Abstract
The incorporation of affinity baits into N-isopropylacrylamide-hydrogel-based nanoparticles offers a novel technology that addresses the major analytical challenges of disease biomarker discovery. In solution in complex biologic fluids (e.g. blood or urine), core-shell bait-containing nanoparticles can perform three functions in one step: (a) sieve molecules according to size, (b) sequestrate and concentrate target analytes, and (c) protect analytes from degradation.
Introduction
There is an urgent need to discover novel biomarkers that provide sensitive and specific disease detection,1–3 since it is widely believed that early detection of disease prior to symptoms will lead to a dramatic improvement in treatment outcome.4 Biomarkers are nucleic acids, proteins, protein fragments or metabolites5,6 indicative of a specific biological state, that are associated with the risk of contraction or presence of disease.7 A serum or plasma sample is thought to contain low-abundance circulating proteins and peptides which can provide a rich source of information regarding the state of the organism as a whole.8 Despite the promise of serum proteomics, there are three fundamental and serious physiologic barriers thwarting biomarker discovery and translation to clinical benefit: 1. Important diagnostic biomarkers may exist in extremely low abundance (concentration) in blood. Early-stage diseased tissue, such as pre-metastatic cancer lesions, may constitute less than a few cubic millimeters. Biomarkers shed into the circulation from such a small tissue volume will become highly diluted in the entire blood volume. Relevant analytes may exist below the detection limits of mass spectrometry and conventional immunoassays. 2. Resident proteins such as albumin and immunoglobulins, accounting for 90% of circulating plasma proteins, confound and mask the isolation of rare biomarkers.9 The vast majority of low abundance biomarkers are non-covalently and endogenously associated with the resident proteins such as albumin and immunoglobulins, which exist in a billionfold excess compared to the biomarker.8 3. Low-abundance biomarkers can be rapidly degraded by endogenous and exogenous proteinases or clotting cascade enzymes immediately after the blood sample is drawn from the patient. Indeed, degradation of candidate biomarkers occurs also during transportation and storage of blood, leading to serious false-positive and false-negative results.10 Over the past five years a variety of new technologies have been applied to address these three fundamental challenges of biomarker discovery.11–14 The technologies focus on improving the sensitivity of detection or the pre-analytical separation and concentration of biomarkers. Two classes of technology that have had significant impact are (a) nanotechnology-based nanosensors, and (b) protein biomarker discovery and sequencing using mass spectrometry (MS).
Nanosensors with clinical applications
Significant advances have been made in building sensitive and reliable nanosensors for diagnostic testing. One widely explored architecture for nanosensors is nanowires, whose working principle relies on the fact that the conductance of a nanosized channel changes upon binding small numbers of biomolecules to probe receptors immobilized on the channel surface.15 Extremely high sensitivity has been reached, e.g. prostate specific antigen (PSA) was detected at concentrations of 1 fg/mL in model solutions. 16 Another example category of nanosensors employs cantilevers coated with receptors specific for biomarkers of interest. When the biomarker binds to the receptor, this causes an increase in mass that is detected by cantilever deflection (static mode) or change in cantilever resonance frequency (dynamic mode).17 Technologies such as beam deflection distance and piezo-resistive read-out, commonly used in atomic force microscopy, are being used to detect the cantilever bending. PSA concentrations of 0.2 ng/mL were detected in a BSA-containing solution with cantilever-based nanosensors. 18 High levels of sensitivity have been achieved with the bio-barcode assay.19 In this muliplex assay, magnetic microparticles and gold nanoparticles are combined in a sandwich immunoassay incorporating a unique DNA sequence specific for the intended analyte. PCR amplification and detection of the DNA bar code via hybridization to a chip allows the identification of the analyte of interest. The detection limit for PSA using this technology was reported to be 1 fg/mL.20 Ultra-sensitive immunosensors for protein biomarkers have been proposed based on surface plasmon resonance (SPR).21 SPR can detect molecules bound to a surface by subtle changes in the SPR angle. Probes are immobilized onto the surface and the analyte-containing solution s passed through the surface. Binding between analyte and probes causes changes in the SPR angle. Amplification methods employing gold nanoparticles generated a linear response to PSA concentration down to 300 fM.22 Despite the high sensitivity of nanosensors for measuring analytes, the critical question remains: What analyte do you measure with the nanosensor? In order to realize the promise of biomarkers for clinical benefit, new analytes must be discovered that provide sensitive and specific association with disease. Thus the discovery of new specific disease-related-biomarkers still remains a critical issue that can not be solved by improving the sensitivity of immunoassay technology alone.23
MS-based biomarker discovery
To address this need for new analytes, mass spectrometry has become the preferred technique for the discovery of candidate biomarkers in biological fluids.24,25 Advances in MS techniques are offering the opportunity to use very complex biological samples to perform de novo analysis of proteins, protein modifications and metabolites present in cells or tissues.26–30 Liquid chromatography coupled with tandem mass spectrometry (LC MS/MS) has reached a sensitivity in the order of attomolar concentration of analytes.31 Nevertheless, when complex solutions are studied, diminished sensitivity of MS/MS can be attributed to its limited dynamic range (3–4 orders of magnitude) compared to the extraordinarily wide concentration range of blood proteins (ten orders of magnitude).1 Moreover, MS analysis accepts only a small input volume, thereby limiting the number of molecules that can be sequenced and limiting the resulting sensitivity. In fact, most low-abundance blood analytes routinely measured using standard clinical laboratory immunoassay machines cannot be detected in the serum or plasma proteome at the current sensitivity of MS. Increasing the dynamic range and the effective sensitivity of current MS technology represents the key technological advance that could lead to discovery of biomarkers previously unknown because of their low abundance in blood and body fluids.
Hydrogel nanoparticles bridge the gap between detection and sensitivity limits
Recently, a new application of nanotechnology has been proposed to bridge the gap between MS-based biomarker discovery and nanosensor-based diagnostic tools. A variety of types of nanoparticles have been applied to capture and separate proteins from complex mixtures prior to MS.32–36 These include solid silica particles and open-structure hydrogel particles. The most promising group of nanoparticles used to date appears to be hydrogel-based. Hydrogel nanoparticle affinity bait sequestration is a successful preprocessing step that can concentrate, purify and protect biomarkers from degradation, capturing them from large volumes of biological fluids and concentrating them in small volumes for analysis either with mass spectrometry or immunoassay systems.35,36 Hydrogels are a class of so-called “smart” materials37 that can perform desired mechanical actions in response to target stimuli, such as changes in temperature, pH, or biological stimuli. Of particular interest, the Ulijn group obtained enzyme-responsive hydrogels by incorporating enzyme-cleavable peptide linkers. These peptides carried a cleavable site and charged arginine residues that caused electrostatic repulsion of polymer chains. In the presence of the specific enzyme, these particles shrank due to the peptide cleavage and consequent loss of the charged arginines.38 Deswelling of particles due to the presence of the enzyme corresponded to a decrease in molecular accessibility of about 30 kDa. Hydrogel materials therefore have the potential to be smart drug carriers able to release their cargo at the desired location following a specific external stimulus.39,40 Hydrogel nanoparticles, integrated with matrices such as silica and ormosil, have been proposed for the intracellular monitoring of small analytes (e.g. H+, Ca2+, Mg2+). These specialized diagnostic microprobes have been named “probes encapsulated by biologically localized embedding” (PEBBLEs).41 The microsensors are fabricated as nanosized particles (20–100 nm) that contain an analyte-specific dye and a reference dye. When the analyte interacts with the specific dye, it elicits a change in the fluorescence that can be compared to the reference dye fluorescence. The inert matrix of the particles protects both the cell and the sensor from mutual chemical perturbation.42
Affinity bait-containing hydrogel particles for solution biomarker harvesting and albumin exclusion
Recently, specialized affinity bait-containing hydrogel particles have shown great promise for biomarker harvesting. The particles simultaneously conduct molecular sieve chromatography and affinity chromatography, in one step, in solution.35 These “smart” nano-particles conduct enrichment and encapsulation of selected classes of proteins and peptides from complex mixtures of biomolecules such as native human serum and urine, purify them from endogenous high abundance proteins such as albumin, and protect them from degradation during subsequent sample handling.35,36 This new class of hydrogel particles has a molecular sieving shell surrounding a specific bait core (Fig. 1). Core-shell hydrogel particles are composed of aN-isopropylacrylamide (NIPAm) shell, that can be modified to alter permeability or porosity, surrounding a NIPAm-bait core which performs protein affinity binding in solutions. 35 The porous structure of the shell excludes molecules above a sharp cut-off size (22–27 kDa, which is the molecular weight of immunoglobulin light chains) from entering the interior of the particle.35 The degree of porosity can be tuned by changing the percentage of cross-linker N,N0-methylenebisacrylamide (BIS) with respect to the monomer (NIPAm). Given their physicochemical properties, hydrogel particles can absorb large amounts of water (over 90% of the particle volume), allowing polypeptides and other small molecules to penetrate the polymer matrix,36,43 thus permitting rapid concentration of rare protein biomarkers. Particles incubated with serum have been shown to trap the target analytes and separate them from carrier albumin.35 The particles can be isolated by centrifugation, and candidate biomarkers can be released from particles by means of electroelution or elution buffers.36 Elution methods are available that are compatible with the current techniques for protein quantification, such as mass spectrometry, 44 gel electrophoresis,45 immunoblotting, 46 ELISA,47 and other immunoassays.48 The ratio of elution buffer volume to the original starting solution establishes the concentration amplification factor. This concentration step is a fundamental point for biomarker measurement and discovery because it provides a means to effectively raise the concentration of rare biomarkers that become the input for a clinical measurement system such as an immunoassay platform36 or mass spectrometry. 35 Protein uptake by the particles is are rapid, with more than 90% of the target protein in solution captured by the particles after 1 minute incubation.35 The extraction of proteins from solution is quantitative and no protein can be detected in the supernatant after 30 minutes’ incubation in the studies performed to date;35,36 thus the efficiency of capture approaches 100%. When applied to human serum and urine samples, the harvesting particles showed low nonspecific binding and exclusion of albumin and other high molecular weight serumderived proteins.35
Fig. 1.
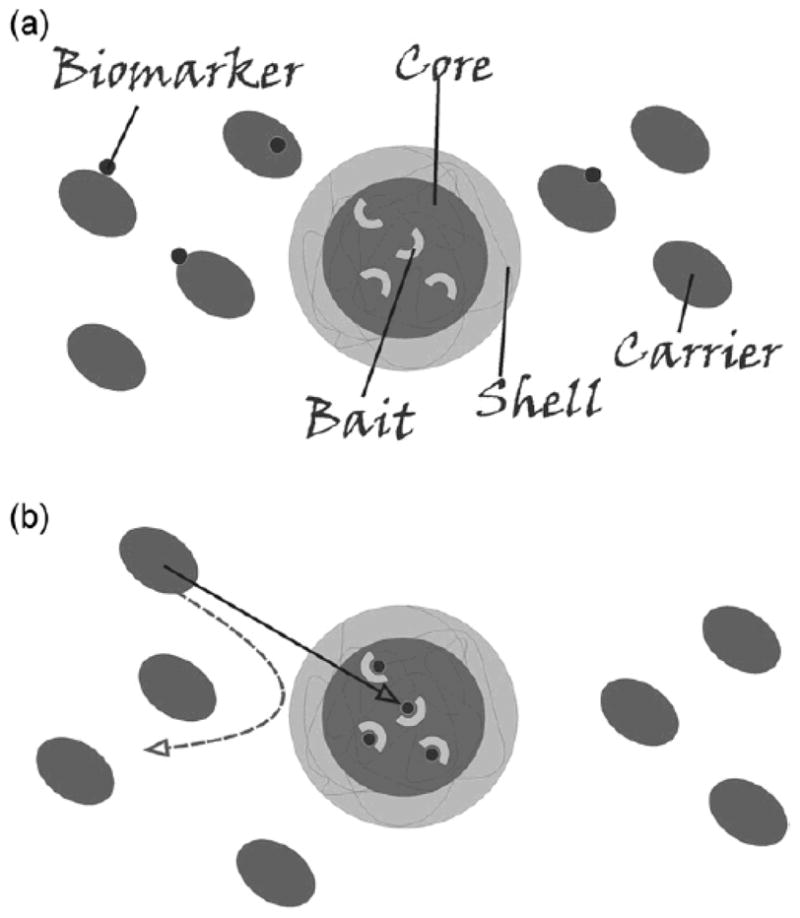
Schematic representation of particle structure and function. (a) Particles are constructed with a bait-containing core, surrounded by a sieving shell. (b) When introduced into a complex solution, such as serum, core-shell particles perform affinity capture of low-molecular-weight proteins from the carrier protein albumin and function as a molecular weight sieve with total exclusion of high molecular weight proteins.
Protection of labile biomarkers from proteolytic degradation
Labile biomarkers, encapsulated into the particles, are also protected against proteolytic degradation, which is an additional serious impediment to biomarker identification. Such stabilization occurs even when the protease is small enough to penetrate the inner space of the particles.35
Bait chemistries are designed to selectively bind diverse classes of biomarkers
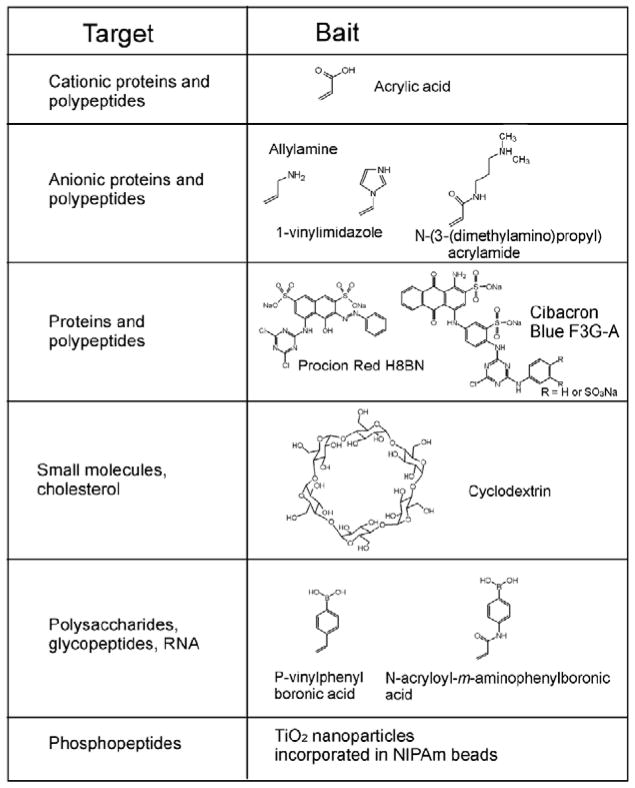
The incorporation of the bait drives the uptake ofmolecules in solution and assures that captured molecules are preserved from degradation. The bait molecule can be incorporated in the hydrogel particle by copolymerization or covalent binding to functionalities present in the particle. A number of bait chemistries have been explored to selectively bind and concentrate different classes of biomarkers such as (a) small proteins and peptides, (b) metabolites, (c) post-translationally modified peptides (e.g. glycosylated and phosphorylated), (d) nucleic acids, and (e) lipids and fatty acids. Bait chemistries comprise charge-based bait (acrylic acid, allylamine co-monomer), triazine-loaded dye (cibacron blue), β-cyclodextrin and boronic acid (Fig 2). At physiological pH, acrylic acid (pK ¼ 3.5) and allylamine (pK ¼ 9.6949) have affinity for cationic and anionic polypeptides and proteins, respectively. An example bait chemistry is the family of triazine-derived textile dyes (e.g., Cibacron blue F3G-A, Procion red H8BN)50 that have been used in affinity chromatography due to their highly specific molecular recognition and low cost.51 The mechanism by which proteins bind to Cibacron blue dye is still unclear, but hypotheses include (a) interaction between the dinucleotide fold structural domain of proteins and the dye, which can assume a conformation that mimics the orientation and anionic groups characteristic of NAD,52 (b) proteins possessing a cluster of apolar residues interact with the aromatic rings of the dye molecule, or with positively charged groups which bind the sulfonate residues, 53 or (c) complex interactions, probably ionic and hydrophobic.54 Dye loaded hydrogel particles have been successfully employed to uptake small proteins (IL-18) and hormones (hGH) from urine and to greatly increase the sensitivity of current diagnostic techniques. 36 Nanoparticles have been designed that contain cyclodextrins. Cyclodextrins are cyclic glucose oligosaccharides that are typically cone-shaped with lipophilic inner cavities and hydrophilic outer surfaces. Because of their structure, cyclodextrins are soluble in aqueous solutions and interact with hydrophobic molecules to form noncovalent complexes via hydrogen bonds, Van der Waals interaction, and electrostatic interactions; 55 these properties make them ideal candidates as drug delivery vectors.56 Cyclodextrins have been shown to bind small hydrophobic molecules, such as cholesterol,57 steroids,58 and DOPA.59 Finally, the capability of boronic acid groups to form complexes with thiol groups has been exploited as a bait strategy for hydrogel nanoparticles. Affinity chromatography based on the -boronate ion has been successfully applied for the selective isolation of nucleotides, RNA, glycated proteins and glycoenzymes.60–65 When an analyte of interest is sought, as reported in Fredolini et al.,36 the first step is choosing a bait that has the highest affinity for the analyte. The second step is optimization of parameters that affect the uptake process: (a) the surface area of the particle, which depends on the dimension of the particle that can be controlled during the synthesis by tuning the concentration of total monomer and initiator; (b) the porosity of the particle, which depends on the percentage of the crosslinking agent with respect to the total monomer during particle synthesis; (c) the characteristics of the solution to be incubated with the particles (pH, ionic force, etc.). Bait strategies include moieties that target classes of analytes. The choice to target a class of analytes instead of a specific molecule has been motivated by the necessity to be as general as possible for biomarker discovery,35 since we do not know ahead of time what we are looking for. Moreover, even when the interest was focused on a specific analyte, class-targeting particles proved to have binding capacity large enough to avoid masking and interference from competing proteins.36 Nevertheless, biologic ligands, receptors, and antibodies are logical as candidate bait molecules for specific ligands. Acrolein-containing particles can be created that carry a free aldehyde group for the covalent immobilization of antibodies and peptides.66
Fig. 2.
Summary of bait chemistries. Chemical formulas of the baits are shown in the right-hand column, and the class of target molecules is shown in the left-hand column. Baits include comonomers (e.g. acrylic acid) or chemical moieties that are covalently bound inside the particle in a second reaction after polymerization (e.g. Cibacron Blue F3G-A).
Properties of hydrogel core-shell bait nanoparticles
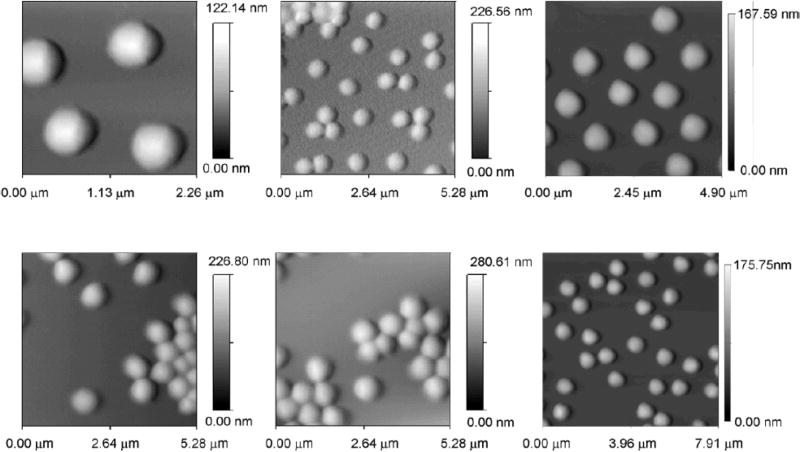
NIPAm-AAc particles have been produced with extremely high yield and reproducibility between and within batches.35,36 The particles have very good colloidal stability at room temperature during the time required for capture, storage and elution of protein analytes (at least 48 hours), and no precipitation was observed, as depicted visually in the report by Fredolini et al.36 The colloidal stability may be very important for rapid uptake. The core-shell nanoparticles have a size of approximately one micron and have been successfully studied and characterized by traditional flow cytometry.35 The particle size was very homogeneous, as shown in Fig. 3.
Fig. 3.
Atomic force microscopy images of NIPAm-AAc particles. Particles suspended in MilliQ water (pH 5.5, 1 μg/mL) were deposited on freshly cleaved mica under a humid atmosphere at room temperature for 15 minutes and dried under nitrogen. The particles exhibit a uniform size distribution (diameter 800 nm). The scale bar for particle height shows a maximum height varying from 100 to 280 nm. The AFM picture was acquired under dry conditions, therefore the particles are distorted (flattened) from their spherical shape due to drying on the mica surface.
Applications of hydrogel nanoparticle harvesting and sieving functions
The envisioned used of this technology is a population of harvesting particles that contain multiple subpopulations specific for individual classes of low molecular weight molecules present in whole blood and other body fluids, such as urine, cerebrospinal fluid, sweat, saliva, nipple aspirates, and amniotic fluid. The particles would simply be stored within the collection tube containing the blood or body fluid. Following introduction of the blood or body fluid, the respective particle populations will remove all of their target molecules, in one step, in solution, from the entire volume of the sample and concentrate, as well as protect the sequestered analyte from degradation. The particle populations carrying different baits can be functionalized with fluorescent dyes and separated by flow cytometry for respective analysis of their harvested biomarker class. The disulfide containing cross-linking agent N,N0-cystaminebisacrylamide (CBAm) provides a means for introducing thiol groups into NIPAm-based hydrogels.41 CBAm can be incorporated as a cross-linking monomer during formation of the particle. Incubating the disulfide-cross-linked hydrogel with aqueous buffer containing mild reagents such as dithiothreitol or tris-2-carboxyethylposphine liberates the sulfurs in the form of thiol groups. These thiol groups can then be used for the chemo-selective covalent attachment of functionalized fluorescent labels such as Alexa Fluor 488 C5 maleimide (Invitrogen). By varying the amount of CBAm monomer incorporated in the hydrogel, it is possible to control the dyeloading capacity of the particle, and thus the degree of labeling.
Conclusion
Future work will be focused on optimizing the separation and washing methods, which are currently based on centrifugation aided by magnetic separation. In this manuscript all applications of the particles are ex vivo, but possible applications in vivo can be envisioned for the future. NIPAm-coated particles have shown low cytotoxicity in previous studies,67 and were not significantly internalized by macrophages, likely due to steric repulsion. Importantly, the repulsion of NIPAm-coated particles did not decrease at 37 °C, above the phase transition temperature at which the polymer chains are expected to shrink and the particle reduces in size.67 Nevertheless, if injection into patients is to be considered in the future, we can also contemplate using FDA-approved polymers such as polyethylene oxide, polylactic acid and polyglycolic acid. Finally, we can envision the extension of the biomarker-harvesting particles to other collection receptacles, such as an adhesive patch for the sampling of skin transudate, exudate, and sweat. The patch configuration would take advantage of the concentration and preservation functions of the particles.
Acknowledgments
The authors appreciate the generous support of Dr. Vikas Chandhoke and the College of Life Sciences at George Mason University. The authors acknowledge financial support from the Italian Istituto Superiore di Sanita in the framework of the Italy/USA cooperation agreement between the U.S. Department of Health and Human Services, George Mason University, and the Italian Ministry of Public Health. This work was partially supported by the U.S. Department of Energy grant number DE-FC52- 04NA25455.
References
- 1.Anderson NL, Anderson NG. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Liotta LA, Ferrari M, Petricoin E. Nature. 2003;425:905. doi: 10.1038/425905a. [DOI] [PubMed] [Google Scholar]
- 3.Hartwell L, Mankoff D, Paulovich A, Ramsey S, Swisher E. Nat Biotechnol. 2006;24:905–908. doi: 10.1038/nbt0806-905. [DOI] [PubMed] [Google Scholar]
- 4.Goldhirsch A, Colleoni M, Domenighetti G, Gelber RD. Ann Oncol. 2003;14:1212–1214. doi: 10.1093/annonc/mdg327. [DOI] [PubMed] [Google Scholar]
- 5.Liotta LA, Petricoin EF. The Journal of Clinical Investigation. 2006;116:26–30. doi: 10.1172/JCI27467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Molecular & Cellular Proteomics. 2003;2:1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Villanueva J, Shaffer DR, Philip J, Chaparro CA, Erdjument-Bromage H, Olshen AB, Fleisher M, Lilja H, Brogi E, Boyd J, Sanchez-Carbayo M, Holland EC, Cordon-Cardo C, Scher HI, Tempst P. J Clin Invest. 2006;116:271–284. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou M, Lucas DA, Chan KC, Issaq HJ, Petricoin EF, 3rd, Liotta LA, Veenstra TD, Conrads TP. Electrophoresis. 2004;25:1289–1298. doi: 10.1002/elps.200405866. [DOI] [PubMed] [Google Scholar]
- 9.Lopez MF, Mikulskis A, Kuzdzal S, Bennett DA, Kelly J, Golenko E, DiCesare J, Denoyer E, Patton WF, Ediger R, Sapp L, Ziegert T, Lynch C, Kramer S, Whiteley GR, Wall MR, Mannion DP, Della Cioppa G, Rakitan JS, Wolfe GM. Clin Chem. 2005;51:1946–1954. doi: 10.1373/clinchem.2005.053090. [DOI] [PubMed] [Google Scholar]
- 10.Ayache S, Panelli M, Marincola FM, Stroncek DF. Am J Clin Pathol. 2006;126:174–184. doi: 10.1309/3WM7-XJ7R-D8BC-LNKX. [DOI] [PubMed] [Google Scholar]
- 11.Viswanathan S, Rani C, Vijan Anand A, Ho JA. Biosens Bioelectron. 2008 doi: 10.1016/j.bios.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Wu Y, Zhuang Y, Liu S, He L. Anal Chim Acta. 2008;630:186–193. doi: 10.1016/j.aca.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Matt P, Fu Z, Fu Q, Van Eyk JE. Physiol Genomics. 2008;33:12–17. doi: 10.1152/physiolgenomics.00282.2007. [DOI] [PubMed] [Google Scholar]
- 14.Whiteaker JR, Zhang H, Eng JK, Fang R, Piening BD, Feng LC, Lorentzen TD, Schoenherr RM, Keane JF, Holzman T, Fitzgibbon M, Lin C, Zhang H, Cooke K, Liu T, Camp DGI, Anderson L, Watts J, Smith RD, McIntosh MW, Paulovich AG. J Proteome Res. 2007;6:828–836. doi: 10.1021/pr0604920. [DOI] [PubMed] [Google Scholar]
- 15.Patolsky F, Zheng G, Lieber CM. Nat Protoc. 2006;1:1711–1724. doi: 10.1038/nprot.2006.227. [DOI] [PubMed] [Google Scholar]
- 16.Kim A, Ah CS, Yu HY, Yang J-H, Baek I-B, Ahn C-G, Park CW, Jun MS, Lee S. Appl Phys Lett. 2007;91:103901–103903. [Google Scholar]
- 17.Nordstroem M, Keller S, Lillemose M, Johansson A, Dohn SR, Haefliger D, Blagoi G, Havsteen-Jakobsen M, Boisen A. Sensors. 2008;8:1595–1612. doi: 10.3390/s8031595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu G, Datar RH, Hansen KM, Thundat T, Cote RJ, Majumdar A. Nat Biotech. 2001;19:856–860. doi: 10.1038/nbt0901-856. [DOI] [PubMed] [Google Scholar]
- 19.Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 20.Muller UR. Mol Biosyst. 2006;2:470–476. doi: 10.1039/b608442g. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y-Y, Hsu H-Y, Huang C-JC. Biosens Bioelectron. 2007;22:980–985. doi: 10.1016/j.bios.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 22.Choi J-W, Kang D-Y, Jang Y-H, Kim H-H, Min J, Oh B-K. Colloids Surf A: Physicochem Eng Asp. 2008;313–314:655–659. [Google Scholar]
- 23.Ahn SM, Simpson RJ. Proteomics Clin Appl. 2007;1:1004–1015. doi: 10.1002/prca.200700217. [DOI] [PubMed] [Google Scholar]
- 24.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 25.Ackermann BL, Hale JE, Duffin KL. Curr Drug Metab. 2006;7:525–539. doi: 10.2174/138920006777697918. [DOI] [PubMed] [Google Scholar]
- 26.Witze ES, Old WM, Resing KA, Ahn NG. Nat Meth. 2007;4:798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 27.Siuti N, Kelleher NL. Nat Meth. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma S, Zhu M. Chem Biol Interact. 2008 [Google Scholar]
- 29.Nita-Lazar A, Saito-Benz H, White FM. Proteomics. 2008;8:4433–4443. doi: 10.1002/pmic.200800231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaia J. Chem Biol. 2008;15:881–892. doi: 10.1016/j.chembiol.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haskins WE, Wang Z, Watson CJ, Rostand RR, Witowski SR, Powell DH, Kennedy RT. Anal Chem. 2001;73:5005–5014. doi: 10.1021/ac010774d. [DOI] [PubMed] [Google Scholar]
- 32.Gaspari M, Ming-Cheng Cheng M, Terracciano R, Liu X, Nijdam AJ, Vaccari L, di Fabrizio E, Petricoin EF, Liotta LA, Cuda G, Venuta S, Ferrari M. J Proteome Res. 2006;5:1261–1266. doi: 10.1021/pr050417+. [DOI] [PubMed] [Google Scholar]
- 33.Geho D, Cheng MM, Killian K, Lowenthal M, Ross S, Frogale K, Nijdam J, Lahar N, Johann D, Herrmann P, Whiteley G, Ferrari M, Petricoin E, Liotta L. Bioconjug Chem. 2006;17:654–661. doi: 10.1021/bc0503364. [DOI] [PubMed] [Google Scholar]
- 34.Terracciano R, Gaspari M, Testa F, Pasqua L, Tagliaferri P, Cheng MM, Nijdam AJ, Petricoin EF, Liotta LA, Cuda G, Ferrari M, Venuta S. Proteomics. 2006;6:3243–3250. doi: 10.1002/pmic.200500614. [DOI] [PubMed] [Google Scholar]
- 35.Luchini A, Geho DH, Bishop B, Tran D, Xia C, Dufour RL, Jones CD, Espina V, Patanarut A, Zhou W, Ross MM, Tessitore A, Petricoin EF, 3rd, Liotta LA. Nano Lett. 2008;8:350–361. doi: 10.1021/nl072174l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fredolini C, Meani F, Reeder KA, Rucker S, Patanarut A, Botterell PJ, Bishop B, Longo C, Espina V, Petricoin EFI, Liotta LA, Luchini A. NanoResearch. 2008;1:502–518. doi: 10.1007/s12274-008-8054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Adv Mat. 2006;18:1345–1360. [Google Scholar]
- 38.Thornton PD, McConnell G, Ulijn RV. Chem Commun. 2005:5913–5915. doi: 10.1039/b511005j. [DOI] [PubMed] [Google Scholar]
- 39.Ehrick JD, Deo SK, Browning TW, Bachas LG, Madou MJ, Daunert S. Nat Mater. 2005;4:298–302. doi: 10.1038/nmat1352. [DOI] [PubMed] [Google Scholar]
- 40.Thornton PD, Mart RJ, Webb SJ, Ulijn RV. Soft Matter. 2008;4:821–827. doi: 10.1039/b714750c. [DOI] [PubMed] [Google Scholar]
- 41.Buck SM, Koo YEL, Park E, Xu H, Philbert MA, Brasuel MA, Kopelman R. Curr Opin Chem Biol. 2004;8:540–546. doi: 10.1016/j.cbpa.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 42.Clark HA, Hoyer M, Philbert MA, Kopelman R. Anal Chem. 1999;71:4831–4836. doi: 10.1021/ac990629o. [DOI] [PubMed] [Google Scholar]
- 43.Pelton R. Adv Colloid Interface Sci. 2000;85:1–33. doi: 10.1016/s0001-8686(99)00023-8. [DOI] [PubMed] [Google Scholar]
- 44.Wu SG, Lai EP, Mayer PM. J Pharm Biomed Anal. 2004;36:483–490. doi: 10.1016/j.jpba.2003.05.001. [DOI] [PubMed] [Google Scholar]
- 45.Bai SF, Cai DZ, Li X, Chen XX. Arch Insect Biochem Physiol. 2008 doi: 10.1002/arch.20279. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 46.Narhi LO, Caughey DJ, Horan T, Kita Y, Chang D, Arakawa T. Anal Biochem. 1997;253:236–245. doi: 10.1006/abio.1997.2375. [DOI] [PubMed] [Google Scholar]
- 47.Singh K, Kaur J, Raje M, Varshney G, Suri C. Anal Bioanal Chem. 2003;377:220–224. doi: 10.1007/s00216-003-2066-z. [DOI] [PubMed] [Google Scholar]
- 48.Croom HA, Richards KM, Best SJ, Francis BH, Johnson EI, Dax EM, Wilson KM. J Clin Virol. 2006;36:68–71. doi: 10.1016/j.jcv.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Braude EA, Nachod FC. Determination of Organic Structures by Physical Methods. Academic Press; New York: 1955. [Google Scholar]
- 50.Denizli A, Piskin E. J Biochem Biophys Methods. 2001;49:391–416. doi: 10.1016/s0165-022x(01)00209-3. [DOI] [PubMed] [Google Scholar]
- 51.Sereikaite J, Bumelis VA. Biomed Chromatogr. 2006;20:195–199. doi: 10.1002/bmc.552. [DOI] [PubMed] [Google Scholar]
- 52.Thompson ST, Cass KH, Stellwagen E. Proc Natl Acad Sci USA. 1975;72:669–672. doi: 10.1073/pnas.72.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian S, Kaufman BT. J Biol Chem. 1980;255:10587–10590. [PubMed] [Google Scholar]
- 54.Arnaud P, Gianazza E. FEBS Lett. 1982;137:157–161. doi: 10.1016/0014-5793(82)80337-2. [DOI] [PubMed] [Google Scholar]
- 55.Dodziuk H. Cyclodextrins and Their Complexes: Chemistry, Analytical Methods, Applications. Wiley-VCH; Hoboken: 2006. [Google Scholar]
- 56.Uekama K, Hirayama F, Arima H. Journal of Inclusion Phenomena and Macrocyclic Chemistry. 2006;56:3–8. [Google Scholar]
- 57.Jin S, Lee Y, Kang H. Archives of Dermatological Research. 300:451–454. doi: 10.1007/s00403-008-0864-z. [DOI] [PubMed] [Google Scholar]
- 58.Cai W, Yao X, Shao X, Pan Z. J Inclusion Phenom Macrocyclic Chem. 2005;51:41–51. [Google Scholar]
- 59.Borst C, Holzgrabe U. J Chromatogr A. 2008;1024:191–196. doi: 10.1016/j.chroma.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 60.Elmas B, Onur M, Sxenel S, Tuncel A. Colloid Polym Sci. 2002;280:1137–1146. [Google Scholar]
- 61.Kataoka K, Miyazaki H, Okano T, Sakurai Y. Macromolecules. 1994;27:1061–1062. [Google Scholar]
- 62.Lorand JP, Edwards JO. J Org Chem. 1959;24:769–774. [Google Scholar]
- 63.Mader HS, Wolfbeis OS. Mikrochim Acta. 2008;162:1–34. [Google Scholar]
- 64.Yamauchi A, Suzuki I, Hayashita T. Glucose Sensing. Springer; US: 2006. pp. 237–258. [Google Scholar]
- 65.Zhang Y, Gao X, Hardcastle K, Wang B. Chemistry. 2006;12:1377–1384. doi: 10.1002/chem.200500982. [DOI] [PubMed] [Google Scholar]
- 66.Markvicheva EA, Bronin AS, Kudryavtseva NE, Rumsh LD, Kirsh YE, Zubov VP. Biotechnol Tech. 1994;8:143–148. [Google Scholar]
- 67.Vihola H, Marttila AK, Pakkanen JS, Andersson M, Laukkanen A, Kaukonen AM, Tenhu H, Hirvonen J. Int J Pharm. 2007;343:238–246. doi: 10.1016/j.ijpharm.2007.04.020. [DOI] [PubMed] [Google Scholar]