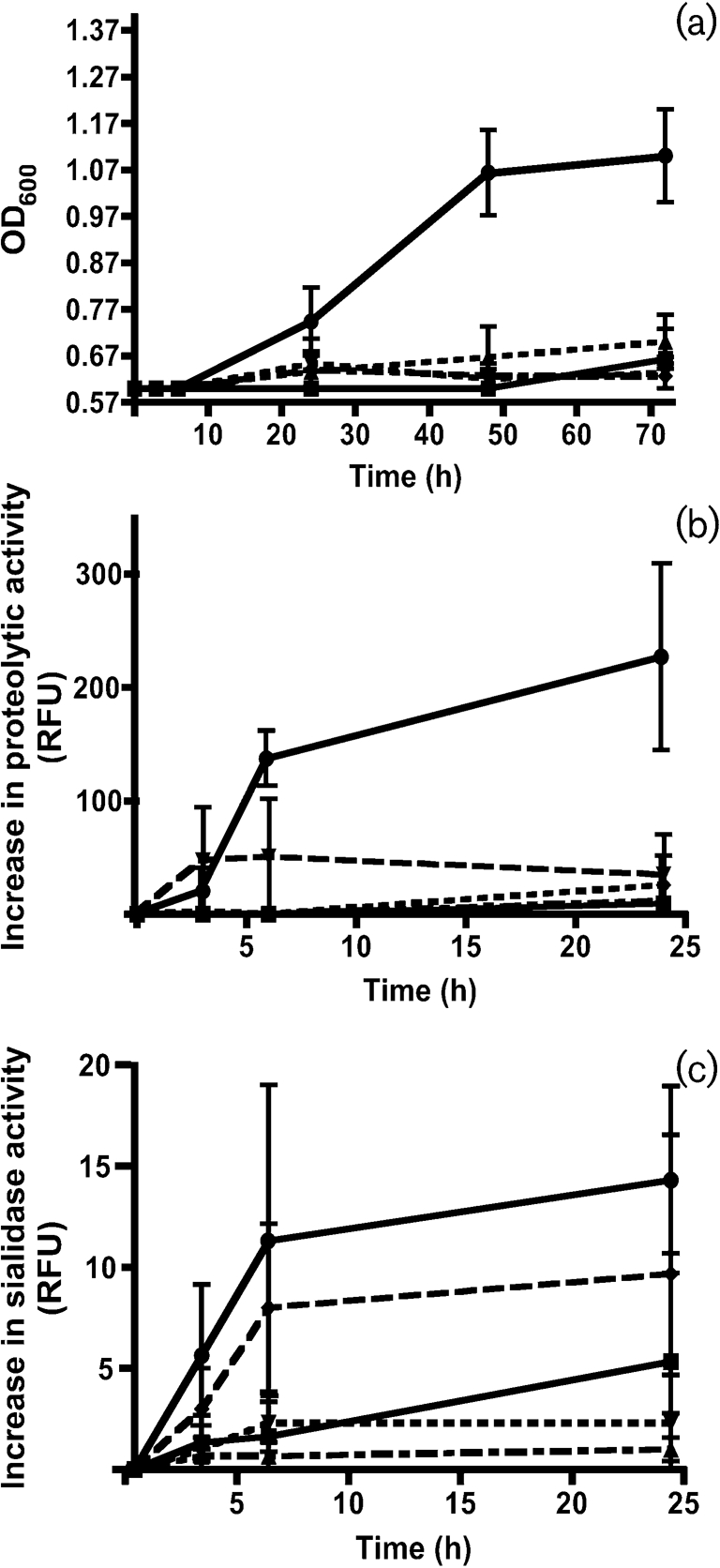
(a) Growth of isolated species and a four-species consortium with MUC5B as sole nutrient. S. mitis biovar 2 (▪), S. gordonii (▴), S. cristatus (▾) and A. naeslundii (⧫), as well as a consortium of the four species (•), were incubated with isolated salivary MUC5B and growth was monitored (OD600) at 3, 6, 24, 48 and 72 h. The experiment was repeated three times and the result shown is the mean±se. (b, c) Induction of proteolytic and sialidase activity during growth. S. mitis biovar 2 (▪), S. gordonii (▴), S. cristatus (▾) and A. naeslundii (⧫), as well as a consortium of the four species (•), were incubated with isolated salivary MUC5B and increase of (b) proteolytic and (c) sialidase activity was monitored (relative fluorescence units, RFU) at 3, 6 and 24 h. The experiment was repeated three times and the result shown is the mean±se.
Induction of proteolytic and sialidase activity
Proteolytic activity was monitored during incubation using FITC-labelled casein as a substrate (Fig. 1b). No increase of proteolytic activity was seen in the single-strain cultures, except for S. cristatus after 6 h. Proteolytic activity was measured in the consortium at 3 h and increased significantly at 6 h (P<0.004). At 24 h, proteolytic activity was evident in all cultures, reflecting the extent of growth in each.
Using 2′-(4-methylumbelliferyl) α-d-N-acetylneuraminic acid as a substrate, sialidase activity increased before or simultaneously with proteolytic activity at 3 and 6 h in all single-strain cultures, with the greatest increase apparent in the A. naeslundii culture (Fig. 1c). A slight increase was also evident after 24 h, especially in the S. mitis biovar 2 culture. The four-species consortium displayed the most prominent increase in sialidase activity after 3 h, continuing from 3 to 6 h (P<0.028), and reaching a plateau at 24 h.
Proteolytic degradation of MUC5B by the bacterial consortium
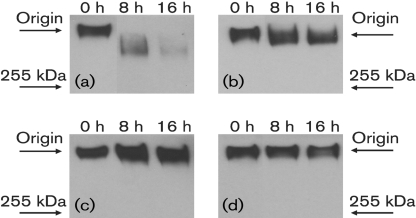
To ascertain whether proteolytic degradation of the MUC5B polypeptide backbone occurred, MUC5B was incubated for 0, 8 and 16 h at 37 °C with the single strains and the four-species consortium. Supernatants were recovered and analysed by SDS-PAGE and Western blotting. MUC5B was detected using an antiserum, LUM5B-2, which recognizes the central domain of the MUC5B polypeptide backbone. When MUC5B was incubated with a consortium of all four isolated species (S. mitis biovar 2, S. gordonii, S. cristatus and A. naeslundii), proteolytic degradation was evident over time and was greater than that expressed by the individual species (Fig. 2).
Fig. 2.
Degradation of the MUC5B polypeptide backbone. MUC5B was incubated with (a) the four-species consortium, (b) S. mitis biovar 2, (c) S. gordonii and (d) a combination of A. naeslundii and S. cristatus for 0, 8 and 16 h at 37 °C. After centrifugation (12 000 g, 2 min), the supernatants were subjected to SDS-PAGE followed by Western blotting. MUC5B was detected using the LUM5B-2 antiserum, recognizing the central domain of the MUC5B polypeptide backbone. The electrophoretic origin of the samples is indicated.
MUC5B degradation and proteolytic activity of dental plaque
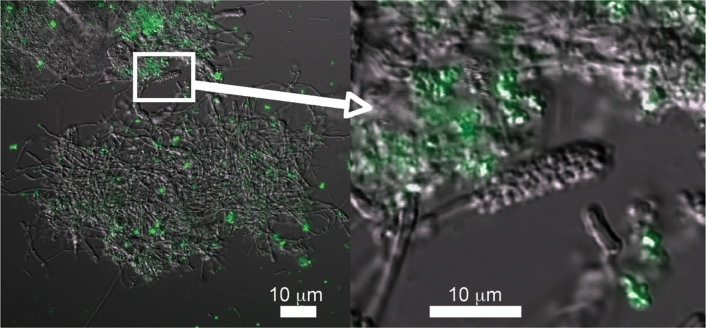
Proteolytic activity in supragingival plaque was visualized by CSLM. In supragingival plaque, protease-expressing cells (fluorescent green) were distributed in a non-uniform manner and appeared to localize in clusters of coccal-shaped bacteria in all the microscopic fields examined (Fig. 3).
Fig. 3.
Distribution of proteolytic activity in supragingival plaque. Freshly harvested dental plaque was incubated with a FITC-conjugated casein substrate and analysed by CSLM. Confocal illumination was provided by an Ar laser (488 nm laser excitation) using a ×60 magnification oil immersion objective.
To establish whether dental plaque degrades MUC5B, whole dental plaque and MUC5B were incubated for 0 and 8 h as above. At 0 h, undegraded MUC5B remained at the origin, failing to enter the SDS-PAGE gel. At 8 h, plaque micro-organisms appeared to degrade MUC5B, as digestion products migrated into the gel and were detected with the LUM5B-2 antiserum in Western blots (Fig. 4).
Fig. 4.
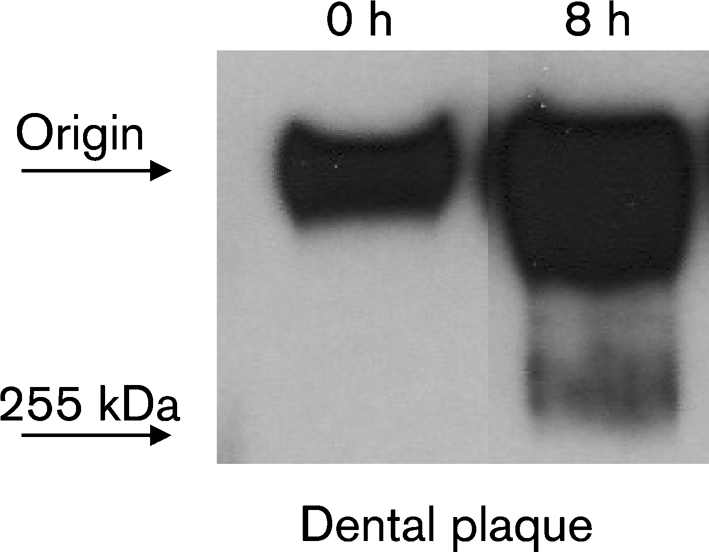
MUC5B degradation by dental plaque. MUC5B was incubated with freshly harvested dental plaque for 0 and 8 h at 37 °C. After centrifugation (12 000 g, 2 min), the supernatants were subjected to SDS-PAGE followed by Western blotting. MUC5B was detected using the LUM5B-2 antiserum, recognizing the central domain of the MUC5B polypeptide backbone.
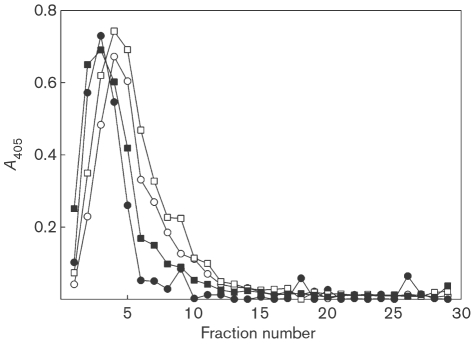
To show that the apparent mass change in MUC5B was associated with proteolytic digestion rather than only deglycosylation, the supernatants from the consortium and dental plaque samples (0 h and 8 h) were subjected to reduction and alkylation, rate-zonal centrifugation and analysis using the LUM5B-2 antiserum (Fig. 5). At 8 h (filled symbols), the mucins from both samples shifted towards a lower molecular mass than at the 0 h time point (open symbols).
Fig. 5.
MUC5B degradation visualized by rate-zonal centrifugation. MUC5B was incubated with freshly harvested dental plaque (○, •) or the four-species consortium (□, ▪) for 0 h (open symbols) and 8 h (filled symbols) at 37 °C. After centrifugation (12 000 g, 2 min), the supernatants were reduced and alkylated, layered on linear 6–8 M guanidinium chloride gradients and subjected to rate-zonal centrifugation (2 h 45 min, Beckman SW 41 Ti rotor). Fractions (400 μl) were collected from the top of the tubes and MUC5B was detected using the LUM5B-2 antiserum, recognizing the central domain of the MUC5B polypeptide backbone.
DISCUSSION
In this report, we show that a naturally occurring substrate, MUC5B, is digested preferentially by cooperation of several members of the dental plaque microbiota. Interspecies cooperation appears to overcome the high level of molecular complexity in MUC5B and makes the digestion products available as a nutrient source. In response to plaque or constituent bacteria, the degradation products were visualized as evidence of proteolysis (Figs 2, 4 and 5). In some experiments, proteolysis was detected using an antiserum recognizing a repetitive epitope located in the central region of the MUC5B molecule, between the heavily glycosylated mucin domains. This approach avoided the appearance of confounding degradation, since both N- and C-terminal protein fragments can be released by post-translational modifications of the MUC5B molecule and become entangled in the mucin network (Wickström et al., 1998; Wickström & Carlstedt, 2001).
To digest the gel-forming mucin, a consortium of species isolated from dental plaque (S. mitis biovar 2, S. gordonii, S. cristatus and A. naeslundii) simulated the degradative activity of the supragingival dental plaque sample, although individual species or the combination of S. cristatus and A. naeslundii did not (Figs 1a and 2). Thus inter-species cooperation facilitates breakdown of this substrate into metabolizable fragments, which could support bacterial growth. Consistent with this conclusion, enzymic activity increased in advance of detectable consortium growth on MUC5B (Fig. 1b, c). Similarly, induction of enzymic activity also preceded limited growth of some members of the consortium when incubated as pure cultures with MUC5B. Enzyme activity and growth of the consortium far exceeded the summed enzyme activity and growth of pure cultures of the individual strains, indicating that the bacteria worked together to induce or upregulate MUC5B-degrading enzymes. This cooperation between bacteria in the consortium is an important example of diversification of function in a native polymicrobial biofilm. Interspecies cooperation is thought to be a factor in maintaining the species diversity of the resident oral microflora (Bradshaw et al., 1994; van der Hoeven & Camp, 1991). As we now show, the degradation of complex substrates, such as salivary mucins, is highly dependent on interactions within consortia of different oral species displaying a complementary range of glycosidase and proteolytic activities. The apparent differences in molecular size (as seen in Figs 2 and 4) between the fragments originating from the consortium and dental plaque degradation of MUC5B, respectively, are not fully understood. In the dental plaque sample, a multitude of oral bacterial species and salivary proteins are present, which may influence the final degradation products.
The microbiota of the oral biofilm express a wide variety of glycosidases necessary to degrade carbohydrate structures found on salivary glycoproteins (e.g. Beighton & Hayday, 1986; Wickström & Svensäter, 2008). Since survival and growth depend upon host-derived nutrients, however, the supragingival flora must also express proteolytic enzymes, and many oral streptococci express endopeptidase activity (Homer et al., 1990). For example, S. oralis has been shown to express proteolytic enzymes and increase in numbers relative to other oral species during fasting, indicating that the ability to digest protein is an advantage in the oral community (Beighton & Hayday, 1986; Homer et al., 1990). Rafay et al. (1996) showed that when porcine gastric mucin was incorporated in the minimal medium used for growth of S. oralis, the cells displayed significantly less protease activity than those that were grown in media rich in glucose. In this report, however, only MUC5B was provided to the cultures as a source of both C and N, forcing the bacteria to enhance their proteolytic activity. Hence, the presence of MUC5B appears to directly induce the range of enzymes involved in its degradation, which indirectly supports the diversity of the oral microbiota.
When incubated with the pooled dental plaque, MUC5B was degraded less efficiently than with the four-species consortium used in this study (Figs 2 and 4). Consistent with the diversified functions of the polymicrobial species in dental plaque, the proteolytic activity in the plaque was distributed in a non-uniform pattern, with activity localized to specific areas. In the plaque, many organisms did not degrade FITC-casein (Fig. 3), which requires expression of endopeptidase activities. Endopeptidase activities in conjunction with full or partial deglycosylation, may be the first step in the degradation of salivary proteins by the oral biofilm. This first step in degradation would increase the availability and number of N- and C-termini to serve as substrates for aminopeptidases, which liberate amino acids and short peptides to support bacterial growth.
MUC5B is also found on mucosal surfaces in the endocervix, respiratory tract, gall bladder and colon (Carlstedt et al., 1983; Wickström et al., 1998; Audie et al., 1993; Porchet et al., 1995; Keates et al., 1997; van Klinken et al., 1998). As in the oral cavity, it could be argued that the commensal microbiota colonizing all epithelial surfaces confer benefit by acting with host defences to prevent colonization by pathogenic microbes. Certainly, the ability of consortia of bacteria resident on different mucosal surfaces to target MUC5B for degradation may be generalizable. Thus, due to the accessibility of the oral cavity, salivary mucins and oral biofilm bacteria may be used as a model system for other mucosal surfaces.
Acknowledgments
This work was supported with research grants from the Crafoord Foundation, the Knowledge Foundation (KK-stiftelsen, Biofilms – Research Centre for Biointerfaces), the Swedish Research Council (grant no. K2005-06X-12266-07A) and NIH/NIDCR R01DE08590 to M. C. H.
We wish to thank Agnethe Henriksson for excellent technical assistance and Dr Luis Chávez de Paz for help with the confocal scanning laser microscope images.
Abbreviations
CSLM, confocal scanning laser microscope/microscopy
References
- Audie, J. P., Janin, A., Porchet, N., Copin, M. C., Gosselin, B. & Aubert, J. P. (1993). Expression of human mucin genes in respiratory, digestive, and reproductive tracts ascertained by in situ hybridization. J Histochem Cytochem 41, 1479–1485. [DOI] [PubMed] [Google Scholar]
- Beighton, D. & Hayday, H. (1986). The influence of dieton the growth of streptococcal bacteria on the molar teeth of monkeys (Macaca fascicularis). Arch Oral Biol 31, 449–454. [DOI] [PubMed] [Google Scholar]
- Beighton, D. & Whiley, R. A. (1990). Sialidase activity of the “Streptococcus milleri group” and other viridans group sterptococci. J Clin Microbiol 28, 1431–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw, D. J., Homer, K. A., Marsh, P. D. & Beighton, D. (1994). Metabolic cooperation in oral microbial communities during growth on mucin. Microbiology 140, 3407–3412. [DOI] [PubMed] [Google Scholar]
- Byers, H. L., Tarelli, E., Homer, K. A. & Beighton, D. (1999). Sequential deglycosylation and utilazation of the N-linked, complex-type glycans of human α1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology 9, 469–479. [DOI] [PubMed] [Google Scholar]
- Byers, H. L., Tarelli, E., Homer, K. A. & Beighton, D. (2000). Isolation and characterisation of sialidase from a strain of Streptococcus oralis. J Med Microbiol 49, 235–244. [DOI] [PubMed] [Google Scholar]
- Carlstedt, I., Lindgren, H. & Sheehan, J. K. (1983). The macromolecular structure of human cervical-mucus glycoproteins. Studies on fragments obtained after reduction of disulphide bridges and after subsequent trypsin digestion. Biochem J 213, 427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman, R. A. & Baron, S. S. (1997). Pathway for uptake and degradation of X-prolyl tripeptides in Streptococcus mutans VA-29R and Streptococcus sanguis ATCC 10556. J Dent Res 76, 1477–1484. [DOI] [PubMed] [Google Scholar]
- Henssge, U., Do, T., Radford, D. R., Gilbert, S. C., Clark, D. & Beighton, D. (2009). Emended description of Actinomyces naeslundii and descriptions of Actinomyces oris sp. nov. and Actinomyces johnsonii sp. nov., previously identified as Actinomyces naeslundii genospecies 1, 2 and WVA 963. Int J Syst Evol Microbiol 59, 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer, K. A. & Beighton, D. (1992). Synergistic degradation of bovine serum albumin by mutans streptococci and other dental plaque bacteria. FEMS Microbiol Lett 69, 259–262. [DOI] [PubMed] [Google Scholar]
- Homer, K. A., Whiley, R. A. & Beighton, D. (1990). Proteolytic activity of oral streptococci. FEMS Microbiol Lett 55, 257–260. [DOI] [PubMed] [Google Scholar]
- Homer, K. A., Kelley, S., Hawkes, D., Beighton, D. & Grootveld, M. C. (1996). Metabolism of glycoprotein-derived sialic acid and N-acetylglucosamine by Streptococcus oralis. Microbiology 142, 1221–1230. [DOI] [PubMed] [Google Scholar]
- Jordan, H. V., Krasse, B. & Möller, A. (1968). A method of sampling human dental plaque for certain “caries-inducing” streptococci. Arch Oral Biol 13, 919–927. [DOI] [PubMed] [Google Scholar]
- Juarez, Z. E. & Stinson, M. W. (1999). An extracellular protease of Streptococcus gordonii hydrolyzes type IV collagen and collagen analogues. Infect Immun 67, 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keates, A. C., Nunes, D. P., Afdhal, N. H., Troxler, R. F. & Offner, G. D. (1997). Molecular cloning of a major human gall bladder mucin: complete C-terminal sequence and genomic organization of MUC5B. Biochem J 324, 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, M. J., Reddy, M. S., Tabak, L. A., Loomis, R. E., Bergey, E. J., Jones, P. C., Cohen, R. E., Stinson, M. W. & Al-Hashimi, I. (1987). Structural aspects of salivary glycoproteins. J Dent Res 66, 436–441. [DOI] [PubMed] [Google Scholar]
- Li, T., Bratt, P., Jonsson, A. P., Ryberg, M., Johansson, I., Griffiths, W. J., Bergman, T. & Strömberg, N. (2000). Possible release of an ArgGlyArgProGln pentapeptide with innate immunity properties from acidic proline-rich proteins by proteolytic activity in commensal Streptococcus and Actinomyces species. Infect Immun 68, 5425–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, P. D. (2003). Are dental diseases examples of ecological catastrophes? Microbiology 149, 279–294. [DOI] [PubMed] [Google Scholar]
- Porchet, N., Pigny, P., Buisine, M. P., Debailleul, V., Degand, P., Laine, A. & Aubert, J. P. (1995). Human mucin genes: genomic organization and expression of MUC4, MUC5AC and MUC5B. Biochem Soc Trans 23, 800–805. [DOI] [PubMed] [Google Scholar]
- Rafay, A. M., Homer, K. A. & Beighton, D. (1996). Effect of mucin and glucose on proteolytic and glycosidic activities of Streptococcus oralis. J Med Microbiol 44, 409–417. [DOI] [PubMed] [Google Scholar]
- Raynal, B. D., Hardingham, T. E., Sheehan, J. K. & Thornton, D. J. (2003). Calcium-dependent protein interactions in MUC5B provide reversible cross-links in salivary mucus. J Biol Chem 278, 28703–28710. [DOI] [PubMed] [Google Scholar]
- Reinholdt, J., Tomana, M., Mortensen, A. B. & Kilian, M. (1990). Molecular aspects of immunoglobulin A1 degradation by oral streptococci. Infect Immun 58, 1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, K. & Beighton, D. (1986). The effects of the availability of diet on the levels of exoglycosidases in the supragingival plaque of macaque monkeys. J Dent Res 65, 1349–1352. [DOI] [PubMed] [Google Scholar]
- Smith, K. & Beighton, D. (1987). Proteolytic activities in the supragingival plaque of monkeys (Macaca fascicularis). Arch Oral Biol 32, 473–476. [DOI] [PubMed] [Google Scholar]
- Thomsson, K. A., Prakobphol, A., Leffler, H., Reddy, M. S., Levine, M. J., Fisher, S. J. & Hansson, G. C. (2002). The salivary mucin MG1 (MUC5B) carries a repertoire of unique oligosaccharides that is large and diverse. Glycobiology 12, 1–14. [DOI] [PubMed] [Google Scholar]
- Thornton, D. J., Khan, N., Mehrotra, R., Howard, M., Veerman, E. C., Packer, N. H. & Sheehan, J. K. (1999). Salivary mucin MG1 is comprised almost entirely of different glycosylated forms of the MUC5B gene product. Glycobiology 9, 293–302. [DOI] [PubMed] [Google Scholar]
- van der Hoeven, J. S. & Camp, P. J. M. (1991). Degradation of mucin by Streptococcus oralis and Streptococcus sanguis in mixed chemostat cultures. J Dent Res 70, 1041–1044. [DOI] [PubMed] [Google Scholar]
- van Klinken, B. J., Dekker, J., van Gool, S. A., van Marle, J., Buller, H. A. & Einerhand, A. W. (1998). MUC5B is the prominent mucin in human gallbladder and is also expressed in a subset of colonic goblet cells. Am J Physiol 274, G871–G878. [DOI] [PubMed] [Google Scholar]
- Veerman, E. C., van den Keybus, P. A., Valentijn-Benz, M. & Nieuw Amerongen, A. V. (1992). Isolation of different high-Mr mucin species from human whole saliva. Biochem J 283, 807–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickström, C. & Carlstedt, I. (2001). N-terminal cleavage of the salivary MUC5B mucin. Analogy with the von Willebrand pro-polypeptide? J Biol Chem 276, 47116–47121. [DOI] [PubMed] [Google Scholar]
- Wickström, C. & Svensäter, G. (2008). Salivary gel-forming mucin MUC5B – a nutrient for dental plaque bacteria. Oral Microbiol Immunol 23, 177–182. [DOI] [PubMed] [Google Scholar]
- Wickström, C., Davies, J. R., Eriksen, G. V., Veerman, E. C. & Carlstedt, I. (1998). MUC5B is a major gel-forming, oligomeric mucin from human salivary gland, respiratory tract and endocervix: identification of glycoforms and C-terminal cleavage. Biochem J 334, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickström, C., Christersson, C., Davies, J. R. & Carlstedt, I. (2000). Macromolecular organization of saliva: identification of 'insoluble' MUC5B assemblies and non-mucin proteins in the gel phase. Biochem J 351, 421–428. [PMC free article] [PubMed] [Google Scholar]
- Wickström, C., Hamilton, I. R. & Svensäter, G. (2009). Differential metabolic activity by dental plaque bacteria in association with two preparations of MUC5B mucins in solution and in biofilms. Microbiology 155, 53–60. [DOI] [PubMed] [Google Scholar]