Abstract
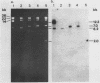
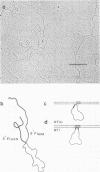
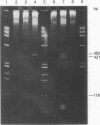
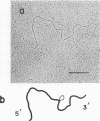
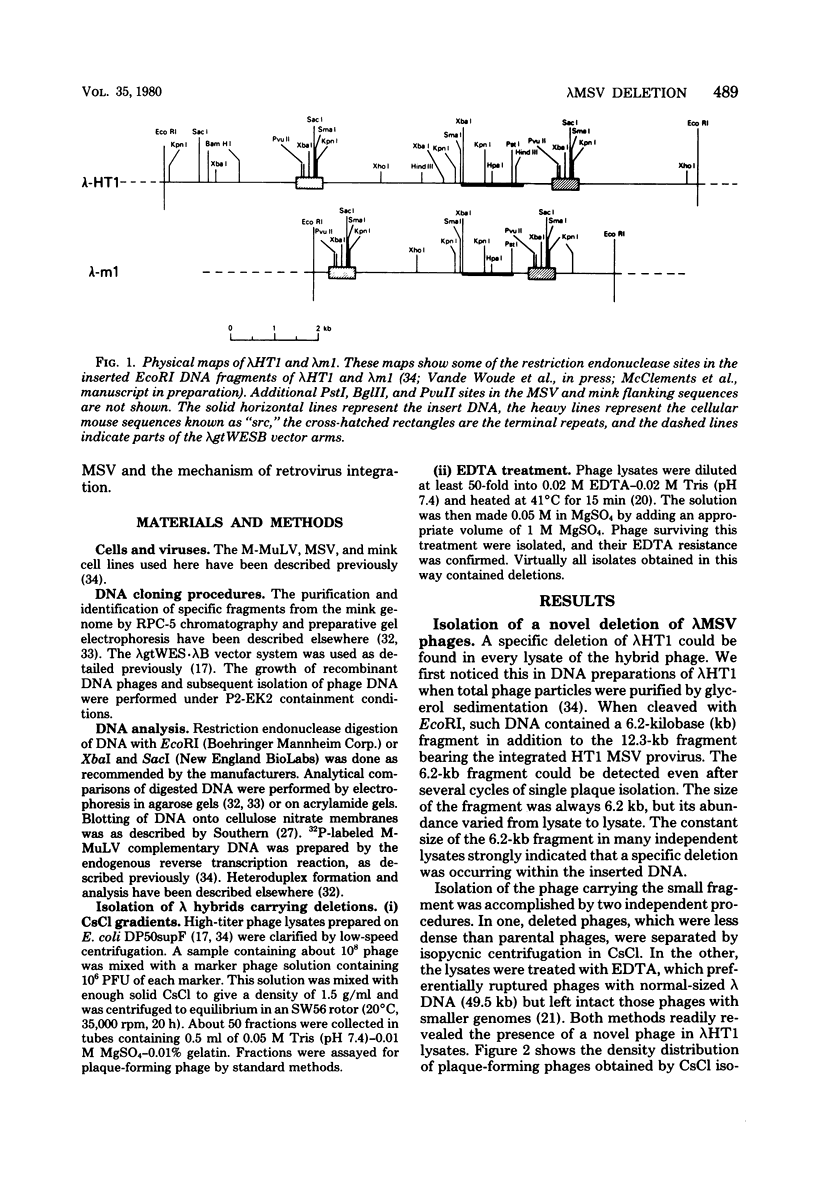
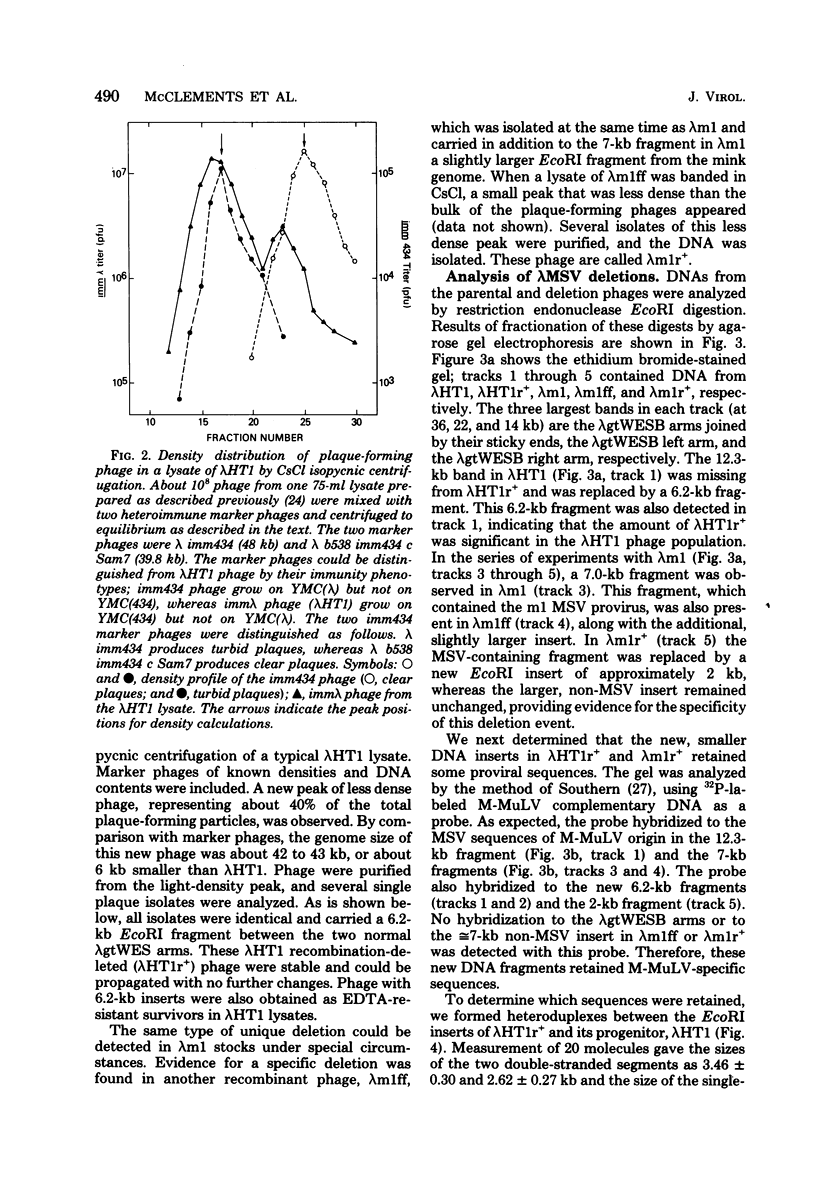
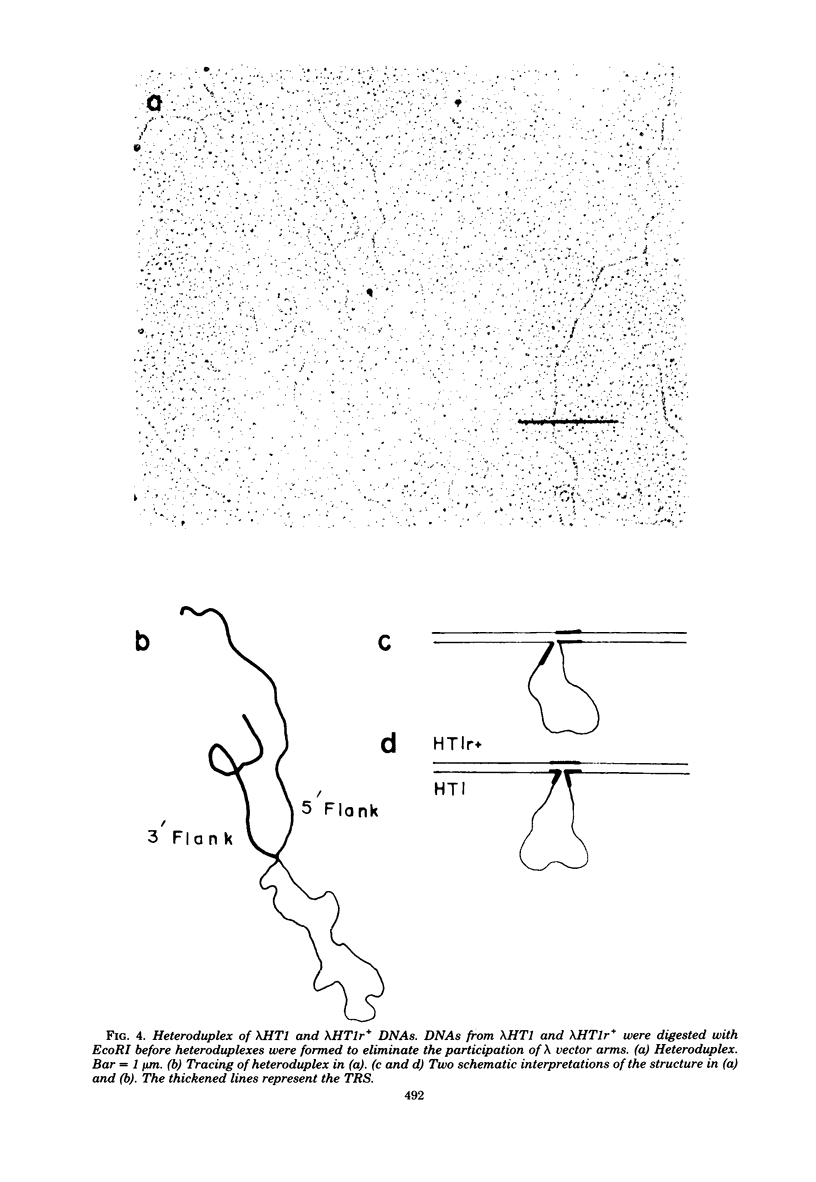
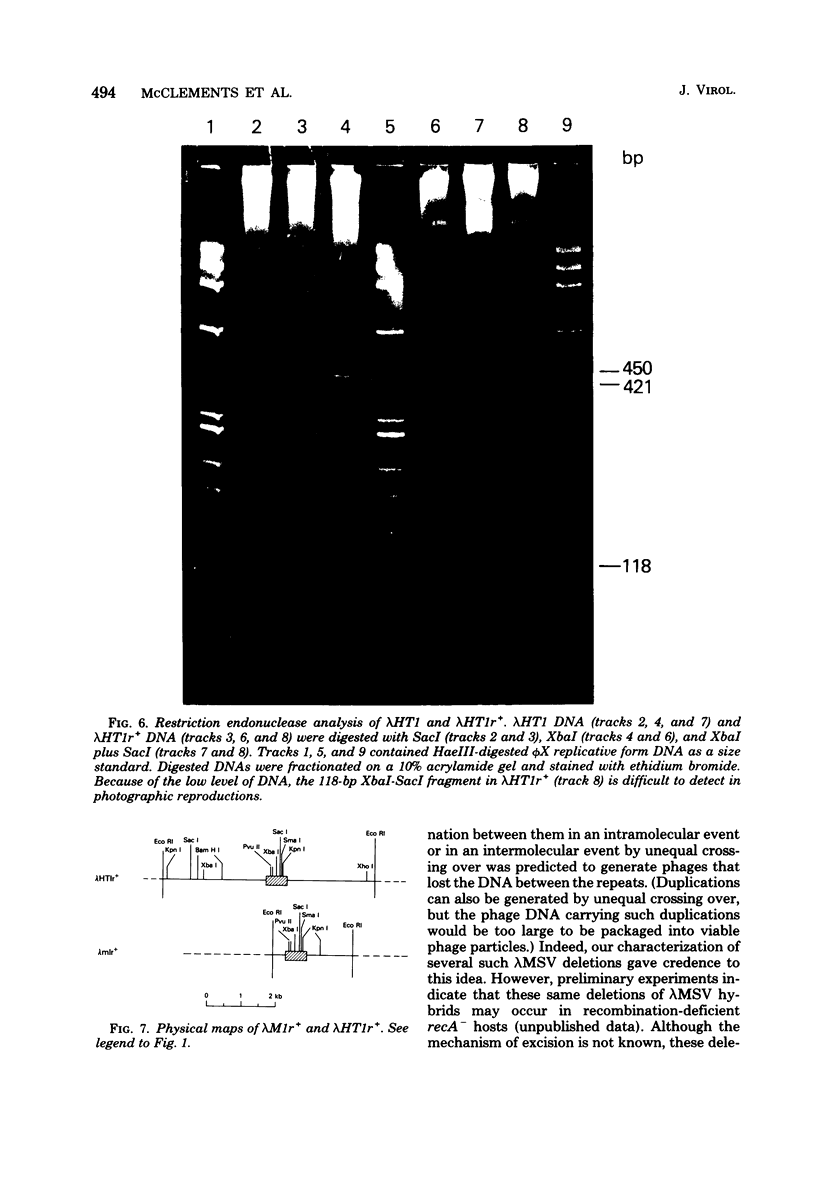
Stocks of hybrid lambda phages carrying the complete integrated provirus of either m1 or HT1 Moloney murine sarcoma virus, as well as flanking host sequences, frequently contain significant numbers of phages carrying a specific deletion. This deletion arises from a recombination event between the terminally repeated sequences in the provirus that deletes the unique Moloney murine sarcoma virus sequences bracketed by the terminally repeated sequences. Physical mapping has shown that the deletion phage retains one complete copy of the terminally repeated sequence and the flanking mink host sequences. One such deletion, lambdaHT1r+, was used to characterize a mink genomic DNA sequence that contains an HT1 Moloney murine sarcoma virus integration site. This integration site sequence from normal mink cells was also cloned into phage lambda. An analysis of the heteroduplexes between the integration site and the lambdaHT1r+ deletion indicated that no major rearrangement of host sequences occurred upon integration of the Moloney murine sarcoma provirus.
Full text
PDF


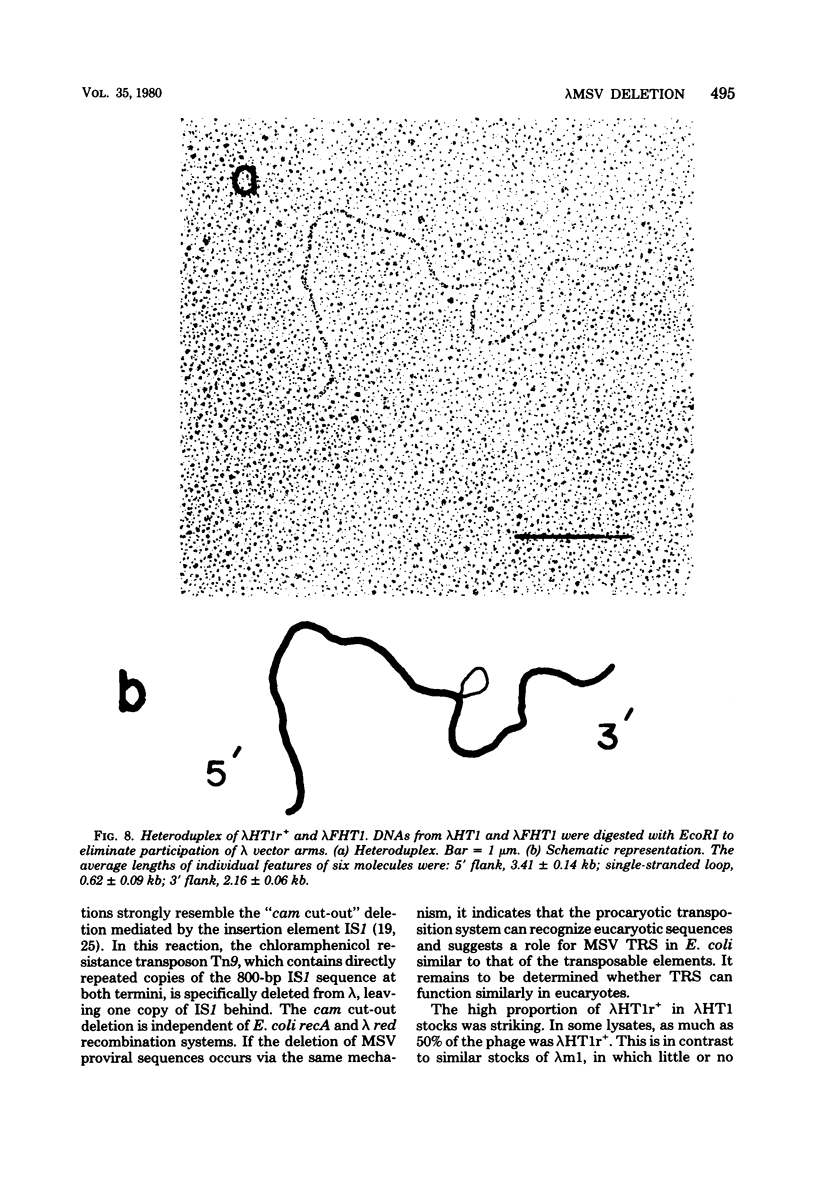


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersson P., Goldfarb M. P., Weinberg R. A. A defined subgenomic fragment of in vitro synthesized Moloney sarcoma virus DNA can induce cell transformation upon transfection. Cell. 1979 Jan;16(1):63–75. doi: 10.1016/0092-8674(79)90188-0. [DOI] [PubMed] [Google Scholar]
- Battula N., Temin H. M. Sites of integration of infectious DNA of avian reticuloendotheliosis viruses in different avian cellular DNAs. Cell. 1978 Feb;13(2):387–398. doi: 10.1016/0092-8674(78)90207-6. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Fry K., Nelson T., Hung P. Synthesis of hybrid bacterial plasmids containing highly repeated satellite DNA. Cell. 1977 Mar;10(3):509–519. doi: 10.1016/0092-8674(77)90038-1. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Johnsrud L., Miller J. H. DNA sequence at the integration sites of the insertion element IS1. Cell. 1978 Mar;13(3):411–418. doi: 10.1016/0092-8674(78)90315-x. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Canaani E., Duesberg P., Dina D. Cleavage map of linear mouse sarcoma virus DNA. Proc Natl Acad Sci U S A. 1977 Jan;74(1):29–33. doi: 10.1073/pnas.74.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A., Ram D. Deletions within E. coli plasmids carrying yeast rDNA. Gene. 1978 Apr;3(2):135–147. doi: 10.1016/0378-1119(78)90057-4. [DOI] [PubMed] [Google Scholar]
- Collins C. J., Parsons J. T. Integration of avian sarcoma virus DNA sequences in transformed mammalian cells. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4301–4305. doi: 10.1073/pnas.74.10.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa E., Goff S., Shields A., Yoshimura F., Mitra S., Baltimore D. In vitro synthesis of a 9 kbp terminally redundant DNA carrying the infectivity of Moloney murine leukemia virus. Cell. 1979 Apr;16(4):863–874. doi: 10.1016/0092-8674(79)90101-6. [DOI] [PubMed] [Google Scholar]
- Grindley N. D. IS1 insertion generates duplication of a nine base pair sequence at its target site. Cell. 1978 Mar;13(3):419–426. doi: 10.1016/0092-8674(78)90316-1. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Sherratt D. J. Sequence analysis at IS1 insertion sites: models for transposition. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1257–1261. doi: 10.1101/sqb.1979.043.01.142. [DOI] [PubMed] [Google Scholar]
- Hughes S. H., Shank P. R., Spector D. H., Kung H. J., Bishop J. M., Varmus H. E., Vogt P. K., Breitman M. L. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell. 1978 Dec;15(4):1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- Huskey R. J. Deletion mutants of bacteriophage lambda. IV. High frequency Int-promoted recombination. Mol Gen Genet. 1973 Dec 14;127(1):39–46. doi: 10.1007/BF00267781. [DOI] [PubMed] [Google Scholar]
- Keshet E., Temin H. M. Sites of integration of reticuloendotheliosis virus DNA in chicken DNA. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3372–3376. doi: 10.1073/pnas.75.7.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder P., Tiemeier D., Enquist L. EK2 derivatives of bacteriophage lambda useful in the cloning of DNA from higher organisms: the lambdagtWES system. Science. 1977 Apr 8;196(4286):175–177. doi: 10.1126/science.322278. [DOI] [PubMed] [Google Scholar]
- Lemons R. S., Nash W. G., O'Brien S. J., Benveniste R. E., Sherr C. J. A gene (Bevi) on human chromosome 6 is an integration site for baboon type C DNA provirus in human cells. Cell. 1978 Aug;14(4):995–1005. doi: 10.1016/0092-8674(78)90353-7. [DOI] [PubMed] [Google Scholar]
- Nash H. A. LambdaattB-attP, a lambda derivative containing both sites involved in integrative recombination. Virology. 1974 Jan;57(1):207–216. doi: 10.1016/0042-6822(74)90121-4. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S., Huskey R. J. Deletion mutants of bacteriophage lambda. I. Isolation and initial characterization. J Mol Biol. 1971 Mar 14;56(2):369–384. doi: 10.1016/0022-2836(71)90471-2. [DOI] [PubMed] [Google Scholar]
- Perricaudet M., Fritsch A., Pettersson U., Philipson L., Tiollais P. Excision and recombination of adenovirus DNA fragments in Escherichia coli. Science. 1977 Apr 8;196(4286):208–210. doi: 10.1126/science.322283. [DOI] [PubMed] [Google Scholar]
- Potter S. S., Brorein W. J., Jr, Dunsmuir P., Rubin G. M. Transposition of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell. 1979 Jun;17(2):415–427. doi: 10.1016/0092-8674(79)90168-5. [DOI] [PubMed] [Google Scholar]
- Shank P. R., Hughes S. H., Kung H. J., Majors J. E., Quintrell N., Guntaka R. V., Bishop J. M., Varmus H. E. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell. 1978 Dec;15(4):1383–1395. doi: 10.1016/0092-8674(78)90063-6. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A., MacHattie L. A. Integration and excision of prophage lambda mediated by the IS 1 element. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1135–1142. doi: 10.1101/sqb.1979.043.01.127. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steffen D., Weinberg R. A. The integrated genome of murine leukemia virus. Cell. 1978 Nov;15(3):1003–1010. doi: 10.1016/0092-8674(78)90284-2. [DOI] [PubMed] [Google Scholar]
- Strobel E., Dunsmuir P., Rubin G. M. Polymorphisms in the chromosomal locations of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell. 1979 Jun;17(2):429–439. doi: 10.1016/0092-8674(79)90169-7. [DOI] [PubMed] [Google Scholar]
- Taylor J. M., Mason W. S., Hsu T. W., Sabran J. L., Yeater C., Mark G. E., Kaji A., Guntaka R. V., Lai M. M. Avian RNA tumor virus DNA before and after integration into the host genome. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):865–867. doi: 10.1101/sqb.1979.043.01.092. [DOI] [PubMed] [Google Scholar]
- Temin H. M., Baltimore D. RNA-directed DNA synthesis and RNA tumor viruses. Adv Virus Res. 1972;17:129–186. doi: 10.1016/s0065-3527(08)60749-6. [DOI] [PubMed] [Google Scholar]
- Tiemeier D. C., Tilghman S. M., Polsky F. I., Seidman J. G., Leder A., Edgell M. H., Leder P. A comparison of two cloned mouse beta-globin genes and their surrounding and intervening sequences. Cell. 1978 Jun;14(2):237–245. doi: 10.1016/0092-8674(78)90110-1. [DOI] [PubMed] [Google Scholar]
- Tilghman S. M., Tiemeier D. C., Polsky F., Edgell M. H., Seidman J. G., Leder A., Enquist L. W., Norman B., Leder P. Cloning specific segments of the mammalian genome: bacteriophage lambda containing mouse globin and surrounding gene sequences. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4406–4410. doi: 10.1073/pnas.74.10.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Woude G. F., Oskarsson M., Enquist L. W., Nomura S., Sullivan M., Fischinger P. J. Cloning of integrated Moloney sarcoma proviral DNA sequences in bacteriophage lambda. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4464–4468. doi: 10.1073/pnas.76.9.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E., Shank P. R., Hughes S. E., Kung H. J., Heasley S., Majors J., Vogt P. K., Bishop J. M. Synthesis, structure, and integration of the DNA of RNA tumor viruses. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):851–864. doi: 10.1101/sqb.1979.043.01.091. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Vogt P. K., Bishop J. M. Integration of deoxyribonucleic acid specific for Rous sarcoma virus after infection of permissive and nonpermissive hosts. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3067–3071. doi: 10.1073/pnas.70.11.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg R. A., Adhya S. Illegitimate recombination in bacteria and bacteriophage. Annu Rev Genet. 1977;11:451–473. doi: 10.1146/annurev.ge.11.120177.002315. [DOI] [PubMed] [Google Scholar]