Abstract
Soluble butane monooxygenase (sBMO), a three-component di-iron monooxygenase complex expressed by the C2–C9 alkane-utilizing bacterium Thauera butanivorans, was kinetically characterized by measuring substrate specificities for C1–C5 alkanes and product inhibition profiles. sBMO has high sequence homology with soluble methane monooxygenase (sMMO) and shares a similar substrate range, including gaseous and liquid alkanes, aromatics, alkenes and halogenated xenobiotics. Results indicated that butane was the preferred substrate (defined by kcat : Km ratios). Relative rates of oxidation for C1–C5 alkanes differed minimally, implying that substrate specificity is heavily influenced by differences in substrate Km values. The low micromolar Km for linear C2–C5 alkanes and the millimolar Km for methane demonstrate that sBMO is two to three orders of magnitude more specific for physiologically relevant substrates of T. butanivorans. Methanol, the product of methane oxidation and also a substrate itself, was found to have similar Km and kcat values to those of methane. This inability to kinetically discriminate between the C1 alkane and C1 alcohol is observed as a steady-state concentration of methanol during the two-step oxidation of methane to formaldehyde by sBMO. Unlike methanol, alcohols with chain length C2–C5 do not compete effectively with their respective alkane substrates. Results from product inhibition experiments suggest that the geometry of the active site is optimized for linear molecules four to five carbons in length and is influenced by the regulatory protein component B (butane monooxygenase regulatory component; BMOB). The data suggest that alkane oxidation by sBMO is highly specialized for the turnover of C3–C5 alkanes and the release of their respective alcohol products. Additionally, sBMO is particularly efficient at preventing methane oxidation during growth on linear alkanes ≥C2, despite its high sequence homology with sMMO. These results represent, to the best of our knowledge, the first kinetic in vitro characterization of the closest known homologue of sMMO.
INTRODUCTION
Aliphatic alkanes are used as a sole source of carbon and energy by a diverse range of aerobic prokaryotes (Shennan, 2006). To initiate growth on C2–C9 alkanes, the Gram-negative β-proteobacterium Thauera butanivorans, formerly called ‘Pseudomonas butanovora’ (Dubbels et al., 2009), expresses a carboxylate-bridged non-haem di-iron monooxygenase, commonly referred to as soluble butane monooxygenase (sBMO) (Sluis et al., 2002; Takahashi et al., 1980). sBMO belongs to a family of bacterial multicomponent monooxygenases which includes soluble methane monooxygenases (sMMOs), phenol hydroxylases (PHs) and aromatic/alkene monooxygenases (TMOs) (Leahy et al., 2003; Sluis et al., 2002). Due to their unusually large substrate ranges, these powerful oxidizers are of particular interest for their potential in bioremediation (Enzien et al., 1994; Halsey et al., 2007; Parales et al., 2002; Smith & Dalton, 2004) and their ability to serve as industrial biocatalysts (Burton, 2003; Parales et al., 2002).
The expression of sBMO is induced by the presence of 1-butanol and butyraldehyde, and is repressed by lactate and succinate (Sayavedra-Soto et al., 2005). When T. butanivorans is grown on butane, sBMO oxidizes butane primarily to 1-butanol (>85 % terminal oxidation), which is then further oxidized to butyrate via butyraldehyde (Arp, 1999). Further metabolism of butyrate probably proceeds through butyryl-CoA prior to β-oxidation (Arp, 1999). The metabolic pathway of odd-chain or subterminally oxidized alkanes has not yet been fully characterized in T. butanivorans; however, it is known that the metabolism of odd- and even-chain alkane growth substrates is differentially controlled (Doughty et al., 2007). Although sBMO is capable of oxidizing methane to methanol (Halsey et al., 2006), T. butanivorans cannot assimilate C1 compounds into biomass.
sBMO is a three-component complex that was recently purified and found to have a similar architecture to that of sMMO: (i) a 250 kDa hydroxylase component (butane monooxygenase hydroxylase; BMOH) with (αβγ)2 subunit composition that, by analogy with sMMO, contains the di-iron active site in the α-subunit; (ii) a 40 kDa flavo-iron–sulfur-containing reductase (butane monooxygenase reductase; BMOR) that shuttles electrons from NADH to the active site of BMOH; and (iii) a 15 kDa regulatory protein (butane monooxygenase regulatory component; BMOB) that enhances the overall activity of the complex (Dubbels et al., 2007). Sequence analysis shows BMOH to be most closely related to sMMO (65, 42 and 38 % amino acid identity in the α-, β- and γ-subunits of BMOH, respectively) (Sluis et al., 2002). Additionally, sequence alignment and structural modelling show that all residues directly lining the active site cavity of BMOH are the same as those in the methane monooxygenase hydroxylase (MMOH). Despite the similarities, several biochemical observations indicate that sBMO represents a class distinct from the sMMO family: (i) methanol accumulation ceases during methane oxidation once ∼20–50 μM has been reached (Halsey et al., 2006), whereas sMMO can continue to accumulate methanol; (ii) sBMO is predominantly a terminal hydroxylator of intermediate-chain-length alkanes (Dubbels et al., 2007), whereas sMMO forms secondary alcohols (Froland et al., 1992); (iii) product regioselectivity is minimally altered by the presence of BMOB (Dubbels et al., 2007), unlike sMMO (Froland et al., 1992); (iv) catalase is required to maintain hydroxylase activity during steady-state turnover (Dubbels et al., 2007) but not to maintain sMMO activity; and (v) the use of peroxide in the ‘peroxide shunt’ mechanism of substrate oxidation is three orders of magnitude less efficient in sBMO than in sMMO (Dubbels et al., 2007).
The structural basis of the catalytic properties of these alkane monooxygenases is still poorly understood, most notably due to a lack of enzyme isolation and subsequent in vitro characterization (van Beilen & Funhoff, 2007). Even though crystal structures of apo-MMOH and product-bound MMOH have been reported (Rosenzweig et al., 1997; Sazinsky & Lippard, 2005), the fully buried active site does not appear to be accessible or large enough to accommodate known substrates such as naphthalene or biphenyl. A few mutational studies of BMOH and MMOH have provided insight into this field, however. Substitution of residue Gly113 in the α-subunit of BMOH with the equivalent residue of MMOHα (Asn) results in a shift of properties to those more characteristic of sMMO, such as increased methanol accumulation and an alteration in product regioselectivity from primary to secondary hydroxylation (Halsey et al., 2006). Alterations in the ‘leucine gate’ residue of MMOHα (Leu110) have shown that it is critical in defining the regioselectivity of sMMO (Borodina et al., 2007).
Given the wide substrate range of alkane monooxygenases, a comprehensive kinetic characterization of sBMO would help provide insights into the mechanism of substrate specificity. Here, we report a characterization of the substrate specificity for sBMO and show that, despite having the same active site residues as sMMO, sBMO is poorly suited for methane yet highly optimized for linear molecules four to five carbons in length. Since measurable substrate turnover is possible without BMOB, we were also able to directly observe a function for BMOB in catalysis using product inhibitors. Such types of experiments have not been possible with sMMO because substrate turnover is too slow without the methane monooxygenase regulatory component (MMOB).
METHODS
Chemicals.
Gaseous alkanes were of reagent grade. Methane was purchased from Airco. Ethane was purchased from Matheson. Propane and butane were purchased from Airgas. Pentane was purchased as reagent grade from VWR. NADH was purchased from Research Organics and residual ethanol was removed by repeated lyophilization in 25 mM PIPES, pH 7.2. Bovine liver catalase was purchased from Sigma. All other chemicals were obtained from Aldrich.
Bacterial cultivation and BMOH/BMOR purification.
Wild-type (WT) T. butanivorans was grown on butane as described previously (Dubbels et al., 2007) with the exception that 100 μM Fe3+-EDTA was used as an iron source instead of 500 μM FeSO4 . 7H2O. Mutant strain G113N, which has residue 113 of the α-subunit of BMOH altered from Gly to Asn (Halsey et al., 2006), was grown identically to WT except that 50 μM MnCl2 and 10 μM Na2CO3 were added to the medium to facilitate the downstream metabolism of secondary alcohols. WT BMOH and BMOR components of sBMO were purified as previously described (Dubbels et al., 2007). The G113N BMOH was purified identically to the WT hydroxylase. The alternative iron source yielded BMOH that exhibited much tighter elution profiles off the Sephacryl S-300 HR gel-filtration column due to significantly reduced self-aggregation, as determined by dynamic light scattering. Additionally, the iron content of BMOH, as measured by the ferrozine spectrophotometric assay (Percival, 1991), contained 2.1–2.4 irons per active site rather than the 1.4–1.8 reported previously (Dubbels et al., 2007). Typical preparations of the enzyme complex resulted in activities ranging from 400 to 700 nmol min−1 mg−1. Protein concentrations were determined by optical absorption at 280 nm using absorption coefficients of 2.2 and 0.56 ml mg−1 cm−1 for BMOH and BMOR, respectively.
Development of a recombinant BMOB expression system.
Purification of BMOH and BMOR from the native host yielded large quantities of highly active protein, and so no recombinant expression system was needed. However, purification of BMOB from the native host required several additional purification steps and the yields were not always satisfactory. We therefore chose to develop a recombinant expression system for BMOB that provided a simpler purification process with significantly increased yields.
To create the recombinant BMOB expression system, genomic and plasmid DNA were first isolated according to established protocols (Ausubel et al., 2003). Primers for the amplification of BMOB were as follows: bmobfNdeI, 5′-CAGGGGCAGACCATATGTCAAACGT-3′, and bmobrBamHI, 5′-CGCACCGGTGTGTGGATCCAAACCT-3′. Bases indicated in bold type are the restriction sites included for subsequent cloning into the expression vector. A standard PCR was carried out with the above primers with Taq (Promega). The PCR product was gel-purified with QIAEX II (Qiagen) and restricted with NdeI and BamHI (Promega). Gel purification of the restriction digest was performed as described above, and the isolated PCR product was ligated into NdeI- and BamHI-digested pT7-7 (Tabor & Richardson, 1985). The ligation reaction was transformed into chemically competent Escherichia coli JM109 (Sambrook et al., 1989). Plasmid DNA was isolated and the sequence of bmob was confirmed by DNA sequencing at the Center for Genome Research and Biocomputing at Oregon State University. The resulting plasmid, pBD400, was transformed into chemically competent E. coli BL21(DE3) (Novagen) for subsequent expression and purification.
Expression and purification of recombinant BMOB.
E. coli BL21(DE3) cells containing the pBD400 plasmid were grown in 3 l LB medium in the presence of 100 μg ampicillin ml−1 at 37 °C to OD600 0.8. Protein expression was stimulated by the addition of 1 mM IPTG. After 3 h further growth, cells were centrifuged at 5000 g for 20 min and stored at −70 °C. Typically, yields of 4–5 g cell paste l−1 were obtained.
All purification steps of recombinant BMOB (rBMOB) were performed at 4 °C. Frozen E. coli BL21(DE3) cell paste (15 g) was resuspended in 25 mM PIPES, pH 7.2, to a total volume of 40 ml containing 1000 U DNase I (Sigma). Cells were lysed by two passes through a French pressure cell disrupter at 52 000 Pa, and then centrifuged at 10 000 g for 20 min. The supernatant was carefully decanted, diluted to 60 ml and centrifuged at 150 000 g for 2 h. The resulting cell-free lysate was decanted, adjusted to pH 7.2, and loaded onto a DEAE-Sepharose FF column (90 mm×30 mm) pre-equilibrated with 25 mM PIPES, pH 7.2, at 2 ml min−1. The column was washed with five column volumes of the same buffer at a linear flow rate of 2 ml min−1, after which a 0–0.4 M KCl linear gradient was applied over four column volumes. Fractions containing rBMOB eluted between 0.2 and 0.3 M KCl. These fractions were pooled, repeatedly dialysed against 25 mM PIPES, pH 7.2, with 150 mM KCl, and concentrated to 3 ml total volume via ultrafiltration. The concentrate was then applied to Sephadex 75 FF gel-filtration columns (550 mm×25 mm) pre-equilibrated with the same buffer at a linear flow rate of 0.5 ml min−1. Fractions containing purified rBMOB were pooled, dialysed against 25 mM PIPES, pH 7.2, concentrated to 2 mM, flash-frozen in liquid nitrogen and stored at −70 °C. From 15 g of cells, 200–250 mg purified rBMOB was obtained. rBMOB was found to display the same enhancement of sBMO activity as native BMOB, alter the product distribution similarly, have the same mobility on SDS-PAGE, and have identical molecular mass as determined by MALDI-TOF MS. rBMOB also displayed the same partial dimerization through intermolecular disulfide linkage as native BMOB (Dubbels et al., 2007). Accordingly, all reactions in this study were performed with the recombinant form of BMOB. Concentrations were determined by A280 measurement using an absorption coefficient of 1.2 ml mg−1 cm−1.
Determination of methane Km.
Vials (7.7 ml) containing a stir bar and a 0.5 ml aliquot of 0.1 μM BMOH, 0.3 μM BMOB, 0.6 μM BMOR and 2400 U catalase ml−1 in 25 mM PIPES, pH 7.2, were crimp-sealed with butyl rubber septa. Methane was added directly as an overpressure. For higher concentrations, however, methane was used to refill the head space, to which additional methane and 20 % (v/v) oxygen were added as an overpressure. This mixture was allowed to equilibrate at 25 °C for 5 min with gentle stirring. To initiate the reaction, NADH was added via a gas-tight syringe to a final concentration of 1 mM. Samples (2 μl) were removed and injected into a Shimadzu GC-8A gas chromatograph equipped with a flame-ionization detector and a stainless steel column packed with Porapak Q (Alltech) (80/100 mesh). Although methanol accumulation halts after the production of ∼30 μM methanol (Halsey et al., 2006), linear rates could be obtained for the first 5–10 min prior to slowing of the reaction. Methane concentrations were based on calculations using a Henry's constant of 0.0014 M atm−1 at 25 °C (Lide & Frederikse, 1995). The Km of methane was determined by fitting the initial rates of reactions as a function of substrate concentration using Origin Pro 7.5 (OriginLab) according to the following equation:
| (Equation 1) |
where v0 is the initial rate of reaction, V is the maximal rate of methanol accumulation under saturating concentrations of substrate, [Methane] is the aqueous concentration of methane and Km is the Michaelis constant. Methanol concentrations were determined based on a standard curve produced from authentic methanol in the same buffered conditions. All reported errors are sds from three independent replicates.
Determination of the Km for C2–C5 alkanes.
Km measurements for longer-chain alkanes could not be performed due to slow diffusion rates and limited detection by GC at submicromolar concentrations of product. Instead, the Km values for alkanes were measured indirectly by competition with nitrobenzene. In a sealed quartz cuvette, 0.5 ml of a mixture containing 0.06 μM BMOH, 0.18 μM BMOB, 0.36 μM BMOR, 2400 U catalase ml−1 and 1 mM nitrobenzene in 25 mM PIPES, pH 7.2, was incubated for 5 min with varying amounts of gaseous alkane added to the head space. Reactions were initiated by the addition of 1 mM NADH, and monitored by the formation of p-nitrophenol (ε404 nm=15 mM−1 cm−1) at 404 nm in a Beckman DU-640 spectrophotometer. The initial linear portion of the reaction curve (∼1–2 min) was taken as the initial reaction rate of nitrobenzene formation when in competition with the alkane. The Km of the alkane was determined by fitting the following equation:
| (Equation 2) |
where v0 is the initial rate of nitrobenzene formation, V is the maximal reaction rate, [S] is the initial concentration of nitrobenzene, Km2 is the Michaelis constant for nitrobenzene (40 μM), [a] is the aqueous concentration of the alkane, and Km is the Michaelis constant for the alkane. Aqueous concentrations of ethane, propane, butane and pentane were determined using Henry's constants of 0.0019, 0.0014, 0.0011 and 0.0008 M atm−1, respectively. To ensure the accuracy of this method, a similar analysis of methane competition with nitrobenzene was performed in an identical manner to that of ethane, propane, butane and pentane, except that only 50 μM nitrobenzene was used instead of 1 mM.
Determination of alcohol inhibition constants.
A sealed quartz cuvette containing 0.25 μM BMOH, 0.75 μM BMOB, 1.5 μM BMOR, 2400 U catalase ml−1 and 1 mM nitrobenzene in 25 mM PIPES, pH 7.2, was incubated with varying amounts of primary and secondary alcohols for 5 min prior to reaction initiation with 1 mM NADH. Three titrations per alcohol were performed, each with different concentrations of nitrobenzene (50, 100 or 200 μM) in order to determine the type of inhibition. Linear rates of p-nitrophenol formation were monitored by the increase in A404 for 2 min. For experiments without BMOB, reactions were monitored for 5 min due to slower rates of product formation. Competitive and uncompetitive inhibition constants were modelled using the mixed inhibitory equation (Cornish-Bowden, 1995):
| (Equation 3) |
where [i] is the concentration of inhibitor and Kic and Kiu are the competitive and uncompetitive inhibition constants, respectively. All observed inhibition data were fitted to Equation 3 with R2 >0.97. Stock nitrobenzene concentrations were determined using a molar absorption coefficient of 7800 M−1 cm−1 at 268 nm (Zhu et al., 2007).
Formaldehyde analysis.
Quantification of formaldehyde was performed by derivatization with acetoacetanilide and subsequent fluorescence detection, as described elsewhere (Li et al., 2007). Concentrations of formaldehyde were based on a standard curve made from a stock solution of known concentration. Formaldehyde stock concentrations were determined by titration of a diluted sample with a molar excess of iodine (I2) in the presence of 0.2 M NaOH. After 15 min, the excess I2 was acidified with H2SO4 and back-titrated with 0.1 M sodium thiosulfate in the presence of 0.02 % (w/v) starch indicator. The same procedure was repeated without formaldehyde. The difference in thiosulfate needed to titrate the two solutions was used to determine the original concentration of formaldehyde.
RESULTS
Oxidation of alkanes by sBMO
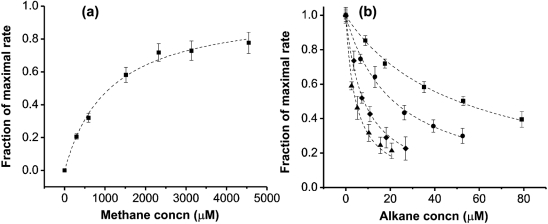
In order to further understand the substrate specificity of sBMO, kinetic parameters Km and kcat were determined for linear C1–C5 alkanes. Although methane is known to be a substrate of sBMO, the accumulation of its product, methanol, has previously been shown to cease once methanol concentrations reach ∼20–30 μM (Halsey et al., 2006). However, at concentrations ranging from 0 to 15 μM, the rates of methanol formation were both constant and dependent on the concentration of aqueous methane. These rates became saturated above 5 mM methane, allowing for a direct measurement of the Km for methane (Fig. 1a). Doubling the enzyme concentration along the first-order region of the titration curve resulted in a doubling of the reaction rate, indicating that the reactions were not diffusion-limited. In contrast to the low micromolar Km of sMMO for methane (3–13 μM) (Green & Dalton, 1986; Nesheim & Lipscomb, 1996), the Km of sBMO for methane was 1.10±0.14 mM (Table 1). Determination of Km values for the C2–C5 growth substrates could not be performed by direct measurement due to limited diffusion rates at low concentrations. Instead, Km values were derived by competition experiments between nitrobenzene and the alkane substrates according to Equation 2 (Fig. 1b). The Km for methane was also measured in this manner, giving similar results to the direct measurement (1.25±0.12 mM, data not shown).
Fig. 1.
Determination of Km values for C1–C5 alkanes. (a) Dependence of the rate of methanol formation on the aqueous concentration of methane. The dashed line is the best fit line according to Equation 1. A fractional rate of 1.0 is equal to 590 nmol min−1 mg−1. (b) Rates of nitrobenzene oxidation with ethane (▪), propane (•), butane (▴) and pentane (⧫) present as a competing substrate. Best fit lines are modelled from the data according to Equation 2. Error bars represent sd from three replicates. A fractional rate of 1.0 is equal to 450 nmol min−1 mg−1.
Table 1.
Kinetic parameters for C1–C5 substrates with WT sBMO
nd, Not detected. Error values represent sd from three replicates.
| Substrate | Km (μM) | kcat (s−1)* | kcat/Km (μM−1 s−1) | Measured specificity† | |||
|---|---|---|---|---|---|---|---|
| Total | 1-OH | 2-OH | 3-OH | ||||
| Methane | 1100±140‡ | 1.3±0.1§ | – | – | – | 0.0012±0.0002 | nd |
| Ethane | 2.2±0.1|| | 0.76±0.05‡ | 0.76±0.05 | – | – | 0.35±0.03 | 1±0.06 |
| Propane | 0.94±0.05|| | 0.65±0.06‡ | 0.55±0.05 | 0.098±0.01 | – | 0.69±0.07 | 2.4±0.2 |
| Butane | 0.24±0.02|| | 0.60±0.03‡ | 0.48±0.02 | 0.12±0.01 | – | 2.5±0.2 | 5.7±0.2 |
| Pentane | 0.34±0.03|| | 0.39±0.03‡ | 0.34±0.02 | 0.039±0.01 | 0.008±0.001 | 1.2±0.1 | 4.4±0.1 |
*The 1-OH column represents the rate of hydroxylation on the primary carbon, 2-OH for the secondary carbon at position 2, 3-OH for the secondary carbon at position 3, and Total represents the sum of all products.
†Direct measurement by competition using an equal concentration of all alkanes in a single reaction, relative to ethane.
‡Direct measurement.
§Extrapolated from Michaelis plot.
||Calculated from competition with nitrobenzene.
The Km values listed in Table 1 indicate a sharp transition of large magnitude from methane to ethane, but less of a difference between propane, butane and pentane. Although preferential binding of substrates is likely to be heavily influenced by the ‘hydrophobic effect’, this cannot explain the drop of nearly three orders of magnitude in Km from methane to ethane (if Km constants are assumed to correlate with binding affinity). Alternatively, given the Bi Uni Uni Bi Ping Pong kinetic mechanism proposed for sMMO (Green & Dalton, 1986), the observed Km for the alkane should be heavily influenced by the rate of alcohol release (Leskovac, 2003). As alcohol release becomes rate limiting, which has been reported for sMMO (Lee et al., 1993; Wallar & Lipscomb, 2001), the Km of the substrate drops below the Kd. Such a scenario would explain the large drop in the Km for ethane. Similar arguments have been proposed for protein kinase A (Werner et al., 1996). The observed maximal turnover rates for methane and ethane listed in Table 1 also support this hypothesis. If the rate-limiting step of the reaction was turnover, the cleavage of a C–H bond for ethane should be three orders of magnitude faster, since its dissociation energy is about 4 kcal mol−1 (16.7 kJ mol−1) weaker than that of methane (Korth & Sicking, 1997; Zheng & Lipscomb, 2006). Such a dramatic increase in ethane turnover was not observed in sBMO, nor in earlier studies with sMMO (Green & Dalton, 1986).
Total turnover rates for C1–C5 substrates were also determined (Table 1). The individual rates of the different alcohol isomers from the oxidation of C3–C5 alkanes were also proportional to previously reported product distributions (Dubbels et al., 2007). The ratio kcat : Km indicated sBMO to be most specific for butane; however, propane and pentane were not substantially different. sBMO was nearly 280-fold more specific for ethane than methane. To confirm the calculated substrate specificities, competition experiments were performed with a mixture of equal concentrations of C1–C5 alkanes. Although methanol formation was not detected, the relative amounts of each C2–C5 alcohol produced were similar to those predicted by the measured kcat : Km ratio (Table 1), providing further evidence that the sBMO enzyme is optimized for butane as a substrate while minimizing C1 oxidation.
Product inhibition of sBMO
Halogenated alcohols have been shown by X-ray crystallography to bind directly in the active site of sMMO, with the oxygen atom of the alcohol group bridging the di-iron centre (Sazinsky & Lippard, 2005). The binding of primary and secondary alcohols to sBMO was characterized in order to gain additional insight into the approximate geometry of the active site. Methanol inhibition of nitrobenzene oxidation was modelled with a mixed inhibitory scenario (Equation 3), which indicated that the inhibition was purely competitive and relatively weak (Kic=1.25±0.06 mM). Previously, our group hypothesized that the plateau of methanol accumulation at ∼30 μM during methane oxidation was the result of strong product inhibition (Halsey et al., 2006). However, the millimolar Kic observed for methanol and similar Km for methane do not support the conclusion that methanol would effectively inhibit methane oxidation at such low concentrations. More insights into the special case of methane oxidation are discussed later in this section.
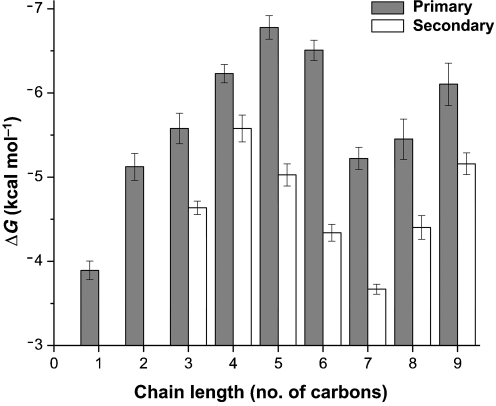
Primary alcohols C1–C6 displayed pure competitive inhibition with nitrobenzene. An average increase of 0.69±0.1 kcal mol−1 (2.9 kJ mol−1) per methylene group was observed for primary C1–C5 alcohols (Fig. 2, shaded bars). Mutational analyses of hydrophobic residues in the interior of proteins have suggested that each methylene group contributes approximately 1.1±0.5 kcal (4.6±2.1 kJ) of stability per mole per methylene group (Pace et al., 1996). It would appear, therefore, that the binding of C1–C5 alcohols to the fully buried, hydrophobic active site of BMOH is heavily influenced by this ‘hydrophobic effect’. After C5, however, binding affinity drops until C7, after which the affinity increases. Interestingly, the tighter inhibition constants for these larger alcohols are accompanied by a distinct change in inhibition from competitive to mixed, suggesting that the alcohol is able to bind to both free enzyme (E) and the enzyme–substrate (ES) complex. Incubation of the enzyme complex with these longer-chain alcohols with and without NADH for 1 h did not yield any significant loss in activity compared with the control after dialysis, indicating that the change in inhibition type is not a result of enzyme inactivation. Given that product-bound structures of MMOH have demonstrated alternative small molecule binding cavities (Sazinsky & Lippard, 2005), it is possible that the uncompetitive nature of primary alcohols larger than C6 is derived from the preferential binding to alternative hydrophobic pockets rather than to the active site.
Fig. 2.
Alcohol binding energies to BMOH. Binding energies of primary alcohols (shaded bars) and secondary alcohols with the hydroxyl group at the second carbon position (white bars) were calculated from the observed competitive inhibition constants using the equation ΔG=−RT ln(Kic−1), where T is the temperature in Kelvin (298 K) and R is the universal gas constant.
Inhibition constants for C1–C9 secondary alcohols were also measured (Fig. 2, white bars). Three distinct observations were made in comparison with primary alcohols. First, all secondary alcohols bound with less affinity than their primary counterparts, indicating that the active site of sBMO is optimized for linear molecules. Second, the break in decreasing Kic values was found between C4 and C5, rather than C5 and C6 as for primary alcohols. Third, the break from competitive to mixed inhibition occurred at C8 rather than C7, indicating that secondary alcohols do not bind to the proposed alternative binding pockets as well as primary alcohols. Lastly, branched alcohols, such as 2-methyl-2-butanol (Kic=1.45±0.08 mM), bound poorly, providing further evidence that the active site of sBMO is structurally optimized for linear molecules.
Effect of BMOB
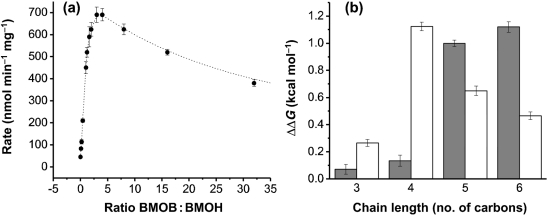
Component B of the sMMO system (MMOB) has profound effects on both the rate of MMOH catalysis and product distribution (Wallar & Lipscomb, 1996). Our previous characterization of purified sBMO revealed that BMOB had little effect on either rate enhancement or product distribution (Dubbels et al., 2007); however, improvements in culturing T. butanivorans have yielded BMOH with catalytic properties more dependent on BMOB (Fig. 3a). At a ratio of 3 : 1 BMOB : BMOH, the turnover rate of nitrobenzene increased 14-fold. Similar effects were seen for ethylene and butane oxidation. Although not as dramatic as the sMMO system, the stimulation of activity by BMOB is more consistent with the role of regulatory components in other di-iron monooxygenases. Unlike MMOB, addition of BMOB had little effect on the product distribution of butane oxidation, generating approximately 80 : 20 and 85 : 15 1-butanol : 2-butanol without and with BMOB, respectively.
Fig. 3.
Influence of BMOB on the sBMO complex. (a) Titration of BMOB into a fixed concentration of BMOH during steady-state nitrobenzene oxidation. (b) The energetic difference (in kcal mol−1) between alcohol binding to the BMOH–BMOB complex and to BMOH alone for primary alcohols (shaded bars) and for secondary alcohols with the hydroxyl group at the second carbon position (white bars). Positive values indicate weaker binding without BMOB. Error bars represent sd from three replicates.
Because substrate turnover by sMMO without MMOB is slow, elucidating its effects on MMOH is difficult and continues to be an active area of research (Mitic et al., 2008). However, because the BMOH–BMOR complex oxidizes substrates at rates of ∼40 nmol min−1 mg−1 without BMOB, we were able to measure the influence of the latter on the inhibition of alcohols during steady-state turnover. Only C3–C6 alcohols were examined, because this was the chain-length range in which breaks in binding affinities were observed. All alcohols bound with less affinity to BMOH when BMOB was not present (Table 2, Fig. 3b). As a consequence, the break in primary alcohol binding affinity was observed between C4 and C5 rather than C5 and C6. C3–C6 secondary alcohols displayed a much less pronounced break between C4 and C5. 2-Methyl-2-butanol could not effectively inhibit sBMO without BMOB present. Lastly, 1-hexanol displayed weak uncompetitive binding characteristics (Kiu >400 μM) without BMOB, implying a possible shift away from active-site binding. The data suggest that BMOB helps to open the active site of BMOH for more efficient substrate access. Similarly, the dissociation of BMOB after substrate turnover may facilitate the release of products. Similar conclusions have been reached in detailed mutational and kinetic analyses in the sMMO system, in which MMOB plays a role in regulating substrate access and product release (Wallar & Lipscomb, 2001; Zheng & Lipscomb, 2006). Whether BMOB enhances the activation of oxygen as MMOB does for MMOH remains to be determined.
Table 2.
Alcohol inhibition constants for WT BMOH
Error values represent sd from three replicates.
| Inhibitor | Alcohol position | With BMOB | Without BMOB | ||
|---|---|---|---|---|---|
| Kic (μM) | Kiu (μM) | Kic (μM) | Kiu (μM) | ||
| Methanol | 1 | 1250±60 | na* | – | – |
| Ethanol | 1 | 150±21 | na | – | – |
| Propanol | 1 | 69±11 | na | 78±5 | na |
| 2 | 349±25 | na | 550±35 | na | |
| Butanol | 1 | 23±3 | na | 29±5 | na |
| 2 | 69±9 | na | 475±27 | na | |
| Pentanol | 1 | 8.8±1.0 | na | 49±3 | na |
| 2 | 180±20 | na | 549±47 | na | |
| Hexanol | 1 | 13±1.4 | na | 89±14 | 414±61 |
| 2 | 580±50 | na | 1290±75 | na | |
| Heptanol | 1 | 127±14 | 102±7 | – | – |
| 2 | 1830±90 | na | – | – | |
| Octanol | 1 | 86±16 | 139±24 | – | – |
| 2 | 521±65 | 1200±175 | – | – | |
| Nonanol | 1 | 28±8 | 29±4 | – | – |
| 2 | 142±16 | 233±21 | – | – | |
| 2-Methyl-1-butanol | 1 | 515±37 | na | – | – |
| 2-Methyl-2-butanol | 2 | 1450±80 | na | na | na |
*na, Not observed. Inhibition constants greater than 10 mM were not considered.
Special case of methane oxidation
Methanol is not an efficient inhibitor of BMOH, even though methanol accumulation during methane oxidation ceases once low micromolar concentrations have been reached both in vivo (Halsey et al., 2006) and in vitro. Even though all the physiologically relevant alcohols were more effective inhibitors than methanol, no stoppage in product accumulation was observed during oxidation of alkanes ≥C2, presumably due to effective competition between substrate and product. Moreover, in vitro characterization of the 1-butanol dehydrogenases BDH and BOH from T. butanivorans showed that the Km for 1-butanol was well below the Kic for sBMO, which further emphasizes that sBMO inhibition by alcohols is unlikely to play an important physiological role in the metabolism of longer-chain alkanes (Vangnai & Arp, 2001; Vangnai et al., 2002). As such, methane oxidation clearly represents a unique case among alkanes.
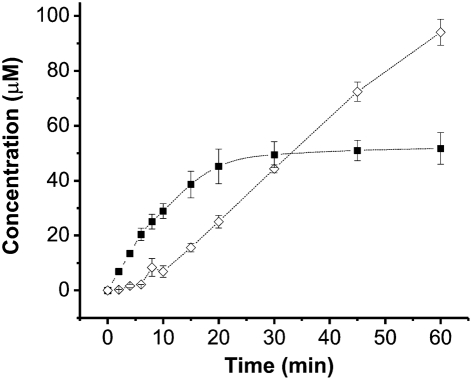
One explanation of the apparent steady level of methanol during methane oxidation is that it is the result of an equilibrium between methane conversion to methanol and methanol conversion to formaldehyde, in which case methane and methanol consumption would be kinetically indistinguishable. Methanol has also been reported to be a substrate for sMMO, but the low micromolar Km for methane and approximately 1 mM Km for methanol make methanol turnover by sMMO physiologically irrelevant (Colby et al., 1977). Kinetic analysis of methane and methanol as substrates of sBMO showed they have nearly identical Km and kcat values (Table 3). As a result, reactions that initially contain only methane generate formaldehyde once methanol begins to accumulate. As the methanol concentration increases, the rate of formaldehyde formation increases until it equals the rate of methanol formation and a steady-state level of methanol is reached (Fig. 4). This non-discrimination of C1 substrates by BMOH is partially alleviated by an alteration in residue 113 from Gly to the corresponding MMOH amino acid, Asn. Kinetic characterization of the G113N variant of BMOH (Table 3) showed that this alteration makes sBMO more specific for methane than methanol, primarily by lowering the Km for methane 3.3-fold. This conserved MMOH Asn residue appears to be critical in maintaining sMMO specificity for methane over methanol, thereby eliminating effective substrate competition between the two.
Table 3.
Kinetic parameters for WT and G113N BMOH oxidation of methane and methanol
Error values represent sd from three replicates.
| Substrate | Km (μM) | kcat (s−1) | kcat/Km (10−4 μM−1 s−1) | |||
|---|---|---|---|---|---|---|
| WT | G113N | WT | G113N | WT | G113N | |
| Methane | 1100±140 | 340±20 | 1.3±0.1 | 0.13±0.01 | 11.8 | 3.8 |
| Methanol | 1250±60 | 750±40 | 1.6±0.1 | 0.21±0.02 | 12.8 | 2.8 |
Fig. 4.
Oxidation of methane by sBMO. Concentrations of both methanol (▪) and formaldehyde (◊) were measured in a reaction initially containing only methane as a substrate. Error bars represent sd from three replicates.
DISCUSSION
The first step in metabolizing an alkane requires the input of energy to cleave the highly stable C–H bond to form an alcohol, which can then be metabolized further in order to provide both the energy and carbon needs for the cell. Attempts to purify alkane monooxygenases from a variety of organisms have proven difficult (Shennan, 2006), limiting in-depth kinetic and structural characterization. In this study, we have characterized an SBMO complex, which is the closest relative to the well-studied sMMO enzymes that has been purified to homogeneity with high activity.
Substrate specificity
With the exception of sMMO, substrate specificity characterizations of alkane monooxygenases are mostly limited to in vivo analyses and only compare relative rates of substrate turnover using saturating concentrations of substrate, thereby neglecting differences in Km values. Our analysis of sBMO shows that even though methane has the highest turnover rate of the alkanes tested, it is clearly a poor substrate due to the high Km relative to C2–C5 alkanes. As such, sBMO is very effective at discriminating between methane and longer alkane substrates. While sMMO is more specific for methane over C2–C5 alkanes, the apparent discrimination for its physiological substrate over longer alkanes is much less striking than that of sBMO. In sMMO, the methane Vmax : Km ratio reported for Methylococcus capsulatus sMMO is only 13-fold higher than that of ethane, and only sevenfold higher than that of propane (Green & Dalton, 1986). For sBMO, the equivalent values for ethane and propane are 290- and 580-fold higher than that for methane, respectively. While it is difficult to compare substrate specificities with those of other alkane monooxygenases due to limited in vitro characterization, in vivo studies with soluble di-iron-containing propane monooxygenases from Gordonia sp. TY-5, Mycobacterium sp. TY-6 and Pseudonocardia sp. TY-7 have suggested that they are similar to sBMO in that they are specific for short-chain C2–C6 alkanes but poorly suited for methane oxidation (Kotani et al., 2003, 2006).
While particulate methane monooxygenase (pMMO) is unusual in that it has a narrow alkane substrate range that includes only linear C1–C5 alkanes (Elliott et al., 1997), wide substrate ranges are not uncommon in membrane-bound alkane monooxygenases. The alkB hydroxylase from Pseudomonas putida GPo1 is capable of oxidizing large (>C12) linear alkanes and substituted cyclic alkanes at similar rates (van Beilen et al., 1994), while also oxidizing the smaller growth substrates propane and butane with high affinity (Ks 66 and 13 μM, respectively) (Johnson & Hyman, 2006). The alkane hydroxylase from propane-utilizing Mycobacterium vaccae JOB5 has also recently been characterized as an alkB hydroxylase (Lopes Ferreira et al., 2007), and has a low reported Ks for propane of 3.3–4.4 μM but high Ks values for branched substrates tert-butyl alcohol and methyl tert-butyl ether (1.36 and 1.18 mM, respectively) (Smith et al., 2003). The soluble haem-containing CYP153A6 alkane hydroxylase from Mycobacterium sp. HXN-1500 has been reported to have low Kd values (20 nM) for C9–C11 growth substrates; however, Kd values for cyclic hydrocarbons are nearly 200-fold higher (Funhoff et al., 2006). Despite the large substrate ranges for these alkane monooxygenases, these data, together with our kinetic analyses of sBMO, emphasize the importance for these alkane monooxygenases to maintain high specificities for physiological substrates in order to out-compete oxidation of molecules that would not provide the carbon and energy needs to sustain cell growth.
Studies have characterized the different iron–oxygen intermediates generated by the hydrocarbon oxidizers toluene/o-xylene monooxygenase (ToMO) and sMMO. While sMMO generates a diamond core bis μ-oxo-(FeIV)2 intermediate (Shu et al., 1997), ToMO generates a weaker oxidizing peroxo-bridged-(FeIII)2 intermediate (Murray et al., 2007), suggesting that this may be a means of substrate selection. This type of selection mechanism, whereby sBMO generates a weaker oxidizing intermediate in order to take advantage of the weaker C–H bonds of longer-chain alkanes compared with methane, is unlikely, given that it oxidizes methane at faster rates than longer-chain alkanes and that it hydroxylates the primary carbon. Therefore, the basis for substrate discrimination is more likely to be based on structural differences in the binding pockets than to be based on different chemical mechanisms. Unfortunately, these structural mechanisms are still poorly understood and are complicated by the observation that naphthalene and methane are both substrates of sMMO and sBMO, despite their fully buried, identical active sites (Halsey et al., 2006; Rosenzweig et al., 1997). However, we have identified one particular residue in the α-subunit of BMOH, Gly-113, which contributes toward defining these specificities. As such, further coordinated mutational analyses of these homologues will certainly provide additional insight into the mechanism of substrate selection.
Component B
Several roles for the small regulatory component have been observed for soluble di-iron-containing multicomponent monooxygenases. Recent studies have demonstrated that in order to enhance the activity of MMOH, MMOB must both (i) induce a geometric rearrangement of a single Fe atom (Fe2) to enhance its reactivity with oxygen, and (ii) cause a more global conformational change within the active site to allow for efficient O2 access (Mitic et al., 2008; Schwartz et al., 2008). The data shown here demonstrate that BMOB also has a large effect on substrate turnover rates (14-fold increase in activity), although this effect is modest compared with that of sMMO. While our data continue to emphasize the importance of these regulatory components in di-iron monooxygenases for maintaining proper substrate selection and efficient product release, one significant difference remains between the sBMO system and both sMMO and other toluene oxidizers: the regulatory component in sBMO does not significantly alter the product regioselectivity for either alkanes or nitrobenzene. The change in regioselectivity caused by MMOB in sMMO was considered to be a consequence of the requirement that a global conformation change within the active site must occur for proper O2 access (Mitic et al., 2008). While it appears that a conformational change within the active site of BMOH must be induced by BMOB, the mechanism must be different from that of sMMO, where product distribution does change substantially. As such, sBMO provides a unique system in which to uncover details about the nature of the active site conformation changes necessary to enhance O2 binding and activation without altering the position of substrate hydroxylation.
The kinetic studies of purified sBMO demonstrate that it is highly specific for linear alkanes ≥C2, while effectively filtering out methane oxidation, even though sBMO and sMMO share identical residues in the active site. While several physiological and enzymic implications can be made, it is clear that sBMO can be a useful tool in helping to elucidate specific factors that influence substrate specificity and activity in bacterial alkane monooxygenases. Efforts to crystallize BMOH are currently under way in our laboratory to address such comparisons from a structural point of view.
Acknowledgments
We thank Michael Schimerlik for his helpful comments and suggestions during the course of this study, and Christine Lastovica and Lisa Roberton for culturing and harvesting of cells. We are grateful for the research support from the National Institutes of Health, grant number 5RO1 GM56128-06.
Abbreviations
BMOB, butane monooxygenase regulatory component
BMOH, butane monooxygenase hydroxylase
BMOR, butane monooxygenase reductase
MMOB, methane monooxygenase regulatory component
MMOH, methane monooxygenase hydroxylase
sBMO, soluble butane monooxygenase
sMMO, soluble methane monooxygenase
WT, wild-type
References
- Arp, D. J. (1999). Butane metabolism by butane-grown ‘Pseudomonas butanovora’. Microbiology 145, 1173–1180. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (2003). Current Protocols in Molecular Biology. New York: Green Publishing & Wiley Interscience.
- Borodina, E., Nichol, T., Dumont, M. G., Smith, T. J. & Murrell, J. C. (2007). Mutagenesis of the “leucine gate” to explore the basis of catalytic versatility in soluble methane monooxygenase. Appl Environ Microbiol 73, 6460–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton, S. G. (2003). Oxidizing enzymes as biocatalysts. Trends Biotechnol 21, 543–549. [DOI] [PubMed] [Google Scholar]
- Colby, J., Stirling, D. I. & Dalton, H. (1977). The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes, n-alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem J 165, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden, A. (1995). Fundamentals of Enzyme Kinetics, p. 108. London: Portland Press Ltd.
- Doughty, D. M., Halsey, K. H., Vieville, C. J., Sayavedra-Soto, L. A., Arp, D. J. & Bottomley, P. J. (2007). Propionate inactivation of butane monooxygenase activity in ‘Pseudomonas butanovora’: biochemical and physiological implications. Microbiology 153, 3722–3729. [DOI] [PubMed] [Google Scholar]
- Dubbels, B. L., Sayavedra-Soto, L. A. & Arp, D. J. (2007). Butane monooxygenase of ‘Pseudomonas butanovora’: purification and biochemical characterization of a terminal-alkane hydroxylating diiron monooxygenase. Microbiology 153, 1808–1816. [DOI] [PubMed] [Google Scholar]
- Dubbels, B. L., Sayavedra-Soto, L. A., Bottomley, P. J. & Arp, D. J. (2009). Thauera butanivorans sp. nov., a C2–C9 alkane oxidizing bacterium previously referred to as ‘Pseudomonas butanovora’. Int J Syst Evol Microbiol in press [DOI] [PMC free article] [PubMed]
- Elliott, S. J., Zhu, M., Tso, L., Nguyen, H. H. T., Yip, J. H. K. & Chan, S. I. (1997). Regio- and stereoselectivity of particulate methane monooxygenase from Methylococcus capsulatus (Bath). J Am Chem Soc 119, 9949–9955. [Google Scholar]
- Enzien, M. V., Picardal, F., Hazen, T. C., Arnold, R. G. & Fliermans, C. B. (1994). Reductive dechlorination of trichloroethylene and tetrachloroethylene under aerobic conditions in a sediment column. Appl Environ Microbiol 60, 2200–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froland, W. A., Andersson, K. K., Lee, S. K., Liu, Y. & Lipscomb, J. D. (1992). Methane monooxygenase component B and reductase alter the regioselectivity of the hydroxylase component-catalyzed reactions. A novel role for protein–protein interactions in an oxygenase mechanism. J Biol Chem 267, 17588–17597. [PubMed] [Google Scholar]
- Funhoff, E. G., Bauer, U., Garcia-Rubio, I., Witholt, B. & van Beilen, J. B. (2006). CYP153A6, a soluble P450 oxygenase catalyzing terminal-alkane hydroxylation. J Bacteriol 188, 5220–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, J. & Dalton, H. (1986). Steady-state kinetic analysis of soluble methane mono-oxygenase from Methylococcus capsulatus (Bath). Biochem J 236, 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsey, K. H., Sayavedra-Soto, L. A., Bottomley, P. J. & Arp, D. J. (2006). Site-directed amino acid substitutions in the hydroxylase α subunit of butane monooxygenase from Pseudomonas butanovora: implications for substrates knocking at the gate. J Bacteriol 188, 4962–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsey, K. H., Doughty, D. M., Sayavedra-Soto, L. A., Bottomley, P. J. & Arp, D. J. (2007). Evidence for modified mechanisms of chloroethene oxidation in Pseudomonas butanovora mutants containing single amino acid substitutions in the hydroxylase α-subunit of butane monooxygenase. J Bacteriol 189, 5068–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E. L. & Hyman, M. R. (2006). Propane and n-butane oxidation by Pseudomonas putida GPo1. Appl Environ Microbiol 72, 950–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth, H.-G. & Sicking, W. (1997). Prediction of methyl C–H bond dissociation energies by density functional theory calculations. J Chem Soc, Perkin Trans 2 715–719.
- Kotani, T., Yamamoto, T., Yurimoto, H., Sakai, Y. & Kato, N. (2003). Propane monooxygenase and NAD+-dependent secondary alcohol dehydrogenase in propane metabolism by Gordonia sp. strain TY-5. J Bacteriol 185, 7120–7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani, T., Kawashima, Y., Yurimoto, H., Kato, N. & Sakai, Y. (2006). Gene structure and regulation of alkane monooxygenases in propane-utilizing Mycobacterium sp. TY-6 and Pseudonocardia sp. TY-7. J Biosci Bioeng 102, 184–192. [DOI] [PubMed] [Google Scholar]
- Leahy, J. G., Batchelor, P. J. & Morcomb, S. M. (2003). Evolution of the soluble diiron monooxygenases. FEMS Microbiol Rev 27, 449–479. [DOI] [PubMed] [Google Scholar]
- Lee, S. K., Nesheim, J. C. & Lipscomb, J. D. (1993). Transient intermediates of the methane monooxygenase catalytic cycle. J Biol Chem 268, 21569–21577. [PubMed] [Google Scholar]
- Leskovac, V. (2003). Comprehensive Enzyme Kinetics. New York: Kluwer Academic/Plenum Publishers.
- Li, Q., Sritharathikhun, P. & Motomizu, S. (2007). Development of novel reagent for Hantzsch reaction for the determination of formaldehyde by spectrophotometry and fluorometry. Anal Sci 23, 413–417. [DOI] [PubMed] [Google Scholar]
- Lide, D. R. & Frederikse, H. P. R. (1995). CRC Handbook of Chemistry and Physics. Boca Raton, FL: CRC Press.
- Lopes Ferreira, N., Mathis, H., Labbe, D., Monot, F., Greer, C. W. & Fayolle-Guichard, F. (2007). n-Alkane assimilation and tert-butyl alcohol (TBA) oxidation capacity in Mycobacterium austroafricanum strains. Appl Microbiol Biotechnol 75, 909–919. [DOI] [PubMed] [Google Scholar]
- Mitic, N., Schwartz, J. K., Brazeau, B. J., Lipscomb, J. D. & Solomon, E. I. (2008). CD and MCD studies of the effects of component B variant binding on the biferrous active site of methane monooxygenase. Biochemistry 47, 8386–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, L. J., Naik, S. G., Ortillo, D. O., Garcia-Serres, R., Lee, J. K., Huynh, B. H. & Lippard, S. J. (2007). Characterization of the arene-oxidizing intermediate in ToMOH as a diiron(III) species. J Am Chem Soc 129, 14500–14510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesheim, J. C. & Lipscomb, J. D. (1996). Large kinetic isotope effects in methane oxidation catalyzed by methane monooxygenase: evidence for C–H bond cleavage in a reaction cycle intermediate. Biochemistry 35, 10240–10247. [DOI] [PubMed] [Google Scholar]
- Pace, C. N., Shirley, B. A., McNutt, M. & Gajiwala, K. (1996). Forces contributing to the conformational stability of proteins. FASEB J 10, 75–83. [DOI] [PubMed] [Google Scholar]
- Parales, R. E., Bruce, N. C., Schmid, A. & Wackett, L. P. (2002). Biodegradation, biotransformation, and biocatalysis (B3). Appl Environ Microbiol 68, 4699–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival, M. D. (1991). Human 5-lipoxygenase contains an essential iron. J Biol Chem 266, 10058–10061. [PubMed] [Google Scholar]
- Rosenzweig, A. C., Brandstetter, H., Whittington, D. A., Nordlund, P., Lippard, S. J. & Frederick, C. A. (1997). Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): implications for substrate gating and component interactions. Proteins 29, 141–152. [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Sayavedra-Soto, L. A., Doughty, D. M., Kurth, E. G., Bottomley, P. J. & Arp, D. J. (2005). Product and product-independent induction of butane oxidation in Pseudomonas butanovora. FEMS Microbiol Lett 250, 111–116. [DOI] [PubMed] [Google Scholar]
- Sazinsky, M. H. & Lippard, S. J. (2005). Product bound structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): protein motion in the α-subunit. J Am Chem Soc 127, 5814–5825. [DOI] [PubMed] [Google Scholar]
- Schwartz, J. K., Wei, P. P., Mitchell, K. H., Fox, B. G. & Solomon, E. I. (2008). Geometric and electronic structure studies of the binuclear nonheme ferrous active site of toluene-4-monooxygenase: parallels with methane monooxygenase and insight into the role of the effector proteins in O2 activation. J Am Chem Soc 130, 7098–7109. [DOI] [PubMed] [Google Scholar]
- Shennan, J. L. (2006). Utilisation of C2–C4 gaseous hydrocarbons and isoprene by microorganisms. J Chem Technol Biotechnol 81, 237–256. [Google Scholar]
- Shu, L., Nesheim, J. C., Kauffmann, K., Munck, E., Lipscomb, J. D. & Que, L., Jr (1997). An Fe2IVO2 diamond core structure for the key intermediate Q of methane monooxygenase. Science 275, 515–518. [DOI] [PubMed] [Google Scholar]
- Sluis, M. K., Sayavedra-Soto, L. A. & Arp, D. J. (2002). Molecular analysis of the soluble butane monooxygenase from ‘Pseudomonas butanovora’. Microbiology 148, 3617–3629. [DOI] [PubMed] [Google Scholar]
- Smith, T. J. & Dalton, H. (2004). Biocatalysis by methane monooxygenase and its implications for the petroleum industry. In Petroleum Biotechnology, Developments and Perspectives, pp. 177–192. Edited by R. Vazquez-Duhalt & R. Qintero-Ramirez. Amsterdam: Elsevier.
- Smith, C. A., O'Reilly, K. T. & Hyman, M. R. (2003). Characterization of the initial reactions during the cometabolic oxidation of methyl tert-butyl ether by propane-grown Mycobacterium vaccae JOB5. Appl Environ Microbiol 69, 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor, S. & Richardson, C. C. (1985). A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A 82, 1074–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, J., Ichikawa, Y., Sagae, H., Komura, I., Kanou, H. & Yamada, K. (1980). Isolation and identification of n-butane assimilating bacterium. Agric Biol Chem 44, 1835–1840. [Google Scholar]
- van Beilen, J. B. & Funhoff, E. G. (2007). Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biotechnol 74, 13–21. [DOI] [PubMed] [Google Scholar]
- van Beilen, J. B., Kingma, J. & Witholt, B. (1994). Substrate specificity of the alkane hydroxylase system of Pseudomonas oleovorans GPo1. Enzyme Microb Technol 16, 904–911. [Google Scholar]
- Vangnai, A. S. & Arp, D. J. (2001). An inducible 1-butanol dehydrogenase, a quinohaemoprotein, is involved in the oxidation of butane by ‘Pseudomonas butanovora’. Microbiology 147, 745–756. [DOI] [PubMed] [Google Scholar]
- Vangnai, A. S., Sayavedra-Soto, L. A. & Arp, D. J. (2002). Roles for the two 1-butanol dehydrogenases of Pseudomonas butanovora in butane and 1-butanol metabolism. J Bacteriol 184, 4343–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallar, B. J. & Lipscomb, J. D. (1996). Dioxygen activation by enzymes containing binuclear non-heme iron clusters. Chem Rev 96, 2625–2658. [DOI] [PubMed] [Google Scholar]
- Wallar, B. J. & Lipscomb, J. D. (2001). Methane monooxygenase component B mutants alter the kinetics of steps throughout the catalytic cycle. Biochemistry 40, 2220–2233. [DOI] [PubMed] [Google Scholar]
- Werner, D. S., Lee, T. R. & Lawrence, D. S. (1996). Is protein kinase substrate efficacy a reliable barometer for successful inhibitor design? J Biol Chem 271, 180–185. [DOI] [PubMed] [Google Scholar]
- Zheng, H. & Lipscomb, J. D. (2006). Regulation of methane monooxygenase catalysis based on size exclusion and quantum tunneling. Biochemistry 45, 1685–1692. [DOI] [PubMed] [Google Scholar]
- Zhu, C. Z., Ouyang, B., Wang, J. Q., Huang, L., Dong, W. B. & Hou, H. Q. (2007). Photochemistry in the mixed aqueous solution of nitrobenzene and nitrous acid as initiated by the 355 nm UV light. Chemosphere 67, 855–861. [DOI] [PubMed] [Google Scholar]