Abstract
Uropathogenic Escherichia coli (UPEC) can grow in environments with significantly elevated osmolarities, such as murine and human urinary tracts. OmpR is the response regulator part of a two-component OmpR–EnvZ regulatory system that responds to osmotic stresses. To determine the role of OmpR in UPEC survival, a ΔompR mutant was created in the UPEC clinical isolate NU149. The ΔompR mutant had a growth defect compared with the wild-type strain under osmotic stress conditions; this defect was complemented by the full-length ompR gene on a plasmid, but not with a mutant OmpR with an alanine substitution for aspartic acid at the phosphorylation site at position 55. Furthermore, the ΔompR mutant displayed up to 2-log reduction in bacterial cell numbers in murine bladders and kidneys compared with wild-type bacteria after 5 days of infection. The ability of the bacteria to survive was restored to wild-type levels when the ΔompR mutant strain was complemented with wild-type ompR, but not when the alanine-substituted ompR gene was used. This study has fulfilled molecular Koch's postulates by showing the pivotal role OmpR plays in UPEC survival within the murine urinary tract.
Escherichia coli is a major cause of urinary tract infections (UTIs) in humans, resulting in billions of dollars in healthcare-related costs each year (Foxman & Brown, 2003; Litwin et al., 2005). The subgroup responsible for these UTIs has been called uropathogenic E. coli (UPEC). For more than two decades, a murine UTI model that mimics UTIs in humans has been used to examine UPEC pathogenesis (Hagberg et al., 1983; Schaeffer et al., 1987). The murine urinary tract (Loeb & Quimby, 1989) has many similarities to the human urinary tract (Ross & Neely, 1983); these similarities include wide ranges in pH and osmolarity.
Several virulence factors have been associated with the ability of UPEC to survive in the urinary tract (Johnson, 1991). Type 1 pili, haemolysins and capsule production are used by UPEC strains causing human and murine UTIs. Molecular Koch's postulates (Falkow, 1988) have been applied to virulence factors expressed by UPEC; these factors include: type 1 pili (Connell et al., 1996); the DegS and DegP proteins that regulate sigma E during extracytoplasmic stress (Redford & Welch, 2006; Redford et al., 2003); the iron transport protein TonB (Torres et al., 2001); PhoU, a negative regulator of the Pho regulon (Buckles et al., 2006); the d-serine deaminase DsdA (Haugen et al., 2007); and flagella (Lane et al., 2007; Schwan, 2008; Wright et al., 2005). Other virulence factors needed by the UPEC strains to maintain a UTI may be those involved in acid tolerance and osmotic stress survival.
To initiate and sustain a UTI, UPEC bacteria must be able to survive in environments with diverse pH values and wide fluctuations in the osmolarity. We have previously reported that the fimB and fimE genes are regulated by OmpR (Schwan et al., 2002), which is a protein that is part of a two-component signal transduction system that serves as a global regulator within E. coli (Oshima et al., 2002). Two-component regulatory systems generally consist of a response regulator, and a sensor kinase that spans the inner membrane. The EnvZ–OmpR tandem represents a two-component regulatory system that is affected by osmotic changes (Mizuno & Mizushima, 1990). During an osmotic shock, some unidentified signal linked to the osmolarity of the external environment triggers autophosphorylation of EnvZ at a conserved histidine residue, using ATP to donate the phosphate group (Forst et al., 1989; Igo & Silhavy, 1988). Once EnvZ is phosphorylated, EnvZ-P donates the phosphate group to OmpR at a conserved aspartic acid residue at position 55 (Forst et al., 1989). OmpR-P can then serve as a transcriptional activator or repressor of various genes within the E. coli cell.
In this study, we deleted the ompR gene from strain NU149, which is a UPEC clinical isolate (Schaeffer et al., 1987), and examined what effect this had on the survival of the isolate in vitro, as well as within a murine UTI model. We demonstrate that, compared with the wild-type, a ΔompR mutation leads to: (i) a growth defect in media with osmolyte concentrations greater than or equal to 400 mM, (ii) attenuated growth in human urine, and (iii) a reduction of 2 logs in viable counts within murine urinary tracts. These results suggest that OmpR is critical for UPEC survival in mammalian urinary tracts.
METHODS
Bacterial strains and plasmids.
The UPEC strain NU149 was used in this study. It has been well characterized, and used extensively in a murine UTI model (Schaeffer et al., 1987; Schwan et al., 2002). Plasmids pFR29* (Russo & Silhavy, 1991) and pD55A (V. K. Tran & L. J. Kenney, unpublished data) were a kind gift from Linda Kenney, the University of Illinois at Chicago, Chicago, IL, USA. The pFR29* plasmid has the full-length ompR gene cloned in front of an IPTG inducible promoter, whereas the pD55A plasmid has a mutated ompR gene, resulting in an alanine substituting for aspartic acid at amino acid position 55 cloned in front of an IPTG-inducible promoter. Strain DH5α MCR was used for transformations. Luria–Bertani (LB) broth and LB agar were used with the addition of the following antibiotics: kanamycin (40 μg ml−1), or ampicillin (100 μg ml−1) (Sigma). The λ Red recombinase system, with plasmids pKD4 (used for amplifying a kanamycin-resistance gene with flanking gene sequences to allow recombination and creation of the deletion mutation), pKD46 (containing the λ Red recombinase), and pCP20 (carrying a Flp recombinase to allow excision of the kanamycin-resistance gene once the deletion mutation had been created), was used for construction of the ΔompR mutation, as described by Datsenko & Wanner (2000). We have previously used this system successfully to create a fliC mutation in strain NU149 (Schwan, 2008).
Construction of the ΔompR mutant strain in a UPEC clinical isolate.
The λ Red recombinase system (Datsenko & Wanner, 2000) was used to construct the ΔompR mutation in the UPEC strain NU149. Primers OmpR1 (5′-ACGCTTACAAATTGTTGCGAACCTTTGGGAGTACAAACATGTGTAGGCTGGAGCTGCTTCG-3′) and OmpR2 (5′-GGGCAAATGAACTTCGTGGCGAGAAGCGCAATCGCCTCATCATATGAATATCCTCCTTAG-3′) were synthesized (IDT DNA Technologies), and used in PCR amplifications, with pKD4 plasmid DNA as a template, under the following conditions: initial denaturation at 95 °C for 5 min, then 35 cycles of 95 °C for 1 min, 55 °C for 1 min, and 72 °C for 2 min. The resulting amplified DNA was concentrated with a Microcon 30 filter, gel purified on a 0.8 % agarose gel, and then electroporated into competent NU149 cells, as described previously (Schwan, 2008), selecting for transformants on LB agar with kanamycin. Several transformants were screened for the lack of ompR gene amplification using a PCR-based system with the OmpR3 (5′-TGCTGACTCGTGAATCTTTCC-3′) and OmpR4 (5′-CGTACCGAGGTTAAGTTTGAA-3′) primer pair (319 bp product). The conditions for the PCR amplifications were as follows: initial denaturation 95 °C for 5 min, 95 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min. One transformant, NU149 OmpR, which did not amplify a 319 bp ompR gene fragment, was chosen for further analysis. Removal of the kanamycin-resistance gene in strain NU149 OmpR using the pCP20 plasmid was done as noted (Datsenko & Wanner, 2000). Transformants were selected on LB agar containing 40 μg kanamycin ml−1. One clone was heated to 42 °C to cure the strain of the pKD46 plasmid. The resulting UPEC strain was labelled NU149 OmpR1. To complement the strain NU149 OmpR1, electroporation of either the pFR29* or the pD55A plasmid was performed, and transformants were selected on LB agar containing 100 μg ampicillin ml−1. One clone representing a successful transformation with each plasmid was chosen for additional analysis.
Growth in buffered LB broth with differences in osmolarity and pH.
To test the strains under different in vitro environmental conditions, the pH of LB medium was adjusted by using 0.1 M sodium phosphate buffer combined with 1 % (v/v) glycerol, as previously described (Schwan et al., 2002). The osmolarity of LB broth was also adjusted, as previously noted (Schwan et al., 2002), using urea, sucrose or NaCl. The broths were labelled low osmolarity (≤200 mM NaCl), moderate osmolarity (400 mM NaCl), and high osmolarity (≥600 mM NaCl). OD600 readings were done after incubation for 24 h.
RT-PCR analysis.
To assess whether there was a difference in envZ expression in the wild-type versus the ompR strain, a RT-PCR procedure was used. Total RNAs were isolated from wild-type and ompR strains grown in pH 7.0 LB broth, with and without the addition of 400 mM NaCl, to mid-exponential phase, using an RNeasy kit (Qiagen) with the addition of a 10 μl volume of 10 mg lysozyme ml−1. After elution, nucleic acid concentrations were determined by spectrophotometry (Shimatzu). The RNAs were treated with RNase-free DNase (Roche) for 30 min at 37 °C to remove residual contaminating DNA. A 6 μg quantity of total RNA was used to create cDNAs using the SuperScript II reverse transcriptase kit (Invitrogen). All of the cDNAs were PCR amplified in a multiplex amplification using the EnvZ1 (5′-ACCACCATATTCGCCACCGC-3′) and EnvZ2 (5′-CTGCGTGAATATGGCGCTTC-3′) primer pair to amplify envZ cDNA, and the EcFtsZ1 and EcFtsZ2 primer pair that has been used to amplify ftsZ (Schwan et al., 2002, 2007). Amplification conditions included an initial denaturation at 95 °C for 5 min followed by 30 cycles of 95 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min. The PCR products were loaded on 1.5 % agarose gels. Expression of the envZ gene was determined by comparing the expression of the envZ product to the ftsZ product, using ImageQuant software (Molecular Dynamics).
Protein extraction and immunoblotting.
SDS-PAGE was performed with 12.5 % separating gels, and 20 μg E. coli cell lysates of NU149, NU149 OmpR1, NU149 OmpR1/pFR29* and NU149 OmpR1/pD55A grown in pH 7.0 LB broth. The lysates were prepared by suspending stationary-phase cultures in cell lysis buffer, as described (Malitschek et al., 1994), and incubating with a 10 μl volume of 10 mg lysozyme ml−1 for 30 min at 37 °C, followed by sonication. Proteins were electrotransferred to nylon membranes (Amersham). Membranes were blocked with blocking buffer [1 % dried milk and 0.1 % BSA (Sigma) in Tris-buffered saline containing 0.05 % Tween 20 (TBS-T)] for 2 h, washed with TBS-T, and then allowed to react with a rabbit polyclonal antibody to OmpR protein (1/500 dilution; provided by Linda Kenney) for 2 h at room temperature. Membranes were washed three times with TBS-T, and horseradish-peroxidase-conjugated anti-rabbit antibody (Amersham) was added at a 1/10 000 dilution, and the membranes were incubated at room temperature for 1 h. After several more washes, bound antibodies were detected by the enhanced chemiluminescence system (Amersham).
Murine UTI model.
A murine UTI model was used to assess the role the ompR mutation may play in E. coli cells infecting murine urinary tracts (Schaeffer et al., 1987). Mid-exponential-phase bacteria at a concentration of 108 c.f.u. ml−1 in PBS were used as the inocula. Female Swiss Webster mice were transurethrally inoculated with either 50 μl per mouse of each E. coli strain to mimic the ascending model (total of five mice per strain), or 200 μl bacteria per mouse to allow infection of more murine kidneys (total of 8–12 mice per strain). For the two injection schemes, the ΔompR mutant (strain NU149 OmpR1), the complemented ompR mutant with the native OmpR (strain NU149 OmpR1/pFR29*), the complemented ompR mutant with the mutated OmpR (strain NU149 OmpR1/pD55A), and wild-type NU149 cells, were used. For the 50 μl injection scheme, mouse organs were collected after 24 h; for the 200 μl injection volume, each organ was collected 1–5 days post-inoculation. Mouse urine was collected before the mouse was killed, and the pH and specific gravity were measured for each sample. Mouse organs were processed by adding PBS (pH 7.4) to each organ. The volume of PBS for each bladder was adjusted based on the mass of each organ, in order to standardize the bladder preparations between mice. Since there was a difference of <10 % in kidney masses, a 1 ml volume of PBS was used for processing kidneys, and no compensation was necessary. The organs were homogenized with a tissue grinder, aliquots were 10-fold serially diluted in PBS, and 100 μl aliquots plated onto LB agar to ascertain viable counts. Both kidneys from an animal were collected at the appropriate time point, and they were processed together as one sample.
Growth in human urine.
Two human urine specimens were collected to perform growth measurements. The pH of each specimen was measured using a pH meter (Orion), and the osmolarity was determined by using specific gravity measurements and freezing point depression (Ross & Neely, 1983).
Statistics.
To determine differences in in vitro growth, Student's t test was used. With the murine UTI model results, an ANOVA, with a Bonferroni correction, was used to assess probability. P values <0.05 were considered significant.
RESULTS
Creation of a ΔompR mutation in the UPEC strain NU149
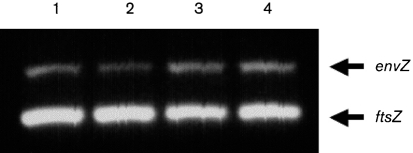
A λ Red recombinase system (Datsenko & Wanner, 2000) was used to create a near-complete deletion mutation in the clinical isolate NU149. Because there is a 3 bp overlap between the ompR and envZ genes, this part of the ompR gene was not deleted to allow proper transcription of the envZ gene. The final deletion mutant, named NU149 OmpR1, was confirmed by PCR to be missing the ompR gene (data not shown). In addition, the mutant strain was shown to have no polar effect on envZ transcription using RT-PCR on cDNA generated from broth-grown cultures of NU149 OmpR1 and NU149 (Fig. 1).
Fig. 1.
Transcriptional analysis of envZ expression in wild-type and ompR strains. The analysis was done on cDNAs from cultures grown in pH 7.0 LB broth, with either no added NaCl or 400 mM added NaCl. Multiplex PCRs were set up with EnvZ1 and EnvZ2 primers to amplify a 424 bp envZ product, and EcFtsZ1 and EcFtsZ2 primers to amplify a 302 bp ftsZ product (Schwan et al., 2002). Each multiplex was run three separate times. The lanes were loaded as follows: 1, NU149 (pH 7.0); 2, NU149 OmpR1 (pH 7.0); 3, NU149 (pH 7.0, 400 mM NaCl); and 4, NU149 OmpR1 (pH 7.0, 400 mM NaCl). Quantification of the data was done using ImageQuant software (Molecular Dynamics), and the number of pixels for each band was quantified. For each lane, the intensities of the envZ product were corrected to the intensity of the ftsZ band.
Confirmation of OmpR expression in the ompR strain transformed with pFR29* or pD55A
To demonstrate that the ompR strain transformed with either pFR29* or pD55A expressed OmpR protein, a Western blot was performed on lysates from NU149, NU149 OmpR1, NU149 OmpR1/pFR29* and NU149 OmpR1/pD55A grown in pH 7.0 LB broth. Strain NU149 OmpR1 did not display an OmpR protein band, whereas the wild-type strain and both plasmid-complemented strains showed an OmpR protein band on the blot (data not shown).
Growth of the ompR mutant strain in buffered LB broth, with differences in osmolarity and pH
Because OmpR is important in osmotically stressed E. coli cells, the ompR strain was compared with the wild-type strain grown in pH 7.0 buffered LB broth, with variations in the osmolarity. The ompR strain grew better than wild-type bacteria in medium with no added NaCl, and in medium with 200 mM added NaCl (Fig. 2a). However, the ompR strain was significantly attenuated for growth in medium supplemented with 400 mM NaCl (OD600 0.38; P<0.003). In medium with 600 mM (OD600 0.05; P<0.01) and 800 mM NaCl (OD600 0.05; P<0.03), the ompR strain failed to grow, whereas the wild-type strain grew. Standard plate counts on selected broth samples following growth confirmed the lower viable bacterial counts for the ompR strain compared with the wild-type (data not shown). When the ompR strain was complemented with the native ompR gene, bacterial growth resembled that of the wild-type strain. On the other hand, complementation with the mutated ompR gene with an amino acid substitution at position 55 produced growth measurements similar to those of the ompR strain.
Fig. 2.
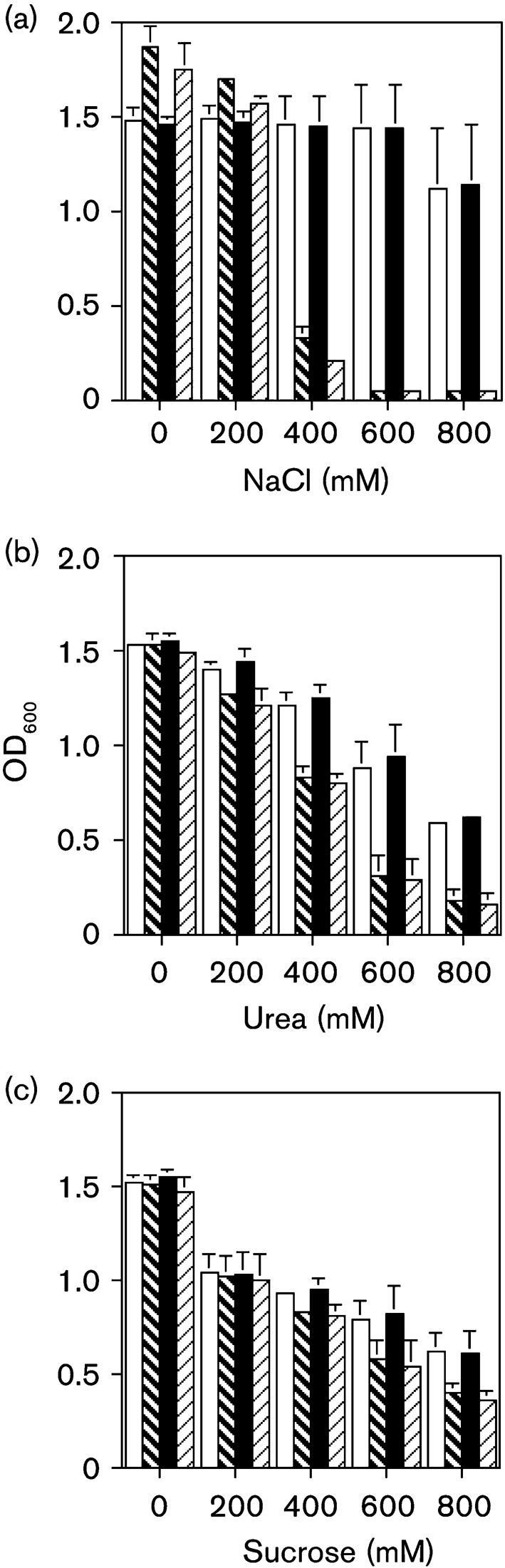
Effects of osmolarity on bacterial growth of wild-type cells (NU149, open bar), an ompR mutant (NU149 OmpR1, bold hatched bar), an ompR mutant complemented with the native ompR gene (NU149 OmpR1/pFR29*, filled bar), and an ompR mutant complemented with a mutated ompR gene (NU149 OmpR1/pD55A, hatched bar). Osmolarity effects were tested by using (a) NaCl, (b) urea and (c) sucrose as the osmolytes, at concentrations ranging from 0 to 800 mM. Growth was measured as the OD at 600 nm, and the values represent the means (±sd) from at least three separate runs.
Growth in medium with variations in the concentration of urea was also tested. In LB broth with no added urea, all four strains had similar OD600 readings (Fig. 2b). When the osmolyte concentration was increased to 200 mM urea, the growth of the ompR strain was lower (OD600 1.269) compared with the wild-type (OD600 1.397; P<0.042). By raising the osmolarity to 400 mM, the ompR strain grew less well (OD600 0.826) compared with the wild-type (OD600 1.204; P<0.035). A further increase in the osmolarity caused a wider separation in the growth of the two strains (OD600: ompR 0.308 versus wild-type 0.875; P<0.009). When growth was compared at the highest urea osmolarity, a greater difference in growth was noted between the mutant and the wild-type strain (OD600: ompR 0.171 versus wild-type 0.586; P<0.006). Complementation of the ompR cells with the pFR29* plasmid resulted in OD600 readings similar to those of the wild-type, in all concentrations of urea, whereas complementation with pD55A failed to restore the growth of the ompR strain to wild-type levels.
A third osmolyte, sucrose, was then tested for its effect on growth of the four strains. Little growth difference was observed between the four strains grown in LB with no added sucrose or when 200 mM sucrose was added (Fig. 2c). In LB with 400 mM sucrose, the ompR strain grew less well than the wild-type (OD600 0.829 versus 0.926, respectively), but the decrease was not significant (P<0.112). An increase in the sucrose concentration to 600 mM resulted in an even lower OD600 reading for ompR (OD600 0.574) compared with the wild-type (OD600 0.781); the difference was significant (P<0.007). Growth of the ompR strain in LB broth containing the highest sucrose concentration showed an OD600 of 0.392 compared with 0.611 for the wild-type (P<0.017). Complementation with the native ompR gene fully restored the growth of the ompR strain, but complementation with the mutated ompR gene did not. Thus, all of the osmolytes that were tested affected growth of the ompR strain at a high concentration.
To further assess how a combination of changes in pH and differences in osmolarity affected in vitro growth, the ompR strain was grown in pH 5.5 and pH 7.0 LB broth, with and without the addition of 400 mM NaCl. The ompR strain grew less well in the pH 5.5 (OD600 0.35; P<0.0002) and pH 7.0 broths (OD600 0.56; P<0.00002) supplemented with 400 mM NaCl compared with the wild-type strain grown in pH 5.5 LB containing 400 mM NaCl (OD600 1.79) or pH 7.0 medium (OD600 1.75) (Fig. 3). Growth was restored to wild-type levels by complementing with the native ompR in pH 5.5 LB broth with 400 mM NaCl (OD600 1.46), and in pH 7.0 LB broth with 400 mM NaCl (OD600 1.63); wild-type levels of growth were not restored by complementing with the mutated ompR gene (pH 5.5 LB broth with NaCl, OD600 0.35; pH 7.0 LB broth with NaCl, OD600 0.49). These results demonstrate that the ompR mutation causes an in vitro growth defect in UPEC cells grown in moderate-to-high osmolarity media.
Fig. 3.
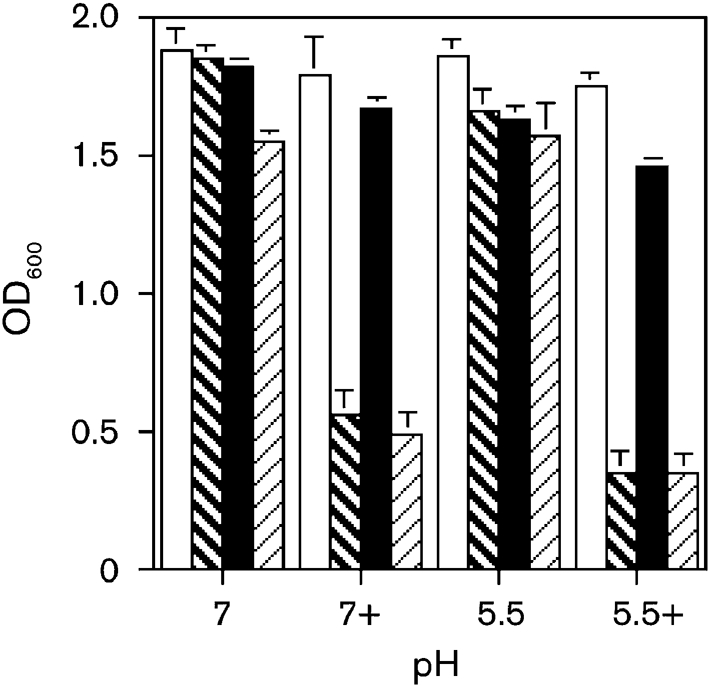
Effects of a combination of osmolarity and pH on bacterial growth of wild-type cells (NU149, open column), an ompR mutant (NU149 OmpR1, bold hatched bar), an ompR mutant complemented with the native ompR gene (NU149 OmpR1/pFR29*, filled bar), and an ompR mutant complemented with a mutated ompR gene (NU149 OmpR1/pD55A, hatched bar). Growth at pH 5.5 and pH 7.0 with 400 mM NaCl (+) and without NaCl was measured at an OD of 600 nm, and the values represent the means (±sd) from at least three separate runs.
Growth in human urine was affected by the ompR mutation
Since there was a difference in growth of the ompR strain compared with the wild-type bacteria in medium with either moderate or high osmolarity, the strains were tested for their ability to grow in human urine. Two human urine samples were collected. Urine no. 1 had a pH of 5.98, and an osmolarity of 300 mM, whereas urine no. 2 had a pH of 6.07, and an osmolarity of 400 mM. The four strains described above were inoculated into aliquots of the human urine, and incubated for 18 h at 37 °C. All of the strains grew in both urine samples. However, the ompR strain did not grow as well as the wild-type strain. The differences in growth were not significant for the first urine specimen [mean (±sd) OD600: ompR 0.400±0.081 versus wild-type 0.529±0.051; P<0.105], but they were significant for urine no. 2 (ompR 0.300±0.03 versus wild-type 4.84±0.012; P<0.045). Complementation of the ompR strain with pFR29* restored the growth to wild-type levels in both urine samples (urine no. 1, 0.567±0.047; urine no. 2, 0.510±0.054), yet complementation with the pD55A plasmid did not restore the growth (urine no. 1, 0.417±0.015; urine no. 2, 0.294±0.03). These results show that ompR mutation is associated with a growth defect in human urine.
Murine urinary tract survival difference is associated with ompR mutation in UPEC
From the in vitro analysis, it was clear that the ompR strain had a growth deficiency when placed in medium with a moderate-to-high osmolarity. To assess whether this ompR mutation caused a similar growth defect in UPEC within the murine urinary tract, the wild-type strain NU149, the ompR strain NU149 OmpR1, and NU149 OmpR1/pFR29* and NU149 OmpR1/pD55A strains, were inoculated into the urinary tracts of female mice using a low-volume (50 μl) or a high-volume (200 μl) inoculum. The mouse urine samples had pH values that ranged from 5.5 to 7.0, and osmolarities that ranged from 350 to 600 mM.
For the low-volume inoculations testing true urinary tract ascension, 4/5 wild-type infected mice had kidneys that were infected. Only 2/5 ompR-infected mice had colonized kidneys. Complementation of the ompR strain with pFR29* resulted in 4/5 mice with infected kidneys, but complementation with pD55A led to 2/5 mice having infected kidneys.
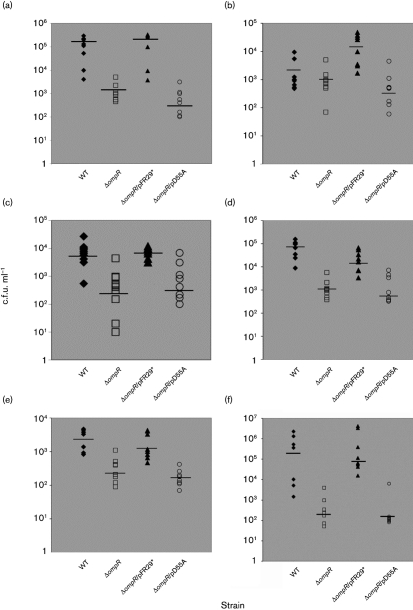
Using a more aggressive UTI model with a larger volume to ensure that a greater number of murine kidneys would be infected, the viable counts were determined for each group after 1, 3 and 5 days post-inoculation. One day post-inoculation, the ompR strain (2.06×103 c.f.u. ml−1, Fig. 4a) displayed a decline of nearly 2 logs in the mean viable count from the bladder as compared with the wild-type strain (1.27×105 c.f.u. ml−1, P<0.015). However, there was no statistical difference when comparing the viable counts in the kidneys from either the ompR strain (1.28×103 c.f.u. ml−1) or the wild-type strain (2.58×103 c.f.u. ml−1, P>0.400) (Fig. 4b). Complementation with the native ompR gene restored the viable counts to wild-type levels in the bladder (2.57×105 c.f.u. ml−1), but complementation with the mutated ompR gene did not.
Fig. 4.
Independent challenges of female BALB/c mice with E. coli strain NU149 (WT, ⧫), an ompR mutant (NU149 OmpR1, □), an ompR mutant strain complemented with the native ompR gene (NU149 OmpR1/pFR29*, ▴), and an ompR mutant strain complemented with a mutated ompR gene (NU149 OmpR1/pD55A, ○). Bacterial counts are shown for: (a) the bladder, 1 day post-inoculation; (b) the kidney, 1 day post-inoculation; (c) the bladder, 3 days post-inoculation; (d) the kidney, 3 days post-inoculation; (e) the bladder, 5 days post-inoculation; and (f) the kidney, 5 days post-inoculation. Each data point represents the c.f.u. ml−1 for one mouse. Horizontal bars represent the median values of the bacterial concentration of the population.
The 3 day post-inoculation data also showed a statistical difference between the ompR strain (4.25×102 c.f.u. ml−1) and the wild-type strain (7.2×103 c.f.u. ml−1; P<0.012) in the murine bladder (Fig. 4c). Moreover, the mean viable count of NU149 OmpR1 in the kidneys (1.03×103 c.f.u. ml−1) was approximately 1.8 logs lower than for the wild-type strain NU149 (8.15×104 c.f.u. ml−1; P<0.006) (Fig. 4d). Again, complementation with the native ompR gene resulted in mean viable counts from the bladders and kidneys that were comparable with those of the wild-type strain, whereas viable counts remained unchanged for the ompR strain that was complemented with the mutated ompR gene.
For the 5 day post-inoculation time point, the mean viable count for the bladder homogenates from the ompR strain infected mice (3.05×102 c.f.u. ml−1) was 1 log less than for the wild-type strain (3.3×103 c.f.u. ml−1, P<0.009) (Fig. 4e). Within the kidneys, day 5 viable count for the wild-type (6.4×105 c.f.u. ml−1) was more than 3 logs higher than for the ompR strain (2.00×102 c.f.u. ml−1; P<0.05) (Fig. 4f). From day 1 to day 5, the mean viable count for strain NU149 in the kidneys rose by approximately 2 logs, whereas the ompR strain displayed a 1 log reduction in the mean viable count in this organ system over the same time period. Successful complementation of the ompR mutation occurred with the native ompR, but not with the mutated ompR gene. The analyses demonstrated that there was a distinct survival advantage for the wild-type strain over the ompR strain in the murine urinary tract.
DISCUSSION
UPEC survival in the murine or human urinary tract is dependent on many variables, including the ability to withstand variations in osmolarity that can reach as high as 800 mM in humans, and 1.5 M in mice (Loeb & Quimby, 1989; Schmidt-Nielsen et al., 1983). Our results show that OmpR is critical for the survival of UPEC within a murine urinary tract. We demonstrated that a UPEC clinical isolate with an ompR deletion was severely crippled in its ability to grow in vitro in medium with moderate-to-high osmolarity as compared with the wild-type parental strain. Growth was attenuated for the NU149 OmpR1 strain in moderate-osmolarity medium (400 mM NaCl) for two out of the three osmolytes tested, and either no growth was seen when the osmolarity was increased with the addition of 600 or 800 mM NaCl (high osmolarity), or the growth was substantially reduced in medium with high concentrations of sucrose or urea. This growth defect was successfully complemented by using the native ompR gene on a plasmid, but not using the mutated ompR gene that resulted in an amino acid substitution at position 55 in the OmpR protein. The change from an aspartic acid to an alanine would negate the transfer of a phosphate group from EnvZ (Delgado et al., 1993), resulting in a potentially unphosphorylated OmpR protein that would be unable to transcriptionally regulate a number of genes within the E. coli cells when the bacteria were grown in an osmotically stressed environment. The phosphorylation of OmpR, in turn, regulates a number of genes within E. coli (Oshima et al., 2002).
Medium with moderate-to-high osmolarity supported the growth of the ompR strain to a lesser extent than it supported growth of the wild-type strain. When moderate osmolarity was combined with an acidic pH medium, an additional growth reduction was shown for the ompR strain. Why are acidity and osmolarity combined important? The two human urine samples had acidic pH values and moderate osmolarities. All of the murine urine samples had moderate-to-high osmolarity, and most were acidic. Most human urine samples are in the acidic range (Ross & Neely, 1983; and personal observations). Certainly, acid conditions combined with moderate osmotic conditions adversely affected growth of the ompR strain more so than they affected growth of the strain in neutral pH medium. Some genes that are regulated by OmpR may play an important role in survival of the bacteria in an acidic/high osmotic environment. In Helicobacter pylori, an OmpR-like protein regulates the acid-adaptation response (Bury-Moné et al., 2004). OmpR is one of the proteins upregulated by acid adaptation in enterohaemorrhagic E. coli (Huang et al., 2007), so there could be a function for OmpR in UPEC growing in an acidified human urinary tract.
In vitro growth attenuation by the ompR strain was also observed when the strain was inoculated into murine urinary tracts. Over a 5 day period of time, viable counts for strain NU149 OmpR1 fell by 1 log in the bladders and kidneys, and this was presumably due to the sensitivity to the high osmotic conditions found in the urinary tract. In contrast, the wild-type strain viable counts in the kidneys rose by over 2 logs. Complementation of the ompR mutation with the native ompR gene enabled the UPEC to grow nearly as well as wild-type bacteria under conditions of high osmolarity conditions; this fulfilled molecular Koch's postulates for the UPEC ompR gene (Falkow, 1988).
Why is the ompR mutation so crippling for UPEC? In terms of overall fitness, one study has shown that an ompR strain grows less well compared with wild-type E. coli in salt water (Darcan et al., 2003). The poor growth in an osmotically stressed environment is likely to be related to OmpR being a global regulator that affects scores of genes within E. coli (Oshima et al., 2002). Mutations in ompR have been shown to cause changes in the outer-membrane-protein profile within E. coli (Forst & Roberts, 1994; Pratt & Silhavy, 1995; Prohinar et al., 2002; Rampersaud et al., 1991). Under an osmotic stress, such as being dropped into a mammalian urinary tract, the bacteria change their outer-membrane proteins (Alteri & Mobley, 2007; Hagan & Mobley, 2007). The loss of OmpR would prevent those changes from occurring. Moreover, OmpR affects expression of surface structures such as type 1 pili (Schwan et al., 2002), curli (Jubelin et al., 2005; May & Okabe, 2008; Römling et al., 1998) and flagella (Shin & Park, 1995). Changes in expression of the surface structures could also have an impact on the ability of UPEC to survive in a mouse or a human urinary tract. In addition, OmpR appears to regulate expression of enterochelin and the aerobactin receptor protein IutA (Oshima et al., 2002; Wooldridge & Williams, 1991). Iron acquisition is critical for UPEC survival in the urinary tract. Alteri & Mobley (2007) demonstrated that IutA expression was induced when the bacteria were grown in human urine. IutA is expressed by UPEC, but not by laboratory strains of E. coli. Thus, an ompR deletion would have an impact on outer-membrane-protein expression, which is a trait shared by all E. coli cells, and it would also affect the expression of virulence factors, such as IutA, that are expressed by UPEC, and are critical for survival in either a murine or a human urinary tract.
Acknowledgments
We wish to thank Linda Kenney for the pFR29* and pD55A plasmids, as well as anti-OmpR antibody. This study was funded by NIH grant AI065432-01A2 to W. R. S.
References
- Alteri, C. J. & Mobley, H. L. (2007). Quantitative profile of the uropathogenic Escherichia coli outer membrane proteome during growth in human urine. Infect Immun 75, 2679–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckles, E. L., Wang, X., Lockatell, C. V., Johnson, D. E. & Donnenberg, M. S. (2006). PhoU enhances the ability of extraintestinal pathogenic Escherichia coli strain CFT073 to colonize the murine urinary tract. Microbiology 152, 153–160. [DOI] [PubMed] [Google Scholar]
- Bury-Moné, S., Thiberge, J. M., Contreras, M., Maitournam, A., Labigne, A. & De Reuse, H. (2004). Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol Microbiol 53, 623–638. [DOI] [PubMed] [Google Scholar]
- Connell, H., Agace, W., Klemm, P., Schembri, M., Marild, S. & Svanborg, C. (1996). Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci U S A 93, 9827–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darcan, C., Ozkanca, R. & Flint, K. P. (2003). Survival of nonspecific porin-deficient mutants of Escherichia coli in Black Sea water. Lett Appl Microbiol 37, 380–385. [DOI] [PubMed] [Google Scholar]
- Datsenko, K. A. & Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado, J., Forst, S., Harlocker, S. & Inouye, M. (1993). Identification of a phosphorylation site and functional analysis of conserved aspartic acid residues of OmpR, a transcriptional activator for ompF and ompC in Escherichia coli. Mol Microbiol 10, 1037–1047. [DOI] [PubMed] [Google Scholar]
- Falkow, S. (1988). Molecular Koch's postulates applied to microbial pathogenicity. Rev Infect Dis 10 (Suppl 2), S274–S276. [DOI] [PubMed] [Google Scholar]
- Forst, S. A. & Roberts, D. L. (1994). Signal transduction by the EnvZ–OmpR phosphotransfer system in bacteria. Res Microbiol 145, 363–373. [DOI] [PubMed] [Google Scholar]
- Forst, S., Delgado, J. & Inouye, M. (1989). Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci U S A 86, 6052–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman, B. & Brown, P. (2003). Epidemiology of urinary tract infections: transmission and risk factors, incidence, and costs. Infect Dis Clin North Am 17, 227–241. [DOI] [PubMed] [Google Scholar]
- Hagan, E. C. & Mobley, H. L. (2007). Uropathogenic Escherichia coli outer membrane antigens expressed during urinary tract infection. Infect Immun 75, 3941–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg, L., Engberg, I., Freter, R., Lam, J., Olling, S. & Svanborg Eden, C. (1983). Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun 40, 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen, B. J., Pellett, S., Redford, P., Hamilton, H. L., Roesch, P. L. & Welch, R. A. (2007). In vivo gene expression analysis identifies genes required for enhanced colonization of the mouse urinary tract by uropathogenic Escherichia coli strain CFT073 dsdA. Infect Immun 75, 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. J., Tsai, T. Y. & Pan, T. M. (2007). Physiological response and protein expression under acid stress of Escherichia coli O157 : H7 TWC01 isolated from Taiwan. J Agric Food Chem 55, 7182–7191. [DOI] [PubMed] [Google Scholar]
- Igo, M. M. & Silhavy, T. J. (1988). EnvZ, a transmembrane environmental sensor of Escherichia coli K-12, is phosphorylated in vitro. J Bacteriol 170, 5971–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J. R. (1991). Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev 4, 80–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubelin, G., Vianney, A., Beloin, C., Ghigo, J. M., Lazzaroni, J. C., Lejeune, P. & Dorel, C. (2005). CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol 187, 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, M. C., Alteri, C. J., Smith, S. N. & Mobley, H. L. (2007). Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A 104, 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin, M. S., Saigal, C. S., Yano, E. M., Avila, C., Geschwind, S. A., Hanley, J. M., Joyce, G. F., Madison, R., Pace, J. & other authors (2005). Urologic Diseases in America Project: analytical methods and principal findings. J Urol 173, 933–937. [DOI] [PubMed] [Google Scholar]
- Loeb, W. F. & Quimby, F. W. (1989). The Clinical Chemistry of Laboratory Animals. New York: Pergamon.
- Malitschek, B., Wittbrodt, J., Fischer, P., Lammers, R., Ullrich, A. & Schartl, M. (1994). Autocrine stimulation of the Xmrk receptor tyrosine kinase in Xiphophorus melanoma cells and identification of a source for the physiological ligand. J Biol Chem 269, 10423–10430. [PubMed] [Google Scholar]
- May, T. & Okabe, S. (2008). Escherichia coli harboring a natural IncF conjugative F plasmid develops complex mature biofilms by stimulating synthesis of colanic acid and curli. J Bacteriol 190, 7479–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, T. & Mizushima, S. (1990). Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol Microbiol 4, 1077–1082. [DOI] [PubMed] [Google Scholar]
- Oshima, T., Aiba, H., Masuda, Y., Kanaya, S., Sugiura, M., Wanner, B. L., Mori, H. & Mizuno, T. (2002). Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol 46, 281–291. [DOI] [PubMed] [Google Scholar]
- Pratt, L. A. & Silhavy, T. J. (1995). Identification of base pairs important for OmpR–DNA interaction. Mol Microbiol 17, 565–573. [DOI] [PubMed] [Google Scholar]
- Prohinar, P., Forst, S. A., Reed, D., Mandic-Mulec, I. & Weiss, J. (2002). OmpR-dependent and OmpR-independent responses of Escherichia coli to sublethal attack by the neutrophil bacteridical/permeability increasing protein. Mol Microbiol 43, 1493–1504. [DOI] [PubMed] [Google Scholar]
- Rampersaud, A., Utssumi, R., Delgado, J., Forst, S. A. & Inouye, M. (1991). Ca2+-enhanced phosphorylation of a chimeric protein kinase involved with bacterial signal transduction. J Biol Chem 266, 7633–7637. [PubMed] [Google Scholar]
- Redford, P. & Welch, R. A. (2006). Role of sigma E-regulated genes in Escherichia coli uropathogenesis. Infect Immun 74, 4030–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redford, P., Roesch, P. L. & Welch, R. A. (2003). DegS is necessary for virulence and is among extraintestinal Escherichia coli genes induced in murine peritonitis. Infect Immun 71, 3088–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling, U., Bian, Z., Hammar, M., Sierralta, W. D. & Normark, S. (1998). Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol 180, 722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, D. L. & Neely, A. E. (1983). Textbook of Urinalysis and Bodily Fluids. Norwalk, VA: Appleton Century Crofts.
- Russo, F. D. & Silhavy, T. J. (1991). EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol 222, 567–580. [DOI] [PubMed] [Google Scholar]
- Schaeffer, A. J., Schwan, W. R., Hultgren, S. J. & Duncan, J. L. (1987). Relationship of type 1 pilus expression in Escherichia coli to ascending urinary tract infections in mice. Infect Immun 55, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Nielsen, B., Graves, B. & Roth, J. (1983). Water removal and solute additions determining increases in renal medullary osmolarity. Am J Physiol 244, F472–F482. [DOI] [PubMed] [Google Scholar]
- Schwan, W. R. (2008). Flagella allow uropathogenic Escherichia coli ascension into murine kidneys. Int J Med Microbiol 298, 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan, W. R., Lee, J. L., Lenard, F. A., Matthews, B. T. & Beck, M. T. (2002). Osmolarity and pH growth conditions regulate fim gene transcription and type 1 pilus expression in uropathogenic Escherichia coli. Infect Immun 70, 1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan, W. R., Shibata, S., Aizawa, S. & Wolfe, A. J. (2007). The two-component response regulator RcsB regulates type 1 piliation in Escherichia coli. J Bacteriol 189, 7159–7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, S. & Park, C. (1995). Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol 177, 4696–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, A. G., Redford, P., Welch, R. A. & Payne, S. M. (2001). TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect Immun 69, 6179–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooldridge, K. G. & Williams, P. H. (1991). Sensitivity of Escherichia coli to cloacin DF13 involves the major outer membrane protein OmpR. J Bacteriol 173, 2420–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, K. J., Seed, P. C. & Hultgren, S. J. (2005). Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun 73, 7657–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]