Abstract
Nitric oxide (NO) regulates biological processes through signaling mechanisms that exploit its unique biochemical properties as a free radical. For the last several decades, the key aspects of the chemical properties of NO relevant to biological systems have been defined, but it has been a challenge to assign these to specific cellular processes. Nevertheless, it is now clear that the high affinity of NO for transition metal centers, particularly iron, and the rapid reaction of NO with oxygen-derived free radicals can explain many of its biological and pathological properties. Emerging studies also highlight a growing importance of the secondary metabolites of NO-dependent reactions in the post-translational modification of key metabolic and signaling proteins. In this minireview, we emphasize the current understanding of the biochemistry of NO and place it in a biological context.
Keywords: Hemoglobin Myoglobin, Mitochondria, Nitric Oxide, Oxidative Stress, Oxygen Radicals
Introduction
Nitric oxide (NO; nitrogen monoxide) and related nitrogen oxides are now known to be critical endogenous regulators of cell and tissue function (1, 2). From the earliest isolation of NO, there has been an intense interest in linking the chemistry of NO to its biological effects, beginning with the near-death experience of the famous physiologist Sir Humphrey Davy after inhaling NO (3). Pharmacological preparations in which NO was the active agent have been used for over 1000 years, although this fact was not appreciated at the time (4). It is even more remarkable that the person commonly credited with the co-discovery of NO, Joseph Priestley, also discovered oxygen. The beneficial and toxic effects of both of these gases and their intermolecular interactions were then a major topic of interest and remain so more than 300 years later.
The biological role of NO was resurrected from the category of a historical side note when its central contributions to fundamental biological processes were identified. In particular, the discovery of NO as a signaling molecule in the cardiovascular system was recognized with the Nobel Prize in Physiology and Medicine in 1998 (4). The notion that such a structurally simple free radical can have a role as a second messenger/effector molecule has resulted in a major paradigm shift in the field of cell signaling and stimulated the now maturing field of redox biology. The purpose of this minireview is to summarize how NO is formed, its relevant chemistry, and how this underpins its biological effects.
Any newcomer to the NO field is confronted with a bewildering nomenclature that uses very similar sounding names to describe very distinct molecules, e.g. nitrous oxide (laughing gas) and nitric oxide (sometimes called crying gas by those working on it!). As an aid to the reader, we provide Table 1, in which we list the nitrogen oxides relevant to biological processes with some key biochemical information (2, 5–7). In addition, we provide in Table 1 the most common adducts that are formed from some of the more reactive nitrogen oxide species. In general, the proper chemical terminology to designate the formation of these adducts is based on the functional group, e.g. nitrosation involves formation of the nitroso group (R-NO), nitration involves the nitro group (R-NO2), and nitrosylation involves the nitrosyl group. Unlike the nitroso and nitro groups, the nitrosyl group is formally an NO radical and is not a covalent functional group; rather, it forms a coordinate complex with a transition metal (2).
TABLE 1.
Nitrogen species occurring from the oxidation or reduction of NO
Selected intracellular targets that are well documented in the literature are listed on the right. For a more in-depth review, the reader is directed to the citations in the text.
| Free molecular species | Abbreviation | Comments | Intracellular targets |
|---|---|---|---|
| Nitric oxide or nitrogen monoxide | NO or ·NO | Uncharged free radical product of NO synthases | sGC, CcOx, NHI, thiyl radicals |
| Nitroxyl | HNO or NO− | Charged reduced state of NO; Angeli's salt product; acts as an electrophile | Transition metals, nucleophiles (e.g. thiols) |
| Nitrosonium ion | NO+ | Formally oxidized NO; does not exist at neutral pH (reacts with water) | Thiolate anion |
| Nitrite | NO2− | Product of NO reaction with O2; can be reduced to NO in presence of iron | Heme |
| Nitrate | NO3− | Non-reactive oxidation product of NO | |
| Dinitrogen trioxide or nitrous anhydride | N2O3 | Nitrosating agent; formed from reaction of NO with O2 and from protonation and further reaction of nitrite | Thiols, amines |
| Nitrous oxide | N2O | Laughing gas | |
| Nitrogen dioxide | NO2 or ·NO2 | Strong oxidizing agent; free radical; nitrating agent | Thiols, phenolics (tyrosine) |
| Peroxynitrite | ONOOH or ONOO− | Formed upon reaction of NO with superoxide; nitrating agent | Thiols, transition metals |
| Peroxynitrate | O2NOO− | Formed upon reaction of NO2 with superoxide | ? |
| Nitrosoperoxocarbonate | ONOOCO2− | Formed upon reaction of CO2 with peroxynitrite; probably very short-lived | Predominantly tyrosine |
| Hydroxylamine | NH2OH | Product of nitroxyl reaction | |
| Ammonia | NH3 | Metabolic waste product |
| Adduct | Name | Type of bond to R |
|---|---|---|
| R-NO | Nitroso | Covalent |
| R-NO2 | Nitro | Covalent |
| R-ONO | Nitrito | Covalent |
| R··NO | Nitrosyl | Coordinate (R = metal) |
A PubMed search based on the keyword “nitric oxide” returns over 99,000 references, and thus, a comprehensive review is clearly not feasible or within the attention span of the authors! For this reason, we have selected predominantly review articles as citations as a guide to the reader for more detailed information regarding key points. As a framework for this article, we have posed a series of major research questions for the mechanisms of NO action, which we then address with particular emphasis on the biochemistry of NO.
What Are the Key Chemical Properties of NO Relevant to Biology?
NO is a free radical and can stabilize its unpaired electron by two mechanisms: reaction with species containing other unpaired electrons (thus pairing up the two lone electrons) and interaction with the d-orbitals of transition metals, particularly iron. Although there are multiple subsequent species generated from these two reactions that result in a panoply of biological effects, the biological reactivity of NO itself is quite limited. In its reaction with iron, NO forms stable high affinity bonds with heme and NHI2 (8). The high affinity and rapid binding to ferrous heme are important biologically for the activation of sGC to generate the second messenger cGMP and the regulation of mitochondrial respiration at CcOx (9–16). NO binding to NHI can also result in significant biological effects, including disruption of iron-sulfur centers and also binding to the so-called chelatable iron pool of cells (17). NO is uncharged and highly soluble in hydrophobic environments. This property may allow free diffusion across biological membranes and the potential to signal many cell diameters distant from its site of generation (18, 19).
A common misconception is that all free radicals are, by definition, unstable and highly reactive, but this is not the case. To place this in perspective, NO is chemically stable and much less reactive with non-radical species than the hydroxyl radical but more reactive than lipid peroxyl radicals. Biologically, this means that the fate of NO is dramatically changed in the presence of other free radicals such as superoxide (O2˙̄) (2, 20–22). Once it has reacted with O2˙̄, it forms peroxynitrite and no longer participates in those signaling actions, which require binding to ferrous heme (23). The rate constant for the reaction of NO and O2˙̄ has been reported to be as high as 1.9 × 1010 m−1 s−1, nearing the diffusion limit for reactions (24). It is important to note that when a reaction is “diffusion-limited,” it means that the rate is determined solely by how fast the two reactants encounter each other, which, in turn, is determined by their diffusion. Factors that affect diffusion (e.g. viscosity, convection, etc.) will therefore influence the rate of reaction, and these factors in vivo are certainly distributed heterogeneously (e.g. variations in cytosolic and tissue viscosity). Thus, the rate of peroxynitrite formation will be spatially heterogeneous and vary from location to location. This, in turn, could result in apparent localized protein and lipid modification mediated by peroxynitrite even though the reaction with NO and O2˙̄ is rapid.
How Is NO Made in Biological Systems?
NO is synthesized intracellularly through the action of a family of NOS enzymes (25). NOS enzymes catalyze the NADPH- and O2-dependent oxidation of l-arginine to l-citrulline and NO. The structures of these proteins and their mechanisms of action have been well defined, and a wide range of pharmacological probes of NOS function have been synthesized (25). Enzymatic synthesis of NO is complex and depends on the availability of prosthetic groups and cofactors such as FAD, FMN, tetrahydrobiopterin, and heme (26). Three distinct isoforms have been identified, all of which have different characteristics and generate NO at different rates (27).
eNOS (NOS3) generates the lowest levels of NO and was originally discovered in the vascular endothelium but is also found in neurons, epithelial cells, and cardiac myocytes (28). Its location in the cell and activity are controlled by Ca2+ and calmodulin, as well as post-translational modifications such as phosphorylation and myristoylation (29, 30). Regulation of eNOS is also intimately related to the physical forces important in vascular function such as shear stress induced by blood flow (27, 31). Neuronal NOS (NOS1) is constitutively present in neurons, skeletal muscle, and epithelial cells. It is also a Ca2+/calmodulin-dependent isoform that, in the brain, is activated physiologically by agonists of the N-methyl-d-aspartate receptor (27). The last major NOS isoform is inducible NOS (NOS2), which has the highest capacity to generate NO. This isoform is expressed in multiple cell types in response to inflammatory stimuli such as that induced by endotoxin and cytokines, e.g. interleukin-1 and -2, tumor necrosis factor-α, and interferon-γ (27, 32). It has also been shown to be constitutively present in some tissues such as lung epithelium. The NOS enzymes are regulated by post-translational modifications, controlled localization within the cell, substrate and cofactor availability, and other proteins with which they form stable complexes (27). The extent to which these regulatory features modify the biochemical actions of NO is an active area of research. For example, several studies have suggested that mitochondrially localized or associated forms of NOS may play a specific role in the regulation of mitochondrial function (33, 34).
Recently, it has been suggested that a major metabolite of NO, nitrite (NO2−), can be reduced back to NO by heme proteins such as hemoglobin (35). Some investigators have considered this an enzymatic activity, i.e. a nitrite reductase (36, 37). The regeneration of NO from one of its metabolites would seem at first glance to be a futile exercise; however, this reaction both changes the location of NO formation and decreases the dependence on oxygen (35, 38). This contrasts with the generation of NO from the NOS enzymes, which require oxygen. For this reason, this mechanism, although presently speculative in details, suggests that NO2− may play an important role in generating NO under hypoxic conditions (35, 38, 39). Whether or not this role for NO2− is a physiological process remains to be seen, but its pharmacological potential is now well recognized and supported by a number of studies (40). One important area, which is an active topic for debate, is the source of NO2− in mammalian systems and its distribution in biological tissues (35, 38). As we have speculated previously (41), reaction with O2 in the biological milieu is almost certainly not important as a source of nitrite formation compared with other much more rapid mechanisms (although it could conceivably be a localized source for reactive oxidative/nitrosative species, especially in hydrophobic environments). Thus, the mechanism(s) of nitrite formation in vivo remains elusive, although there is evidence that it may occur mainly from NO formation from eNOS compared with other sources (35, 38). Interestingly, it has been shown that the copper protein ceruloplasmin can catalyze the conversion of NO to NO2− (38).
Recent interest in nitrite's therapeutic potential has focused on its dietary or pharmacological applications (35, 38). This is supported by several studies describing improvements in ischemia/reperfusion injury following acute myocardial infarction, pulmonary hypertension, and organ transplantation following dietary supplementation with nitrite (35).
How Does the Biochemistry of NO Change with Concentration?
As mentioned above, the different NOS isoforms generate NO at different rates. An interesting aspect of the biochemistry of NO is that both concentration and location are key determinants of its ability to activate different cell signaling pathways. Perhaps surprisingly, controversy remains regarding the levels of NO that can be achieved biologically (42). With the widespread availability of NO donors, most biochemical experiments are essentially unconstrained in terms of the amounts of NO that can be added to a cell culture or in vitro system. Therefore, it is possible that NO will not achieve the concentrations necessary in vivo to exhibit all of its biochemical properties defined in vitro.
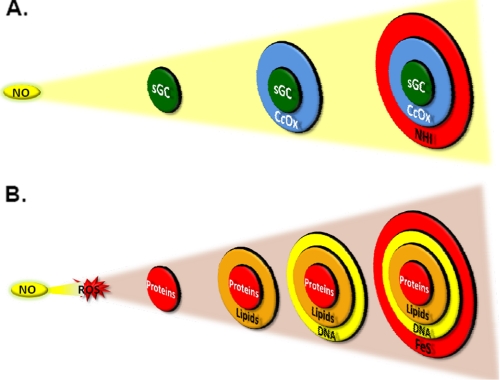
To illustrate this important aspect of NO biology, we have exploited the concept of the “bullseye” with reference to increasing concentrations of NO (Fig. 1). At the center of the bullseye is sGC, which is the most sensitive “receptor” or target for NO (Fig. 1A). In the next ring is the mitochondrial enzyme CcOx (43). As the concentration of NO increases, modification of NHI occurs, which can also regulate cell signaling (44). The concentration at which the endogenous NO-dependent formation of S-nitrosothiols occurs is not clear and is likely to be both protein- and cellular compartment-specific (45). It is important to note that this model does not fully account for the effects of NO generation at different sites in the cell. In addition, augmenting the rate of formation of NO increases the distance over which NO diffuses, which also increases the target (bullseye) size.
FIGURE 1.
Model for the cellular targets of NO and associated nitrogen oxides. A, the most sensitive target of NO is sGC, followed by CcOx and NHI. B, reaction of NO with O2 or ROS changes target susceptibility. Under high O2 tensions or conditions of increased free radical production, NO forms oxidation products that react with proteins, lipids, DNA, and FeS centers predominantly over the targets shown in A.
An interesting effect occurs if we now include ROS such as O2˙̄ in the same compartment where NO is generated (Fig. 1B). In this case, oxidative modification of proteins, lipids, and DNA occurs, the target composition changes, and (Fig. 1B). For sGC to be activated under this condition, the levels of NO must exceed those of superoxide. This is clearly a simplified model, but it begins to illustrate the biological necessity to have NOS isoforms that are capable of generating NO at such different levels and sites within the cell. For example, the control of vascular tone by eNOS requires a localized transient effect, whereas the modulation of synaptic transmission by neuronal NOS requires a more persistent and long-range effect. At the extreme of this spectrum, it is known that for inducible NOS to kill microbes, parasites, or cancer cells, high levels of NO are needed to destroy FeS centers and respiratory proteins in the target organism (28, 31, 32, 46).
What Is the Fate of NO, and How Does That Change Its Biochemistry?
Once generated, NO has a number of potential biological fates. The intra- and extravascular half-lives of NO, which have been measured to be in the range of 2 ms to >2 s, appear to be dependent on the availability of intracellular reactants of NO (47). The most specific and highest affinity interactions of NO with biological targets are those with metalloproteins such as sGC, CcOx, and hemoglobin. It is the individual interactions of NO with each of these metalloproteins that lead to some of the best understood biological effects of NO.
NO reacts rapidly with other free radicals such as O2˙̄ or lipid peroxyl radicals (5, 48). The best studied of these reactions is the very rapid reaction of NO with superoxide to form peroxynitrite (2, 21, 22, 24). For example, peroxynitrite (or reactive species derived from it such as nitrogen dioxide and carbonate anion radical) can mediate the oxidation and nitration of proteins, lipids, or DNA, whereas NO cannot directly mediate any of these effects. Since the realization that peroxynitrite is produced in biological systems, it has been proposed to be a major mediator of the pathological effects associated with NO, particularly inflammation (22). Interestingly, the nitroproteome appears to be quite specific, and pathological responses may be due to modification of particularly sensitive proteins, including those that regulate signaling and cell metabolism (49–52). Peroxynitrite reacts directly with sulfur-containing amino acids (cysteine and methionine), as well as those with aromatic structure (e.g. tryptophan and tyrosine). Furthermore, secondary products of peroxynitrite oxidation can also modify proteins. Although the rate constant for reaction of peroxynitrite with free cysteine is 5.9 × 103 m−1 s−1, reaction with protein thiols is much slower. This may then contribute to the selectivity in protein targets. Additionally, because NO is freely diffusible and O2˙̄ is not, one factor controlling peroxynitrite reactivity may be the location of O2˙̄ production. It is important to also realize that protein nitration can occur through other mechanisms not directly involving the reaction of NO and O2˙̄ such as that of nitrite with the enzyme myeloperoxidase (53). In addition, nitration could be mediated by nitrogen dioxide (NO2˙), which is an important environmental toxin and a byproduct of the reaction of NO with O2. If solely dependent on NO2˙, this mechanism would require two molecules of the oxidant and is therefore less likely to occur biologically. However, there are many ROS that can generate a phenoxyl radical on a tyrosine residue, which could then react with NO2˙, resulting in nitration. A major challenge in understanding the biochemistry of NO has been to determine why the NO-dependent proteomes such as the S-nitrosoproteome and nitroproteome involve limited subsets of proteins within the cell.
How Does NO Act as a Signaling Molecule?
There are essentially two major biochemical mechanisms under current investigation through which NO can mediate signal transduction, which we will discuss in turn.
Binding to Ferrous Heme
The reaction of NO with metalloproteins involves nitrosylation of ferrous iron (Fe2+). However, ligand discrimination between metalloproteins differs widely because of the effects of the protein structure. For instance, sGC does not bind O2 but does form 5-coordinate Fe2+·NO complexes, which subsequently activate the enzyme and are responsible for the cGMP-dependent effects of NO. In contrast, hemoglobin and CcOx have different affinities for NO but also bind O2 (15). Of these three interactions of NO with ferrous heme proteins, it is widely accepted that binding sGC satisfies all of the requirements for cell signaling, including regulation and amplification of the signaling pathway (14, 15).
Notably, CcOx is considerably less sensitive to NO-mediated inhibition than sGC is to NO-dependent activation (14, 43). However, unlike sGC, the interaction of NO with CcOx is competitive with O2 (13, 54). In addition, modulation of mitochondrial respiration by CcOx is dependent on the state of oxidative phosphorylation (55). NO is a better inhibitor of respiration in mitochondria that are actively respiring and at low oxygen tensions (13, 54). The conjunction of these properties has led to the idea that NO may control oxygen gradients in organs such as the heart and liver (54, 56).
Heme-containing O2 transport proteins are found in both intracellular and extracellular compartments and are exposed to varying levels of both NO and O2. Unlike CcOx, interaction of NO with hemoglobin or myoglobin typically results in its consumption. The extremely rapid reaction with HbO2 results in conversion of the NO to nitrate, and NO binds very rapidly to the deoxyheme to form a stable complex (35, 36). These reactions should effectively prevent NO from activating sGC. This results in a true conundrum: how could nanomolar levels of NO generated by the endothelium be capable of relaxing vascular smooth muscle cells in the presence of millimolar HbO2? Thus, a great deal of effort has been expended by researchers to define the interactions of NO with hemoglobin and its biological significance. Although this remains a contentious and controversial area, some key insights are slowly emerging on three fronts. (a) There are significant diffusional barriers between the endothelium and the red blood cell that decrease the inactivation of NO by HbO2. These include the static fluid layer generated by blood flow and a diffusional barrier at the red blood cell membrane (47). (b) NO may be generated within the red blood cell by metabolism of nitrite or, intriguingly, through the presence of an erythrocytic NOS (57). (c) Another possibility is that although the vast majority of NO that enters the red blood cell is rapidly inactivated, a small proportion may be converted to metabolites such as nitrite, which, through as yet unidentified mechanisms, restores NO-dependent activation of sGC. Recent studies also suggest a role for S-nitrosothiol formation, although the mechanism whereby this occurs is unclear. These reactions may be particularly important under conditions of hypoxia in restoring NO-dependent vascular function (35, 58, 59). There is, however, no general consensus on these possibilities. We await further developments in this area for the insights they may offer into NO-dependent regulation of vascular function and the basic biochemistry of NO reactions with heme proteins.
NO-dependent Protein Modifications
The reaction of NO with O2 is also an area of research critical to our understanding of how NO can lead to nitrosation, nitration, or oxidation of protein side chains. Cysteinyl thiols are targets of NO, and NO from exogenous and endogenous sources has been shown to increase intracellular low molecular weight S-nitrosothiols (e.g. S-nitrosoglutathione), promote protein glutathiolation (protein S-glutathione), and increase thiol oxidation (protein oxysulfur acids, PSOxH) (60). However, the different modifications induced by NO appear to be dependent on the RNS, the protein itself, and the subcellular localization of the protein.
Recently, RSNOs have attracted interest because of their potential roles as intermediates in the transport, storage, and delivery of NO; as post-translational protein modifications in cell signaling and inflammation; and as molecular markers of RNS. Well over 100 proteins have been shown to be S-nitrosated, and as the list of S-nitrosated proteins grows, the potential relevance of this modification and other NO-induced protein modifications in redox biology and signaling is of increasing interest (61, 62).
NO can promote S-nitrosation by four basic mechanisms (45). 1) In radical-radical reactions, thiyl radicals on glutathione and proteins can react with NO to form S-nitrosoglutathione and S-nitrosated proteins. 2) In transnitrosation, once formed, the nitroso group can be transferred among thiols on proteins and low molecular weight peptides such as glutathione. 3) Proteins may also be S-nitrosated by N2O3, which is formed from the reaction of NO with O2. Hence, the steady-state concentrations of O2, thiyl radicals, and antioxidants can interact to modulate the formation of RSNOs. 4) The fourth mechanism of RSNO formation is transfer of the nitroso group from metal nitrosyls in a reaction that may not involve O2. The relative contributions of these mechanisms to biological nitrosation are not clear; however, recent evidence seems to implicate cellular non-heme metal nitrosyls as important contributors to nitrosation reactions (63). One of the controversial aspects of the S-nitrosothiol field involves the accurate measurement of NO-induced protein modifications. Initially, the application of nonspecific techniques resulted in estimates of S-nitrosothiols several orders of magnitude higher than the nanomolar amounts now thought to be present in most biological samples. The S-nitrosoproteome would now appear to be much more exclusive than previously thought.
It is important to recognize that S-nitrosation is likely networked with other thiol modifications, including S-glutathiolation and S-oxidation. Several proteins have cysteines that react with RNS and are nitrosated, glutathiolated, and/or oxidized. This ability to incur multiple types of NO-induced modifications underscores a novel yet fundamental facet of redox biology, i.e. the redox switch (64).
NO-induced thiol modifications of proteins can lead to transient changes in enzyme activity, and such modifications could be regulated by enzymes such as glutathione S-transferase Pi and glutaredoxin, which can catalyze the binding of glutathione to oxidized protein thiols or restore thiols to their reduced state (65). Hence, redox switches may represent pathways in which thiol proteomes could be modulated in a well regulated manner.
Despite major advances in this field, it remains extremely challenging to determine whether an NO-dependent post-translational protein modification is a bystander effect or an obligatory step in a cell signaling pathway. The solution to this question will likely lie in the ability to selectively introduce into cellular proteins a defined modification such as protein nitration and observe the effects (66). Quantitative mass spectrometry techniques capable of determining the extent of protein modification are now also being developed and will aid in this quest (67).
How Do We Translate Our Understanding of the Biochemistry of NO to Understanding Its Physiology?
As mentioned above, the biochemistry of NO is governed by 1) its interaction with metalloproteins, oxygen, and ROS and 2) its concentration. In a biological setting, location of both the source of NO production and the targets provides an additional layer of complexity. We will now discuss how the integration of the key biochemical properties of NO outlined above can interact in physiological and pathological processes. To illustrate this, we will use emerging concepts from both the hepatology and cardiovascular fields.
Importantly, mitochondrial dysfunction and altered NO bioavailability, down-regulation of the sGC pathway, and increased protein modifications play a key role in the pathogenesis of liver disease. As discussed above, NO regulates mitochondrial function through reversible binding at the heme site in CcOx, which inhibits O2 consumption and can influence cross-talk mechanisms between the mitochondrion and sGC. Recently, we have shown that both alcoholic steatohepatitis and nonalcoholic steatohepatitis are associated with increased sensitivity of the respiratory chain to inhibition by NO, increased hypoxia, and protein nitration (54, 68, 69). It is proposed that these NO-dependent changes result in excessive inhibition or altered regulation of the respiratory chain. This leads to bioenergetic dysfunction, reductive stress, and ROS production, all of which are key features of mitochondrial dysfunction in diabetes/obesity and alcohol-mediated liver disease.
One key function of NO-mediated inhibition of respiration is to modulate O2 gradients and hypoxia-responsive targets in cells via inhibiting O2 consumption in respiring mitochondria (47). A consequence of this inhibition would be to extend the O2 gradient within tissues and prevent hypoxia. Indeed, a cardioprotective mitochondrially targeted S-nitrosothiol was shown recently to enhance oxygen levels under hypoxic conditions and to promote S-nitrosation of mitochondrial protein thiols (63). These findings suggest that, like its deleterious actions, the protective effects of NO also involve its binding to ferrous iron and its regulation of protein thiol modifications.
Summary
In this minireview, we hope to have demonstrated how the understanding of the biochemistry of NO, gained over the last 30 years, can be used to illuminate its biological properties. It is now becoming clear that these biochemical mechanisms can operate simultaneously within the context of any specific organ or cell. Thus far, the study of these specific properties of NO, i.e. its binding to metalloproteins, reaction with free radicals, and protein modifications, has proceeded in parallel. The challenge in future research will be to integrate these characteristics into a more comprehensive model of NO biology.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AA15172, AA18841, and DK73775 (to S. M. B.); CA131653 (to J. R. L.); AA13395, ES/HL10167, and DK75865 (to V. M. D.-U.); T32 HL007918 (to B. P. D.); and T32 HL07457 (to B. G. H.). This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- NHI
- non-heme iron
- sGC
- soluble guanylate cyclase
- CcOx
- cytochrome c oxidase
- NOS
- nitric-oxide synthase
- eNOS
- endothelial NOS
- ROS
- reactive oxygen species
- RNS
- reactive nitrogen species
- RSNO
- S-nitrosothiol.
REFERENCES
- 1.Moncada S., Palmer R. M., Higgs E. A. (1991) Pharmacol. Rev. 43, 109–142 [PubMed] [Google Scholar]
- 2.Pacher P., Beckman J. S., Liaudet L. (2007) Physiol. Rev. 87, 315–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sprigge J. S. (2002) Anaesthesia 57, 357–364 [DOI] [PubMed] [Google Scholar]
- 4.Butler A. R. (2006) J. R Coll. Physicians Edinb. 36, 185–189 [PubMed] [Google Scholar]
- 5.Patel R. P., McAndrew J., Sellak H., White C. R., Jo H., Freeman B. A., Darley-Usmar V. M. (1999) Biochim. Biophys. Acta 1411, 385–400 [DOI] [PubMed] [Google Scholar]
- 6.Darley-Usmar V., Halliwell B. (1996) Pharm. Res. 13, 649–662 [DOI] [PubMed] [Google Scholar]
- 7.Bruckdorfer R. (2005) Mol. Aspects Med. 26, 3–31 [DOI] [PubMed] [Google Scholar]
- 8.Thomas D. D., Miranda K. M., Colton C. A., Citrin D., Espey M. G., Wink D. A. (2003) Antioxid. Redox Signal. 5, 307–317 [DOI] [PubMed] [Google Scholar]
- 9.Brookes P. S., Levonen A. L., Shiva S., Sarti P., Darley-Usmar V. M. (2002) Free Radic. Biol. Med. 33, 755–764 [DOI] [PubMed] [Google Scholar]
- 10.Brown G. C. (2001) Biochim. Biophys. Acta 1504, 46–57 [DOI] [PubMed] [Google Scholar]
- 11.Denninger J. W., Marletta M. A. (1999) Biochim. Biophys. Acta 1411, 334–350 [DOI] [PubMed] [Google Scholar]
- 12.Erusalimsky J. D., Moncada S. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 2524–2531 [DOI] [PubMed] [Google Scholar]
- 13.Cooper C. E., Giulivi C. (2007) Am. J. Physiol. Cell Physiol. 292, C1993–C2003 [DOI] [PubMed] [Google Scholar]
- 14.Garthwaite J. (2010) Mol. Cell. Biochem. 334, 221–232 [DOI] [PubMed] [Google Scholar]
- 15.Derbyshire E. R., Marletta M. A. (2009) Handb. Exp. Pharmacol. 191, 7–31 [DOI] [PubMed] [Google Scholar]
- 16.Moncada S., Higgs E. A. (2006) Br. J. Pharmacol. 147, Suppl. 1, S193–S201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler A. R., Megson I. L. (2002) Chem. Rev. 102, 1155–1166 [DOI] [PubMed] [Google Scholar]
- 18.Lancaster J. R., Jr. (2004) Science 304, 1905 (Author Reply 1905) [DOI] [PubMed] [Google Scholar]
- 19.Lancaster J. R., Jr. (1997) Nitric Oxide. 1, 18–30 [DOI] [PubMed] [Google Scholar]
- 20.Trujillo M., Ferrer-Sueta G., Radi R. (2008) Antioxid. Redox Signal. 10, 1607–1620 [DOI] [PubMed] [Google Scholar]
- 21.Szabó C., Ischiropoulos H., Radi R. (2007) Nat. Rev. Drug Discov. 6, 662–680 [DOI] [PubMed] [Google Scholar]
- 22.Beckman J. S. (2009) Arch. Biochem. Biophys. 484, 114–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McAndrew J., Patel R. P., Jo H., Cornwell T., Lincoln T., Moellering D., White C. R., Matalon S., Darley-Usmar V. (1997) Semin. Perinatol. 21, 351–366 [DOI] [PubMed] [Google Scholar]
- 24.Koppenol W. H. (2001) Redox Rep. 6, 339–341 [DOI] [PubMed] [Google Scholar]
- 25.Alderton W. K., Cooper C. E., Knowles R. G. (2001) Biochem. J. 357, 593–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Poulos T. L. (2005) J. Inorg. Biochem. 99, 293–305 [DOI] [PubMed] [Google Scholar]
- 27.Kone B. C., Kuncewicz T., Zhang W., Yu Z. Y. (2003) Am. J. Physiol. Renal Physiol. 285, F178–F190 [DOI] [PubMed] [Google Scholar]
- 28.Dudzinski D. M., Michel T. (2007) Cardiovasc. Res. 75, 247–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oess S., Icking A., Fulton D., Govers R., Müller-Ester W. (2006) Biochem. J. 396, 401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sessa W. C. (2004) J. Cell Sci. 117, 2427–2429 [DOI] [PubMed] [Google Scholar]
- 31.Balligand J. L., Feron O., Dessy C. (2009) Physiol. Rev. 89, 481–534 [DOI] [PubMed] [Google Scholar]
- 32.Kleinert H., Pautz A., Linker K., Schwarz P. M. (2004) Eur. J. Pharmacol. 500, 255–266 [DOI] [PubMed] [Google Scholar]
- 33.Finocchietto P. V., Franco M. C., Holod S., Gonzalez A. S., Converso D. P., Arciuch V. G., Serra M. P., Poderoso J. J., Carreras M. C. (2009) Exp. Biol. Med. 234, 1020–1028 [DOI] [PubMed] [Google Scholar]
- 34.Kato K., Giulivi C. (2006) Front. Biosci. 11, 2725–2738 [DOI] [PubMed] [Google Scholar]
- 35.Lundberg J. O., Gladwin M. T., Ahluwalia A., Benjamin N., Bryan N. S., Butler A., Cabrales P., Fago A., Feelisch M., Ford P. C., Freeman B. A., Frenneaux M., Friedman J., Kelm M., Kevil C. G., Kim-Shapiro D. B., Kozlov A. V., Lancaster J. R., Jr., Lefer D. J., McColl K., McCurry K., Patel R. P., Petersson J., Rassaf T., Reutov V. P., Richter-Addo G. B., Schechter A., Shiva S., Tsuchiya K., van Faassen E. E., Webb A. J., Zuckerbraun B. S., Zweier J. L., Weitzberg E. (2009) Nat. Chem. Biol. 5, 865–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gladwin M. T., Grubina R., Doyle M. P. (2009) Acc. Chem. Res. 42, 157–167 [DOI] [PubMed] [Google Scholar]
- 37.Gladwin M. T., Kim-Shapiro D. B. (2008) Blood 112, 2636–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundberg J. O., Weitzberg E., Gladwin M. T. (2008) Nat. Rev. Drug Discov. 7, 156–167 [DOI] [PubMed] [Google Scholar]
- 39.Gladwin M. T. (2006) Adv. Exp. Med. Biol. 588, 189–205 [DOI] [PubMed] [Google Scholar]
- 40.Kim-Shapiro D. B., Schechter A. N., Gladwin M. T. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 697–705 [DOI] [PubMed] [Google Scholar]
- 41.Möller M. N., Li Q., Lancaster J. R., Jr., Denicola A. (2007) IUBMB Life 59, 243–248 [DOI] [PubMed] [Google Scholar]
- 42.Hall C. N., Garthwaite J. (2009) Nitric Oxide 21, 92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodríguez-Juárez F., Aguirre E., Cadenas S. (2007) Biochem. J. 405, 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Demple B. (2002) Mol. Cell. Biochem. 234–235, 11–18 [PubMed] [Google Scholar]
- 45.Zhang Y., Hogg N. (2005) Free Radic. Biol. Med. 38, 831–838 [DOI] [PubMed] [Google Scholar]
- 46.Kleppisch T., Feil R. (2009) Handb. Exp. Pharmacol. 191, 549–579 [DOI] [PubMed] [Google Scholar]
- 47.Thomas D. D., Liu X., Kantrow S. P., Lancaster J. R., Jr. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Donnell V. B., Freeman B. A. (2001) Circ. Res. 88, 12–21 [DOI] [PubMed] [Google Scholar]
- 49.Alvarez B., Radi R. (2003) Amino Acids 25, 295–311 [DOI] [PubMed] [Google Scholar]
- 50.Xu S., Ying J., Jiang B., Guo W., Adachi T., Sharov V., Lazar H., Menzoian J., Knyushko T. V., Bigelow D., Schöneich C., Cohen R. A. (2006) Am. J. Physiol. Heart Circ. Physiol. 290, H2220–H2227 [DOI] [PubMed] [Google Scholar]
- 51.Koeck T., Stuehr D. J., Aulak K. S. (2005) Biochem. Soc. Trans. 33, 1399–1403 [DOI] [PubMed] [Google Scholar]
- 52.Aulak K. S., Miyagi M., Yan L., West K. A., Massillon D., Crabb J. W., Stuehr D. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12056–12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brennan M. L., Wu W., Fu X., Shen Z., Song W., Frost H., Vadseth C., Narine L., Lenkiewicz E., Borchers M. T., Lusis A. J., Lee J. J., Lee N. A., Abu-Soud H. M., Ischiropoulos H., Hazen S. L. (2002) J. Biol. Chem. 277, 17415–17427 [DOI] [PubMed] [Google Scholar]
- 54.Shiva S., Oh J. Y., Landar A. L., Ulasova E., Venkatraman A., Bailey S. M., Darley-Usmar V. M. (2005) Free Radic. Biol. Med. 38, 297–306 [DOI] [PubMed] [Google Scholar]
- 55.Antunes F., Boveris A., Cadenas E. (2007) Antioxid. Redox Signal. 9, 1569–1579 [DOI] [PubMed] [Google Scholar]
- 56.Shiva S., Brookes P. S., Patel R. P., Anderson P. G., Darley-Usmar V. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7212–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ozüyaman B., Grau M., Kelm M., Merx M. W., Kleinbongard P. (2008) Trends Mol. Med. 14, 314–322 [DOI] [PubMed] [Google Scholar]
- 58.Stamler J. S., Singel D. J., Piantadosi C. A. (2008) Nat. Med. 14, 1008–1009 (Author Reply 1009–1010) [DOI] [PubMed] [Google Scholar]
- 59.Angelo M., Hausladen A., Singel D. J., Stamler J. S. (2008) Methods Enzymol. 436, 131–168 [DOI] [PubMed] [Google Scholar]
- 60.Hogg N. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 585–600 [DOI] [PubMed] [Google Scholar]
- 61.Lopez-Sanchez L. M., Muntane J., de la Mata M., Rodriguez-Ariza A. (2009) Proteomics 9, 808–818 [DOI] [PubMed] [Google Scholar]
- 62.Foster M. W., Hess D. T., Stamler J. S. (2009) Trends Mol. Med. 15, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bosworth C. A., Toledo J. C., Jr., Zmijewski J. W., Li Q., Lancaster J. R., Jr. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4671–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cooper C. E., Patel R. P., Brookes P. S., Darley-Usmar V. M. (2002) Trends Biochem. Sci. 27, 489–492 [DOI] [PubMed] [Google Scholar]
- 65.Mieyal J. J., Gallogly M. M., Qanungo S., Sabens E. A., Shelton M. D. (2008) Antioxid. Redox Signal. 10, 1941–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Neumann H., Hazen J. L., Weinstein J., Mehl R. A., Chin J. W. (2008) J. Am. Chem. Soc. 130, 4028–4033 [DOI] [PubMed] [Google Scholar]
- 67.Schöneich C., Sharov V. S. (2006) Free Radic. Biol. Med. 41, 1507–1520 [DOI] [PubMed] [Google Scholar]
- 68.Venkatraman A., Shiva S., Wigley A., Ulasova E., Chhieng D., Bailey S. M., Darley-Usmar V. M. (2004) Hepatology 40, 565–573 [DOI] [PubMed] [Google Scholar]
- 69.Mantena S. K., Vaughn D. P., Andringa K. K., Eccleston H. B., King A. L., Abrams G. A., Doeller J. E., Kraus D. W., Darley-Usmar V. M., Bailey S. M. (2009) Biochem. J. 417, 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.