Abstract
ATP-citrate lyase (Acly) is one of two cytosolic enzymes that synthesize acetyl-coenzyme A (CoA). Because acetyl-CoA is an essential building block for cholesterol and triglycerides, Acly has been considered a therapeutic target for hyperlipidemias and obesity. To define the phenotype of Acly-deficient mice, we created Acly knockout mice in which a β-galactosidase marker is expressed from Acly regulatory sequences. We also sought to define the cell type-specific expression patterns of Acly to further elucidate the in vivo roles of the enzyme. Homozygous Acly knockout mice died early in development. Heterozygous mice were healthy, fertile, and normolipidemic on both chow and high fat diets, despite expressing half-normal amounts of Acly mRNA and protein. Fibroblasts and hepatocytes from heterozygous Acly mice contained half-normal amounts of Acly mRNA and protein, but this did not perturb triglyceride and cholesterol synthesis or the expression of lipid biosynthetic genes regulated by sterol regulatory element-binding proteins. The expression of acetyl-CoA synthetase 1, another cytosolic enzyme for producing acetyl-CoA, was not up-regulated. As judged by β-galactosidase staining, Acly was expressed ubiquitously but was expressed particularly highly in tissues with high levels of lipogenesis, such as in the livers of mice fed a high-carbohydrate diet. β-Galactosidase staining was intense in the developing brain, in keeping with the high levels of de novo lipogenesis of the tissue. In the adult brain, β-galactosidase staining was in general much lower, consistent with reduced levels of lipogenesis; however, β-galactosidase expression remained very high in cholinergic neurons, likely reflecting the importance of Acly in generating acetyl-CoA for acetylcholine synthesis. The Acly knockout allele is useful for identifying cell types with a high demand for acetyl-CoA synthesis.
ATP-citrate lyase (Acly)1 is a cytosolic enzyme that catalyzes the formation of acetyl-coenzyme A (CoA) and oxaloacetate from citrate and CoA, with the hydrolysis of ATP to ADP and phosphate. Acetyl-CoA is the key building block for de novo lipogenesis. Acly is not, however, the sole source of acetyl-CoA in the cytosol. Another enzyme, acetyl-CoA synthetase 1 (Acs1), generates acetyl-CoA from acetate and CoA, with the hydrolysis of ATP to AMP and diphosphate (1). Whether Asc1 might be sufficient in the setting of a genetic deficiency in Acly is not known. The expression of both Acly and Acs1 is controlled by the sterol regulatory element-binding protein (SREBP)-1c, a transcriptional regulator of fatty acid synthesis (2, 3).
An important role for Acly in lipid biosynthesis in the liver has been suggested by studies with the Acly inhibitor hydroxycitrate. In rats treated with hydroxycitrate, the rate of fatty acid and cholesterol synthesis in the liver fall (4). Those experiments and studies with other inhibitors (5–10) have suggested that Acly could represent a therapeutic target for elevated plasma lipid levels. As with other genes involved in lipid synthesis, the expression of Acly in the liver is regulated, at least in part, at the transcriptional level by SREBP-1 (3, 11, 12). When lipid biosynthesis levels in the liver are low, as in fasted mice, Acly mRNA levels are low. However, when fasted mice are fed a carbohydrate-rich diet, lipid biosynthesis rates and Acly mRNA levels increase (3, 12).
Lipid synthesis is important in the developing brain, particularly for myelination (13). Northern blots have shown that Acly mRNA levels increase in the rat brain during the first week of suckling, when de novo lipogenesis is high (14). However, those experiments have not revealed which regions of the brain, or which cell types in the brain, express Acly at high levels. Acly is also highly expressed in the kidney (15, 16), and moderate amounts of the Acly mRNA are found in the adrenals, intestine, and lung (14).
In this study, we explored the physiologic importance of Acly in mice. We had three goals. First, we were interested in determining if a complete absence of Acly would be compatible with embryonic development. Second, we wanted to determine whether half-normal levels of Acly, as in heterozygous Acly knockout mice, would be associated with alterations in lipid synthesis, plasma lipid levels, or body weight. Third, we sought to define the cell type-specific expression of Acly, both during development and in adult mice, to better understand the in vivo roles of Acly. To address these questions, we produced and characterized Acly-deficient mice.
EXPERIMENTAL PROCEDURES
Acly-deficient Mice
A mouse embryonic stem cell line (cell line NPX098, strain 129/Ola) containing an insertional mutation in Acly was identified within BayGenomics, a gene-trapping resource (17). The gene-trap vector used (pGT1δTMpfs) contains a splice-acceptor sequence upstream of a reporter gene, βgeo (a fusion of β-galactosidase and neomycin phosphotransferase II) (17). As judged by 5′ rapid amplification of cDNA ends (18), the insertional mutation in NPX098 occurred in the first intron of Acly, following a noncoding first exon. Thus, the mutation results in the production of a fusion transcript consisting of exon 1 sequences from Acly and βgeo. The mutation created a new EcoRV site in intron 1, which was useful for genotyping mice by Southern blots (see below).
NPX098 was injected into C57BL/6 blastocysts to generate chimeric mice, which were bred to establish Acly knockout mice. All mice described herein had a mixed genetic background (~50% C57BL/6 and ~50% 129/Ola). The mice were weaned at 21 days of age, housed in a barrier facility with a 12-h light/12-h dark cycle, and fed a chow diet containing 4.5% fat (Ralston Purina, St. Louis, MO). In some experiments, the mice were fed a Western diet (21% fat, 50% carbohydrate, and 20% protein) from Research Diets (New Brunswick, NJ) or a fat-free, carbohydrate-rich diet (60.2% sucrose and 20% casein) from ICN (Irvine, CA).
Mice were genotyped by Southern blots with EcoRV-digested genomic DNA and a 5′ Acly flanking probe. The probe was amplified from mouse genomic DNA with primers 5′-CGCTGGTCAAGAATGGGTGTTACAA-3′ and 5′-CTCCCTCCCCCAACCTCAAACTAAG-3′.
Mouse Primary Embryonic Fibroblasts
Timed matings were established between Acly+/− mice, and pregnant females were sacrificed 13.5–17.5 days post-coitus (dpc). Embryos were incubated overnight in 5 ml of 0.25% trypsin-EDTA (Invitrogen, Carslbad, CA) at 4 °C. The next morning, embryos were mechanically disrupted in 5 ml of Dulbecco’s modified Eagle’s medium supplemented with fetal bovine serum, l-glutamine, nonessential amino acids, penicillin, streptomycin, Fungizone (Invitrogen), and 2-mercaptoethanol (Sigma). After removal of debris, cells were plated in 100-mm Petri dishes.
Mouse Primary Hepatocytes
Mice were anesthetized with 2.5% avertin, and a catheter was introduced into the inferior vena cava via the right atrium. The liver was perfused with liver perfusion media (Invitrogen) and then collagenase-lipase media (Invitrogen). The digested liver was then removed and mechanically disrupted. The cell suspension was filtered through sterile gauze and centrifuged at 50 × g. Hepatocytes were then washed three times with hepatocyte wash media (Invitrogen), and 5 × 105 cells/well were plated in 6-well collagen-coated tissue culture plates. Cells were allowed to attach to the plate for 4 h before starting the labeling experiments (see below). Cells were grown at 37 °C under 5% CO2 in 3 ml of Dulbecco’s modified Eagle’s medium supplemented with fetal bovine serum, sodium pyruvate, l-glutamine, nonessential amino acids, penicillin, streptomycin, and Fungizone.
Analysis of Cholesterol and Triglyceride Synthesis by Thin-layer Chromatography
Acly+/+ and Acly+/− fibroblasts were grown to 90% confluency in 100-mm Petri dishes. Cells were then labeled with 50 µCi of [1(3)-3H]glycerol (Amersham Biosciences), 10 µCi of [1,5-14C]citric acid (American Radiolabeled Chemicals, St. Louis, MO), or 25 µCi of [1-14C]acetic acid (Amersham Biosciences). Primary hepatocytes were labeled with 10 µCi of [1(3)-3H]glycerol. Lipids from fibroblasts and hepatocytes were extracted with hexane:isopropyl alcohol (3:2 (v/v)), dried under nitrogen, and resuspended in 100 µl of chloroform. Triglycerides and free cholesterol were separated on thin-layer chromatography silica plates (60 Å) in hexane:ethyl ether:acetic acid (80:20:1 (v/v/v)). Samples were assayed in duplicates (40 µl/lane). After lipid extraction, cell protein was quantified with the Bradford assay (Bio-Rad).
Blood samples were taken from the retroorbital sinus under anesthesia (2.5% avertin). Plasma cholesterol, triglycerides, and free fatty acids were measured with enzymatic assays (Abbott Spectrum (Abbott, Abbott Park, IL), triglycerides/glycerol blanked (Roche/Hitachi, Indianapolis, IN), and free fatty acids half-micro test (Roche Applied Science), respectively). Liver lipids were extracted (19), and the amounts of cholesterol and triglycerides were measured (20). Before harvesting of liver tissue, mice were anesthetized and perfused with saline.
RNA Isolation and Northern Blot Analysis
Total RNA was isolated from 50 to 150 mg of mouse tissue or from fibroblasts grown on 100-mm Petri dishes with Tri-Reagent (Sigma). Total RNA (25 µg) was separated by electrophoresis on a 1% agarose/formaldehyde gel and transferred onto a Nytran SuPerCharge membrane (Schleicher & Schuell). A mouse multiple-tissue poly(A)+ RNA blot and a mouse embryo poly(A)+ RNA blot (Clontech, Palo Alto, CA) were used to determine the tissue pattern of Acly expression in adult mice and to examine the temporal expression of Acly during embryogenesis. Bands on Northern blots were visualized by autoradiography (Hyperfilm ECL, Amersham Biosciences) and quantified by densitometry (Molecular Imager FX, Bio-Rad). We used the same Acly cDNA probe that has been described previously (11). A β-galactosidase cDNA was amplified by reverse transcriptase-PCR from Acly+/− mouse liver RNA with primers 5′-TTTTCGATGAGCGTGGTGGTTATGC-3′ and 5′-GCGCGTACATCGGGCAAATAATATC-3′; an Asc1 cDNA was amplified with oligonucleotides 5′-TGCTGAGGACCCACTCTTCATCTTG-3′ and 5′-AAGCAGAACCAGGTTTCATGGGTGT-3′. A glyceraldehyde-3-phosphate dehydrogenase (Gapdh) probe and an 18 S cDNA were used as controls for RNA integrity and loading. [α-32P]dCTP-labeled cDNA probes were prepared with All-in-One random prime labeling mixture (Sigma). Standard prehybridization, hybridization, and washing procedures were used (21).
Quantitative Real-time PCR
Real-time PCR conditions and oligonucleotides to measure mRNA levels for Acly, Acs2, acetyl-CoA carboxylase, fatty acid synthase, stearoyl-CoA desaturase 1, glycerol-3-phosphate-acyltransferase, long-chain fatty acyl elongase, glucose-6-phosphatase, glucokinase, glucose-6-phosphate dehydrogenase, malic enzyme, 3-hydroxy-3-methylglutaryl-CoA synthase, and 3-hydroxy-3-methylglutaryl-CoA reductase were described previously (22). The level of Gapdh mRNA was used as a control.
Western Blot Analysis
Fresh mouse tissues (liver, white adipose tissue, kidney, and brain) (0.5–1 g), or embryonic fibroblasts (grown to 80–90% confluency in 100-mm Petri dishes) were homogenized in RIPA buffer containing a mixture of protease inhibitors (Complete Mini EDTA-free, Roche Applied Science). Samples were incubated on ice for 30 min and centrifuged at 14,000 rpm for 10 min at 4 °C. The protein content of the supernatant fluid was determined with the Bradford assay (Bio-Rad). Denatured proteins (60–100 µg) were loaded onto precast 4–15% polyacrylamide gels (Bio-Rad). After electrotransfer onto polyvinylidene difluoride membrane (Amersham Biosciences), blots were blocked with phosphate-buffered saline containing 0.1% Tween and 3% bovine serum albumin for 4 h at 4 °C and incubated for 1 h at room temperature with a rabbit antibody against rat Acly (14) at a dilution of 1:500 in phosphate-buffered saline containing 0.10% bovine serum albumin. Blots were then incubated with a horseradish peroxidase-linked donkey anti-rabbit IgG antibody (Amersham Biosciences) at a dilution of 1:50,000 in phosphate-buffered saline containing 0.1% bovine serum albumin for 45 min at room temperature. Antibody binding was detected with the ECL Plus Western blotting kit (Amersham Biosciences); the blots were exposed to Hyperfilm ECL (Amersham Biosciences) and quantified by densitometry as described above.
Immunohistochemistry and β-Galactosidase Staining
Mice were anesthetized with avertin and perfusion-fixed with 4% paraformaldehyde. Tissues were harvested and fixed in 10% formalin for 4 h at 4 °C, immersed in 30% glucose for ~16 h at 4 °C, and frozen in O.C.T. (Sakura Finetek, Torrance, CA) for cryostat sectioning. β-Galactosidase activity was assessed by incubating 10-µm thick sections with a staining solution containing 1.3 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) (Invitrogen) for 16 h at 37 °C (23). After staining, slides were counterstained with Eosin Y (Sigma). In some experiments, β-galactosidase staining was followed by immunohistochemistry. The following primary antibodies were used: rabbit anti-human glial fibrillary acidic protein (Sigma, 1:1,000), mouse monoclonal anti-neuronal nuclei (Chemicon, Temecula, CA, 1:1,000), goat anti-human choline acetyltransferase (Chemicon, 1:1,000), and goat anti-rat vesicular acetylcholine transporter (Chemicon, 1:5,000). Sections were subsequently incubated with the appropriate biotinylated secondary antibody (anti-rabbit or anti-goat IgG), and antigen-antibody complexes were detected after incubation with horseradish peroxidase and diaminobenzidine substrate (ABC kit, Vector Laboratories, Burlingame, CA). Sections were examined and photographed on an Eclipse 6600 microscope (Nikon, Japan) equipped with a Spot RT Slider digital camera (Diagnostic Instruments, Burlingame, CA).
RESULTS
The insertional mutation in the first intron of Acly introduced a new EcoRV site, making it possible to use Southern blots to distinguish between a 29.2-kilobase (kb) EcoRV fragment in the wild-type allele and a shorter (~6.2-kb) EcoRV fragment in the mutant allele (Fig. 1A). The mutation results in the production of a fusion transcript containing sequences from exon 1 joined to βgeo (Fig. 1B). There is little doubt that the insertional mutation inactivated Acly. Acly mRNA levels were decreased by ~50% in Acly+/− mice (Fig. 1B), as were Acly protein levels (Fig. 1C).
Fig. 1. Insertional mutation in Acly.
A, schematic of the insertional mutation in Acly. Cell lines and mice were genotyped by Southern blot with a 5′ flanking probe. Black boxes represent exons. SA, splice acceptor; SV40pA, simian virus poly adenylation signal sequence. B, Northern blot of total RNA from liver, heart, white adipose tissue (WAT), kidney, and brain of Acly+/+ and Acly+/− mice. Three probes were used: an Acly cDNA, a β-galactosidase cDNA, and an 18S cDNA (for normalization). C, Western blot of protein extracts from liver, WAT, kidney, and brain of Acly+/+ and Acly+/− mice with an Acly-specific antiserum.
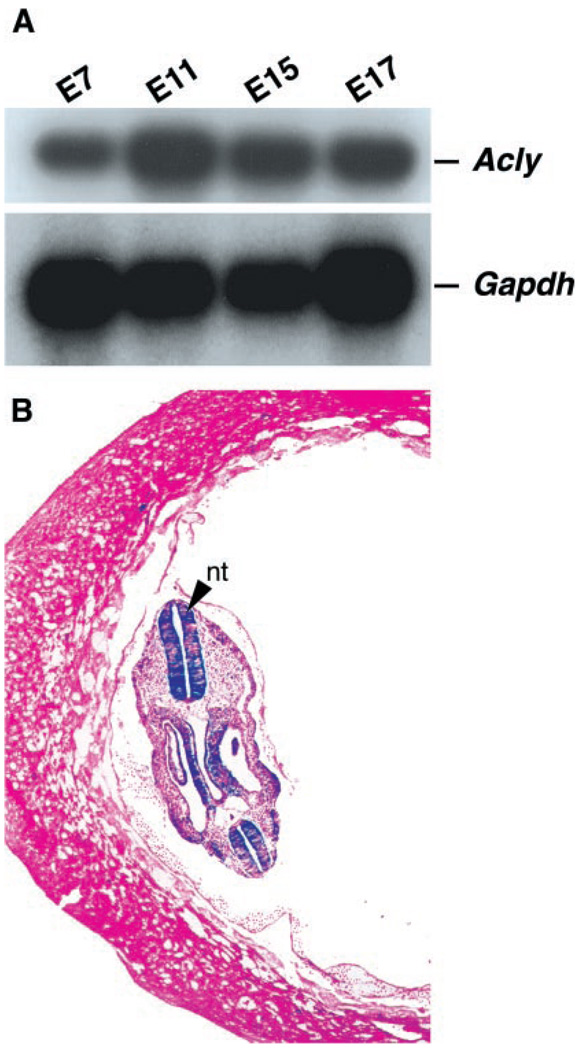
Acly+/− mice were intercrossed in an attempt to obtain homozygous knockout mice. However, genotyping of more than 60 litters did not yield any homozygous mice. Moreover, we genotyped more than 80 embryos from 8.5 to 17.5 dpc, and found no homozygous knockout embryos. Acly is expressed at high levels throughout embryonic development (Fig. 2A), and β-galactosidase staining revealed that Acly is expressed at high levels in the neural tube at 8.5 dpc (Fig. 2B). We concluded that Acly expression in embryos is required for development.
Fig. 2. Expression of Acly during development.
A, mouse embryo poly(A)+ RNA blot demonstrating that Acly is expressed throughout development. B, β-galactosidase staining of an 8.5-dpc Acly+/− embryo showing very high levels of Acly expression in the neural tube (nt). Original magnification, ×4.
We predicted that Acly+/− mice would have reduced body weight and lower plasma lipid levels as a consequence of a reduced capability for lipid synthesis. However, the levels of cholesterol, triglycerides, and free fatty acids in the plasma were no different in chow-fed Acly+/+ and Acly+/− mice (Table I). Moreover, feeding the mice a Western diet did not yield any significant difference in plasma lipid levels (Table I) or in hepatic cholesterol and triglyceride levels (not shown). Body weights of Acly+/+ and Acly+/− mice were virtually identical (Table I).
Table I.
Plasma levels of cholesterol, triglycerides, and free fatty acids (FFA) in Acly+/+ and Acly+/− mice fed a Western diet
| Mice were 3 weeks old at baseline. | ||||
|---|---|---|---|---|
| Baseline | 1 month | |||
| +/+ (n = 18) | +/− (n = 14) | +/+ (n = 18) | +/− (n = 14) | |
| Body weight (g) | 20.6 ± 0.63 | 19.1 ± 1.07 | 33.0 ± 0.70 | 31.1 ± 1.13 |
| Cholesterol (mg/dl) | 83.4 ± 3.41 | 82.1 ± 3.61 | 148 ± 9.28 | 137 ± 8.34 |
| Triglycerides (mg/dl) | 67.4 ± 18.4 | 52.4 ± 14.6 | 129 ± 18.7 | 113 ± 14.2 |
| FFA (µM) | 411 ± 33.7 | 428 ± 70.6 | 688 ± 78.7 | 668 ± 106 |
We suspected that Acly+/− mice might exhibit reduced weight gain and lower plasma lipid levels when fed a high-carbohydrate diet, which results in high levels of de novo lipogenesis in the liver. Contrary to our hypothesis, however, body weights and plasma levels of cholesterol, triglycerides, free fatty acids, and glucose were similar in Acly+/+ and Acly+/− mice over 3 months of observation (Table II). Also, feeding the carbohydrate-rich diet to Acly+/− mice was not associated with reduced liver stores of cholesterol and triglycerides (Table III).
Table II.
Plasma levels of cholesterol, triglycerides, and free fatty acids (FFA) in Acly+/+ and Acly+/− mice fed a fat-free, carbohydrate-rich diet
| Mice were 3 weeks old at baseline. | ||||
|---|---|---|---|---|
| Baseline | 1 month | |||
| +/+ (n = 8) | +/− (n = 8) | +/+ (n = 8) | +/− (n = 8) | |
| Body weight (g) | 21.2 ± 0.80 | 18.5 ± 1.26 | 29.2 ± 1.24 | 26.4 ± 1.31 |
| Cholesterol (mg/dl) | 95.3 ± 5.00 | 85.3 ± 3.88 | 83.3 ± 16.4 | 78.8 ± 3.78 |
| Triglycerides (mg/dl) | 49.8 ± 8.25 | 42.6 ± 6.21 | 12.8 ± 2.37 | 14.1 ± 2.55 |
| FFA (µM) | 435 ± 50.1 | 428 ± 70.7 | 413 ± 61.4 | 318 ± 39.7 |
Table III.
Hepatic cholesterol and triglyceride contents in Acly+/+ and Acly+/− mice fed a fat-free, carbohydrate-rich diet
| Mice were 3 weeks old at baseline. | ||||
|---|---|---|---|---|
| Baseline | 3 months | |||
| +/+ (n = 6) | +/− (n = 14) | +/+ (n = 6) | +/− (n = 14) | |
| Cholesterol (mg/g protein) | 7.88 ± 1.16 | 5.97 ± 0.77 | 32.3 ± 17.0 | 49.1 ± 7.21 |
| Triglycerides (mg/g protein) | 395 ± 39.0 | 361 ± 19.1 | 702 ± 207 | 628 ± 53.2 |
We also examined lipid synthesis in primary fibroblasts from Acly+/+ and Acly+/− embryos. As expected, Acly mRNA (Fig. 3A) and protein levels (Fig. 3B) were reduced by ~50% in heterozygous cells. However, the amount of cholesterol synthesis in Acly+/+ and Acly+/− fibroblasts was similar, as judged by [14C]citrate metabolic labeling experiments (Fig. 3C). It was not possible to assess the impact of reduced Acly expression on triglyceride synthesis, because very little [14C]citrate was incorporated into triglycerides. However, Acly+/+ and Acly+/− fibroblasts displayed similar amounts of triglyceride synthesis in [3H]glycerol labeling experiments (Fig. 3C). Similarly, cholesterol and triglyceride synthesis rates were entirely normal in primary hepatocytes from Acly+/− mice (Fig. 4).
Fig. 3. Acly mRNA, protein, and activity levels in Acly+/+ and Acly+/− primary fibroblasts.
A, Northern blot of total RNA from embryonic fibroblasts. Three probes were used: an Acly cDNA, an Acs1 cDNA, and an 18S cDNA (for normalization). B, Western blot of protein extracts from embryonic fibroblasts with an Acly-specific antiserum. C, cholesterol and triglyceride synthesis in embryonic fibroblasts. Cholesterol and triglyceride synthesis rates in Acly+/− cells are expressed as a percentage of those in Acly+/+ cells. The levels of incorporation into cholesterol in wild-type cells were: 2,289 cpm/mg of protein for [1,5-14C]citric acid and 108,626 cpm/mg of protein for [1-14C]acetic acid. The levels of incorporation into triglycerides in wild-type cells were: 34,560 cpm/mg of protein for [1(3)-3H]glycerol and 26,882 cpm/mg of protein for [1-14C]acetic acid. Data represent the average of duplicate determinations, and each experiment was repeated twice. D, relative amount of mRNAs coding for a variety of enzymes involved in triglyceride, cholesterol, and glucose metabolism in Acly+/+ and Acly+/− embryonic fibroblasts. Total RNA was prepared from three cell lines per genotype. Equal aliquots were pooled according to genotype and analyzed by quantitative reverse transcription- PCR. Gapdh was used as a control in this experiment, although a cyclophilin control yielded virtually identical results. Values represent the relative amount of mRNA in relation to that measured in Acly+/+ fibroblasts. Acc, acetyl-CoA carboxylase; Fas, fatty acid synthase; Scd1, stearoyl-CoA desaturase 1; Gpat, glycerol-3-phosphate-acyltransferase; Lce, long-chain fatty acyl elongase; G6pc, glucose-6-phosphatase, catalytic; Gck, glucokinase; G6pd, glucose-6-phosphate dehydrogenase; Me, malic enzyme; Hmgcs, 3-hydroxy-3-methylglutaryl-CoA synthase; Hmgcr, 3-hydroxy-3-methylglutaryl-CoA reductase.
Fig. 4. Levels of cholesterol and triglyceride synthesis in primary hepatocytes from Acly+/+ and Acly+/− mice.
Cholesterol and triglyceride synthesis rates in Acly+/− cells are expressed as a percentage of those in Acly+/+ cells. The levels of incorporation of [1(3)-3H]glycerol into cholesterol and triglycerides in wild-type cells were 583 ± 33 cpm/µg of protein and 56,085 ± 2,919 cpm/µg of protein, respectively. Data represent the average of triplicate determinations, and each experiment was repeated twice.
The fact that the Acly+/− mice and Acly+/− cells did not manifest detectable perturbations in lipid levels or lipid biosynthesis suggested that half-normal levels of Acly have little impact on cellular lipid homeostasis. Indeed, this appeared to be the case. As judged by quantitative reverse transcriptase-PCR, Acly mRNA levels were reduced by 50% in Acly+/− cells, but the expression of a host of other enzymes involved in triglyceride, cholesterol, and glucose metabolism was not perturbed (Figs. 3D and 5A). Of note, the half-normal levels of Acly expression did not lead to an up-regulation of the expression of Asc1, the other acetyl-CoA-synthesizing enzyme in the cytosol (Figs. 3A and 5B). In keeping with the latter finding, the synthesis of cholesterol and triglycerides from [14C]acetate (a measure of Acs1 activity (2)) was identical in Acly+/+ and Acly+/− cells (Fig. 3C).
Fig. 5. Expression of Acly, Acs1, and lipid biosynthetic genes in liver from Acly+/+ and Acly+/− mice.
A, relative amount of mRNAs coding for a variety of enzymes involved in triglyceride, cholesterol, and glucose metabolism in the livers of Acly+/+ and Acly+/− mice. Reverse transcriptase-PCR was performed as described in the legend to Fig. 3D. B, Northern blot of total RNA from liver. Three probes were used: Acly cDNA, Acs1 cDNA, and 18S cDNA (for normalization).
Northern blots showed that Acly is expressed in multiple tissues (Fig. 6A). To explore the cell-type specific expression patterns, sections of tissues from Acly+/− mice were stained for β-galactosidase expression. Strong β-galactosidase expression was identified in multiple cell types, including Leydig cells in testis (Fig. 6B) and renal tubular cells (Fig. 6C). In the livers of chow-fed mice, β-galactosidase staining was largely confined to the periportal hepatocytes (Fig. 6D). In mice fed a fat-free, carbohydrate-rich diet, β-galactosidase staining of the liver was much more widespread and more intense (Fig. 6E), reflecting increased levels of Acly expression (Fig. 6G). On the Western diet, β-galactosidase staining of the liver was only modestly increased, compared with the chow diet (Fig. 6F).
Fig. 6. Tissue pattern of Acly expression in adult mice and regulation of Acly expression in liver by diet.
A, mouse multiple-tissue poly(A)+ RNA blot showing Acly expression in the tissues of wild-type mice. A Gapdh cDNA was used for normalization. B, β-galactosidase staining of testis from a 4-week-old Acly+/− mouse, showing intense staining in Leydig cells. Original magnification, ×20. At higher power, less-intense staining of the Sertoli cells was also apparent. The same pattern of cellular expression was observed in testis from 7- and 36-week-old mice. C, β-galactosidase staining of a section of Acly+/− kidney. Original magnification, ×10. Arrows indicate tubules. D–F, β-galactosidase staining of liver from Acly+/− adult mice fed a chow diet (D), a fat-free, carbohydrate-rich diet (E), or a Western diet (F). Original magnification, ×10. G, Northern blot analysis of total RNA from the liver of Acly+/+ mice on the three different diets. Two probes were used: Acly cDNA and 18S cDNA (for normalization).
β-Galactosidase expression was intense throughout the brains of 15.5 dpc Acly+/− embryos (Fig. 7A) and 1-day-old Acly+/− pups (Fig. 7B), consistent with the presumed role of Acly in the de novo lipogenesis required for myelin formation (14). We suspected that Acly expression in the brain would decrease in older animals, mirroring a reduced requirement for lipid synthesis (24–28). This suspicion was confirmed. In 3-week-old pups, β-galactosidase staining was less intense than in the 1-day-old mice, although staining was still observed in the hippocampus, thalamus, medulla, and the white matter of cerebellum (Fig. 7C). In 6-month-old mice, Acly was expressed at far lower levels in the brain than in the younger mice (Fig. 7D). In the adult mice, β-galactosidase was expressed in glial cells (Fig. 8A) and in neurons (Fig. 8B).
Fig. 7. Evolution of Acly expression in the maturing brain.
A–D, β-galactosidase staining of sagittal sections of brains from a 15-dpc Acly+/− embryo (A), 1-day-old Acly+/− mouse (B), a 3-week-old Acly+/− mouse (C), and a 6-month-old Acly+/− mouse (D). Original magnifications, ×2 (A and B) and ×1 (C and D). Arrows indicate hippocampus (h), granular layer of the cerebellum (gr), nuclei of pons (p), and facial nucleus (f).
Fig. 8. Acly is expressed in glial cells and in neurons in the adult mouse nervous system.
A and B, β-galactosidase and immunohistochemical staining of adult Acly+/− thalamus (A) and cortex (B). Specific antibodies against glial fibrillary acidic protein (A) and neuronal nuclei (B) were used. C, β-galactosidase staining of adult Acly+/− spinal cord (cervical). Arrow indicates ventral horn. D, immunohistochemical staining of adult Acly+/− spinal cord (cervical) with a specific antibody against neuronal nuclei. Original magnifications, ×40 (A and B) and ×4 (C and D).
Although the overall level of β-galactosidase expression was much lower in adult brains than in embryos, discrete regions of the adult brain showed very intense β-galactosidase staining. For example, relatively high levels of Acly expression persisted in hippocampus, the granular layer of the cerebellum, and other specific regions within the cerebellum, pons, medulla, and spinal cord (Fig. 7D). The facial nucleus in the medulla stained intensely for β-galactosidase (Fig. 7D), as did the gray matter of the spinal cord, particularly the ventral horns (Fig. 8C).
Initially, we hypothesized that the regions of the adult brain that stained highly for β-galactosidase were simply a subset of neurons with a high metabolic requirement for new lipid synthesis. Later, however, we remembered that acetyl-CoA is a key substrate for choline acetyltransferase in the synthesis of acetylcholine, and we became attracted to the hypothesis that those cells might be cholinergic neurons. To test this hypothesis, we stained histological sections for Acly expression (i.e. β-galactosidase) and two markers of cholinergic neurons, choline acetyltransferase and vesicular acetylcholine transporter. Indeed, Acly (Fig. 9A) and choline acetyltransferase (Fig. 9B) displayed strikingly overlapping patterns of expression in the hippocampus, as did Acly and vesicular acetylcholine transporter (Fig. 9C). Acly and vesicular acetylcholine transporter also colocalized in Purkinje cells of the cerebellum (Fig. 9E) and in several cerebellar nuclei (Fig. 9F). Finally, Acly was expressed very highly in vesicular acetylcholine transporter-expressing neurons in the facial nucleus within the medulla (Fig. 9H).
Fig. 9. Acly is expressed in cholinergic neurons in the adult mouse brain.
A, β-galactosidase staining of a sagittal section of adult Acly+/− hippocampus. B, immunohistochemical staining of adult Acly+/− hippocampus with a specific antibody against choline acetyltransferase. C–H, β-galactosidase and immunohistochemical staining of adult Acly+/− hippocampus (C), pons nuclei (D), cerebellum (E), cerebellar nuclei (F), and medulla (facial nucleus) (G and H). Specific antibodies against vesicular acetylcholine transporter (C, E, and H), choline acetyltransferase (F), or neuronal nuclei (D and G) were used. Original magnifications, ×10 (A–C and E), ×20 (F), and ×40 (D, G, and H).
DISCUSSION
In the current study, we created Acly knockout mice in which a β-galactosidase marker is expressed from Acly regulatory sequences, with the goal of further elucidating the physiologic roles of Acly in mammals. Acly is clearly required for embryonic development: no viable homozygous Acly knockout embryos were identified. The other enzyme capable of producing acetyl-CoA in the cytosol, Acs1, was evidently not sufficient to support development. Knockout mouse experiments frequently offer up surprises, but the lethal developmental defect in the Acly knockout homozygotes probably makes perfect sense, given the central role of Acly in energy metabolism. Acly is important for generating acetyl-CoA from glycolysis and, in particular, is required for obtaining acetyl-CoA from mitochondria. Acetyl-CoA can also be generated within mitochondria, either by pyruvate dehydrogenase or by the β-oxidation of fatty acids. To be used for lipid synthesis in the cytosol, however, acetyl-CoA must be exported from the mitochondria. To accomplish the transfer, mitochondrial acetyl-CoA condenses with oxaloacetate to form citrate, which is then transported to the cytosol, where it is used by Acly to create acetyl-CoA. Hence, the elimination of Acly effectively prevents the export of lipid building blocks from mitochondria.
The fact that lipid homeostasis appeared to be totally unaffected by half-normal levels of Acly was unexpected. One possibility is that Acly is synthesized in abundant quantities and that half-normal levels of the enzyme are simply not limiting for acetyl-CoA production. Another possibility is suggested by the intrinsic enzymatic properties of Acly. Acly activity in mammals and lower organisms is allosterically regulated by one of its cosubstrates, citrate (29). It is therefore likely that even evanescent increases in intracellular citrate levels would stimulate enzyme activity and increase the conversion of citrate into acetyl-CoA, thereby preventing measurable metabolic effects (e.g. reduced lipid synthesis or perturbed expression of SREBP-regulated genes).
The β-galactosidase marker in the mutant Acly allele provides novel information on the cell type-specific expression of Acly. For example, the high levels of Acly expression in the testis revealed by Northern blot (14) are largely explained by high levels of β-galactosidase activity in Leydig cells. One could argue that this pattern of expression is not surprising, given the role of Leydig cells in the synthesis of a steroid hormone. On the other hand, we would maintain that this pattern was not entirely predictable a priori. Two other lipid biosynthetic genes that we have studied, phosphatidylserine synthase 2 and 1-acyl-sn-glycerol-3-phosphate acyltransferase 4, are expressed highly in testis but mainly in Sertoli cells (23),2 which likely have a role in the delivery of lipid nutrients to the germ cells.
The expression of β-galactosidase in Acly+/− mice faithfully mirrored Acly expression. In mice fed a high-carbohydrate diet, high levels of Acly expression were detected by Northern blot in the liver, and the hepatocytes of those livers manifested a high level of β-galactosidase activity. Interestingly, Acly was clearly expressed predominantly in a periportal distribution. This study, as far as we are aware, is the first to allow the direct visualization of different levels of de novo lipogenesis in distinct zones of the liver.
The up-regulation of hepatic Acly expression on a high-carbohydrate diet is controlled at the transcriptional level by SREBP-1 (3, 11, 12). (In transgenic mice expressing a truncated form of SREBP-1c, Acly was one of the most up-regulated of all of the SREBP-controlled genes (11).) We suggest that the β-galactosidase marker in the mutant Acly allele could be a useful tool for visualizing SREBP-1c gene regulation in vivo in response to changing metabolic conditions. As far as we are aware, the mutant Acly allele reported here is the only such “SREBP activity marker” currently available.
β-Galactosidase staining proved to be useful in understanding the role of Acly in the brain. First, β-galactosidase expression was high in the developing brain and was much lower in older mice. That temporal expression pattern undoubtedly reflects different rates of de novo lipogenesis at different stages of development (26–28). Acly was expressed in both glial cells and neurons. Second, in adult mice, we identified very high levels of β-galactosidase expression in cholinergic neurons, particularly in the brain stem and spinal cord. Whereas one might conceivably argue that this β-galactosidase expression reflects some peculiar requirement of cholinergic neurons for high levels of lipid synthesis, we think it is more likely that the very intense staining of cholinergic neurons reflects a role for Acly in acetylcholine synthesis. This conclusion is not farfetched: acetyl-CoA is a cosubstrate for choline acetyltransferase, which produces acetylcholine. Moreover, citrate and hydroxycitrate stimulate and inhibit, respectively, the in vitro synthesis of acetylcholine in rat caudate nuclei (30) and perfused rat phrenic nerve (31). Also, acetyl-CoA levels in cells have been reported to correlate directly with the rate of in vitro acetylcholine synthesis in rat caudate nuclei (32) and SN56 hybrid cholinergic cells (33).
It has been suggested that Asc1 could play a role in the regeneration of acetyl-CoA from acetate that is taken up after the breakdown of acetylcholine by acetylcholinesterase in nerve synapses (34). Based on published Northern blot experiments, the level of Asc1 expression in the brain is low (2, 35). However, if Asc1 is truly important for acetate metabolism in cholinergic neurons, we would predict that Asc1, like Acly, might be expressed highly in the cholinergic neurons of the brain stem and spinal cord. Finally, although both Acly and Asc1 expression are unequivocally regulated by SREBP-1c (2, 11), we speculate that the genetic regulation of Acly and Asc1 expression in cholinergic neurons might be more complex, taking into account the additional requirement of those cells for acetyl-CoA for acetylcholine synthesis.
In summary, our data show that Acly is critically important for embryonic development of the mouse, but there were no defects in lipid synthesis in the setting of half-normal levels of Acly. Our studies clearly support a major role for Acly in lipid synthesis, and suggest that Acly expression in the adult brain is important for the production of acetyl-CoA for acetylcholine synthesis.
Acknowledgments
We thank Dr. Nabil A. Elshourbagy (SmithKline Beecham, King of Prussia, PA) for the antibody against rat Acly; Dr. Lorenzo Arnaboldi for technical advice and lipid analyses; and S. Ordway and G. Howard for comments on the manuscript. We thank Dr. Jacquelyn J. Maher, Gene Lee, and Claudio Villanueva for help in preparing primary hepatocytes.
Footnotes
This work was supported by National Institutes of Health NHLBI Program in Genomics Applications Grants “BayGenomics” HL66621, HL66600, and HL66590.
The abbreviations used are: Acly, ATP-citrate lyase; Acs1, acetyl-CoA synthetase 1; SREBP, sterol regulatory element-binding protein; βgeo, a fusion of β-galactosidase and neomycin phosphotransferase II; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; dpc, days post-coitus.
A. P. Beigneux, unpublished observations.
REFERENCES
- 1.Knowles SE, Jarrett IG, Filsell OH, Ballard FJ. Biochem. J. 1974;142:401–411. doi: 10.1042/bj1420401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luong A, Hannah VC, Brown MS, Goldstein JL. J. Biol. Chem. 2000;275:26458–26466. doi: 10.1074/jbc.M004160200. [DOI] [PubMed] [Google Scholar]
- 3.Liang G, Yang J, Horton JD, Hammer RE, Goldstein JL, Brown MS. J. Biol. Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan AC, Triscari J, Hamilton JG, Miller ON, Wheatley VR. Lipids. 1974;9:121–128. doi: 10.1007/BF02532136. [DOI] [PubMed] [Google Scholar]
- 5.Bray GA, Greenway FL. Clin. Endocrinol. Metab. 1976;5:455–479. doi: 10.1016/s0300-595x(76)80031-x. [DOI] [PubMed] [Google Scholar]
- 6.Chee H, Romsos DR, Leveille GA. J. Nutr. 1977;107:112–119. doi: 10.1093/jn/107.1.112. [DOI] [PubMed] [Google Scholar]
- 7.Greenwood MR, Cleary MP, Gruen R, Blase D, Stern JS, Triscari J, Sullivan AC. Am. J. Physiol. 1981;240:E72–E78. doi: 10.1152/ajpendo.1981.240.1.E72. [DOI] [PubMed] [Google Scholar]
- 8.Saxty BA, Novelli R, Dolle RE, Kruse LI, Reid DG, Camilleri P, Wells TN. Eur. J. Biochem. 1991;202:889–896. doi: 10.1111/j.1432-1033.1991.tb16448.x. [DOI] [PubMed] [Google Scholar]
- 9.Pearce NJ, Yates JW, Berkhout TA, Jackson B, Tew D, Boyd H, Camilleri P, Sweeney P, Gribble AD, Shaw A, Groot PH. Biochem. J. 1998;334:113–119. doi: 10.1042/bj3340113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leonhardt M, Hrupka B, Langhans W. Physiol. Behav. 2001;74:191–196. doi: 10.1016/s0031-9384(01)00547-9. [DOI] [PubMed] [Google Scholar]
- 11.Shimomura I, Shimano H, Korn BS, Bashmakov Y, Horton JD. J. Biol. Chem. 1998;273:35299–35306. doi: 10.1074/jbc.273.52.35299. [DOI] [PubMed] [Google Scholar]
- 12.Shimano H, Yahagi N, Amemiya-Kudo M, Hasty AH, Osuga J, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, Harada K, Gotoda T, Ishibashi S, Yamada N. J. Biol. Chem. 1999;274:35832–35839. doi: 10.1074/jbc.274.50.35832. [DOI] [PubMed] [Google Scholar]
- 13.Dietschy JM, Turley SD. Curr. Opin. Lipidol. 2001;12:105–112. doi: 10.1097/00041433-200104000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Elshourbagy NA, Near JC, Kmetz PJ, Sathe GM, Southan C, Strickler JE, Gross M, Young JF, Wells TN, Groot PH. J. Biol. Chem. 1990;265:1430–1435. [PubMed] [Google Scholar]
- 15.Foster DW, Srere PA. J. Biol. Chem. 1968;243:1926–1930. [PubMed] [Google Scholar]
- 16.Melnick JZ, Srere PA, Elshourbagy NA, Moe OW, Preisig PA, Alpern RJ. J. Clin. Invest. 1996;98:2381–2387. doi: 10.1172/JCI119051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stryke D, Kawamoto M, Huang CC, Johns SJ, King LA, Harper CA, Meng EC, Lee RE, Yee A, L’Italien L, Chuang PT, Young SG, Skarnes WC, Babbitt PC, Ferrin TE. Nucleic Acids Res. 2003;31:278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Townley DJ, Avery BJ, Rosen B, Skarnes WC. Genome Res. 1997;7:293–298. doi: 10.1101/gr.7.3.293. [DOI] [PubMed] [Google Scholar]
- 19.Björkegren J, Beigneux A, Bergo MO, Maher JJ, Young SG. J. Biol. Chem. 2002;277:5476–5483. doi: 10.1074/jbc.M108514200. [DOI] [PubMed] [Google Scholar]
- 20.Shimano H, Horton JD, Hammer RE, Shimomura I, Brown MS, Goldstein JL. J. Clin. Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beigneux A, Withycombe SK, Digits JA, Tschantz WR, Weinbaum CA, Griffey SM, Bergo M, Casey PJ, Young SG. J. Biol. Chem. 2002;277:38358–38363. doi: 10.1074/jbc.M205183200. [DOI] [PubMed] [Google Scholar]
- 22.Moon YA, Horton JD. J. Biol. Chem. 2003;278:7335–7343. doi: 10.1074/jbc.M211684200. [DOI] [PubMed] [Google Scholar]
- 23.Bergo MO, Gavino BJ, Steenbergen R, Sturbois B, Parlow AF, Sanan DA, Skarnes WC, Vance JE, Young SG. J. Biol. Chem. 2002;277:47701–47708. doi: 10.1074/jbc.M207734200. [DOI] [PubMed] [Google Scholar]
- 24.Spady DK, Dietschy JM. J. Lipid Res. 1983;24:303–315. [PubMed] [Google Scholar]
- 25.Dietschy JM, Kita T, Suckling KE, Goldstein JL, Brown MS. J. Lipid Res. 1983;24:469–480. [PubMed] [Google Scholar]
- 26.Jurevics H, Morell P. J. Neurochem. 1995;64:895–901. doi: 10.1046/j.1471-4159.1995.64020895.x. [DOI] [PubMed] [Google Scholar]
- 27.Jurevics HA, Kidwai FZ, Morell P. J. Lipid Res. 1997;38:723–733. [PubMed] [Google Scholar]
- 28.Turley SD, Burns DK, Dietschy JM. Am. J. Physiol. 1998;274:E1099–E1105. doi: 10.1152/ajpendo.1998.274.6.E1099. [DOI] [PubMed] [Google Scholar]
- 29.Barnum TE. Enzyme Handbook. New York: Springer-Verlag; 1969. [Google Scholar]
- 30.Ricny J, Tucek S. J. Neurochem. 1982;39:668–673. doi: 10.1111/j.1471-4159.1982.tb07944.x. [DOI] [PubMed] [Google Scholar]
- 31.Endemann G, Brunengraber H. J. Biol. Chem. 1980;255:11091–11093. [PubMed] [Google Scholar]
- 32.Ricny J, Tucek S. Biochem. J. 1980;188:683–688. doi: 10.1042/bj1880683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szutowicz A, Jankowska A, Blusztajn JK, Tomaszewicz M. J. Neurosci. Res. 1999;57:131–136. doi: 10.1002/(SICI)1097-4547(19990701)57:1<131::AID-JNR14>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 34.Carroll PT. Brain Res. 1997;753:47–55. doi: 10.1016/s0006-8993(96)01485-0. [DOI] [PubMed] [Google Scholar]
- 35.Fujino T, Kondo J, Ishikawa M, Morikawa K, Yamamoto TT. J. Biol. Chem. 2001;276:11420–11426. doi: 10.1074/jbc.M008782200. [DOI] [PubMed] [Google Scholar]