Abstract
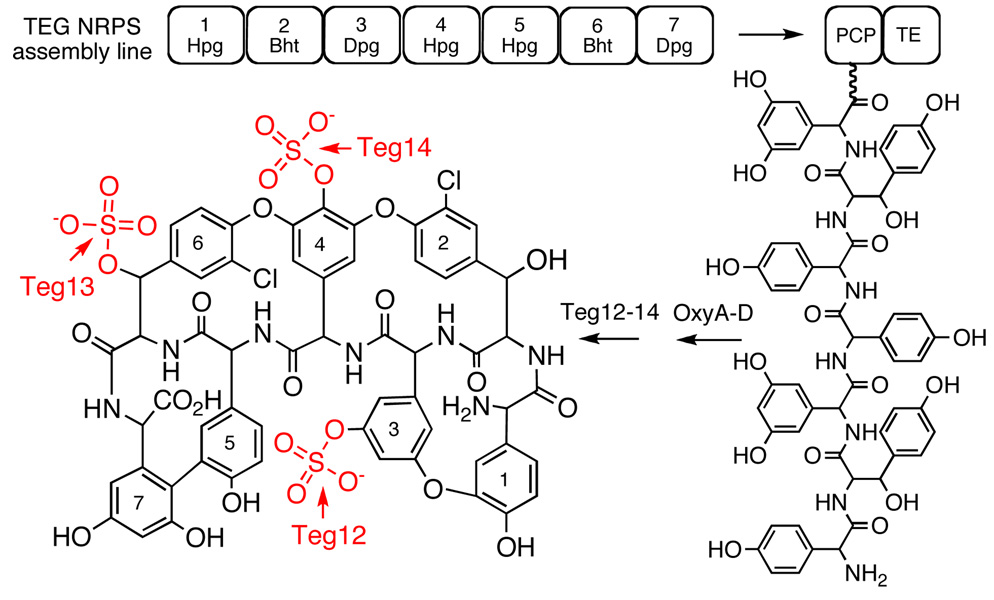
The TEG gene cluster, a glycopeptide biosynthetic gene cluster that is predicted to encode the biosynthesis of a polysulfated glycopeptide congener, was recently cloned from DNA extracted directly from desert soil. This predicted glycopeptide gene cluster contains three closely related sulfotransferases (Teg12, 13, and 14) that sulfate teicoplanin-like glycopeptides at three unique sites. Here we report a series of structures including: an apo structure of Teg12, Teg12 bound to the desulfated co-substrate 3'-phosphoadenosine 5'-phosphate and Teg12 bound to the teicoplanin aglycone. Teg12 appears to undergo a series of significant conformational rearrangements during glycopeptide recruitment, binding and catalysis. Loop regions that exhibit the most conformational flexibility show the least sequence conservation between TEG sulfotransferases. Site directed mutagenesis guided by our structural studies confirmed the importance of key catalytic residues as well as the importance of residues found throughout the conformationally flexible loop regions.
During the 20th century, widespread use of antibiotics significantly reduced the threat of many once lethal infectious diseases. However, the success of these wonder drugs may soon become their Achilles’ heel. Bacterial pathogens that have developed resistance to most widely used antibiotics are now regularly seen in clinical settings. Vancomycin and teicoplanin are glycopeptide antibiotics used in the treatment of many gram-positive bacterial infections, including methicillin-resistant Staphylococcus aureus (MRSA). With the appearance of vancomycin resistant Enterococci in the late 1980s and resistant Staphylococci in the early 1990s, these traditional antibiotics of last resort are in danger of becoming clinically compromised (1–3). As with many bacterial natural products, the discovery of additional glycopeptide congeners that might combat the growing problem of antibiotic resistance has slowed as it has become increasingly difficult to identify new biodiversity from which novel molecules might be characterized.
The vast majority of bacteria present in the environment remain recalcitrant to culturing (4). This uncultured majority no doubt contains previously inaccessible glycopeptide biosynthetic gene clusters, many of which could encode the biosynthesis of novel glycopeptide congeners. Although metabolites produced by bacteria that are difficult to culture in the laboratory cannot be characterized using standard microbiological methods, it is possible to extract DNA directly from environmental samples and then analyze this DNA for sequences that might encode the biosynthesis of new natural products. In a recent analysis of DNA extracted directly from desert soil, we uncovered a new glycopeptide biosynthetic gene cluster (the TEG gene cluster) that is predicted to encode the biosynthesis of the first polysulfated glycopeptide congeners (5). The TEG gene cluster contains three closely related 3'-phosphoadenosine 5'-phosphosulfate (PAPS) dependent sulfotransferases (Teg12, 13 and 14). In vitro, these three sulfotransferase finishing enzymes sulfate teicoplanin-like glycopeptides at three unique sites and, in combination, can be used to produce seven different glycopeptide sulfation patterns (Figure 1).
Figure 1.
Predicted structures for the glycopeptide congeners produced by the TEG gene cluster are shown. The seven nonribosomal peptide synthetase modules encoded by the TEG cluster are predicted to biosynthesize a heptapeptide that is oxidatively cross-linked by OxyA-D homologs and then sulfated by Teg12, 13, and 14.
Vancomycin- and teicoplanin-like glycopeptides are structurally defined by the presence of an oxidatively cross-linked heptapeptide core (6, 7). The peptide core is initially produced as a linear polymer by nonribosmal peptide synthetases, which is then oxidatively cross-linked into either three or four large macrocycles by conserved cytochrome P450 oxidases. During their biosynthesis, each member of this family of antibiotics is functionalized by a unique collection of finishing enzymes that includes glycosyl transferases, halogenases, acyl transferases and sulfotransferases. Although glycopeptides show some variation in the sequence of the heptapeptide core, the bulk of the structural diversity seen within this class of antibiotics arises from the functionality added by finishing enzymes. Over 150 different glycosylated, halogenated, and alkylated glycopeptide congeners have been characterized from cultured bacteria (8). Only three naturally occurring sulfated congeners have been identified to date from studying this same pool of bacteria (8, 9). While anionic glycopeptides have rarely been reported as natural products, telavancin, a semisynthetic phosphono congener that proved to be a very effective antibiotic in clinical trials, was recently approved for use in humans by the FDA (10, 11). Increasing the hydrophilicity with the addition of the negatively charged phosphono group was found to significantly improve the adsorption, distribution, metabolism, and excretion profile of this class of antibiotics (12). The enzymatic synthesis of anionic glycopeptides may provide a facile means to access additional anionic congeners with improved pharmacological properties.
Here we report a series of Teg12 sulfotransferase structures, including an apo structure, a binary structure complexed with the teicoplanin aglycone substrate, as well as a ternary structure containing both PAP and the teicoplanin aglycone. In the binary and ternary structures, the glycopeptide substrate is observed bound at two different locations. Teg12 appears to undergo a series of conformational rearrangements during glycopeptide recruitment, binding and catalysis. These studies provide insight into the sulfotransferase mechanism, as well as insights into how this rarely seen class of finishing enzymes might be engineered to produce novel anionic glycopeptides.
Materials and Methods
Teg12 Expression and Purification
Teg12 was cloned and expressed as previously described (5). Briefly, teg12 was amplified (30 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 90 s; FailSafe system from Epicentre) from eDNA cosmid clone D30 using the following primers: Teg12FWD(BclI):GCGCTGATCAATGAACGGAATTCGATGG, Teg12REV(HindIII):GCGCAAGCTTTCCTTAACCGGCATACCCGTA. Restriction enzyme sites used for cloning are shown in bold. The resulting product was doubly digested with BclI and HindIII and subsequently ligated into pET28a, which had been BamHI/HindIII doubly digested. The resulting construct was then transformed into E. coli BL21(DE3) for protein expression. Expression cultures were grown to OD600=0.6, followed by IPTG induction, and overnight growth at 20 °C. The culture was pelleted by centrifugation (3,200 × g for 30 min), the supernatant was discarded, and the cell pellet was resuspended in 40 mL lysis buffer [50 mM HEPES, pH 7.5, 0.5 M NaCl, 5% (vol/vol) glycerol, 20 mM imidazole, pH 8, 10 mM β-mercaptoethanol and 0.5% (vol/vol) Triton X-100]. The resuspended cell pellet was lysed by sonication, and the insoluble portion was removed by centrifugation (15,000 × g for 30 min). The cleared cell lysate was incubated with 1 mL Ni-NTA resin for 15 min. The slurry was loaded onto a column, allowed to empty by gravity flow, washed with 40 mL lysis buffer, and finally washed with 40 mL wash buffer [50 mM HEPES, pH 7.5, 0.5 M NaCl, 5% (vol/vol) glycerol, 20 mM imidazole, pH 8.0, and 10 mM β-mercaptoethanol]. The protein was eluted by the addition of 15 mL of elution buffer [50 mM HEPES, pH 7.5, 0.5 M NaCl, 5% (vol/vol) glycerol, 125 mM imidazole, pH 8, and 10 mM β-mercaptoethanol]. No attempt was made to remove the vector derived 6X-histidine tag, resulting in a Teg12 protein plus 34 additional residues N-terminal to the start methionine. Protein was concentrated using Vivascience Vivaspin 30,000 MWCO ultrafiltration concentrators, and was buffer exchanged 3 times into protein buffer [200 mM NaCl, 20 mM HEPES, pH 7.5, 5% glycerol, and 1 mM DTT].
Teg12 Crystallization
Concentrated protein was centrifuged at 14,000 rpm (4 °C, 30 min) in a microcentrifuge to remove any insoluble material prior to crystallization. All crystals were grown using the hanging drop vapor diffusion method. Initial Teg12-apo crystals were obtained by mixing 1 µl of protein (7.5 mg/ml in protein buffer) with 1 µl of reservoir solution (1.0 M sodium citrate, 0.1 M sodium cacodylate, pH 6.5, JCSG core III-48, Qiagen) over a 500 µl reservoir. Blade-like crystals grew overnight at 22 °C and reached a maximal size of 400 µm × 50 µm × 20 µm in approximately one week. To improve crystal thickness, Teg12-apo crystals were optimized by microseeding, in addition to mixing 1 µl of protein at 7.5 mg/ml with 0.5 µl of reservoir and 0.5 µl of Silver Bullets™ reagent 29 (Hampton Research). A component of the Silver Bullets™ screen, aspartame, was modeled into the PAPS binding site of one of the monomers of the Teg12 dimer. Crystals were soaked in a 20 µl drop containing reservoir solution plus 10% ethylene glycol. The drop was allowed to dehydrate by exposure to open air at room temperature for approximately 5 hours before flash cooling the crystals in liquid ethane.
Teg12-ternary crystals were co-crystallized at 4 °C in the presence of 2 mM PAP and 1 mM teicoplanin aglycone. Teg12 was first concentrated to 20 mg/ml in protein buffer. The protein was then diluted 1:1 with 50 mM CHES, pH 9.1, 2 mM teicoplanin aglycone, 4 mM PAP, achieving a final concentration of 10 mg/ml Teg12 in 0.5× protein buffer, 25 mM CHES, pH 9.1, 1 mM teicoplanin aglycone, 2 mM PAP. 1 µl of protein was mixed 1:1 with reservoir solution (0.2 M ammonium acetate and 20% w/v PEG 3350, JCSG, core I-25, Qiagen). Crystals appeared in 2–3 days and grew to a maximal size of 100 µm × 50 µm × 50 µm in approximately 1 week. These crystals were of an irregular chunk-like morphology and had cracks throughout. Crystals were cryo-protected by quickly dunking in reservoir solution plus 15% ethylene glycol and were flash cooled in liquid nitrogen.
Teg12-binary crystals were co-crystallized at 4 °C in the presence of 1 mM teicoplanin aglycone. Similar to Teg12-ternary crystallization, the protein was first concentrated to 20 mg/ml in protein buffer, then diluted to 10 mg/ml with 50 mM CHES, pH 9.1, 2 mM aglycone (final 0.5× protein buffer, 25 mM CHES, 1 mM teicoplanin aglycone). 1 µl of protein solution was mixed with 1 µl of reservoir solution (2.0 M sodium formate, 0.1 M sodium acetate, pH 4.6, JCSG core III-85, Qiagen). Cubic crystals grew between 2 and 3 weeks, and were approximately 50 µm × 50 µm × 50 µm. Crystals were soaked in 6.0 M sodium formate, 0.1 M sodium acetate, pH 4.6, 1 mM teicoplanin aglycone overnight, prior to flash cooling in liquid nitrogen.
Data Collection and Structure Solving
All data sets were reduced and scaled using the HKL2000 package (18). Data for Teg12-apo crystals were collected at the NSLS, beamline X29A. All but one of the crystals screened diffracted poorly to approximately 4 Å resolution. The crystal from which the 2.91 Å dataset was collected rotated briefly out of the cryostream, and thereby had gone through a room temperature annealing cycle of several seconds. Diffraction from this crystal was dramatically improved compared with other crystals taken from the same drop. Data for Teg12-apo was reduced and scaled in space group C2221. Phase information was obtained by molecular replacement using the program Phaser and StaL (GenBank accession number AAM80529, PDB code 2OV8), devoid of all flexible loops, as the search model (19). The initial molecular replacement model was refined against the Teg12-apo dataset using rigid body refinement in Refmac (20). Additional features of the map were enhanced through density modification, and 2-fold ncs averaging in CNS (21, 22). The model was rebuilt manually using the program Coot (23). Full restrained refinement was carried out using the translation/libration/screw model in Refmac, with the addition of hydrogen atoms, converging to a final Rwork and Rfree of 21.96 and 27.19, respectively (24). NCS restraints were not used during refinement. The final model comprises residues 1–129, 136–203, and 251–285 for monomer A, and 1–27, 42–128, 137–210, 247–285 for monomer B. The Teg12-apo model was used as a molecular replacement model for all subsequent structures.
Teg12-ternary and Teg12-binary data sets were collected at the APS, microfocus beamline 24-IDE. Teg12-ternary data was reduced and scaled in space group P212121. Teg12-binary was scaled in space group I212121. The crystal structure of glycopeptide aglycone A-40926 was used as a starting point to generate a restraint definition file for teicoplanin aglycone using the program Phenix Elbow (25). Geometry optimization was achieved using the semi-empirical quantum mechanical AM1 method. Teg12-binary and Teg12-ternary models were refined using the translation/libration/screw model in Phenix Refine to a final Rwork and Rfree of 17.30 and 22.61, and 17.12 and 22.47, respectively (26). The final Teg12-binary model comprises residues 1–129, 135–216, and 247–285. The final Teg12-ternary model comprises residues 1–215, 220–224, and 231–285 for monomer A, and residues 1–129, 136–224, and 240–285. All structures were validated using the Molprobity server from the Richardson laboratory at Duke University (27).
Site-Directed Mutagenesis
Teg12 point mutants were generated using the “megaprimer” method, with slight modifications (28). Oligonucleotide primers were designed for each mutant (Table 2), and a megaprimer was generated by PCR amplification from the Teg12/pET28a construct using the Pfx Accuprime System (Invitrogen), the relevant mutant oligonucleotide primer, and either the T7 promoter (for mutations at residues 9–108) or the T7 terminator (for mutations at residues 167–251) as the second oligonucleotide primer, (30 rounds of amplification: 95 °C for 30 s, 55°C for 30 s, 68 °C for 30 s). The full length mutant Teg12 gene was amplified from the Teg12/pET28a construct, using the megaprimer, which then contained the bases that code for the specific mutant residue, and either the T7 terminator (for mutations at residues 9–108) or the T7 promoter (for mutations at residues 167–251) as the second oligonucleotide primer (30 rounds of amplification: 95 °C for 30 s, 55 °C for 30 s, 68 °C for 80 s). Full-length mutant amplicons were then sequentially digested with BamHI and HindIII, and subsequently ligated into BamHI/HindIII doubly digested pET28a. Ligated constructs were transformed into E. coli EC100 (Epicentre), and sequenced to identify successfully mutated constructs. Mutant constructs containing the desired point mutation were then transformed into E. coli BL21 (DE3) for protein expression.
Table 2.
Oligonucleotide primers for site-directed mutagenesis
| Mutant | Primer (5’-3’) |
|---|---|
| S9A | GTTTCCAGCCTTTGGATACGCTGCGATCCATCGAATTCC |
| K12A | CCACGTGTTTCCAGCCGCTGGATACGATGC |
| T16A | CACCTGACCCACGCGTTTCCAGCCTTTGG |
| W17A | CAACATGCACCTGACCGCCGTGTTTCCAGC |
| K65A | CATCGGCCTTGAGGTGCGTCGCCACCAGCACCGGTTC |
| H67A | CACATCGGCCTTGAGGGCCGTCTTCACCAG |
| R90A | CATATCCCGCGGGTTCGCCACGAGATAGAG |
| S98A | GGCCATGCGATCGAGGCGAGCAGCATATC |
| R101A | TATCGAGGCCATGGCCATCGAGCTGAG |
| S106A | CTACGTCGTCGCGCGCTATCGAGGCCATGCG |
| R107A | GCTTTTTTCTACGTCGTCGGCCGATATCGAGGC |
| D108A | GCTTTTTTCTACGTCGGCGCGCGATATCGAGGCC |
| Y167A | GTGCTGACGATGCGTGCTGAGGACCTGAAGGGC |
| K171A | CGTTATGAGGACCTGGCGGGCGATCCGGTCGCACGG |
| E206A | GCTGCCTCCACGCTGGCGCGGATGCGTGAACTG |
| R207A | GCTGCCTCCACGCTGGAGGCGATGCGTGAACTG |
| E210A | GAGCGGATGCGTGCACTGGAGAAACGGAG |
| E212A | CGGATGCGTGAACTGGCGAAACGG |
| K213A | CGGATGCGTGAACTGGAGGCACGGAGCGAGCAGCAG |
| R214A | CGGATGCGTGAACTGGAGAAAGCGAGCGAGCAGCAG |
| M232A | GGTGATGCGAGAATGGCGAAAGGG |
| K233A | GATGCGAGAATGATGGCAGGGGG |
| G234A | GCGAGAATGATGAAAGCGGGACC |
| G235A | AGAATGATGAAAGGGGCACCTGGTG |
| P236A | ATGATGAAAGGGGGAGCTGGTGG |
| G238A | AAAGGGGGACCTGGTGCCGCGAGG |
| A239R | AAAGGGGGACCTGGTGGCCGGAGG |
| R240A | GGACCTGGTGGCGCGGCGCCCCAG |
| P241A | CCTGGTGGCGCGAGGGCCCAGTTC |
| Q242A | GGTGGCGCGAGGCCCGCGTTCGTG |
| R248A | CAGTTCGTGGGCGAGGGCGCGTACGACCAGTCCCTG |
| Q251A | GAGGGCAGGTACGACGCGTCCCTGTCCTTCTTG |
Mutant Teg12 Expression and Purification
Mutant proteins were expressed and purified in a manner similar to the native Teg12, except on a reduced scale. 100 mL overnight expression cultures were pelleted and resuspended in 4 mL lysis buffer. After sonication to lyse the cells, the crude lysates were centrifuged to remove insoluble material (10 min at 15,000 × g). The cleared lysates were incubated with 100 µl Ni-NTA resin for 15 min. The slurry was then loaded onto a column, allowed to empty by gravity flow, washed with 4 mL lysis buffer, followed by a second wash with 4 mL wash buffer. The protein was eluted by the addition of 1.5 mL elution buffer. All Teg12 mutants used in activity assays appeared to be homogeneous by polyacrylamide gel electrophoresis
Teg12 Activity Assays
All soluble Teg12 mutants were assayed for activity using the teicoplanin aglycone as a substrate. 50 µL reactions were run in duplicate, as follows: 15 mM HEPES, pH 7.5, 1 mM 3’-phosphoadenosine-5’-phosphosulfate (PAPS), 0.1 mM DTT, 1.2 mM teicoplanin aglycone (in DMSO), and 500 ng purified protein in elution buffer. Reactions were carried out at 30 °C for each of the four time points (10, 15, 20, 25 min), followed by heat inactivation at 99 °C for 10 min, and a further 10 min in an ice water bath. Vmax and Km values were determined under the same reaction conditions using the teicoplanin aglycone as substrate (5 µM to 100 µM). 25 µL of each reaction was run on a Waters analytical HPLC system (C18 (4.6 × 150 mm)). A linear gradient (1.5 ml/min) was run from an initial condition of 95:5 20 mM ammonium acetate:acetonitrile to 70:30 20 mM ammonium acetate:acetonitrile over twenty minutes. The area under the UV peak (Diode Array, 240 nm–400 nm) was determined for both the monosulfated product and the teicoplanin aglycone substrate at each time point. The percent substrate conversion for duplicate time points was averaged. The slope of the graph derived from the four time points for Teg12 and each mutant was then determined. Relative activity of each mutant is reported as a percent of the slope for wild-type Teg12.
Results
Teg12 apo structure
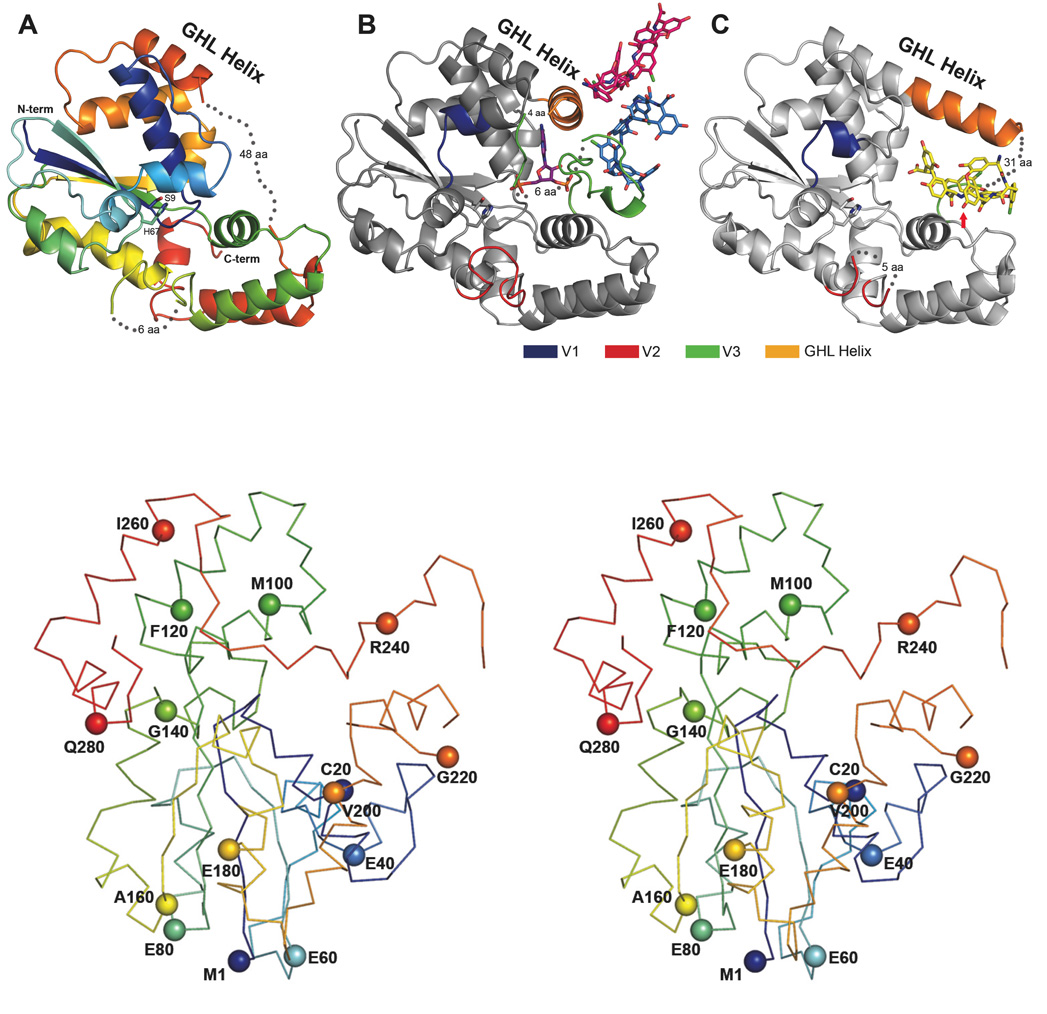
N-terminally His tagged Teg12 was affinity purified using nickel NTA resin and crystallized without the need for further purification. The apo structure was solved to 2.91 Å resolution by molecular replacement using StaL (PDB code 2OV8) as a search model (13). StaL is a related sulfotransferase involved in the biosynthesis of the monosulfated glycopeptide congener A47934 (9, 14). StaL has 52.9% sequence identity with Teg12. The Teg12 structure superimposes well on StaL (rmsd of 1.053 Å over 210 Cα atoms). Teg12 crystallizes as a dimer and the overall structure resembles that of StaL. Running Teg12 through the DALI server shows that its closest eukaryotic sulfotransferase relative is the human cytosolic sulfotransferase 1C1 (SULT1C1, pdb code 1zhe), with an rmsd of 2.8 Å over all Cα atoms (15). As with other sulfotransferases in this family, Teg12 consists of a single globular α/β domain composed of a parallel beta sheet core surrounded by alpha helices (Figure 2A). The beta sheet core of both StaL and Teg12 contains four strands. The dimer interface, which resembles that seen in StaL, consists of a symmetrical interaction between a short helix-loop motif from one monomer and the same helix-loop motif from the other monomer. In addition to the hydrophobic contacts that exist between the two helices there is a hydrogen bond between the carbonyl oxygen of Gly51 at the end of the helix from one monomer and the backbone nitrogen of Val74 of the other monomer. The active site cavity of each monomer faces the dimer interface.
Figure 2.
The Three Teg12 structures are presented. Only a single monomer from the dimer is shown for clarity. (A) Teg12-Apo is colored using a rainbow scheme, from blue (N-term) to red (C-term). Regions of disorder are connected with grey dots, with the number of disordered residues indicated. The flexible GHL Helix is shown in orange. Side chains for the proposed active site residues His67 and Ser9 are also shown. (B) Teg12-ternary complex containing PAP and the teicoplanin aglycone. The protein has been colored grey in order to accentuate regions of the structure that differ from Teg12-Apo. Specifically, these regions are the variable loops V1–V3, colored blue, red, and green, respectively. Also shown is a molecule of PAP in the active site, a molecule of teicoplanin aglycone that interacts with the V3 loop (colored sky blue), and an additional molecule of teicoplanin aglycone involved in crystal packing interactions (colored hot pink). (C) Teg12-binary structure complexed with teicoplanin aglycone in the active site cavity. Again, the protein has been colored grey with V1–V3 colored as in the ternary structure. The active site is shown as a stereo close-up image in figure 6. Teg12 sulfates the hydroxyl of residue 3 of the teicoplanin aglycone, which has been indicated in the figure with a red arrow. Omitted from the binary structure is a second molecule of teicoplanin aglycone bound to the outside of the protein on the opposite side of the GHL helix. (D) Stereo view of a Cα trace of the Teg12-ternary structure represented as a ribbon diagram with the Cα carbon of every 20th amino acid shown as a sphere.
Minor differences exist between the two monomers of the apo structure. The pET28a vector derived N-terminal tail, including the 6X-His, thrombin cleavage site and T7 tags, from monomer A is ordered and involved in crystal packing interactions. The arrangement of these residues within the crystal causes distortions in the region Gly28 through Ser41 when compared with StaL, and subsequent Teg12 complex structures. In monomer B, residues Gly28 through Ser41 could not be modeled in the electron density map, further underscoring the conformational flexibility of this region. From our co-crystal structures and that of StaL, it is known that residues Glu216 through Asp250 constitute a largely disordered loop. In the Teg12 apo structure, this disorder extends even further N-terminally into what is observed as a helix in the Teg12 binary and ternary structures and StaL. Electron density falls off in monomer A after Ser203, but extends through Glu210 in monomer B. Ser203 corresponds to Cys196 in StaL. In StaL Cys196 makes a disulfide bond with Cys20 (residue 20 is also a cysteine in Teg12). The absence of a disulfide in Teg12 could impart a greater flexibility in this region and explain the additional observed disorder. In subsequent Teg12 co-crystal structures, portions of this helix-loop region (Thr204-Asp250) play key roles in binding the glycopeptide substrate. Because of its role in binding the glycopeptide, we have termed this entire conformationally flexible region the GHL (glycopeptide-helix-loop).
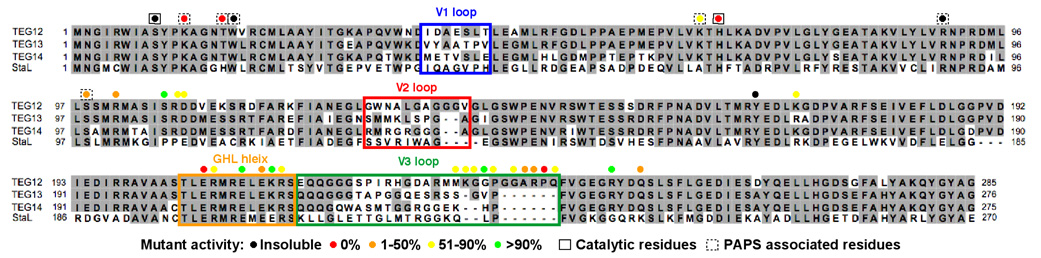
ClustalW alignment of the three TEG sulfotransferases shows that most of the sequence variability seen within this family of sulfotransferases is concentrated on three short loops (variable regions V1, V2, V3) that surround the predicted active site (Figure 3). The large disordered loop from the GHL corresponds to the longest of these three variable regions, V3. V2 encompasses residues Gly127 through Gly137. Much of V2 (Ala130 to Gly135 in monomer A and Asn129 through Gly136 in monomer B) is disordered in the Teg12 apo structure. V1, the shortest of the three variable sequences, corresponds to an ordered region (though disordered in monomer B) that spans Ile37 through Thr43.
Figure 3.
ClustalW alignment of glycopeptide sulfotransferases. Highly variable sequences (V1, V2 and V3) and the flexible part of the GHL helix appear in colored boxes that match the coloration seen in Figures 2B and 2C. Results from alanine exchange mutagenesis experiments are color coded by percent activity. Results for catalytic and PAPS associated residues are shown in solid or dashed boxes, respectively. All other mutated residues are thought to interact with the glycopeptides. The H67A mutant was insoluble and therefore H67Q data is reported.
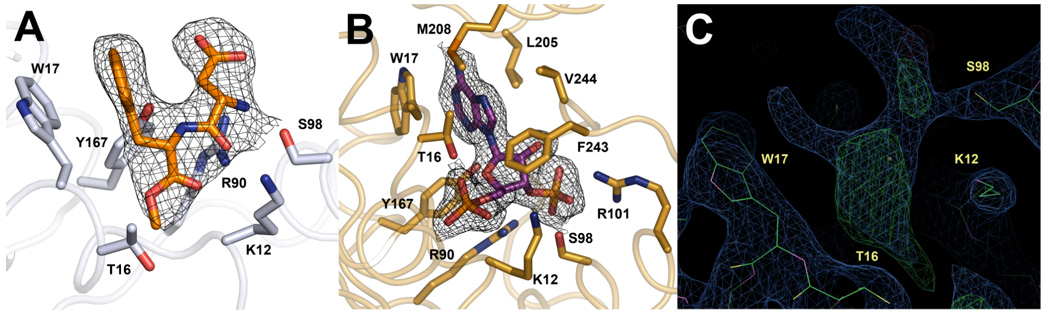
Electron density was observed in the PAPS binding site of monomer A (this binding site was empty in monomer B). However, neither PAP nor PAPS could be modeled into the density. We were able to model the dipeptide aspartame (N-(l-a-aspartyl)-l-phenylalanine,1-methyl ester) into this density (Figure 4). Aspartame is a component of the Silver Bullets™screen (Hampton) used during crystallization. The orientation of aspartame in the structure mimics that of PAP. Its phenylalanine ring stacks with Trp17, as does the adenine ring of PAP, and its N-terminal nitrogen hydrogen bonds with the hydroxyl of Ser98. Eukaryotic sulfotransferases are known to tightly bind ribose, adenine and other nucleotides in the PAPS binding pocket (16).
Figure 4.
Comparison of the ligands bound in the PAPS binding sites from Teg12-Apo and Teg12-ternary. Electron density from the 2fo-fc map surrounding each ligand and contoured at 1σ is shown. (A) The PAPS binding site of monomer A from Teg12-Apo contained a molecule of the dipeptide aspartame (N-(L-a-aspartyl)-L-phenylalanine,1-methyl ester). Several Teg12 residues important for coordinating PAP also make contacts with aspartame. Their side chains are shown. (B) PAP bound in the Teg12-ternary complex. Leu205 and Met208 from the GHL helix and Phe243 and Val244 from the V3 loop make significant Van der Waals contacts with PAP. (C) Electron density in the PAPS binding site of the Teg12-apo monomer A, before the addition of aspartame to the model is shown. Both the 2fo-fc (contoured at 1 sigma) (green) and the fo-fc (contoured at 3 sigma) (blue) maps are shown. Aspartame was added to the model only after the protein had been fully built, at the second to last refinement step (before the addition of waters).
Teg12 co-crystal structures
The TEG gene cluster is predicted to encode the biosynthesis of a heptapeptide (hydroxyphenylglycine (Hpg)-betahydroxytyrosine (Bht)-dihydroxyphenylglycine (Dpg)-Hpg-Hpg-Bht-Dpg) that is oxidatively cross-linked into the four macrocycles seen in teicoplanin-like glycopeptides (Figure 1). This TEG derived, oxidatively cross-linked heptapeptide skeleton only differs from the teicoplanin aglycone by the substitution of Bht for Tyr at the 2nd position in the peptide core. All three TEG sulfotranferases can use the teicoplanin aglycone as a substrate. We therefore used this molecule in Teg12 co-crystallization experiments. The Vmax and Km values for the teicoplanin aglycone were determined to be 215.3 ± 25.2 nmol/min mg and 59.6 ± 1.1 uM, respectively. Attempts to obtain co-crystals by soaking Teg12 apo crystals with either the teicoplanin aglycone or the co-substrate PAP were unsuccessful. The addition of PAP to drops containing apo crystals caused the crystals to rapidly dissolve into the mother liquor. Therefore, we co-crystallized Teg12 and the teicoplanin aglycone with and without PAP. PAP is the desulfated byproduct of PAPS from the sulfonation reaction. Our co-crystallization experiments led to a Teg12 binary structure bound to teicoplanin, and a ternary structure bound to PAP and teicoplanin. These structures represent the first examples of sulfotransferase structures containing glycopeptide substrates.
Teg12 Ternary Structure
We obtained a 2.05 Å structure of a Teg12-PAP-teicoplanin aglycone ternary complex by co-crystallization (Figure 2B and 2D). Several regions of the Teg12 sequence not seen in the other Teg12 structures could be modeled into the electron density. Loop region Gly127 to Val137, which is primarily disordered in the other Teg12 structures, is fully ordered in one of the monomers of the ternary complex. This short loop contains the V2 region, one of the three regions that are highly variable within the three TEG sulfotransferase sequences. The V2 loop in Teg12 is extended by four amino acids compared with StaL, and by two amino acids when compared with Teg13 and Teg14. A much larger portion of the GHL helix and loop could be modeled in the ternary structure. The GHL helix extends through Ser15 in one monomer. In the opposite monomer the helix extends to Gln218 and transitions uninterrupted to the GHL loop.
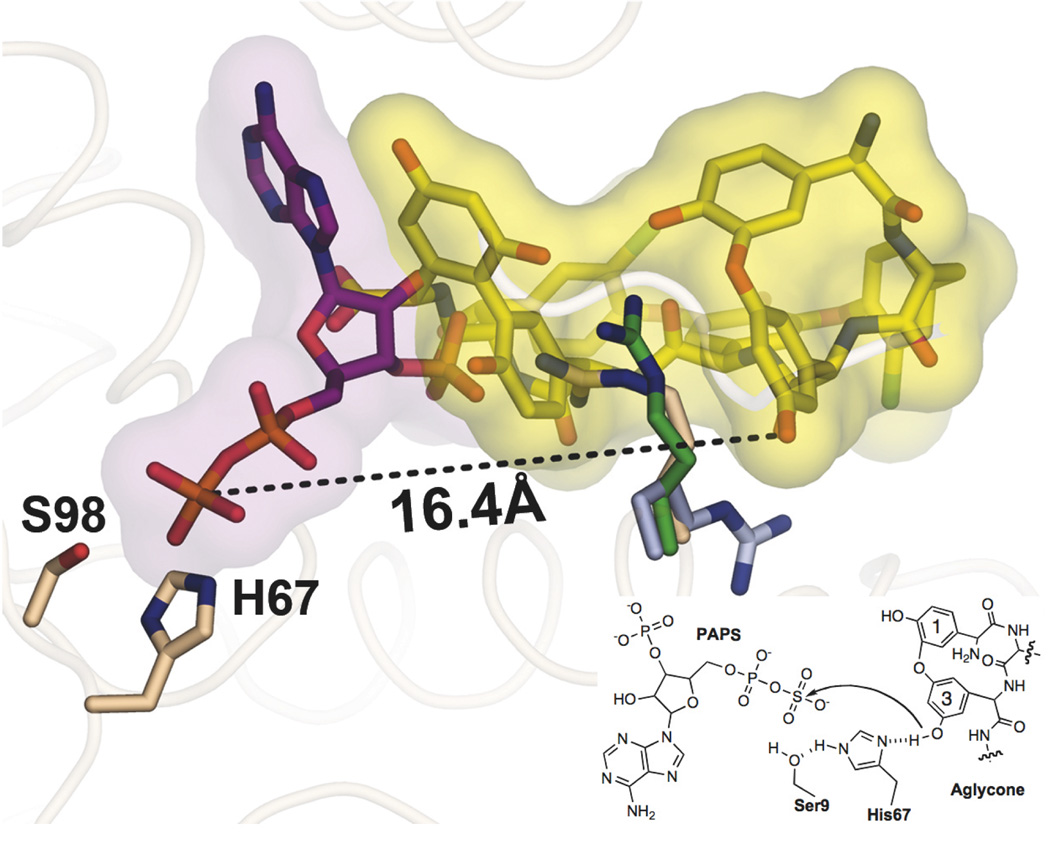
Both monomers of the ternary structure contain a single molecule of PAP bound in the predicted active site location. Three highly conserved structural motifs in sulfotransferases are known to play a role in binding PAPS, all of which are seen in Teg12. Lys12 from the 5’-phosphate binding loop (PSB) (Pro11-Thr16) hydrogen bonds with the 5’-phosphate of PAP, Ser98 from the 3’-phosphate binding loop (PB) (Val89-Ser99) hydrogen bonds with the 3’-phosphate of PAP, and Trp17 parallel stacks with the adenine base. A number of additional residues from the PSB loop region, the PB loop region and the conformationally flexible GHL region are within hydrogen bonding distance of PAP (Figure 4B). Thr16 is only ~2.6 Å from the 5’ phosphate, to which it hydrogen bonds. This residue corresponds to a histidine in StaL, although threonine is the more common side chain at this position in eukaryotic sulfotransferases (13). A general hydrogen bond donor at position 16 is likely to be important for properly coordinating PAPS in the active site. Indeed, an alanine replacement mutant of Thr16 was inactive for the sulfation reaction. A K12A mutant was also inactive, while S98A was partially active (27% compared with wild-type). A W17A mutant did not express as soluble protein, indicating a role not only in binding PAP, but also in protein folding and stability.
PAPS dependent sulfotransferases are believed to use a histidine as a general base to activate the hydroxyl or amine used in the sulfate exchange reaction. Teg12 contains three histidine residues, His67, His226 and His269. Of these, only His67 is located near the co-substrate binding site. The 5’-phosphate of PAP is ~5 Å from the imidazole of His67, and when PAPS is modeled in its place, the sulfate is positioned directly adjacent to His67 (Figure 7). In this model, both Lys12 and Lys65 are close enough to coordinate with the sulfate. In place of the sulfate from PAPS, a water molecule occupies this space in the ternary structure. Ser9, which is within hydrogen bonding distance of the imidazole ring, was proposed to help activate the catalytic histidine in StaL. Attempts to mutate the predicted catalytic histidine in StaL resulted in an insoluble protein and therefore its role in catalysis had not been previously confirmed (13). Alanine exchange mutants were generated for His67, Ser9, and Lys65. H67A and S9A did not express as soluble proteins, while K65A was partially active (65%). Additional glutamate and glutamine mutants of His67 were generated to potentially provide better expression. H67E also expressed as an insoluble protein. However, H67Q, which was soluble, produced a completely inactive sulfotransferase.
Figure 7.
A composite of the active site from the Teg12 binary and ternary structures. PAPS was modeled into the ternary structure in place of PAP. In this aligned view, PAPS and teicoplanin aglycone would not be able to occupy their respective positions simultaneously, as there would be strong clashes between the two substrates. Residue 3 of the teicoplanin aglycone, the site of sulfation, is more than 16 angstroms from the sulfate of PAPS when the ternary and binary structures are overlapped. A substantial rearrangement of teicoplanin aglycone within the active site would have to occur in order for sulfation to proceed by the proposed in-line attack mechanism. The positions of Arg101 in the binary (yellow) and ternary (purple) structures appear to preclude the movement of teicoplanin aglycone towards His67. Arg101’s position in the context of the apo structure creates a more open active site, where the aglycone could pivot towards His67 more easily.
Teicoplanin aglycone bound in the ternary structure
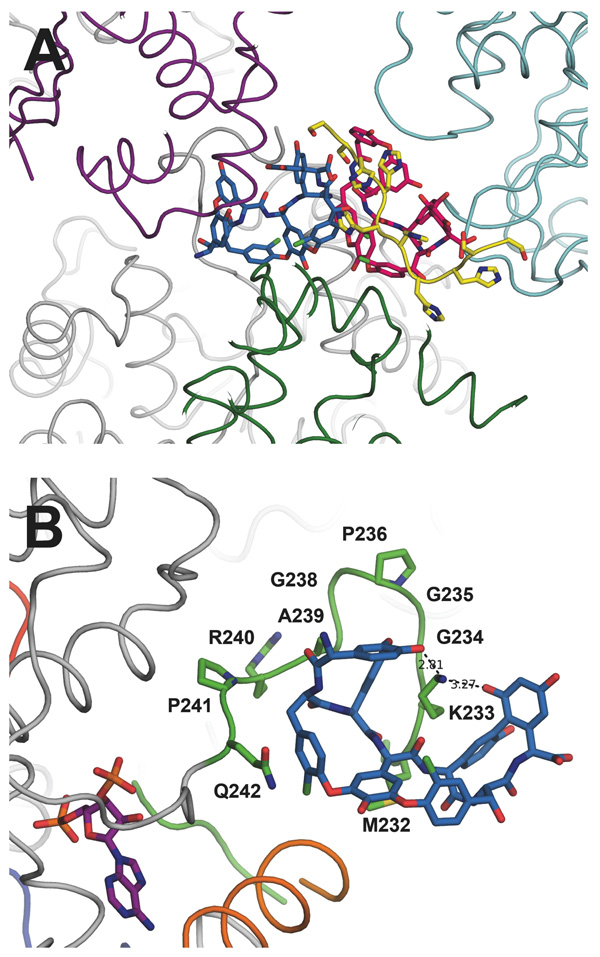
One of the monomers in the ternary structure, in addition to PAP, contains two molecules of teicoplanin aglycone. Both molecules of the aglycone are located outside of the predicted active site pocket, and appear to be organized primarily by crystallographic forces (Figure 5A). One of the molecules of aglycone makes numerous contacts with the GHL loop (Figure 5B). Beginning with the N-terminal section of the GHL loop, residues Gly220 through Ile224 fold back towards the dimer interface. Pro223 stacks with Trp134 and helps to stabilize this region. The subsequent six amino acids (Arg 225-Arg230) are disordered, before density picks up again at Met231. However, these residues occupy a region of space where they could easily make additional contacts with the glycopeptides. Lys233 through Gln242 closely trace the outer surface of the N-terminal three amino acids of the aglycone. These ten amino acids immediately precede the five residues (Phe243-Gly247) that interact with PAP. Lys233 intercalates between the Dpg at position 3 and the Hpg at position 5, and it hydrogen bonds with hydroxyls from the Hpg at position 1 and the Dpg at position 7. The carboxylate from Asp271 of a symmetry mate makes several contacts with backbone nitrogens in the concave cleft of the glycopeptides. The C-terminal end of the teicoplanin aglycone extends out from the surface of the protein and interacts with a second glycopeptide molecule that is bound to the His-tag from an adjacent monomer. Despite the assumption that these aglycone-GHL interactions are mediated primarily by crystal packing, we generated a series of mutants in GHL loop residues that contact the glycopeptides. All mutants in this loop region, with the exception of G235A, had deleterious effects on the rate of substrate conversion, supporting the notion that the V3 loop interacts with the substrate and is involved in recruitment. The P241A mutation abolished Teg12’s activity completely. While the observed glycopeptide-GHL interactions observed in the ternary structure are clearly stabilized by crystal packing interactions, our mutagenesis experiments make it so we cannot rule out the possibility that this represents a snapshot of the substrate recruitment path. A summary of all mutations made in Teg12 and their corresponding activities is given in Table 3.
Figure 5.
(A) Two molecules of teicoplanin aglycone were modeled in the Teg12-ternary complex, colored sky blue and hot pink. Their location within the crystal appears to mediate the packing of several Teg12 monomers, as shown in purple, cyan, dark green, and grey. The two teicoplanins belong to the grey Teg12 in the crystal. Also shown in yellow is the 6X-His portion of the N-terminal derived tag from the pET28a expression vector. The tag-aglycone interaction is proposed to mimic the binding of the aglycone to its cellular target, D-Ala-D-Ala of the bacterial cell wall. (B) A close-up of the GHL loop-aglycone complex from the Teg12-ternary structure. Side chains for all alanine replacement experiments are shown. Lys233 intercalates into the glycopeeptide makes hydrogen bond contacts with residues 1 and 7 of the aglycone. For orientation purposes, the molecule of PAP is shown in purple. Also shown in orange is the GHL helix.
Table 3.
Relative activity of TEG12 mutant sulfotransferases1
| Catalytic or PAP binding |
Activity (%) |
V3 aglycone binding |
Activity (%) |
Non-V3 aglycone binding |
Activity (%) |
|---|---|---|---|---|---|
| S9A | N/A | M232A | 74 | R101A | 29 |
| K12A | 0 | K233A | 79 | S106A | 92 |
| T16A | 0 | G234A | 82 | R107A | 82 |
| W17A | N/A | G235A | 100 | D108A | 82 |
| K65A | 61 | P236A | 73 | Y167A | N/A |
| H67A | N/A | G238A | 56 | K171A | 69 |
| H67E | N/A | A239R | 37 | E206A | 0 |
| H67Q | 0 | R240A | 48 | R207A | 79 |
| R90A | N/A | P241A | 0 | E210A | 97 |
| S98A | 27 | Q242A | 64 | E212A | 26 |
| K213A | 99 | ||||
| R214A | 67 | ||||
| R248A | 196 | ||||
| Q251A | 41 |
N/A indicates a mutation that produced an insoluble protein
Teg12 binary structure and the Teg12 active site
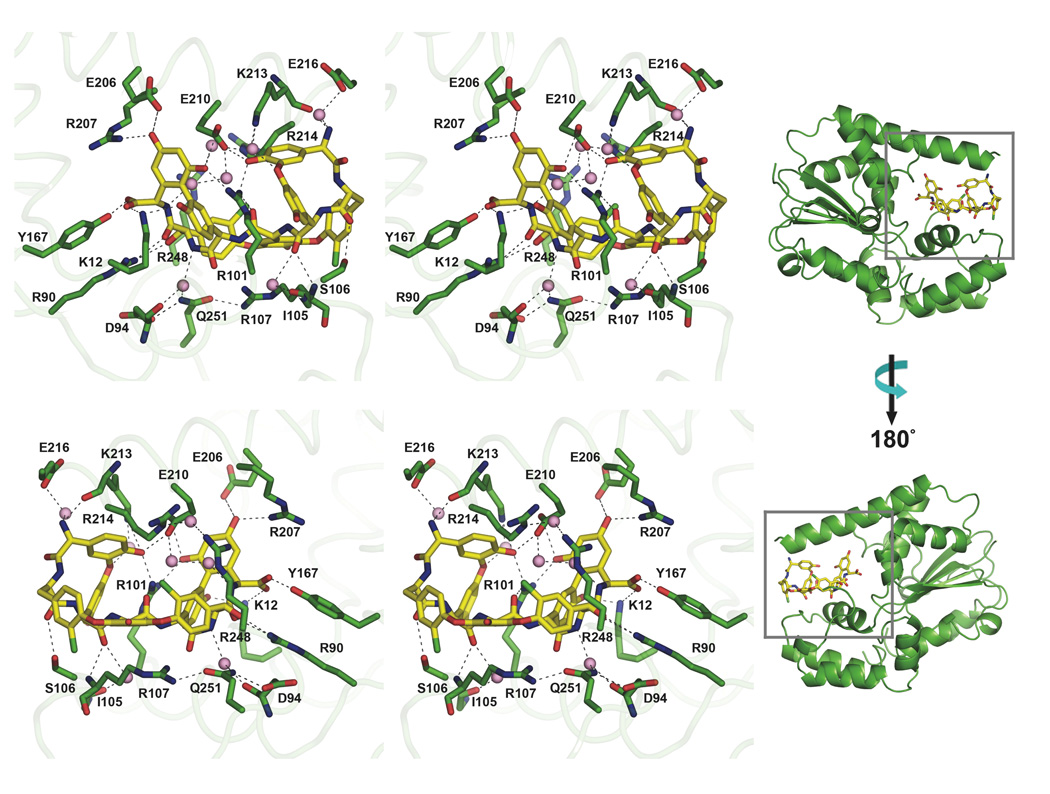
We solved a Teg12-teicoplanin aglycone structure to 2.27 Å resolution (Figure 2C). A single monomer constitutes the asymmetric unit and the dimer is generated by crystallographic symmetry. The active site contains a single molecule of teicoplanin aglycone. Two sets of side chain hydrogen bonds play key roles in creating the glycopeptide binding cavity. The base of the binding pocket is largely defined by the outstretched side chain of Arg107 coordinating with the side chain of Gln251. The back of the cavity is largely dSefined by two outstretched arginine side chains (Arg214 and Arg248) that interact with each other via a mediating water molecule. Numerous hydrogen bonds exist between amino acid side chains from residues found in the GHL helix (Glu206, Arg207, Glu210 and Lys213) and the glycopeptide (Figure 6). Several of the residues that make contacts with PAP also interact with teicoplanin (Lys12, Arg90, Arg101, and Tyr167). Side chains from Tyr167 and Lys12 coordinate the C-terminal carboxylate, while a backbone carbonyl oxygen from Lys213 and water hydrogen bonded to Glu216 coordinate the N-terminal amine.
Figure 6.
Stereo views of the front and back sides of the Teg12-aglycone complex in the active site of the Teg12-binary structure. Protein residues are colored green, and the aglycone is colored yellow. Hydrogen bonds are indicated with dashed lines. Water molecules that make important hydrogen bonds are shown as pink spheres.
The GHL helix and the helix immediately N-terminal (residues Ile193 to Ser203) form a single contiguous helix that runs along the top surface of the aglycone (Figure 2C). In the ternary structure with an empty active site pocket, these two helices adopt a bent conformation that results from unwinding of the long helix at Thr204 (compare orange GHL helix, Figures 2B and 2C). This bent conformation is also observed in StaL. In the bent conformation, the helices are at a nearly perpendicular angle to one another. The long, largely disordered loop region of the GHL that interacts with PAP in the ternary structure is completely displaced by the bound glycopeptide in the binary structure. This disordered loop is now positioned at the back of Teg12, behind the bound glycopeptide. The front surface of the glycopeptide, where the sulfotransferase reaction is predicted to occur on residue 3 (see red arrow in Figure 2C), is largely exposed to solvent. With the exception of an additional small compensatory movement of a short loop (Gln31 through Ile37) into the void left by the straightening of the flexible helix from the GHL, the remainder of this Teg12 binary structure is essentially identical to that seen in the other Teg12 structures.
The C-terminus of the glycopeptide is buried in the PSB loop from the strand-loop-helix motif and the helix from the PB loop containing the strand-turn-helix motif runs along the under side of the glycopeptide. While most residues only contact the outer surface of the glycopeptide, one residue, Arg101, extends into the core of the molecule. The side chain of Arg101 intercalates between the Dpg at position 3 and the Hpg at position 5. The guanidinium from Arg101 hydrogen bonds with the backbone carbonyl from the Hpg at position 4 as well as with hydroxyls from the Dpg at position 3 and the Dpg at position 7. In this respect, Arg101 behaves in much the same way as Lys233 in the ternary structure. We generated a series of alanine replacement mutants for residues contacting the aglycone to determine which are important for substrate conversion. The results are summarized in Table 3. R101A showed a reduction of activity to 27% compared with wild-type. Interestingly, an E212A mutation also showed a reduction of activity to 26%. Glu212 is part of the GHL helix, but does not make contacts with the aglycone in the binary structure. Instead, this residue is found on the upper, solvent exposed face of the helix. Arg248, along with Arg214 forms the back of the active site cavity. An alanine placed at position 248 resulted in a two-fold increase in sulfation. Removing the bulky side chain at this position may impart more flexibility on the active site, allowing the glycopeptide to shift more easily towards His67 (see discussion).
Although the glycopeptide substrate interacts with different sets of GHL residues in the binary and ternary structures, the general glycopeptide binding motif seen in these structures is very similar. In both cases, a long positively charged amino acid side chain (Lys233 in the ternary, Arg101 in the binary) extends into the core of the glycopeptide, where it coordinates with the same set of side chain hydroxyls. All other glycopeptide-protein contacts observed in these structures involve the outer surface of the glycopeptide.
In addition to the glycopeptide bound in the active site cavity, there is a second glycopeptide molecule bound on the surface of the protein (not shown in Figure 2C). The location of the second teicoplanin aglycone in the crystal is likely not biologically relevant. Instead, it appears to mediate crystal contacts between symmetry partners. A stretch of electron density, large enough to contain four residues, is located in the D-ala-D-ala binding cleft of the second aglycone. Although it is presumed that this stretch of residues comes from the N-terminal vector derived His-tag of an adjacent symmetry mate, we were not able to determine its sequence, and hence it was modeled as polyalanine.
Discussion
Teg12 sulfates the hydroxyl on the dihydroxyphenyl glycine at position 3 of the teicoplanin aglycone (Figure 2C, red arrow). In an alignment of the binary and ternary structures, this hydroxyl is 16.4 Å from where the sulfate of PAPS would be and there is significant overlap between the C-terminus of the glycopeptide and PAP (Figure 7). Before the sulfation reaction can proceed, there must be a substantial conformational rearrangement in the active site. In the binary structure, the interaction of the glycopeptide with Arg101 appears to preclude the substrate from shifting to accommodate PAPS or from its shifting into the vicinity of His67. Arg101 makes mutually exclusive contacts with the substrates present in the binary and ternary structures. In the binary structure it is inserted into the peptide core and hydrogen bonds with residues 1 and 7, while in the ternary structure it forms a key salt bridge with the 3-phosphate of PAP. If, when both substrates are present in the active site, Arg101 were to adopt the conformation seen in the apo structure, the glycopeptide would largely be free to move out of the PAPS binding pocket and towards His67. By pivoting towards His67, the glycopeptide would occupy the same general location that substrates bind in eukaryotic sulfotransferases (17), and it would also be positioned so that it could interact with all three variable loops.
The movement the glycopeptide must undergo to reach the PAPS sulfate traces a path similar to that which the GHL helix undergoes when it bends. This conformational rearrangement may serve not only as a mechanism to allow large glycopeptide substrates into the active site, but it could also play a role in positioning a glycopeptide for catalysis once it enters the active site. Alanine exchange mutagenesis of residues found on both the lower glycopeptide associated face (E206A, R207A, R214A) and the upper solvent exposed face of the helix (E212A) led to decreases in substrate conversion. Residues around the entire helix likely make key contacts with the glycopeptide at different points in time as a result of the rotation in the helix as it bends. In fact, all residues that contact the glycopeptide in the binary structure face out from the active site cavity in the ternary structure. One alanine exchange mutant, R248A, resulted in a two-fold increase in activity. R248, along with R214 of the GHL helix, and a mediating water molecule, forms a cage-like enclosure over the top of the glycopeptide (Figure 6). Introduction of an alanine side chain at this position would open up the back side of the active site to the solvent, and could provide substrates in the active site more room for positional rearrangement. The sequence of the GHL helix is conserved throughout the TEG sulfotransferases and a large portion of it is also conserved in StaL. The mechanics of the conformational change from the straight to bent structures may therefore be conserved throughout this family of finishing enzymes.
Sequence and structural similarities between eukaryotic and prokaryotic sulfotransferases indicate that all members have likely arisen from one common ancestor. In eukaryotic structures the loop that would correspond to the V3 loop of the GHL behaves in much the same way as it does in Teg12; it is disordered in apo structures and associated with PAP and the substrate in co-crystal structures (17). The open active site conformation that results from the straightening of the GHL helix was not observed in previous StaL structures. While a GHL-like helix-loop region is present in eukaryotic sulfotransferases, the helix from these structures does not appear to exhibit the same conformational rearrangements from a single straight helix to two helices with a bent conformation during substrate binding as it does in Teg12. At over 1000 daltons, glycopeptides are significantly larger than the substrates used by most eukaryotic sulfotransferases. The increased conformational plasticity that results from the ability of the GHL helix to easily flex may help TEG-like sulfotransferases accommodate larger substrates.
In addition to the conformational flexibility seen in the V3 loop, both the V1 and V2 loops adopt different conformations in this series of structures, suggesting they could easily reorganize to accommodate an incoming glycopeptide substrate. The sequence differences seen between Teg12, 13, and 14 suggest that these three short variable regions likely control the selection and orientation of the glycopeptide substrate bound in the active site. Systematically altering the residues found in the three TEG variable regions may provide a means to generate new glycopeptide finishing enzymes that sulfate a broader collection of glycopeptide congeners than is currently possible with the small number of sulfotransferases that have been identified naturally. The sulfated teicoplainin aglycone derivatives produced by the native TEG sulfotransferase retain potent in vitro antibacterial activity (5).
The cloning and characterization of biosynthetic gene clusters derived from uncultured bacteria provides a means to access both novel small molecules and new biosynthetic enzymes. Teg12 is one of the first enzymes discovered using culture independent methodologies to be characterized structurally, and the Teg12-teicoplanin aglycone co-crystal structures are the first examples of a substrate complexed with a member of this family of glycopeptide finishing enzymes. This series of Teg12 structures provides key insights into how sulfotransferases might be engineered to generate additional anionic glycopeptides that could be evaluated against clinically relevant drug resistant bacteria.
Table 1.
Diffraction statistics for Teg12 structures
| Teg12 apo | Teg12 ternary | Teg12 binary | |
|---|---|---|---|
| Data collection | |||
| Space group | C2221 | P212121 | I212121 |
| Cell dimensions | |||
| a, b, c (Å) | 79.39, 126.09, 145.13 | 77.12, 78.56, 100.54 | 66.12, 80.15, 132.94 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| No. reflections (observed/unique) |
55,339/15,372 | 177,647/38,619 | 66,588/16,647 |
| Resolution (Å) | 50.00 − 2.93 (3.03 − 2.93) |
50.00 − 2.05 (2.12 − 2.05) |
50.00 − 2.28 (2.36 − 2.28) |
| Rmerge/Rsym | 0.063 (0.396) | 0.084 (0.464) | 0.079 (0.542) |
| I/σI | 19.10 (2.04) | 16.43 (2.62) | 15.50 (2.17) |
| Completeness (%) | 95.5 (98.0) | 97.7 (96.6) | 99.9 (100.0) |
| Redundancy | 3.6 (3.6) | 4.6 (4.6) | 4.0 (4.1) |
| Refinement | |||
| Resolution (Å) | 72.57 − 2.91 | 32.95 − 2.04 | 34.32 − 2.27 |
| No. reflections | 14,562 | 36,951 | 15,761 |
| Rwork / Rfree | 21.96 / 27.19 | 17.12 / 22.47 | 17.30 / 22.61 |
| No. atoms | 3,628 | 4,820 | 2,222 |
| Protein atoms | 3,550 | 4,247 | 1,945 |
| Ligand/ion | 44 | 233 | 170 |
| Waters | 34 | 340 | 95 |
| B factors | 36.2 | 35.7 | 43.0 |
| Protein | 35.9 | 35.6 | 40.3 |
| Ligand/ion | 61.7 | 30.1 | 69.5 |
| Water | 39.1 | 40.5 | 47.4 |
| Solvent Content | 52.4 | 40.9 | 46.5 |
| Rms bond (Å) | 0.014 | 0.007 | 0.006 |
| Rms angle (°) | 1.495 | 1.183 | 1.038 |
| Rotamer Outlier (%) | 3.6 | 0.7 | 1.0 |
| Ramachandran (favored/outlier) (%) |
94.6 / 0.6 | 98.7 / 0.0 | 100.0 / 0.0 |
| PDB code |
Acknowledgment
We thank Wuixan Shi of beamline X29 at the NSLS, Kanagalaghatta Rajashankar of NE-CAT 24-ID beamline at the APS, and Deena Oren of the Rockefeller University Structural Biology Resource Center. We also thank Nigel Moriarty for his assistance with Phenix.elbow and generation of a teicoplanin restraints file for use in refinement. The teicoplanin aglycone was generously provided by Supelco.
This work was supported by NIH GM077516, the Howard Hughes Medical Institute, the Beckman Foundation and the Searle Foundation.
Abbreviations
- TEG
teicoplanin-like eDNA derived gene cluster
- PAP
3’ – phosphoadenosine 5’ – phosphate
- PAPS
3’ – phosphoadenosine 5’ – phosphosulfate
- Hpg
hydroxyphenylglycine
- Bht
betahydroxytyrosine
- Dpg
diydroxyphenylglycine
- HEPES
2-[4-(2-hydroxyethyl)piperazin-1-1yl]ethanesulfonic acid
- CHES
2-(Cycolhexylamino)ethanesulfonic acid
- DTT
dithiothreitol
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
REFERENCES
- 1.Linden PK. Vancomycin resistance: are there better glycopeptides coming? Expert Rev. Anti-Infect. Ther. 2008;6:917–928. doi: 10.1586/14787210.6.6.917. [DOI] [PubMed] [Google Scholar]
- 2.Uttley AH, Collins CH, Naidoo J, George RC. Vancomycin-resistant enterococci. Lancet. 1988;1:57–58. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 3.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 4.Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu. Rev. Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 5.Banik J, Brady SF. Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc. Nat. Acad. Sci. U.S.A. 2008;105:17273–17277. doi: 10.1073/pnas.0807564105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kahne D, Leimkuhler C, Lu W, Walsh C. Glycopeptide and lipoglycopeptide antibiotics. Chem. Rev. (Washington, DC, U. S.) 2005;105:425–448. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]
- 7.Hubbard BK, Walsh CT. Vancomycin assembly: nature's way. Angew. Chem. Int. Ed. 2003;42:730–765. doi: 10.1002/anie.200390202. [DOI] [PubMed] [Google Scholar]
- 8.Nicolaou KC, Boddy CNC, Brase S, Winssinger N. Chemistry, biology, and medicine of the glycopeptide antibiotics. Angew. Chem. Int. Ed. 1999;38:2096–2152. doi: 10.1002/(sici)1521-3773(19990802)38:15<2096::aid-anie2096>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 9.Boeck LD, Mertz FP. A47934, a novel glycopeptide-aglycone antibiotic produced by a strain of Streptomyces toyocaensis taxonomy and fermentation studies. Jpn. J. Antibiot. 1986;39:1533–1540. doi: 10.7164/antibiotics.39.1533. [DOI] [PubMed] [Google Scholar]
- 10.Higgins DL, Chang R, Debabov DV, Leung J, Wu T, Krause KM, Sandvik E, Hubbard JM, Kaniga K, Schmidt DE, Jr, Gao Q, Cass RT, Karr DE, Benton BM, Humphrey PP. Telavancin, a multifunctional lipoglycopeptide, disrupts both cell wall synthesis and cell membrane integrity in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2005;49:1127–1134. doi: 10.1128/AAC.49.3.1127-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stryjewski ME, O'Riordan WD, Lau WK, Pien FD, Dunbar LM, Vallee M, Fowler VG, Jr, Chu VH, Spencer E, Barriere SL, Kitt MM, Cabell CH, Corey GR. Telavancin versus standard therapy for treatment of complicated skin and soft-tissue infections due to gram-positive bacteria. Clin. Infect. Dis. 2005;40:1601–1607. doi: 10.1086/429914. [DOI] [PubMed] [Google Scholar]
- 12.Leadbetter MR, Adams SM, Bazzini B, Fatheree PR, Karr DE, Krause KM, Lam BM, Linsell MS, Nodwell MB, Pace JL, Quast K, Shaw JP, Soriano E, Trapp SG, Villena JD, Wu TX, Christensen BG, Judice JK. Hydrophobic vancomycin derivatives with improved ADME properties: discovery of telavancin (TD-6424) Jpn. J. Antibiot. 2004;57:326–336. doi: 10.7164/antibiotics.57.326. [DOI] [PubMed] [Google Scholar]
- 13.Shi R, Lamb SS, Bhat S, Sulea T, Wright GD, Matte A, Cygler M. Crystal structure of StaL, a glycopeptide antibiotic sulfotransferase from Streptomyces toyocaensis. J. Biol. Chem. 2007;282:13073–13086. doi: 10.1074/jbc.M611912200. [DOI] [PubMed] [Google Scholar]
- 14.Lamb SS, Patel T, Koteva KP, Wright GD. Biosynthesis of sulfated glycopeptide antibiotics by using the sulfotransferase StaL. Chem. Biol. 2006;13:171–181. doi: 10.1016/j.chembiol.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Dombrovski L, Dong A, Bochkarev A, Plotnikov AN. Crystal structures of human sulfotransferases SULT1B1 and SULT1C1 complexed with the cofactor product adenosine-3'-5'-diphosphate (PAP) Proteins. 2006;64:1091–1094. doi: 10.1002/prot.21048. [DOI] [PubMed] [Google Scholar]
- 16.Lin ES, Yang YS. Nucleotide binding and sulfation catalyzed by phenol sulfotransferase. Biochem. Biophys. Res. Commun. 2000;271:818–822. doi: 10.1006/bbrc.2000.2722. [DOI] [PubMed] [Google Scholar]
- 17.Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, Loppnau P, Martin F, Thornton J, Edwards AM, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith CH. Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol. 2007;5:e97. doi: 10.1371/journal.pbio.0050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276A:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 21.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 25.Schafer M, Schneider TR, Sheldrick GM. Crystal structure of vancomycin. Structure. 1996;4:1509–1515. doi: 10.1016/s0969-2126(96)00156-6. [DOI] [PubMed] [Google Scholar]
- 26.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 27.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarkar G, Sommer SS. The "megaprimer" method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]