Abstract
Nitric oxide synthases (NOS) are modular, calmodulin (CaM)-dependent, flavo-heme enzymes that catalyze oxidation of L-arginine to generate nitric oxide (NO) and citrulline. During catalysis, the FMN subdomain cycles between interaction with an NADPH-FAD subdomain to receive electrons, and interaction with an oxygenase domain to deliver electrons to the NOS heme. This process can be described by a three-state, two equilibrium model for the conformation of the FMN subdomain, in which it exists in two distinct bound states (FMN-shielded), and one common unbound state (FMN-deshielded). We studied how each partner subdomain, the FMN redox state, and CaM binding may regulate the conformational equilibria of the FMN module in rat neuronal NOS (nNOS). We utilized four nNOS protein constructs of different subdomain composition, including the isolated FMN subdomain, and determined changes in the conformational state by measuring the degree of FMN shielding by fluorescence, electron paramagnetic resonance, or stopped-flow spectroscopic techniques. Our results suggest: (i) The NADPH-FAD subdomain has a far greater capacity to interact with the FMN subdomain than does the oxygenase domain. (ii) CaM binding has no direct effects on the FMN subdomain. (iii) CaM destabilizes interaction of the FMN subdomain with the NADPH-FAD subdomain but does not measurably increase its interaction with the oxygenase domain. Our results imply that a different set point and CaM regulation exists for either conformational equilibrium of the FMN subdomain. This helps to explain the unique electron transfer and catalytic behaviors of nNOS, relative to other dual-flavin enzymes.
Nitric oxide (NO)1 has important functions in the cardiovascular, immune, and neuronal systems during normal and disease states (1–4). NO synthases (NOS; EC1.14.13.39) are a family of homodimeric enzymes that catalyze a two-step oxidation of l-Arginine (l-Arg) to generate NO and citrulline (5). Mammals express three NOS isoforms that are known as neuronal (nNOS), endothelial (eNOS), and inducible NOS (iNOS). All three are modular enzymes comprised of an N-terminal oxygenase domain (NOSoxy) that contains binding sites for iron protoporphyrin IX (heme), (6R)-5,6,7,8,-tetrahydro-L-biopterin (H4B), and l-Arg, a C-terminal flavoprotein domain that contains binding sites for flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), and nicotinamide adenine dinucleotide phosphate (NADPH), and an intervening calmodulin (CaM) binding sequence(6). The NOSoxy, flavoprotein, and CaM binding domains can be expressed independently and have been subject to detailed structural, kinetic, and regulatory studies (7–23).
The flavoprotein domain of NOS shares structural and catalytic features with a family of NADPH-utilizing dual-flavin enzymes, including cytochrome P450 reductase (CPR), methionine synthase reductase, sulfite reductase flavoprotein, and novel reductase-1 (24–32). These enzymes are all comprised of a NADP+/FAD-binding subdomain that is related to ferredoxin-NADP+ reductase (FNR) (33–35) and a FMN-binding subdomain that is homologous to flavodoxin (36). During catalysis, NADPH reduces bound FAD via hydride transfer and then electrons are passed individually from FAD to FMN to eventually form FMN hydroquinone (FMNH2). During catalysis the reduced FMN subdomain (containing FMNH2) is thought to undergo a large conformational motion that frees it from the FNR subdomain and enables it to transfer an electron to either an attached heme domain (as in NOS) or to an exogenous electron acceptor protein (like cytochrome P450 or cytochrome c). Once this is accomplished, the FMN subdomain (containing FMN semiquinone, FMNsq) must then recombine with the FNR subdomain in order to receive another electron and continue catalysis. A simple 3-state, 2-equilibrium model describing FMN subdomain function is illustrated in Fig. 1. The model stipulates that electron input into FMN and output from FMN are mutually exclusive steps that involve two distinct binding events of the FMN subdomain.
FIGURE 1.
A 3-state, 2-equilibrium model of FMN subdomain function in electron transfer in NOS. Two protein hinge elements (H1 and H2) connect the FMN subdomain to the FNR and NOSoxy subdomains. The CaM binding site is located on the H2 hinge. Interaction of the FMN subdomain with either partner is described by equilibrium A and B. The model assumes that the same face of the FMN subdomain (red) interacts with specific surface regions (blue) on the FNR and NOSoxy partners in order to receive and give electrons, and that the FMN subdomain of one NOS subunit (green) interacts exclusively with the NOSoxy subdomain of the partner subunit (black) in a NOS dimer.
Molecular details of FMN subdomain interactions and how they are regulated in dual-flavin enzymes are topics of ongoing interest (18;21;27;37–39). Recent studies have focused on either one of the two FMN subdomain interactions (depicted as equilibrium A or B in Fig. 1) (13;18;27;37;38;40–44). Evidence suggests that NADPH binding site occupancy (45;46) and flavin redox state (15;20) may be two facets that regulate equilibrium A in the dual flavin reductases. Besides these, NOS enzymes display unique additional aspects because interactions of their FMN subdomains are also controlled by CaM binding (12;18;19;41;47;48). CaM binding destabilizes the FMN-shielded conformation in eNOS and nNOS flavoprotein domain (shifts equilibrium A to the right in Fig. 1), thereby increasing rates of cytochrome c reduction in either steady-state or single turnover reactions (20;39;41). CaM also impacts equilibrium B in NOS, because CaM binding is required for electron transfer from FMNH2 to the NOSoxy domain heme (49–53).
The molecular mechanisms by which CaM impacts equilibria A and B in NOS are under active investigation. Intriguingly, evidence suggests that CaM effects on equilibrium A and B may be largely independent of one another, and may even involve different lobes of CaM (21;54–56). CaM binding alters the behavior of at least one, and perhaps as many as three, unique protein regulatory elements that are inserted within the NOS flavoprotein domain and act to repress electron transfer from the FMN subdomain in the absence of CaM (18;19;57). Comparatively, our understanding of how CaM binding impacts equilibrium B has lagged, although valuable investigations have been initiated recently using NOS protein constructs consisting of the attached FMN, CaM, and NOSoxy subdomains (FMNCaMoxy) (42;58;59). Questions that remain include: How do the interactions of each partner subdomain (FNR and NOSoxy) influence FMN subdomain conformational states in equilibrium A and B (Fig. 1)? How does CaM binding impact these distributions? To what extent do the partner subdomains (FNR and NOSoxy) mediate CaM effects on the FMN subdomain, versus CaM having a direct effect on the FMN subdomain itself? Finally, does the flavin redox state influence these aspects?
To address these questions, we took advantage of the modular structure of NOS and studied FMN subdomain function using four protein constructs that are each a fragment of the holo-nNOS enzyme (Fig. 2). The constructs allowed us to study the free FMN subdomain itself, and the separate impact of each partner subdomain (FNR and NOSoxy) on the conformation and function of the FMN subdomain, in the presence or absence of bound CaM, and with FMN poised in each of its three redox states (FMNox, FMNsq, FMNH2). Our results provide insight into the regulation of equilibrium A and B in nNOS (Fig. 1), and can serve as a blueprint to study these aspects in other dual-flavin reductases.
FIGURE 2.
Schematic illustration of the rat nNOS polypeptide with its subdomains, hinge elements (purple), and a C-terminal control element (CT, red) as marked. The nNOS protein constructs used in the study: nNOSr (amino acids 695–1429); FNR (amino acids 969–1383); FMNCaM (amino acids 695–956); and FMNCaMoxy (amino acids 295–951).
Materials and Methods
General
All reagents and materials were obtained from Sigma, Amersham Biosciences, or other resources as previously reported (46). Absorption, fluorescence, EPR, and stopped-flow spectroscopic experiments were done as previously described (39). For all experiments and protein purification, the buffer contained 40 mM EPPS (pH 7.6), 10 % glycerol, and 250 mM NaCl (buffer A), unless noted otherwise.
Generation of rat nNOS FMNCaM and FMNCaMoxy constructs
The FMNCaM construct (amino acids 695–956) was created using PCR. The PCR primers introduced a Ndel and a start codon at the 5’ end (5’-TTTTCATATGTCGGG CAGCATCACCCCTG-3’) and stop codon at 956 followed by a XbaI sites at the 3’ end (5’-TTTTGGATCCTCTAGATCACTCGATGTTGACGTCATCCCC-3’). The PCR-product fragment was gel-purified and inserted into the TOPO vector (pSC-b) using the StrataClone Blunt PCR Cloning Kit (Stratagene). This was then digested using NdeI and XbaI and the FMNCaM fragment was inserted into the pCWori vector and transformed into JM109 cells. After confirmation of the DNA insert sequence, the expression vector was transformed into E. coli BL21 (DE3) cells that contained the pACYC human calmodulin (CaM) plasmid and selected with ampicillin and chloramphenicol.
The FMNCaMoxy construct (amino acids 295–951) was generated by PCR. A fragment was generated using oligo CaM_OxyF (5’-AACGGGGAGAAATTCGGCTGTGC-3’) and CaM_OxyR (5’-AAAAAGCTTTCTAGATCATTACTCGATGTTGACGTCATCCCCCAC-3’) with the latter creating a translational stop site at residue 952 and a new XbaI site. PCR was done using nNOS full-length as a template. The PCR product was gel-purified and digested with HindIII and XbaI. The resulting 66 base-pair fragment was subcloned into a pCWori/Δ296nNOS vector (60) that had been digested with HindIII and XbaI. After confirming the construct DNA sequence, the expression vector for FMNCaMoxy was transformed into E. coli Rosetta cells and selected using ampicillin and chloramphenicol.
Protein Expression and Purification
The FMNCaM or FMNCaMoxy proteins were expressed in E. coli BL21 (DE3) cells carrying the pACYC plasmid encoding human Calmodulin or in E. coli Rosetta cells, respectively. The transformed bacteria were grown at 37 °C in 4 liters of terrific broth supplemented with 125 mg/ml ampicillin and 50 mg/ml chloramphenicol. Protein expression was induced when the cultures reached an OD600 of 0.8 to 1 by adding 1 mM isopropyl-β-D-thiogalactoside. After further growth at room temperature for 24 h, the cells were harvested. The FMNCaM cell pellets were resuspended in buffer B (40 mM EPPS, and 50 mM NaCl) containing 1 mM EDTA, 0.5 mg/ml each of leupeptin and pepstatin, 1 mg/ml lysozyme, and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cells were lysed by sonication with five 30 s pulses with a 1 min rest on ice between pulses. Cell lysates were centrifuged at 4 °C for 30 min to remove debris and the cell-free supernatant was loaded at 4 °C onto a Q-sepharose column that had been equilibrated with buffer B on an FPLC. The loaded column was washed with 5 column volumes of buffer B. The FMNCaM protein was eluted with buffer C (40 mM EPPS, 150 mM NaCl) containing 1 mM EDTA. The eluted protein sample was buffer-exchanged with 40 mM EPPS, 10% glycerol, 250 mM NaCl and 2 mM CaCl2 and applied to a column of CaM-Sepharose pre-equilibrated with buffer A plus 2 mM CaCl2. The bound protein was washed extensively with the equilibration buffer and then eluted with buffer A containing 4 mM EDTA. The pure protein was concentrated, dialyzed against buffer A to remove the EDTA and stored frozen in aliquots at −80 °C. The purity of protein was verified by SDS-PAGE and absorption spectroscopy. The concentration of FMNCaM proteins was estimated using an extinction coefficient of 10.4 mM−1 cm−1 at 457 nm for the fully oxidized form (15). To quantify protein-bound FMN, a known amount of FMNCaM was boiled in a sealed vial shielded from light to release the FMN, followed by centrifugation to remove the denatured protein. The concentration of FMN in the supernatant was determined by absorbance spectroscopy (λmax = 447 nm; ε447nm = 12.2 mM−1 cm−1) (61).
Cell pellets that contained FMNCaMoxy were resuspended in buffer A containing 4 µM FMN, 0.5 mg/ml each of leupeptin and pepstatin, 1 mg/ml lysozyme, 1mM PMSF, 1 mM l-Arg and 10 µM H4B. Cells were lysed and centrifuged at 4 °C as described above. 60 mM imidazole was added to the supernatant before loading to a Ni2+-NTA column that had been equilibrated with buffer A containing 60 mM imidazole. The protein in the column was washed with 5 column volumes of equilibration buffer and then bound protein was eluted with buffer A containing 250 mM imidazole. 2 mM CaCl2 was added into the elutate, which was then loaded onto a CaM-Sepharose column, and eluted as described above. The eluted FMNCaMoxy protein was concentrated, dialyzed against buffer A to remove the EDTA and stored frozen in aliquots at −80 °C. The concentration of FMNCaMoxy protein was determined based on heme content by the reduced CO difference spectra (ε = 74 mM−1 cm−1 for ΔA445–500). The bound FMN to heme ratio was obtained by boiling known amounts of FMNCaMoxy protein and analyzing for free FMN by absorbance spectroscopy as described above. The nNOSr was expressed and purified as previously described (46). The FNR fragment of nNOSr was generated by limited trypsin proteolysis of nNOSr and purified as previously described (61).
Redox Potentiometry
Protein sample manipulations and redox titrations were carried out in an anaerobic glove box (Belle Technology) at 15 ±1 °C under N2 atmosphere with oxygen levels below 5 ppm as previously described (39). The FMNCaM protein concentration was 40–50 µM containing either EDTA (1 mM) or CaCl2 (2 mM) + CaM (80–100 µM) in buffer A. The six redox mediators we added (0.5–1 µM) and their midpoint potentials (parenthesis) were 2-hydroxy-1,4-naphthaquinone (−152 mV), anthraquinone-2,6-disulfide (−184 mV), phenosafranine (−252 mV), safranine O (−280 mV), benzylviologen (−348 mV), and methyl viologen (−443 mV). The absorption changes at 457 nm and at 600 nm were plotted with electrochemical potentials (mV). The midpoint potentials for the FMNox/FMNsq and FMNsq/FMNH2 redox couples were calculated using the two-electron Nernst equation (eq.1):
| (Eq. 1) |
where A is the absorbance, a − c are the relative absorbance values contributed by the FMN in each of three nondegenerate oxidation states, E is the observed system potential, and are the two midpoint potentials for FMN.
Fluorescence Spectroscopy
Flavin fluorescence emission was measured at room temperature using a Hitachi model F-2000 spectrofluorometer as described in (39). Briefly, different concentrations from 0.5 to 3 µM of the oxidized nNOSr, FNR, FMNCaM, FMNCaMoxy proteins and free FMN were dissolved in buffer A. The samples were placed in a 1-ml quartz cuvette and were excited at 457 nm wavelength and their fluorescence emission spectra were monitored from 480 to 700 nm. In some cases, nNOSoxy was added (0 to 3µM) to samples of the FMNCaM protein to investigate possible quenching by the nNOSoxy heme. The FAD fluorescence of the FNR protein was subtracted from the total flavin fluorescence intensity of the nNOSr to obtain the FMN-specific fluorescence value, as described previously (39).
EPR Spectroscopy
EPR sample preparation and power saturation experiments were carried out essentially as previously described (39;46). Briefly, solutions (approximately 40 µM) of nNOSr, FMNCaM, or FMNCaMoxy were prepared in 20 mM HEPES buffer (pH=7.4) containing 25% glycerol (v/v). The FMNCaMoxy sample contained 1 mM N-Nitro-L-Arginine Methyl Ester (L-NAME), 90 µM of H4B, and 1 mM DTT. CaM-bound samples of nNOSr, FMNCaM and FMNCaMoxy also contained CaM (80 µM) and CaCl2 (2 mM). The nNOSr samples were given a slight molar excess of NADPH while the FMNCaM and FMNCaMoxy samples were treated with a small excess of sodium dithionite, and were all allowed to air oxidize to generate the bound, air stable FMN semiquinone radical. Varying amounts (0, 3.3, 6.7, 10, or 13 mM) of dysprosium (III)-HEDTA complex (DyIII-HEDTA) were added to the samples followed by immediate freezing and were then kept in liquid N2 for EPR measurements. The EPR parameters and data analysis were previously described in detail (39). In Ohio State University (OSU), EPR spectra were recorded on a Bruker ESP 300 EPR spectrometer equipped with an ER 035 NMR gauss meter and a Hewlett-Packard 5352B microwave frequency counter. The EPR data from Kent State University (KSU) were recorded on a Bruker EMX-A spectrometer equipped with an ER 041XG microwave X-band solid state bridge an ER 4119HS-W1 high sensitivity resonator and a Bruker BVT 3000 temperature controller. All spectra were obtained at a temperature of 150 K and a microwave frequency of 9.45 GHz, modulation amplitude of 4.01 G at 100 kHz modulation frequency, and a center field of 3375 G (OSU) or microwave frequency of 9.19 GHz, modulation amplitude of 10.0 G at 100 kHz modulation frequency, and a center field of 3275 G (KSU). Sweep width was 400 G. Five scans were accumulated and averaged for each spectrum. A total of seventeen power settings, ranging from 0.020 to 200.0 mW, were used for the saturation experiments.
Anaerobic Pre-Steady-State Cytochrome c Reduction
Anaerobic solutions of nNOSr, FMNCaM, or FMNCaMoxy proteins were initially fully reduced by titration with sodium dithionite solution in a spectrophotometer. CaM-free proteins contained EDTA (1 mM) while the CaM-bound samples had CaCl2 (2mM) and CaM (3 times the concentration of the nNOS protein). For the FMNCaMoxy sample, 1 mM l-Arg, 50 mM of H4B, and 1mM DTT were added. Each fully-reduced protein at different initial concentrations (9, 15, 21, and 27 mM) was rapidly mixed with a fixed solution of cytochrome c (3 µM) in the stopped-flow instrument at 10 °C. The reaction was monitored by absorbance changes from 300 to 700 nm and the kinetic traces of absorbance gain at 550 nm were fit to a single-exponential function. The observed rate constants (s−1) were plotted against the corresponding protein concentrations. The data points were subject to a linear fit through the origin and we obtained the second order rate constants from the slope of the line (µM−1 s−1).
Solvent accessible surface calculations
The solvent accessible surface of each protein residue was calculated using the Surface racer 5.0 software (62) on the 1TLL structure of rat nNOSr (only one monomer was used, and the part of the FMN-FNR linker visible in the structure (residues 943–951) was removed for the calculations) or on the designed oxy-FMN complexes described below. A probe diameter of 10 Å mimicking the Dy(III)-HEDTA complex (63) was used. The values obtained for the FMN domain alone (1TLL, residues 750–942) were compared to those obtained for the various FMN-oxy or FMN-FNR complexes to identify which residues have a decreased in solvent accessibility in the complexes.
Models of oxy-FMN domain interaction
Four different models of the oxy-FMN domain interaction were constructed manually using the structures 1ZVL (nNOSoxy domain dimer) and 1TLL (nNOSr FMN domain, residues 750–942). The restrictions used were as follows: no overlapping of the two structures; FMN to heme distance was equal or less than 15 Å Lys423 (oxy domain) and Glu762 (FMN domain) must be in the interface (both have been shown to be essential for FMN to heme electron transfer) (38;64); residues 716 (oxy domain) and 750 (FMN domain) must remain within a reasonable distance to reflect their being linked by a 34 amino acid hinge (the structure of part of this linker (20 amino acids) in complex with CaM is available as PDB 2O60). Two of the oxy-FMN complexes we designed are shown in Fig. S1.
Results
Generation and purification of nNOS FMNCaM and FMNCaMoxy proteins
The FMNCaM protein was purified by tandem Q-sepharose and CaM affinity chromatography. Co-expressed CaM was removed from the FMNCaM protein in the Q-sepharose step by adding EDTA. Following elution from the CaM-agarose column, we obtained for the first time a FMNCaM protein that was stable in the absence of CaM. It was more stable at a higher salt concentration (250 mM NaCl) than at 150 mM NaCl. The FMNCaMoxy protein was purified using Ni2+-NTA column followed by a CaM affinity column, similar to the nNOS full-length purification (65). The typical yield of purified FMNCaM and FMNCaMoxy proteins was 3–4 mg/L culture. The FMNCaM protein preparations typically contained a 1:1 stoichiometric FMN-to-protein ratio, while the FMNCaMoxy preparations contained approximately 0.8 FMN per heme (data not shown). Gel-filtration chromatography was used to determine the ratio of monomer and dimer. The FMNCaM protein was predominantly monomeric, while the FMNCaMoxy protein was predominantly dimeric (Fig. S2). Dithiothreitol (DTT) had to be present to prevent the reversible formation of FMNCaMoxy multimers cross-linked via intermolecular disulfide bonds.
FMN midpoint potential
We investigated if an attached FNR subdomain or if CaM binding would affect the thermodynamic properties of the FMN cofactor. We performed a reductive titration using sodium dithionite followed by an oxidative titration of the same protein sample using potassium ferricyanide. Representative spectra collected during titration of the CaM-free FMNCaM protein are shown in Fig. 3A. We obtained similar data when titrating the CaM-bound protein (data not shown). In the reductive direction there was an initial absorbance decrease around 457 nm and a broad absorbance increase around 600 nm (FMNsq formation), with an isosbestic point near 510 nm as the first electron equivalent was added. A second phase then took place that was characterized by a decrease in absorbance from 450 to 700 nm until the fully reduced state (FMNH2) was reached. Plots of the absorbance at 457 and 600 nm versus the recorded potential for the CaM-free and CaM-bound FMNCaM protein are shown in Fig. 3Band C. The plots at 457 nm showed a sigmoidal shape while at 600 nm produced a bell-shaped curve, reflecting the appearance and disappearance of the neutral FMNsq. The two sets of data obtained at both wavelengths were fit simultaneously to the two-electron Nernst equation as described under Experimental Procedures. The equilibrium midpoint potentials (Em) of the FMN redox couples (oxidized/semiquinone, ox/sq and semiquinone/hydroquinone, sq/hq) obtained from the fitting are reported in Table 1. The data show that the FMN midpoint potentials in the FMNCaM protein are not altered by bound CaM.
FIGURE 3.
Potentiometric titration of the FMNCaM protein in the presence and absence of CaM. (A) Representative set of visible spectra collected during titration of CaM-free FMNCaM with sodium dithionite. (B & C) Plots of absorbance at 457 nm (solid squares) and at 600 nm (open squares) versus the electrochemical potential (mV) for CaM-free and CaM-bound FMNCaM protein along with the lines of best-fit as calculated using the two-electron Nernst equation described in Materials and Methods. Data were collected at 15 °C and are representative of at least two experiments.
Table 1.
Equilibrium midpoint potentials (Em) of NOS constructs vs standard hydrogen electrode
| Sample | Condition | FMN | FAD | ||
|---|---|---|---|---|---|
| ox/sq | sq/hq | ox/sq | sq/hq | ||
| FMNCaM | − CaM | − 121 ± 2 | − 252 ± 3 | ||
| + CaM | − 117 ± 2 | − 258 ± 3 | |||
| FNR | − CaM | − 264 ± 2 | − 279 ± 3 | ||
| nNOSr | − CaM | − 49 ± 2 | − 273 ± 6 | − 269 ± 23 | − 286 ± 13 |
The midpoint potentials of FAD and FMN cofactors (ox/sq, and sq/hq) were determined for the NOS constructs by potentiometric titration at 15 ± 1 C° under a N2 atmosphere as described in Materials and Methods.
For comparison we carried out potentiometric titrations of the nNOS FAD-containing FNR subdomain and of the CaM-free nNOSr. Flavin midpoint potentials that we obtained for these proteins (Table 1) are in good agreement with previously published data (15;61;66). The midpoint potentials of the FAD (ox/sq, sq/hq) cofactor in the FNR protein are essentially identical to those for FAD in CaM-free nNOSr (Table 1). In contrast, the FMNox/sq midpoint potential in the FMNCaM protein is approximately 70 mV lower than its value in CaM-free nNOSr. This indicates an attached FNR subdomain alters the redox properties of the FMN cofactor in nNOSr, as previously seen (15), such that the FNR subdomain tends to thermodynamically stabilize the FMNsq.
FMN shielding
We next used our four nNOS protein constructs to study what regulates FMN shielding in nNOS when the FMN is poised in each of its 3 redox states. We reasoned that data obtained with the FMNCaM subdomain should provide lower boundary values for FMN shielding (i.e., it will represent the least-shielded form of FMN that is possible within nNOS) in each redox state, while the nNOSr and FMNCaMoxy constructs would indicate how each individual partner subdomain (FNR and NOSoxy, respectively) can influence FMN shielding in the presence or absence of bound CaM.
Shielding of fully-oxidized FMN
Previous work showed that the intensity of flavin fluorescence is inversely proportional to the degree of FMN shielding in NOSr proteins whose flavins are in the fully-oxidized state (20;39). We therefore measured the fluorescence intensities (per mole) of the oxidized FMNCaM, FMNCaMoxy, nNOSr, FNR, and free FMN in buffer to compare their levels of FMN shielding (Fig. 4). We estimated the fluorescence emission that is solely due to the bound FMN cofactor within nNOSr by subtracting2 the fluorescence emission obtained for corresponding concentrations of the FNR subdomain (39), whose FNR fluorescence emission is only due to its bound FAD cofactor. FAD fluorescence typically accounts for a minor portion (6 to 15 %) of the total fluorescence of nNOSr due to the fluorescence of FAD being quenched by its adenine ring (67). In Fig. 4B, the fluorescence of the FNR subdomain (black bar) is superimposed onto the bar value for nNOSr, and was then subtracted to obtain an estimate of the FMN-specific fluorescence for nNOSr as shown in Fig. 4B, inset. The fluorescence intensity of the FMNCaM protein was about 4-fold less than that of free FMN, similar to but somewhat less than the quenching that is observed in other FMN-containing flavoproteins (67). The FMNCaMoxy protein exhibited slightly lower fluorescence than the FMNCaM protein. Control experiments showed that this difference was not due to a through-solution quenching of FMN fluorescence by the NOSoxy heme (data not shown). In comparison, the nNOSr protein exhibited much less FMN fluorescence. CaM binding to each construct did not detectably alter their FMN fluorescence (data not shown). Our data imply the following hierarchy exists for FMN shielding in the three NOS proteins (with or without bound CaM) when the FMN is in the fully-oxidized state: FMNCaM ≤ FMNCaMoxy < nNOSr.
FIGURE 4.
Fluorescence intensities of the FMN subdomain in various nNOS protein constructs. (A) Fluorescence intensities of fully-oxidized FMNCaM, FMNCaMoxy, and nNOSr proteins, and free FMN were determined at the indicated concentrations by excitation at 457 nm and monitoring emission at 525 nm. The fluorescence intensities of the FNR subdomain were also obtained to subtract its contribution from the total flavin fluorescence intensity in the nNOSr. (B) Fluorescence intensity versus the concentration slope value for each protein obtained by linear regression analysis of the data in panel A. The slope of the FNR subdomain is depicted as a black bar that is superimposed onto the bar value for nNOSr. It was then subtracted to obtain an FMN-specific fluorescence value for nNOSr. Inset, scale-up plot of the FMN-specific fluorescence values for each nNOS protein. Data are representative of at least two experiments.
Shielding of the FMNsq
The FMNCaM, FMNCaMoxy, and nNOSr proteins can all form airstable FMNsq radicals (10;15;47;59). This enabled us to measure FMNsq shielding by determining the effect of an added spin-relaxing agent DyIII-HEDTA on the EPR microwave power saturation characteristics of the FMNsq radical (68;69). We have previously used this method to compare FMNsq shielding in various NOSr proteins (39;46). Here we treated the CaM-free and CaM-bound FMNCaM and FMNCaMoxy proteins with a slight molar excess of sodium dithionite and then allowed them to air-oxidize to their FMNsq state, as judged by their visible spectra (data not shown). Known amounts of DyIII-HEDTA were then added and the protein solutions were immediately frozen in liquid nitrogen for EPR measurements. A representative set of EPR spectra that was recorded for each of the three proteins is shown in Fig. S3.
The power saturation of the FMNsq radical in the nNOSr, FMNCaM, and FMNCaMoxy proteins were affected by DyIII-HEDTA in a concentration-dependent manner (Fig. S4), similar to what we observed previously for nNOSr (39;46). We obtained P1/2 values for each sample by fitting the power saturation curves (Fig. S4), which were converted to ΔP1/2 values as previously described (46). The ΔP1/2 values for a set of nNOSr and FMNCaM samples are plotted versus the DyIII-HEDTA concentration in Fig. 5 A and B. Linear regression analysis of the data gave the slopes of the lines, m (mW/mM) as noted in the Figure. A larger slope indicates an FMNsq that is more solvent-accessible and therefore less shielded. Our data confirm that FMNsq shielding in nNOSr is significantly diminished upon CaM binding, as reported previously (39;46). In contrast, CaM binding to the FMNCaM protein did not significantly alter its FMNsq shielding. Also, the degree of FMNsq shielding that was observed in the CaM-bound nNOSr was similar to the level of FMNsq shielding observed in the FMNCaM protein with or without CaM. This indicates a near minimal degree of FMNsq shielding exists in nNOSr when CaM is bound.
FIGURE 5.
Comparative shielding of the FMNsq in nNOSr, FMNCaM, and FMNCaMoxy as measured by EPR spectroscopy. EPR power saturation measurements were recorded at 150 K for nNOS constructs in the presence of the indicated concentrations of DyIII-HEDTA, and the ΔP1/2 values were calculated as described in Materials and Methods and are plotted versus [DyIII-HEDTA] in the presence (filled circles) or absence (open circles) of bound CaM. EPR data for samples of (A) nNOSr and (B) FMNCaM were obtained at the Ohio State University and EPR data for samples of (C) FMNCaMoxy and (D) FMNCaM were obtained at Kent State University. The lines of best fit were calculated by linear regression. Data are representative of at least two experiments.
The ΔP1/2 values for a distinct set of FMNCaM and FMNCaMoxy proteins are plotted versus the DyIII- HEDTA concentration in Fig. 5C and D3. FMNsq shielding in the FMNCaMoxy protein was not significantly affected by CaM binding, and its absolute degree of FMNsq shielding was equivalent to that observed in the FMNCaM protein. These results indicate that a near minimal degree of FMNsq shielding exists in the FMNCaMoxy protein irrespective of bound CaM. The results also suggest the following hierarchy exists for FMNsq shielding in the nNOSr proteins: FMNCaM = FMNCaMoxy = CaM-bound nNOSr < CaM-free nNOSr.
Shielding of FMNH2
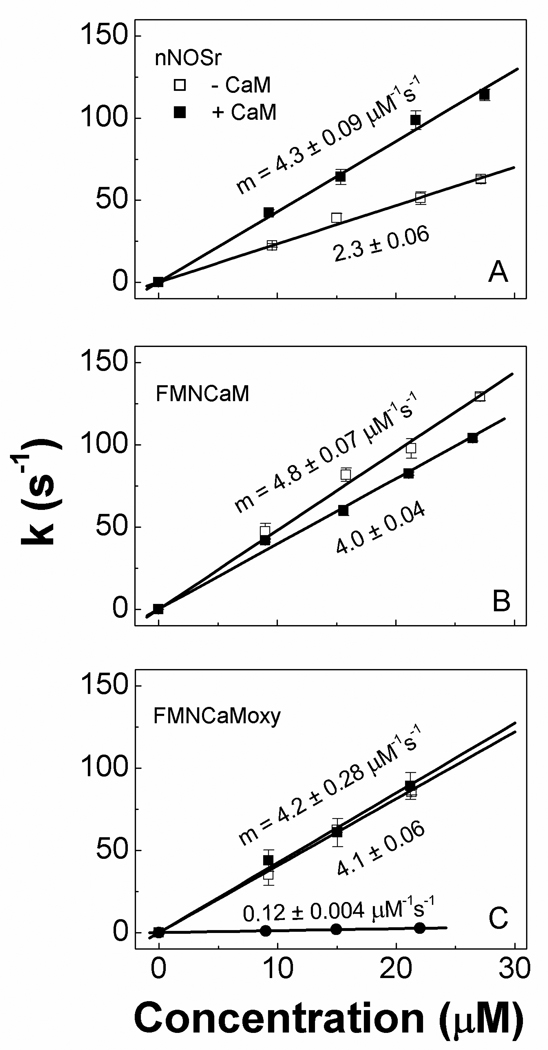
We measured FMNH2 shielding in our nNOS proteins by an established stopped-flow spectroscopic method that monitors reaction of the FMNH2 with cytochrome c as an external electron acceptor (39;41;46). Pseudo-first order rate constants for cytochrome c reduction were determined after rapid-mixing a 3, 5, 7, or 9-fold molar excess of each dithionite-reduced nNOS protein with cytochrome c in a stopped-flow spectrometer and fitting each absorbance increase at 550 nm to a single-exponential function. We also ran replica reactions using nNOSoxy protein as a control for possible reduction of cytochrome c by the ferrous heme in our FMNCaMoxy construct. Representative spectra recorded during reactions of each nNOS protein, along with averaged kinetic traces obtained at 550 nm, are shown in Figs. S5 & S6. The derived rate constants were plotted versus NOS protein concentration and found to be linearly dependent in all cases (Fig. 6A–C). Regarding the FMNCaMoxy data, our control nNOSoxy reactions showed that cytochrome c reduction by the NOS ferrous heme was kinetically insignificant (Fig. 6C). From these plots we calculated the second order rate constants for FMNH2-based cytochrome c reduction by linear regression analysis. The values are reported in Fig. 6AC. We observed that CaM binding to nNOSr4 approximately doubled the second order rate constant relative to that observed for the CaM-free enzyme. In contrast, CaM binding to the FMNCaM protein diminished its second order rate constant by 17%, and CaM caused no significant change on the rate constants obtained for the FMNCaMoxy protein. The second order rate constant for CaM-bound nNOSr was of similar magnitude to the rate constants we obtained for both the FMNCaM and FMNCaMoxy proteins. Together, the data indicate that CaM binding diminished FMNH2 shielding in the NOSr protein, and appeared to do so to the full extent possible, whereas CaM binding to the FMNCaMoxy protein did not increase its FMNH2 shielding.
FIGURE 6.
Second order rate constants for pre-steady state cytochrome c reduction by excess nNOS proteins in the presence and absence of bound CaM. The indicated initial concentrations of fully-reduced (A) nNOSr, (B) FMNCaM, and (C) FMNCaMoxy proteins were rapidly mixed with cytochrome c (3 µM initial concentration) in the stopped-flow instrument under anaerobic conditions at 10 °C. Data obtained from reactions of fully-reduced nNOSoxy and cytochrome c is also indicated in panel C (solid circle). The kinetic traces at 550 nm were fit to a single-exponential function and the measured rate constants are plotted in the panels versus the initial concentrations (pre-mixing) of the various nNOS proteins. The second order rate constants (µM−1s−1) were obtained by linear regression analysis. Data are representative of at least two experiments.
Discussion
The FMN subdomain has coordinate electron-accepting and electron-donating roles in nNOS and in related dual-flavin enzymes. Here we examined how the conformational properties of the FMN subdomain in nNOS are influenced by its two partner subdomains, the FMN redox state, and CaM binding. Investigating these facets within a single study has advantages over studies that typically focused only on individual facets (14;39;45;46;58;61). Our findings help to discern regulation in the NOS enzymes and also provide a foundation for studies on related dual-flavin enzymes. We discuss our results below in terms of the three-state, two-equilibrium model for FMN function that was introduced in Fig.1.
Effects of CaM on the isolated FMN subdomain
Given that CaM is of similar size to the FMN subdomain (17 vs 28 kDa, respectively) and binds to an adjacent helical hinge element on the NOS polypeptide (H2 in Fig.1), it is conceivable that CaM could directly affect the properties of the FMN subdomain. Previously this possibility could not be examined because the expressed FMNCaM proteins were found to be unstable in the absence of CaM (15). However, in our hands, CaM binding was not required for protein stabilization. We found that CaM binding had little or no discernable impact on the properties of the isolated FMN subdomain when the FMN was poised in each of its three redox states. This establishes that CaM affects the behavior of the FMN subdomain in nNOS (redox potential, FMN shielding, electron transfer activities) through a coordinate effect involving the partner subdomains (FNR and NOSoxy). By excluding a direct effect of CaM on the FMN subdomain, our findings also help to discern how CaM may impact equilibrium A and B in nNOS (Fig. 1).
Equilibrium A: interaction of the FNR and FMN subdomains
Measuring the degree of FMN shielding is one approach to study equilibrium A in NOSr proteins, as done previously for nNOSr and eNOSr proteins poised in the zero (FAD/FMN), one-electron (FAD/FMNsq), and 4-electron (FADH2/FMNH2) reduced states (39). However, data interpretation had been limited due to the uncertainty of a direct CaM effect on the FMN subdomain, which we have now ruled out (see above), and by the unavailability of boundary values for the isolated FMN subdomain (i.e., providing the maximally and minimally FMN-shielded values). Our measures of FMN shielding in the FMNCaM protein now provide the minimal FMN shielding boundary values for nNOS, and can be used to extend the previous and current findings. Our measures indicate that FMN shielding in CaM-free nNOSr is 4×, 2.5×, and 2× greater compared to the minimum FMN shielding that we observed for the FMNCaM protein, when nNOSr is poised in the 0, 1, and 4 electron-reduced states, respectively. This establishes that the attached FNR subdomain causes significant FMN shielding in CaM-free nNOSr, in each of the three FMN redox states.
The boundary values for a maximally-shielded FMN in nNOSr are not yet available, and are expected to be non-zero when the FMN is poised in a 0- or 1-electron reduced state (if FMN shielding in these two redox states is measured by FMN fluorescence emission and FMNsq paramagnetic interaction with DyIII-HEDTA, respectively). This means that we cannot yet estimate the equilibrium A distribution of CaM-free nNOSr when the FMN is in the 0 or 1-electron reduced states. In comparison, the maximal boundary value for FMNH2 shielding in nNOSr is expected to be zero (i.e., it should display no measurable reduction of cytochrome c)5. If we assume the maximal boundary value is zero, then a comparison of our FMN shielding measures for the fully-reduced nNOSr and FMNCaM proteins imply that the equilibrium A distribution of the reduced, CaM-free nNOSr is an approximate 50:50 mixture of the shielded and deshielded conformations depicted in Fig. 1 (i.e., KeqA = 1). We recently derived a similar KeqA estimate for a fully-reduced CaM-free nNOSr using a somewhat different technique (39). Thus, our data indicate that a significant fraction (50%) of fully-reduced CaM-free nNOSr exists in an FMN-deshielded conformation in solution. In principle, this distribution should enable a significant interaction in unit time between the FMNH2 of nNOSr and electron acceptor proteins (or partner subdomains), which in turn would be consistent with CaM-free nNOSr catalyzing a robust cytochrome c reductase activity. However, the analysis also points out that despite the fully-reduced FMN subdomain being in a conformation that is able to interact with electron acceptors, the electron transfer to the heme in the nNOS oxygenase domain still does not occur in the absence of bound CaM (49;52). Thus, we conclude that heme reduction in nNOS is not primarily controlled at the level of equilibrium A.
CaM binding diminishes FMN shielding in nNOSr (20;39;41). Here we found that FMN shielding in CaM-bound nNOSr dropped to a level that was nearly equivalent to the FMN shielding we observed for the FMNCaM protein, which represents the lower boundary value. This implies that CaM binding to nNOSr shifts equilibrium A to the right such that nearly all of the nNOSr molecules exist in an FMN deshielded conformation (Fig. 1). In addition, our data imply that CaM caused equilibrium A to shift in this way regardless of whether nNOSr is in a 1- or 4-electron reduced state. A shift in equilibrium A to favor the FMN deshielded conformation would seem desirable for a 4-electron reduced nNOSr, because it is an obligate electron-donating species whose FMNH2 needs to interact with an electron-accepting partner or domain. But this same shift in the FMN conformation may not be desirable for the 1-electron reduced nNOSr, because in this case the FMN subdomain contains FMNsq, which is an obligate electron acceptor that must interact with the FNR subdomain in order to receive an electron to continue catalysis. The apparent dilemma could be resolved if NADPH binding and/or FAD reduction in nNOSr causes equilibrium A to shift to the left, thereby facilitating reduction of the FMNsq by the reduced FNR subdomain. How NADP+/NADPH binding and combinations of FAD or FMN redox states other than those studied heres might impact equilibrium A in a dual-flavin enzyme is not clear and is being investigated in several labs (20;41;43;61;70;71).
Remarkably, for the related dual-flavin enzyme CPR, measures of its FMN subdomain interaction with cytochrome c by isothermal titration calorimetry (43) imply that the equilibrium A for the 1- electron reduced CPR (containing FMNsq) lies heavily toward the left. The basis for this apparent difference between nNOSr and CPR is unclear, but it could certainly involve inherent differences in the measurement techniques. Indeed, we measured FMNsq shielding in 1-electron reduced nNOSr based on the through-space paramagnetic interaction of FMNsq with DyIII-HEDTA, the latter which has an estimated molecular diameter of 10–12 Åin solution (63;69) (Fig. S7). The crystal structure of nNOSr depicts the protein in one of the tightest possible “FMN-shielded” conformations, such that the FNR and FMN subdomain interface is tight enough to exclude water or DyIII-HEDTA and thus prevent its close approach to the bound FMNsq (Fig. S8 A–C). However, because interactions between subdomain redox partners can be multiple and highly dynamic (72), the conformer that is depicted by the nNOSr crystal structure is likely to be only one of many similar conformations that are being sampled in unit time in solution, and therefore are present as an ensemble during our EPR measurement. Because of the small diameter of DyIII-HEDTA, relatively small movements of the FMN subdomain could loosen up the FNR-FMN subdomain interface enough to allow a closer approach of DyIII-HEDTA to the bound FMNsq. Thus, our measurements based on DyIII-HEDTA are almost certain to score a greater percentage nNOSr molecules to be in an “FMN-deshielded” conformation as compared to measures that are based on an interaction between the FMN subdomain and cytochrome c. This is partly because cytochrome c has a much larger molecular diameter (at least 35 Å) that is similar to the diameter of the FMN subdomain itself (Fig. S7), and so would require that nNOSr undergo a more significant structural perturbation before its FMN subdomain could interact and consequently be scored as being in an FMN-deshielded state.
This caveat underscores the need to consider how a given measurement technique might score the degree of FMN shielding in a given dual-flavin enzyme, and how the scoring may relate to the subset of protein conformations that are relevant to the catalytic function of interest. For nNOSr, the relevant reactions include electron transfer to FMNsq (via the FNR-FMN subdomain interaction, equilibrium A), and electron transfer from FMNH2 to either cytochrome c or NOSoxy (equilibrium B), or to O2 (i.e., flavin autoxidation). Because cytochrome c is of similar size to the FMN subdomain, measuring their interaction is likely to be a good indicator for the subset of nNOSr conformers whose FMN subdomain is positioned far enough from the FNR subdomain to deliver electrons to a partner protein or domain. In comparison, we expect that DyIII-HEDTA would score these conformers as FMN-deshielded, along with an additional subset of nNOSr conformers whose FMN subdomain may be loosened from the FNR subdomain but not yet in a position to deliver electrons to a large acceptor like NOSoxy or to cytochrome c.
Equilibrium B: interaction of the FMN and NOSoxy subdomains
The model in Fig. 1 suggests that the FNR subdomain, when present, might compete with and antagonize NOSoxy domain interaction with the FMN subdomain. Because our FMNCaMoxy protein is missing the FNR subdomain, it conceivably provides the best chance for maximum complex formation between NOSoxy and the FMN subdomain as determined by equilibrium B (Fig. 1). We therefore surmised that the FMNCaMoxy protein would display greater FMN shielding than the FMNCaM protein, and might further increase its FMN shielding in the presence of bound CaM. However, this was not the case. Instead, our measures show that the extent of FMN shielding in the CaM-free FMNCaMoxy protein was similar to that in the FMNCaM protein, irrespective of whether the FMN was in the 0, 1, or 2-electron reduced state. In addition, CaM binding to the FMNCaMoxy protein did not significantly increase our measures of FMN shielding in any circumstance. Given that the FMN shielding level in the FMNCaMoxy protein always remained close to the minimal boundary value as indicated by the FMNCaM protein, we conclude that the attached NOSoxy domain has little or no capacity to influence FMN shielding in the CaM-free or CaM-bound FMNCaMoxy protein. This contrasts sharply with how the attached FNR subdomain can significantly increase FMN shielding in CaM-free nNOSr. Together, these results distinguish the two protein partners of the FMN subdomain (FNR and NOSoxy) in nNOS, and imply that there are fundamental differences in the protein interactions that define equilibrium A and B.
What can our FMN shielding measures tell us about equilibrium B in nNOS? Any increase in FMN shielding in the FMNCaMoxy protein would have been straightforward to interpret, because it could only have been caused by an increased interaction between the FMN subdomain and NOSoxy. Instead, our measures indicated that FMN shielding in the FMNCaMoxy protein nearly matched the minimum boundary value for FMN shielding, irrespective of CaM or the reduction state of the FMN. This could either mean: (i) FMN-NOSoxy complexes form infrequently and/or transiently, and so exist only to a minor extent at equilibrium (i.e., equilibrium B is set far to the left), or (ii) stable FMN-NOSoxy complexes form and are a major conformer (i.e., equilibrium B is set to the right), but do not cause the measured FMN shielding level to increase. Possibility (i) is the simplest interpretation of our data, and will be further discussed below. Possibility (ii) hinges on whether FMN shielding is measurably increased when a complex forms between the FMN subdomain and the NOSoxy domain. Although a definitive answer is not yet possible, we may have enough information to consider this question. Despite there being no crystal structure for a FMN-NOSoxy complex, a model for the interaction was proposed, based on the existing crystal structures of nNOSr and NOSoxy (13;73), point mutagenesis studies of protein surface residues (38;64), and known distance constraints for electron transfer (74). Specifically, an electronegative surface patch on the FMN subdomain that surrounds the bound FMN cofactor is thought to dock against a complementary electropositive patch on the surface of the NOSoxy dimer near the heme (13;75). This complex would enable the FMN and heme to achieve a 12 to 17 Å edge-to-edge distance that is sufficiently close for electron transfer (13). But would such an FMN-NOSoxy complex, if formed, measurably increase FMN shielding in our FMNCaMoxy protein? At this point, we know too little about how the complex formation might affect the FMN fluorescence emission to know whether complex formation in the FMNCaMoxy protein would be detectable by our fluorescence measures. However, this is not likely the case for our other measurements of flavin shielding when the FMNCaMoxy protein was poised to contain the reduced forms of FMN (FMNsq or FMNH2). For these cases, we used the existing information as a guide to build models of the docking between the FMN subdomain and NOSoxy, and then calculated the corresponding surface region on the FMN subdomain that would be shielded from a direct interaction with DyIII-HEDTA or water molecules. As shown in Fig. S8, panels D-F, the docking interfaces we modeled would shield at minimum a surface area on the FMN subdomain of 400 to 900 Å2 (water excluded surface, 1.2 Å probe radius) that surrounds the FMN cofactor in a non-symmetric manner. The docking interface would shield a larger surface area (2200 to 2500 Å2) from DyIII-HEDTA, due to its larger radius (5 Å probe radius). In general, this modeled FMN-NOSoxy interface is somewhat smaller and less protective of the bound FMN than is the known interface formed by the FMN-FNR subdomain complex in the crystal structure of nNOSr (around 2500 Å2 (13) for the water excluded surface or 4100 Å2 when calculated with a 5 Å probe radius; FMN-NOSoxy and FMN-FNR surfaces are compared in Fig. S8). On the basis of our analysis, we expect that if a stable FMN-NOSoxy complex were present at equilibrium, it might be somewhat more difficult, but not impossible, to detect with DyIII-HEDTA as a probe to measure FMNsq shielding in the FMNCaMoxy protein. On the other hand, our analysis suggests that a FMN-NOSoxy complex would be easily detected by using the cytochrome c reaction as a probe to detect shielding of bound FMNH2 in FMNCaMoxy. Thus, the cytochrome c and EPR data we obtained for our FMNCaMoxy protein leads us to favor interpretation (i) above. Namely, FMNsq or FMNH2 shielding in the FMNCaMoxy protein, with or without bound CaM, remains near the minimal boundary level because FMN-NOSoxy complexes either form infrequently or are transient and do not build up. This suggests that equilibrium B is poised far to the left in nNOS, and is little affected by CaM binding.
Our current results, when combined with previous information, can update the model for FMN subdomain function in nNOS. The model has FMN subdomain interactions with its FNR and NOSoxy partners being mutually exclusive and under their own regulation. Equilibrium A, which defines the FNR-FMN subdomain interaction, has a range of set points that include KeqA values both below and above unity. KeqA appears to be influenced by NADPH binding site occupancy (41;46), flavin redox state (15;20), and CaM binding (12;18;48). When the FMN subdomain is in an obligate electron-donating redox state (in our case, FMNH2 in 4-electron reduced nNOSr), equilibrium A is an approximate 50:50 distribution between FMN-shielded (non-reactive) and FMN-deshielded (reactive) conformers in the CaM-free nNOSr. Subsequent CaM binding shifts equilibrium A almost completely to the right so that nearly all the nNOSr molecules are in a reactive, FMN-deshielded conformation. Equilibrium B, which defines the FMN-NOSoxy interaction, appears to have a more narrow range of KeqB values that are all below unity, such that the FMN deshielded conformer predominates. KeqB does not appear to be significantly influenced by CaM binding or possibly by the FMN redox state. Thus, equilibrium B is set in nNOS such that FMN-NOSoxy complex formation may be infrequent and/or transient in practically all circumstances.
The model described for equilibrium A and B can help to explain the catalytic behaviors of nNOS. For example, it is consistent with (i) cytochrome c reduction being much faster than is heme reduction in NOS (39;76); (ii) CaM increasing the cytochrome c reductase activity of nNOS; (iii) cytochrome c blocking NO synthesis by “stealing” electrons from the NOSr domain (77); (iv) the poor capacity of isolated nNOSr and nNOSoxy proteins to interact and catalyze heme reduction or NO synthesis when they are mixed together (78); and (v) NOS enzymes having slow rates of heme reduction compared to other flavo-heme proteins (76).
If one accepts that CaM binding does not cause significant FMN-NOSoxy complex formation, how might it trigger heme reduction in nNOS? Although our data indicate that CaM binding to NOS deshields the fully-reduced FMN module almost completely, we know this alone is insufficient. Perhaps without CaM bound to the hinge linker (H2 in Fig. 1), a deshielded FMN subdomain has too much freedom of motion and cannot “find” the NOSoxy docking site, or perhaps it is physically blocked from forming a complex with NOSoxy that is productive for electron transfer. Accordingly, bound CaM might lower the degrees of freedom by restricting the motions of the FMN subdomain, and might also eliminate a steric block to productive complex formation with nNOSoxy, perhaps manifested through an effect on inhibitory control elements present in nNOSr (18;19). Even so, when CaM binds it appears to enable only a low yield of productive FMN-NOSoxy complexes. This is consistent with a dynamic conformational sampling model as described by Scrutton and colleagues for the electron transfer flavoprotein (72), where rapid sampling of many conformations yields a range of transient protein donor-acceptor complexes, only a subset of which are competent for electron transfer. Under this circumstance, the macroscopic rate of heme reduction that is observed in a population of nNOS molecules is limited by the relatively infrequent formation of competent FMN-NOSoxy complexes. Related work suggests that productive FMN-NOSoxy complex formation in nNOS is influenced by several structural factors including the composition and length of inter-domain hinges (79), charge pairing at subdomain interfaces(38), and also by an overriding selective pressure to keep NOS heme reduction slow enough to allow the enzyme to release the NO that it makes (76). This latter facet may explain why NOS enzymes would have a low frequency formation of productive FMN-NOSoxy complexes, despite their bidomain structure potentially enabling a much more efficient and rapid rate of heme reduction as occurs in the related bi-domain enzyme cytochrome P450BM3 (80).
Relationship to similar work
Feng and colleagues have used laser flash techniques to show that electron transfer can occur between the FMNsq and ferrous heme groups in various NOS FMNCaMoxy proteins (42;81). Their data suggest that a small percentage of the protein molecules are in a conformation that allows the electron transfer, even in the absence of CaM. This is consistent with our data suggesting that equilibrium B is set far to the left in nNOS. For the related dual-flavin enzyme CPR, recent FRET and surface plasmon resonance studies have measured binding of the oxidized reductase or its FMN subdomain to a physiologic partner protein, heme oxygenase 1. The data gave binding Kd values of 0.5 to 2.5 µM, depending on NADP+ binding site occupancy(82;83). In addition, a significant degree of FRET was observed at equilibrium, implying a significant amount of stable protein complexes can form in the system. For the related dual-flavin enzyme methionine synthase reductase, flavin fluorescence and isothermal titration calorimetry were used to measure binding of the oxidized reductase or its isolated FMN subdomain to the activation domain of methionine synthase (27). Besides reporting Kd values that ranged from 1.5 to 5 µM, the study provided evidence for stable protein complex formation, as indicated by their observing a significant decrease in FMN fluorescence at equilibrium and a significant extent of chemical cross-linking across the protein-protein interface. In general, we surmise that these types of dual-flavin enzymes have maintained a relatively stronger binding affinity between their FMN subdomains and their exogenous protein partners (44;78;84;85) because their equilibrium B involves a through-solution bimolecular reaction instead of a unimolecular reaction as occurs in the NOS enzymes. Clearly, there is still more to be learned about what regulates the conformational equilibrium of the FMN subdomain, its binding interactions, and electron transfer within this diverse family of enzymes. Our current study provides insight into these issues for nNOS and may provide a useful strategy to study the related dual-flavin enzymes.
Supplementary Material
Computer models of the FMN-NOSoxy complexes, gel filtration chromatography results for nNOSr, FMNCaM and FMNCamoxy proteins, representative EPR spectra sets, power saturation profiles, presteady state cytochrome c reduction data, a comparison of the relative sizes of DyIII-HEDTA, FMN domain and cytochrome c and solvent accessibility of the surface residues of the FMN domain. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
We thank Drs. Mahfuzul Haque and Ashis Biswas for helpful advice and discussions, and Deborah Durra for helping with protein purification. We also thank the Department of Chemistry at the Kent State Tuscarawas campus for access to their EPR instrument.
Footnotes
This work was supported by National Institutes of Health Grants (CA53914, GM51941, & HL76491 to D.J.S.; HL63744, HL65608, & HL38324 to J. L. Z.) and by American Heart Association Postdoctoral Fellowships (0725341B to R.P.I. and 0625632B to J.T.).
Abbreviations: NO, nitric oxide; NOS, nitric oxide synthase; NOSr, NOS reductase domain, nNOSr, neuronal NOSr; eNOSr, endothelial NOSr; NOSoxy, NOS oxygenase domain; CaM, calmodulin; FMNCaM, FMN subdomain and CaM domain of neuronal NOS; FMNCaMoxy, FMN subdomain, CaM and oxygenase domains of neuronal NOS; FNR, ferredoxin NADP+ reductase; FADH2, FAD hydroquinone; FMNH2, FMN hydroquinone; FMNsq, FMN semiquinone; CPR, cytochrome P450 reductase; Dy(III)-HEDTA, dysprosium-hydroxy-ethylethylenediaminetriacetic acid complex; NADPH, reduced β-nicotinamide adenine dinucleotide phosphate; DTT, dithiothreitol.
The flavin fluorescence intensity of nNOSr may not be a simple sum of the individual FNR and FMN subdomain fluorescence values. However, this potentially confounding factor is lessened here by the FNR fluorescence being much weaker than that of the FMN subdomain.
Due to instrument availability, two sets of protein samples (one representing the nNOSr versus the FMNCaM protein, and the other representing FMNCaMoxy protein versus the FMNCaM protein) were each run on a different EPR instrument, and this resulted in the nNOSr protein samples having different absolute slope values for either set (see Fig. 5).
NADPH-free enzyme
In this circumstance, the FNR and FMN subdomains would be permanently associated with one another.
References
- 1.Ricciardolo FL, Sterk PJ, Gaston B, Folkerts G. Nitric oxide in health and disease of the respiratory system. Physiol Rev. 2004;84:731–765. doi: 10.1152/physrev.00034.2003. [DOI] [PubMed] [Google Scholar]
- 2.Schulz R, Rassaf T, Massion PB, Kelm M, Balligand JL. Recent advances in the understanding of the role of nitric oxide in cardiovascular homeostasis. Pharmacol. Ther. 2005;108:225–256. doi: 10.1016/j.pharmthera.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Sessa WC. Regulation of endothelial derived nitric oxide in health and disease. Mem. Inst. Oswaldo Cruz. 2005;100 Suppl 1:15–18. doi: 10.1590/s0074-02762005000900004. [DOI] [PubMed] [Google Scholar]
- 4.Knott AB, Bossy-Wetzel E. Nitric oxide in health and disease of the nervous system. Antioxid. Redox. Signal. 2008 doi: 10.1089/ars.2008.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stuehr DJ. Mammalian nitric oxide synthases. Biochim. Biophys. Acta. 1999;1411:217–230. doi: 10.1016/s0005-2728(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Poulos TL. Structure-function studies on nitric oxide synthases. J. Inorg. Biochem. 2005;99:293–305. doi: 10.1016/j.jinorgbio.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Crane BR, Arvai AS, Ghosh DK, Wu C, Getzoff ED, Stuehr DJ, Tainer JA. Structure of nitric oxide synthase oxygenase dimer with pterin and substrate. Science. 1998;279:2121–2126. doi: 10.1126/science.279.5359.2121. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Martasek P, Paschke R, Shea T, Masters BSS, Kim JJ. Crystal structure of the FAD/NADPH-binding domain of rat neuronal nitric-oxide synthase.Comparisons with NADPH-cytochrome P450 oxidoreductase. J. Biol. Chem. 2001;276:37506–37513. doi: 10.1074/jbc.M105503200. [DOI] [PubMed] [Google Scholar]
- 9.Panda K, Rosenfeld RJ, Ghosh S, Meade AL, Getzoff ED, Stuehr DJ. Distinct dimer interaction and regulation in nitric-oxide synthase types I, II, and III. J. Biol. Chem. 2002;277:31020–31030. doi: 10.1074/jbc.M203749200. [DOI] [PubMed] [Google Scholar]
- 10.Knight K, Scrutton NS. Stopped-flow kinetic studies of electron transfer in the reductase domain of neuronal nitric oxide synthase: re-evaluation of the kinetic mechanism reveals new enzyme intermediates and variation with cytochrome P450 reductase. Biochem. J. 2002;367:19–30. doi: 10.1042/BJ20020667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roman LJ, Martasek P, Masters BS. Intrinsic and extrinsic modulation of nitric oxide synthase activity. Chem. Rev. 2002;102:1179–1190. doi: 10.1021/cr000661e. [DOI] [PubMed] [Google Scholar]
- 12.Daff S. Calmodulin-dependent regulation of mammalian nitric oxide synthase. Biochem. Soc. Trans. 2003;31:502–505. doi: 10.1042/bst0310502. [DOI] [PubMed] [Google Scholar]
- 13.Garcin ED, Bruns CM, Lloyd SJ, Hosfield DJ, Tiso M, Gachhui R, Stuehr DJ, Tainer JA, Getzoff ED. Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J. Biol. Chem. 2004;279:37918–37927. doi: 10.1074/jbc.M406204200. [DOI] [PubMed] [Google Scholar]
- 14.Dunford AJ, Marshall KR, Munro AW, Scrutton NS. Thermodynamic and kinetic analysis of the isolated FAD domain of rat neuronal nitric oxide synthase altered in the region of the FAD shielding residue Phe1395. Eur. J. Biochem. 2004;271:2548–2560. doi: 10.1111/j.1432-1033.2004.04185.x. [DOI] [PubMed] [Google Scholar]
- 15.Garnaud PE, Koetsier M, Ost TW, Daff S. Redox properties of the isolated flavin mononucleotide- and flavin adenine dinucleotide-binding domains of neuronal nitric oxide synthase. Biochemistry. 2004;43:11035–11044. doi: 10.1021/bi049312v. [DOI] [PubMed] [Google Scholar]
- 16.Jachymova M, Martasek P, Panda S, Roman LJ, Panda M, Shea TM, Ishimura Y, Kim JJ, Masters BS. Recruitment of governing elements for electron transfer in the nitric oxide synthase family. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15833–15838. doi: 10.1073/pnas.0506522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ost TW, Daff S. Thermodynamic and kinetic analysis of the nitrosyl, carbonyl, and dioxy heme complexes of neuronal nitric-oxide synthase. The roles of substrate and tetrahydrobiopterin in oxygen activation. J. Biol. Chem. 2005;280:965–973. doi: 10.1074/jbc.M411191200. [DOI] [PubMed] [Google Scholar]
- 18.Roman LJ, Masters BS. Electron transfer by neuronal nitric oxide synthase is regulated by concerted interaction of calmodulin and two intrinsic regulatory elements. J. Biol. Chem. 2006;281:23111–23118. doi: 10.1074/jbc.M603671200. [DOI] [PubMed] [Google Scholar]
- 19.Tiso M, Tejero J, Panda K, Aulak KS, Stuehr DJ. Versatile regulation of neuronal nitric oxide synthase by specific regions of its C-terminal tail. Biochemistry. 2007;46:14418–14428. doi: 10.1021/bi701646k. [DOI] [PubMed] [Google Scholar]
- 20.Dunford AJ, Rigby SEJ, Hay S, Munro AW, Scrutton NS. Conformational and thermodynamic control of electron transfer in neuronal nitric oxide synthase. Biochemistry. 2007;46:5018–5029. doi: 10.1021/bi7001339. [DOI] [PubMed] [Google Scholar]
- 21.Spratt DE, Taiakina V, Palmer M, Guillemette JG. Differential binding of calmodulin domains to constitutive and inducible nitric oxide synthase enzymes. Biochemistry. 2007;46:8288–8300. doi: 10.1021/bi062130b. [DOI] [PubMed] [Google Scholar]
- 22.Berka V, Yeh HC, Gao D, Kiran F, Tsai AL. Redox function of tetrahydrobiopterin and effect of L-arginine on oxygen binding in endothelial nitric oxide synthase. Biochemistry. 2004;43:13137–13148. doi: 10.1021/bi049026j. [DOI] [PubMed] [Google Scholar]
- 23.Berka V, Wu G, Yeh HC, Palmer G, Tsai AL. Three different oxygen-induced radical species in endothelial nitric-oxide synthase oxygenase domain under regulation by L-arginine and tetrahydrobiopterin. J. Biol. Chem. 2004;279:32243–32251. doi: 10.1074/jbc.M404044200. [DOI] [PubMed] [Google Scholar]
- 24.Finn RD, Basran J, Roitel O, Wolf CR, Munro AW, Paine MJ, Scrutton NS. Determination of the redox potentials and electron transfer properties of the FAD- and FMN-binding domains of the human oxidoreductase NR1. Eur. J. Biochem. 2003;270:1164–1175. doi: 10.1046/j.1432-1033.2003.03474.x. [DOI] [PubMed] [Google Scholar]
- 25.Munro AW, Leys DG, McLean KJ, Marshall KR, Ost TW, Daff S, Miles CS, Chapman SK, Lysek DA, Moser CC, Page CC, Dutton PL. P450 BM3:the very model of a modern flavocytochrome. Trends Biochem. Sci. 2002;27:250–257. doi: 10.1016/s0968-0004(02)02086-8. [DOI] [PubMed] [Google Scholar]
- 26.Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolthers KR, Scrutton NS. Protein interactions in the human methionine synthase-methionine synthase reductase complex and implications for the mechanism of enzyme reactivation. Biochemistry. 2007;46:6696–6709. doi: 10.1021/bi700339v. [DOI] [PubMed] [Google Scholar]
- 28.Davydov DR, Kariakin AA, Petushkova NA, Peterson JA. Association of cytochromes P450 with their reductases: opposite sign of the electrostatic interactions in P450BM-3 as compared with the microsomal 2B4 system. Biochemistry. 2000;39:6489–6497. doi: 10.1021/bi992936u. [DOI] [PubMed] [Google Scholar]
- 29.Kitazume T, Haines DC, Estabrook RW, Chen B, Peterson JA. Obligatory intermolecular electron-transfer from FAD to FMN in dimeric P450BM-3. Biochemistry. 2007;46:11892–11901. doi: 10.1021/bi701031r. [DOI] [PubMed] [Google Scholar]
- 30.Zeghouf M, Fontecave M, Coves J. A simplifed functional version of the Escherichia coli sulfite reductase. J. Biol. Chem. 2000;275:37651–37656. doi: 10.1074/jbc.M005619200. [DOI] [PubMed] [Google Scholar]
- 31.Eschenbrenner M, Coves J, Fontecave M. The flavin reductase activity of the flavoprotein component of sulfite reductase from Escherichia coli. A new model for the protein structure. J.Biol.Chem. 1995;270:20550–20555. doi: 10.1074/jbc.270.35.20550. [DOI] [PubMed] [Google Scholar]
- 32.Olteanu H, Banerjee R. Human methionine synthase reductase, a soluble P-450 reductase-like dual flavoprotein, is sufficient for NADPH-dependent methionine synthase activation. J. Biol. Chem. 2001;276:35558–35563. doi: 10.1074/jbc.M103707200. [DOI] [PubMed] [Google Scholar]
- 33.Karplus PA, Daniels MJ, Herriott JR. Atomic structure of ferredoxin-NADP+reductase: prototype for a structurally novel flavoenzyme family. Science. 1991;251:60–66. [PubMed] [Google Scholar]
- 34.Arakaki AK, Ceccarelli EA, Carrillo N. Plant-type ferredoxin-NADP+reductases: a basal structural framework and a multiplicity of functions. FASEB J. 1997;11:133–140. doi: 10.1096/fasebj.11.2.9039955. [DOI] [PubMed] [Google Scholar]
- 35.Carrillo N, Ceccarelli EA. Open questions in ferredoxin-NADP+ reductase catalytic mechanism. Eur. J. Biochem. 2003;270:1900–1915. doi: 10.1046/j.1432-1033.2003.03566.x. [DOI] [PubMed] [Google Scholar]
- 36.Sancho J. Flavodoxins: sequence, folding, binding, function and beyond. Cell Mol. Life Sci. 2006;63:855–864. doi: 10.1007/s00018-005-5514-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welland A, Garnaud PE, Kitamura M, Miles CS, Daff S. Importance of the domain-domain interface to the catalytic action of the NO synthase reductase domain. Biochemistry. 2008;47:9771–9780. doi: 10.1021/bi800787m. [DOI] [PubMed] [Google Scholar]
- 38.Panda K, Haque MM, Garcin-Hosfield ED, Durra D, Getzoff ED, Stuehr DJ. Surface charge interactions of the FMN module govern catalysis by nitric-oxide synthase. J. Biol. Chem. 2006;281:36819–36827. doi: 10.1074/jbc.M606129200. [DOI] [PubMed] [Google Scholar]
- 39.Ilagan RP, Tiso M, Konas DW, Hemann C, Durra D, Hille R, Stuehr DJ. Differences in a conformational equilibrium distinguish catalysis by the endothelial and neuronal nitric-oxide synthase flavoproteins. J. Biol. Chem. 2008;283:19603–19615. doi: 10.1074/jbc.M802914200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiso M, Konas DW, Panda K, Garcin ED, Sharma M, Getzoff ED, Stuehr DJ. C-terminal tail residue Arg1400 enables NADPH to regulate electron transfer in neuronal nitric-oxide synthase. J. Biol. Chem. 2005;280:39208–39219. doi: 10.1074/jbc.M507775200. [DOI] [PubMed] [Google Scholar]
- 41.Craig DH, Chapman SK, Daff S. Calmodulin activates electron transfer through neuronal nitric-oxide synthase reductase domain by releasing an NADPH-dependent conformational lock. J. Biol. Chem. 2002;277:33987–33994. doi: 10.1074/jbc.M203118200. [DOI] [PubMed] [Google Scholar]
- 42.Feng CJ, Tollin G, Hazzard JT, Nahm NJ, Guillemette JG, Salerno JC, Ghosh DK. Direct measurement by laser flash photolysis of intraprotein electron transfer in a rat neuronal nitric oxide synthase. Journal of the American Chemical Society. 2007;129:5621–5629. doi: 10.1021/ja068685b. [DOI] [PubMed] [Google Scholar]
- 43.Grunau A, Paine MJ, Ladbury JE, Gutierrez A. Global effects of the energetics of coenzyme binding: NADPH controls the protein interaction properties of human cytochrome P450 reductase. Biochemistry. 2006;45:1421–1434. doi: 10.1021/bi052115r. [DOI] [PubMed] [Google Scholar]
- 44.Sevrioukova IF, Li H, Zhang H, Peterson JA, Poulos TL. Structure of a cytochrome P450-redox partner electron-transfer complex. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1863–1868. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gutierrez A, Munro AW, Grunau A, Wolf CR, Scrutton NS, Roberts GC. Interflavin electron transfer in human cytochrome P450 reductase is enhanced by coenzyme binding. Relaxation kinetic studies with coenzyme analogues. Eur. J. Biochem. 2003;270:2612–2621. doi: 10.1046/j.1432-1033.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- 46.Konas DW, Zhu K, Sharma M, Aulak KS, Brudvig GW, Stuehr DJ. The FAD-shielding residue Phe1395 regulates neuronal nitric-oxide synthase catalysis by controlling NADP+ affinity and a conformational equilibrium within the flavoprotein domain. J. Biol. Chem. 2004;279:35412–35425. doi: 10.1074/jbc.M400872200. [DOI] [PubMed] [Google Scholar]
- 47.Guan ZW, Iyanagi T. Electron transfer is activated by calmodulin in the flavin domain of human neuronal nitric oxide synthase. Arch. Biochem. Biophys. 2003;412:65–76. doi: 10.1016/s0003-9861(03)00009-2. [DOI] [PubMed] [Google Scholar]
- 48.Matsuda H, Iyanagi T. Calmodulin activates intramolecular electron transfer between the two flavins of neuronal nitric oxide synthase flavin domain. Biochim. Biophys. Acta. 1999;1473:345–355. doi: 10.1016/s0304-4165(99)00193-2. [DOI] [PubMed] [Google Scholar]
- 49.Abu-Soud HM, Yoho LL, Stuehr DJ. Calmodulin controls neuronal nitric-oxide synthase by a dual mechanism. Activation of intra- and interdomain electron transfer. J. Biol. Chem. 1994;269:32047–32050. [PubMed] [Google Scholar]
- 50.Daff S, Sagami I, Shimizu T. The 42-amino acid insert in the FMN domain of neuronal nitric-oxide synthase exerts control over Ca(2+)/calmodulin-dependent electron transfer. J. Biol. Chem. 1999;274:30589–30595. doi: 10.1074/jbc.274.43.30589. [DOI] [PubMed] [Google Scholar]
- 51.Kobayashi K, Tagawa S, Daff S, Sagami I, Shimizu T. Rapid calmodulin-dependent interdomain electron transfer in neuronal nitric-oxide synthase measured by pulse radiolysis. J. Biol. Chem. 2001;276:39864–39871. doi: 10.1074/jbc.M102537200. [DOI] [PubMed] [Google Scholar]
- 52.Panda K, Ghosh S, Stuehr DJ. Calmodulin activates intersubunit electron transfer in the neuronal nitric-oxide synthase dimer. J. Biol. Chem. 2001;276:23349–23356. doi: 10.1074/jbc.M100687200. [DOI] [PubMed] [Google Scholar]
- 53.Roman LJ, Martasek P, Miller RT, Harris DE, de La Garza MA, Shea TM, Kim JJ, Masters BS. The C termini of constitutive nitric-oxide synthases control electron flow through the flavin and heme domains and affect modulation by calmodulin. J. Biol. Chem. 2000;275:29225–29232. doi: 10.1074/jbc.M004766200. [DOI] [PubMed] [Google Scholar]
- 54.Spratt DE, Taiakina V, Guillemette JG. Calcium-deficient calmodulin binding and activation of neuronal and inducible nitric oxide synthases. Biochim. Biophys. Acta. 2007;1774:1351–1358. doi: 10.1016/j.bbapap.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 55.Gachhui R, Abu-Soud HM, Ghosh DK, Presta A, Blazing MA, Mayer B, George SE, Stuehr DJ. Neuronal nitric-oxide synthase interaction with calmodulin-troponin C chimeras. J. Biol. Chem. 1998;273:5451–5454. doi: 10.1074/jbc.273.10.5451. [DOI] [PubMed] [Google Scholar]
- 56.Newman E, Spratt DE, Mosher J, Cheyne B, Montgomery HJ, Wilson DL, Weinberg JB, Smith SM, Salerno JC, Ghosh DK, Guillemette JG. Differential activation of nitric-oxide synthase isozymes by calmodulin-troponin C chimeras. J. Biol. Chem. 2004;279:33547–33557. doi: 10.1074/jbc.M403892200. [DOI] [PubMed] [Google Scholar]
- 57.Knudsen GM, Nishida CR, Mooney SD, Ortiz de Montellano PR. Nitric-oxide synthase (NOS) reductase domain models suggest a new control element in endothelial NOS that attenuates calmodulin-dependent activity. J. Biol. Chem. 2003;278:31814–31824. doi: 10.1074/jbc.M303267200. [DOI] [PubMed] [Google Scholar]
- 58.Feng C, Tollin G, Holliday MA, Thomas C, Salerno JC, Enemark JH, Ghosh DK. Intraprotein electron transfer in a two-domain construct of neuronal nitric oxide synthase: the output state in nitric oxide formation. Biochemistry. 2006;45:6354–6362. doi: 10.1021/bi060223n. [DOI] [PubMed] [Google Scholar]
- 59.Ghosh DK, Holliday MA, Thomas C, Weinberg JB, Smith SM, Salerno JC. Nitric-oxide synthase output state. Design and properties of nitric-oxide synthase oxygenase/FMN domain constructs. J. Biol. Chem. 2006;281:14173–14183. doi: 10.1074/jbc.M509937200. [DOI] [PubMed] [Google Scholar]
- 60.Panda K, Adak S, Aulak KS, Santolini J, McDonald JF, Stuehr DJ. Distinct influence of N-terminal elements on neuronal nitric-oxide synthase structure and catalysis. J. Biol. Chem. 2003;278:37122–37131. doi: 10.1074/jbc.M304456200. [DOI] [PubMed] [Google Scholar]
- 61.Konas DW, Takaya N, Sharma M, Stuehr DJ. Role of Asp(1393) in catalysis, flavin reduction, NADP(H) binding, FAD thermodynamics, and regulation of the nNOS flavoprotein. Biochemistry. 2006;45:12596–12609. doi: 10.1021/bi061011t. [DOI] [PubMed] [Google Scholar]
- 62.Tsodikov OV, Record MT, Jr, Sergeev YV. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 2002;23:600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 63.Oliver ME, Hales BJ. Using dysprosium complexes to probe the nitrogenase paramagnetic centers. Biochemistry. 1993;32:6058–6064. doi: 10.1021/bi00074a017. [DOI] [PubMed] [Google Scholar]
- 64.Shimanuki T, Sato H, Daff S, Sagami I, Shimizu T. Crucial role of Lys(423) in the electron transfer of neuronal nitric-oxide synthase. J. Biol. Chem. 1999;274:26956–26961. doi: 10.1074/jbc.274.38.26956. [DOI] [PubMed] [Google Scholar]
- 65.Wu C, Zhang J, Abu-Soud H, Ghosh DK, Stuehr DJ. High-level expression of mouse inducible nitric oxide synthase in Escherichia coli requires coexpression with calmodulin. Biochem. Biophys. Res. Commun. 1996;222:439–444. doi: 10.1006/bbrc.1996.0763. [DOI] [PubMed] [Google Scholar]
- 66.Noble MA, Munro AW, Rivers SL, Robledo L, Daff SN, Yellowlees LJ, Shimizu T, Sagami I, Guillemette JG, Chapman SK. Potentiometric analysis of the flavin cofactors of neuronal nitric oxide synthase. Biochemistry. 1999;38:16413–16418. doi: 10.1021/bi992150w. [DOI] [PubMed] [Google Scholar]
- 67.Munro AW, Noble MA. Fluorescence analysis of flavoproteins. Methods Mol. Biol. 1999;131:25–48. doi: 10.1385/1-59259-266-X:25. [DOI] [PubMed] [Google Scholar]
- 68.Galli C, MacArthur R, AbuSoud HM, Clark P, Stuehr DJ, Brudvig GW. EPR spectroscopic characterization of neuronal NO synthase. Biochemistry. 1996;35:2804–2810. doi: 10.1021/bi9520444. [DOI] [PubMed] [Google Scholar]
- 69.Innes JB, Brudvig GW. Location and magnetic relaxation properties of the stable tyrosine radical in photosystem II. Biochemistry. 1989;28:1116–1125. doi: 10.1021/bi00429a028. [DOI] [PubMed] [Google Scholar]
- 70.Jones RJ, Gao YT, Simone TM, Salerno JC, Smith SM. NADPH analog binding to constitutive nitric oxide activates electron transfer and NO synthesis. Nitric. Oxide. 2006;14:228–237. doi: 10.1016/j.niox.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 71.Murataliev MB, Feyereisen R. Functional interactions in cytochrome P450BM3.Evidence that NADP(H) binding controls redox potentials of the flavin cofactors. Biochemistry. 2000;39:12699–12707. doi: 10.1021/bi001068u. [DOI] [PubMed] [Google Scholar]
- 72.Toogood HS, van Thiel A, Scrutton NS, Leys D. Stabilization of non-productive conformations underpins rapid electron transfer to electron-transferring flavoprotein. J. Biol. Chem. 2005;280:30361–30366. doi: 10.1074/jbc.M505562200. [DOI] [PubMed] [Google Scholar]
- 73.Matter H, Kumar HS, Fedorov R, Frey A, Kotsonis P, Hartmann E, Frohlich LG, Reif A, Pfleiderer W, Scheurer P, Ghosh DK, Schlichting I, Schmidt HH. Structural analysis of isoform-specific inhibitors targeting the tetrahydrobiopterin binding site of human nitric oxide synthases. J. Med. Chem. 2005;48:4783–4792. doi: 10.1021/jm050007x. [DOI] [PubMed] [Google Scholar]
- 74.Page CC, Moser CC, Dutton PL. Mechanism for electron transfer within and between proteins. Current Opinion in Chemical Biology. 2003;7:551–556. doi: 10.1016/j.cbpa.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 75.Crane BR, Rosenfeld RJ, Arvai AS, Ghosh DK, Ghosh S, Tainer JA, Stuehr DJ, Getzoff ED. N-terminal domain swapping and metal ion binding in nitric oxide synthase dimerization. EMBO J. 1999;18:6271–6281. doi: 10.1093/emboj/18.22.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stuehr DJ, Santolini J, Wang ZQ, Wei CC, Adak S. Update on mechanism and catalytic regulation in the NO synthases. J. Biol. Chem. 2004;279:36167–36170. doi: 10.1074/jbc.R400017200. [DOI] [PubMed] [Google Scholar]
- 77.Klatt P, Heinzel B, John M, Kastner M, Bohme E, Mayer B. Ca2+/calmodulin-dependent cytochrome c reductase activity of brain nitric oxide synthase. J. Biol. Chem. 1992;267:11374–11378. [PubMed] [Google Scholar]
- 78.Rozhkova EA, Fujimoto N, Sagami I, Daff SN, Shimizu T. Interactions between the isolated oxygenase and reductase domains of neuronal nitric-oxide synthase: assessing the role of calmodulin. J. Biol. Chem. 2002;277:16888–16894. doi: 10.1074/jbc.M200642200. [DOI] [PubMed] [Google Scholar]
- 79.Haque MM, Panda K, Tejero J, Aulak KS, Fadlalla MA, Mustovich AT, Stuehr DJ. A connecting hinge represses the activity of endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9254–9259. doi: 10.1073/pnas.0700332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Munro AW, Daff S, Coggins JR, Lindsay JG, Chapman SK. Probing electron transfer in flavocytochrome P-450 BM3 and its component domains. Eur. J. Biochem. 1996;239:403–409. doi: 10.1111/j.1432-1033.1996.0403u.x. [DOI] [PubMed] [Google Scholar]
- 81.Feng C, Thomas C, Holliday MA, Tollin G, Salerno JC, Ghosh DK, Enemark JH. Direct measurement by laser flash photolysis of intramolecular electron transfer in a two-domain construct of murine inducible nitric oxide synthase. J. Am. Chem. Soc. 2006;128:3808–3811. doi: 10.1021/ja0578606. [DOI] [PubMed] [Google Scholar]
- 82.Higashimoto Y, Sakamoto H, Hayashi S, Sugishima M, Fukuyama K, Palmer G, Noguchi M. Involvement of NADPH in the interaction between heme oxygenase-1 and cytochrome P450 reductase. J. Biol. Chem. 2005;280:729–737. doi: 10.1074/jbc.M406203200. [DOI] [PubMed] [Google Scholar]
- 83.Wang J, Ortiz de Montellano PR. The binding sites on human heme oxygenase-1 for cytochrome p450 reductase and biliverdin reductase. J. Biol. Chem. 2003;278:20069–20076. doi: 10.1074/jbc.M300989200. [DOI] [PubMed] [Google Scholar]
- 84.Chen PF, Tsai AL, Berka V, Wu KK. Endothelial nitric-oxide synthase. Evidence for bidomain structure and successful reconstitution of catalytic activity from two separate domains generated by a baculovirus expression system. J. Biol. Chem. 1996;271:14631–14635. [PubMed] [Google Scholar]
- 85.Ghosh DK, Abu-Soud HM, Stuehr DJ. Reconstitution of the second step in NO synthesis using the isolated oxygenase and reductase domains of macrophage NO synthase. Biochemistry. 1995;34:11316–11320. doi: 10.1021/bi00036a003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Computer models of the FMN-NOSoxy complexes, gel filtration chromatography results for nNOSr, FMNCaM and FMNCamoxy proteins, representative EPR spectra sets, power saturation profiles, presteady state cytochrome c reduction data, a comparison of the relative sizes of DyIII-HEDTA, FMN domain and cytochrome c and solvent accessibility of the surface residues of the FMN domain. This material is available free of charge via the Internet at http://pubs.acs.org.