Abstract
γ-Secretase is a multimeric membrane protein complex composed of presenilin (PS), nicastrin, Aph-1, and Pen-2, which mediates intramembrane proteolysis of a range of type I transmembrane proteins. We previously analyzed the functional roles of the N-terminal transmembrane domains (TMDs) 1–6 of PS1 in the assembly and proteolytic activity of the γ-secretase using a series of TMD-swap PS1 mutants. Here we applied the TMD-swap method to all the TMDs of PS1 for the structure-function analysis of the proteolytic mechanism of γ-secretase. We found that TMD2- or -6-swapped mutant PS1 failed to bind the helical peptide-based, substrate-mimic γ-secretase inhibitor. Cross-linking experiments revealed that both TMD2 and TMD6 of PS1 locate in proximity to the TMD9, the latter being implicated in the initial substrate binding. Taken together, our data suggest that TMD2 and the luminal side of TMD6 are involved in the formation of the initial substrate-binding site of the γ-secretase complex.
Keywords: Alzheimers Disease, Amyloid, Aspartic Protease, Membrane Enzymes, Presenilin, Secretases
Introduction
γ-Secretase is an unusual aspartic protease that mediates an intramembrane proteolysis of a number of type I transmembrane proteins, including amyloid precursor protein (APP)3 and Notch receptor (1, 2). A body of evidence revealed that γ-secretase is a high molecular weight (HMW) complex, composed of presenilin (PS), nicastrin (Nct), Aph-1, and Pen-2 (3). Mutations in PS genes are linked to the early onset familial Alzheimer disease. Familial Alzheimer disease-linked PS mutations influence the processing of APP, leading to an overproduction of amyloid peptide ending at the 42nd residue (Aβ42) that is more prone to forming amyloid deposits. Thus, PS-γ-secretase complex is a plausible therapeutic target for the treatment of Alzheimer disease (1, 2).
PS is a highly conserved polytopic membrane protein with nine transmembrane domains (TMDs), and two conserved aspartates in PS (i.e. Asp257 in TMD6 and Asp385 in TMD7) comprise the catalytic site of the γ-secretase (2). Chemical biological approaches using γ-secretase inhibitors (GSIs) revealed its unusual enzymatic characters; immobilized transition state analogue-type GSIs directly target PS NTF/CTF heterodimer and co-purify with γ-secretase substrates (4). In contrast, designed helical peptide-type GSI binds to PS irrespective of the presence or absence of transition state analogue-type GSIs that occupy the catalytic site (5, 6). These findings strongly suggested that γ-secretase has an “initial substrate-binding site” that is distinct from the catalytic site (2).
During the biosynthetic process of the γ-secretase complex, Nct and Aph-1 form a heterodimeric subcomplex and bind to PS (7, 8). Pen-2 then is assembled to the heterotrimeric complex and triggers the endoproteolysis of PS to generate N- and C-terminal fragments (NTF and CTF), which represent the catalytically active form (3). PS forms a catalytic pore structure in which catalytic aspartates face a hydrophilic environment, enabling the proteolysis of hydrophobic substrates within the lipid bilayer (9–11). However, information regarding the initial substrate-binding site within PS at the molecular level is still lacking. Using TMD-swap mutants of PS1 that are replaced at each one of the TMDs of the NTF with the TMDs of irrelevant proteins, we have shown that all the PS1 TMDs in NTF (i.e. TMDs 1–6), except for TMD3, are required for the acquisition of the γ-secretase activity (12). We further showed that PS1 TMD4 represents the direct binding region of Pen-2. Here we further systematically analyzed the TMD-swap mutants of PS1 and revealed the stepwise formation of the functional γ-secretase complex. Notably, we found that TMD2 and the luminal side of TMD6 of PS1 contribute to the formation of the initial substrate-binding site of the γ-secretase complex.
EXPERIMENTAL PROCEDURES
Plasmid Construction, Cell Culture, Transfection, and Retroviral Infection
cDNAs encoding PS1, APP carrying Swedish mutation (APPNL), and NotchΔE were inserted into pMXs-puro (9, 11–13). cDNAs encoding mutant PS1 were generated by long PCR-based QuikChangeTM strategy (Stratagene). All constructs were sequenced using Thermo Sequenase (USB Corp., Cleveland, OH) on an automated sequencer (Li-Cor, Lincoln, NE) (see Table 1). Maintenance of DKO cells, viral packaging in Plat-E cells, retroviral infection, and generation of stable infectant pools were done as described previously (9, 11–14).
TABLE 1.
Amino acid alignment of swapped region in TMD-swap PS1 mutants
Alignments of amino acid sequences of the TMDs of PS1 and CD4 or CLAC-P are shown. Numbers indicate the region for TMD-swap mutant in PS1. Amino acid sequences remained as those of PS1 in TM6mt are underlined.
| TM1mt | 82VIMLFVPVTLCMVVVVATI100 | PS1 TMD1 |
| 82CAVLAALLSVVAVVSCLYL100 | CLAC-P | |
| TM2mt | 135NAAIMISVIVVMTILLVVLY154 | PS1 TMD2 |
| 135ALIVLGGVAGLLLFIGLGIF154 | CD4 | |
| TM3mt | 164AWLIISSLLLLFFFSFIYLG183 | PS1 TMD3 |
| 164CAVLAALLSVVAVVSCLYLG183 | CLAC-P | |
| TM4mt | 190NVAVDYITVALLIWNFGVVGMISI213 | PSI TMD4 |
| 190MALIVLGGVAGLLLFIGLGIFFCV213 | CD4 | |
| TM5mt | 219LRLQQAYLIMISALMALVFI238 | PS1 TMD5 |
| 219CAVLAALLSVVAVVSCLYLG238 | CLAC-P | |
| TM6mt | 244WTAWLILAVISVYDLVAVL255 | PS1 TMD6 |
| 244MALIVLGGVAGLYDLVAVL255 | CD4/TMD6 | |
| TM8mt | 408IACFVAILIGLCLTLLLLAIF428 | PS1 TMD8 |
| 408MALIVLGGVAGLLLFIGLGIF428 | CD4 | |
| TM9mt | 434ALPISITFGLVFYFATDYLV453 | PS1 TMD9 |
| 434CAVLAALLSVVAVVSCLYLG453 | CLAC-P |
Antibodies and Immunochemical Analyses
Anti-G1Nr2, G1L3, and PNT3 antibodies against glutathione S-transferase-fused human PS1 N terminus, glutathione S-transferase-fused human PS1 loop, or a synthetic peptide corresponding to the N-terminal 26 amino acids of human/mouse Pen-2, respectively, have been previously described (3, 15, 16). Anti-PS1NT, anti-PS-C3, and anti-SPPct antibodies were kindly provided by Drs. G. Thinakaran (University of Chicago, Chicago, IL), A. Takashima (RIKEN, Saitama, Japan), and T. E. Golde (Mayo Clinic, Jacksonville, FL), respectively (17–19). The monoclonal antibody anti-α-tubulin AA4.3 developed by Dr. C. Walsh was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD, National Institutes of Health, and maintained by The University of Iowa, Department of Biology, Iowa City, IA. Other antibodies were purchased from Chemicon (anti-PS1 loop (MAB5232)), Cell Signaling Technology (anti-cleaved Notch1 (V1744)), Covance (anti-Aph-1aL (O2C2)), Santa Cruz Biotechnology (anti-Nct (N19)), or Sigma (anti-Nct (N1660)). Membrane fractionation, immunoblot analysis, immunoprecipitation of CHAPSO-solubilized lysates, in vitro γ-secretase assay, cycloheximide treatment, trypsin digestion, or quantitation of Aβ by two-site ELISAs using BNT77 as a capture antibody were performed as described previously (3, 8, 9, 11–16, 20).
Blue Native-PAGE (BN-PAGE) Analysis
BN-PAGE was performed according to the manufacturer's protocol (Invitrogen). Briefly, membrane fractions were suspended in Native PAGETM sample buffer containing 1% digitonin (Wako Biochemicals). The mixtures were centrifuged for 10 min at 15,000 × g, and Coomassie Brilliant Blue was added to the supernatant to give a final concentration of 0.25% w/v. NativeMarkTM unstained protein standard (Invitrogen) was used as a molecular weight standard. After electrophoresis, the gel was transferred to polyvinylidene difluoride membranes. The membranes were destained briefly in methanol before being incubated with specific antibodies.
Compounds, Photoaffinity Labeling, Substituted Cysteine-scanning Method (SCAM), and Cross-linking Experiments
L-685,458, peptide 11 (pep.11) and pep.11-Bt were purchased from Bachem (Torrance, CA), Ito Life Sciences (Moriya, Japan), and BEX (Tokyo, Japan), respectively. L-852,646 was kindly provided by Dr. Yueming Li (21). All methanethiosulfonate (MTS) reagents (Toronto Research Chemicals) were dissolved in dimethyl sulfoxide (DMSO) at 200 mm prior to use or stored at −80 degree until use. Photoaffinity labeling, SCAM, and cross-linking experiments were performed as described previously (6, 9, 11, 22, 23).
RESULTS
Transmembrane Domains 8 and 9 of Presenilin 1 are Required for the γ-Secretase Activity
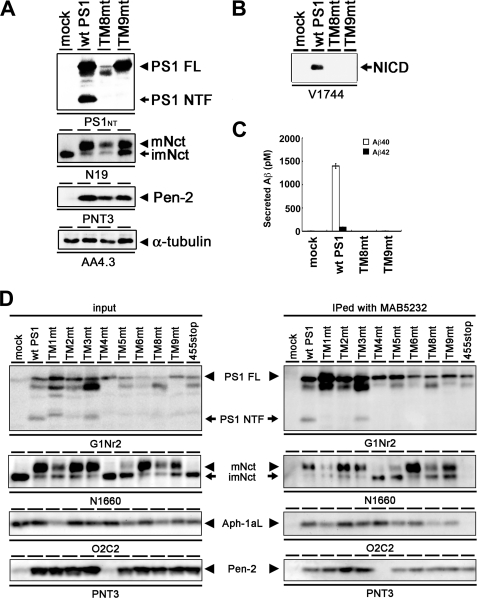
We have previously analyzed the functional role(s) of the PS TMDs using TMD-swap mutants (12). We applied this strategy to PS1 CTF and constructed cDNAs encoding PS1 mutants (i.e. TM8mt, TM9mt) in which TMD8 (408–428 amino acids) or TMD9 (434–453 amino acids) was replaced with TMDs of functionally unrelated transmembrane proteins, i.e. CD4 (type I transmembrane protein) or CLAC-P (type II transmembrane protein), respectively (Table 1). As reported previously, overexpression of wild-type (WT) PS1 in DKO cells resulted in the generation of PS fragments and recovered the levels of mature Nct and the accumulation of Pen-2 (Fig. 1A) (12). Both TM8mt and TM9mt also rescued the accumulation of mature Nct and Pen-2, although the level of mature Nct in DKO cells expressing TM8mt was lower than that in cells expressing WT PS1. However, endoproteolysis of TM8mt or TM9mt PS1 was not observed. We next analyzed the γ-secretase activities of the TMD-swap mutants in DKO cells. Unexpectedly, neither TM8mt nor TM9mt recovered the γ-secretase activity to generate NICD or Aβ in cell-based assays (Fig. 1, B and C). These TMD-swap mutants also displayed no proteolytic activity in in vitro assays (data not shown).
FIGURE 1.
Complementation of maturation of Nct and accumulation of Pen-2 by expression of TM8mt and TM9mt in DKO cells. A, immunoblot analysis of DKO cells stably transfected with WT or mutant PS1 (as indicated above the panel). Cell lysates were analyzed by immunoblotting with each antibody (as indicated below the panel). FL, full length; mNct, mature nicastrin; imNct, immature nicastrin. B, immunoblot analysis of the generation of NICD from NotchΔE-stable DKO cells. C, sandwich ELISA analysis of secreted Aβ from APPNL-stable DKO cells transiently expressing TMD-swap PS1 mutants. The levels of Aβ40 and Aβ42 are shown by white and black bars, respectively. Error bars indicate S.E. D, co-immunoprecipitation analysis of 1% CHAPSO-solubilized fractions prepared from DKO cells transiently transfected with WT or mutant PS1. Soluble fractions were precipitated (IPed) by MAB5232 and then analyzed by immunoblotting with each antibody as indicated below the panel.
To address whether replacement of the PS1 TMDs interferes with the formation of γ-secretase complex, we analyzed the CHAPSO lysates of DKO cells expressing WT or mutant PS1 by co-immunoprecipitation (Fig. 1D). Upon overexpression of WT PS1, all other γ-secretase components (i.e. Nct, Aph-1aL, and Pen-2) were co-precipitated with WT PS1. In contrast, TM4mt and the C-terminal deletion mutant PS1 that lacks the C-terminal 12 amino acids (i.e. PS1/455stop) failed to bind the Pen-2 or Nct-Aph-1 subcomplex, respectively, in accordance with previous reports (12, 24, 25). Notably, all TMD-swap mutants including TM8mt and TM9mt interacted with Nct, Aph-1aL, and Pen-2, indicating that the primary amino acid sequences of TMD8 or -9 are dispensable to the interaction with γ-secretase components. Taken together, these results indicate that the primary structures of PS1 TMD8 and -9 contribute to the acquisition of the conformation of active γ-secretase or its enzymatic activity but not to the interactions with other essential components of γ-secretase.
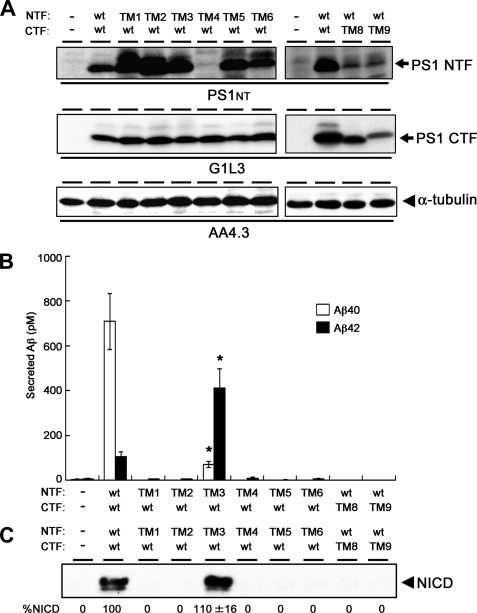
Intramolecular Complementation Assay by Co-expression of NTF and CTF Individually in DKO Cells
Several lines of evidence suggest that the biologically active form of PS is the NTF/CTF heterodimer, although the functional significance of endoproteolysis remains to be elucidated. To ask whether ineffective endoproteolysis inhibited the proteolytic activity of the TMD-swap PS mutant, we further examined the effects of TMD-swaps in an intramolecular complementation analysis by co-expression of NTF and CTF in DKO cells (Fig. 2A). The level of protein expression of TM4mt NTF was relatively low, supporting the notion that the interaction of PS1 with Pen-2 is involved in the stabilization of the γ-secretase complex (12, 26) (see below). As reported, expression of WT NTF and WT CTF from separate vectors was able to restore the γ-secretase activity (25) (Fig. 2B). Interestingly, co-expression of TM3mt NTF with WT CTF improved the total Aβ-generating as well as the NICD-generating activities of γ-secretase more than those in cells expressing the TM3mt holoprotein (12). In addition, TM3mt showed a significant decrease in the production of Aβ40 and a reciprocal increase in that of Aβ42, respectively. Of note, the same trend in the generation of Aβ40 and Aβ42 had been observed in holoprotein-based TM3mt PS1 (12). In fact, the total levels of Aβ and NICD were almost similar to those in cells expressing WT NTF/WT CTF (n = 3, p = 0.12 (for Aβ) and p = 0.30 (for NICD) by Student's t test). This result suggests that the TMD3 of PS1 harbors as yet unknown function to promote the γ-secretase activity after endoproteolysis regardless of the substrates. Further analysis on the role of TMD3 on substrates other than APP and Notch would be required. Intriguingly, some residues on TMD3 have been implicated in the determination of PS1/PS2 specificity of a subset of the γ-secretase inhibitors (26). Nevertheless, these data support the idea that TMD3 is involved in the acquisition of proper cleavage activity of the γ-secretase after the assembly of the complex. However, none of the other TMD-swap mutants expressed in an endoproteolyzed form rescued any γ-secretase activity, suggesting that the structures of each of these TMDs of PS1, other than TMD4, are indispensable to the acquisition of γ-secretase activity, independently from their roles in PS endoproteolysis.
FIGURE 2.
Intramolecular complementation analysis in DKO cells. A, immunoblot analysis of DKO cells transiently co-expressing NTF and CTF of PS1. B, sandwich ELISA analysis of secreted Aβ from APPNL-stable DKO cells transiently co-expressing mutant NTFs and CTFs. The levels of Aβ40 and Aβ42 are shown by white and black bars, respectively. TM3mt exhibited a significantly decreased Aβ40 activity and increased Aβ42-generating activities (*, statistically significant (n = 3, p < 0.001) when compared with those in cells co-expressing WT NTF/WT CTF by Student's t test). Error bars indicate S.E. C, immunoblot analysis of the generation of NICD from NotchΔE-stable DKO cells using anti-V1744 antibody. Numbers shown below each lane denote the relative band intensities of NICD (%, n = 3) when compared with those in cells co-expressing WT NTF/WT CTF.
TMD1, -5, -8, and -9 Are Required for the Stability, but Not the Formation, of the HMW Complex
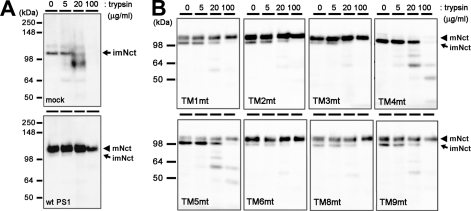
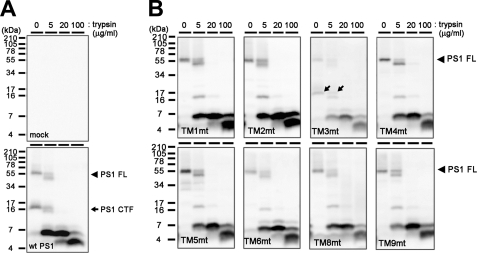
During the assembly of the functional γ-secretase complex, Nct acquires resistance against trypsin and undergoes the complex-type N-glycosylation (27). These posttranslational modifications represent conformational maturation of the extracellular domain of Nct and the proper trafficking of the γ-secretase complex (28–31). Mature Nct proteins were detected in all TMD-swap-expressing cell lysates, except for TM4mt, suggesting that the trafficking of the γ-secretase complex was not significantly altered by the swapping of TMDs (Fig. 3). Moreover, the maturated form of Nct proteins expressed with TMD-swap PS1 acquired the resistance against trypsin treatment independent of the PS1 genotype. However, the levels of mature Nct were significantly lower in cells expressing TM1mt, TM5mt TM8mt, and TM9mt PS1 when compared with WT PS1. Altogether, the maturation of Nct was unaffected by TMD-swap mutation in PS1 TMDs, except for the TM4mt.
FIGURE 3.
Trypsin digestion of Nct polypeptides in DKO cells expressing TMD-swap PS1 mutants. 1% CHAPSO-solubilized lysates prepared from DKO cells transiently transfected with WT PS1 (A) or TMD-swap PS1 mutants (B) were divided into four samples and incubated with trypsin as indicated by concentrations above the panels and analyzed by immunoblotting using anti-Nct antibody (N1660). imNct, immature nicastrin; mNct, mature nicastrin.
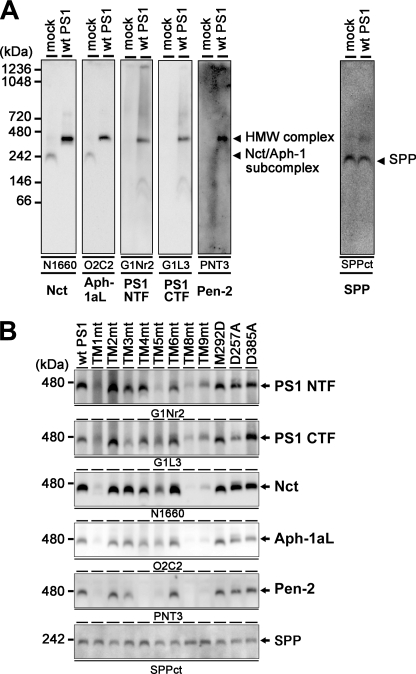
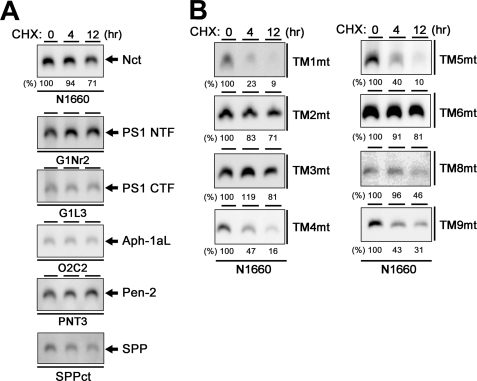
The formation of stabilized HMW complex containing PS is tightly related to its enzymatic function (15, 18). Upon BN-PAGE analysis, all components of the γ-secretase complex were detected at the position of the HMW complex with an apparent molecular mass of 440 kDa (Fig. 4A). We used the endogenous signal peptide peptidase (SPP) as a loading control, which was mainly detected as a 200-kDa complex irrespective of the overexpression of PS1.4 Mutant PS1 carrying M292D, D257A, or D385A mutation was also detected as the ∼440-kDa HMW complex, suggesting that the HMW complex formation was independent from the endoproteolysis or the catalytic activity of PS1 (Fig. 4B). Consistent with the results of the immunoprecipitation assay, all TMD-swap PS1 mutants were also detected as the HMW complex together with other γ-secretase components. Notably, TM4mt was detected as an ∼440-kDa complex, too, indicating that Pen-2 is dispensable for the HMW complex formation. In good agreement with our previous result that TM4mt was unstable (12), the amount of the TM4mt-containing HMW complex was smaller than that of WT PS1. In addition, the steady state levels of TM1mt-, TM5mt-, TM8mt-, or TM9mt-containing HMW complex were significantly lower than that of the HMW complex containing WT PS1. To further analyze the effects of swap of TMDs on the γ-secretase complex, we examined the stability of the HMW complex by cycloheximide treatment. Complex containing WT PS1 was highly stable at the chase of 12 h (Fig. 5A). In contrast, TM1mt-, TM4mt-, TM5mt-, TM8mt-, or TM9mt-containing HMW complexes were rapidly degraded (Fig. 5B), whereas those containing TM2mt, TM3mt, or TM6mt were highly stabilized similarly to that containing WT PS1. We then examined the major conformational change of TMD-swap PS1 mutants by trypsin digestion (Fig. 6). Unexpectedly, the patterns of digested bands of the TMD-swap PS1 mutants were almost similar to those of WT PS1, suggesting that the TMD-swap mutations did not cause significant defects in the structural integrity of the PS1 polypeptides. Taken together, these results indicate that the TMD1, -5, -8, and -9 of PS1 are critical regions for the stabilization of the γ-secretase subsequent to the formation of the HMW complex.
FIGURE 4.
HMW complex formation of TMD-swap PS1 mutants. A, BN-PAGE analysis of digitonin-solubilized lysate prepared from DKO cells expressing mock or WT PS1. Antibodies used as well as target proteins are indicated below the panel. Note that the overexpression of PS1 in DKO cells restored the formation of the HMW complex that migrated at 440 kDa. B, BN-PAGE analysis of mutant PS1. SPP was used as loading control as SPP was detected at 200 kDa in this analysis irrespective of PS1 overexpression.
FIGURE 5.
Stability of the HMW complex harboring TMD-swap PS1 mutants. A, analysis of stability of the HMW complex containing WT PS1 in DKO cells incubated in culture medium containing cycloheximide (CHX) (30 mg/ml). DKO cells after various incubation periods (0–12 h) were solubilized in BN-PAGE lysis buffer and analyzed by BN-PAGE analysis with each antibody (as indicated below the panel). In immunoblot using anti-Nct antibody, numbers shown above or below each lane denote the incubation period and the relative band intensity, respectively. Note that all components were stable over 12 h. B, analysis of stability of the HMW complex harboring TMD-swap PS1 mutant probed by anti-Nct antibody.
FIGURE 6.
Trypsin digestion of TMD-swap PS1 polypeptides in DKO cells. 1% CHAPSO-solubilized lysates prepared from DKO cells transiently transfected with WT PS1 (A) or TMD-swap PS1 mutants (B) were divided into four samples and incubated with trypsin at the indicated concentrations above the panels and analyzed by immunoblotting using the anti-PS1 C terminus antibody (anti-PS-C3). Full-length PS1 (PS1 FL) and CTF are shown by black arrowheads and arrows, respectively. Note that the digestion patterns of mutant PS1 are almost similar, whereas the stability of HMW complex is different (see Fig. 5B).
Photoaffinity Labeling Experiments Using TMD-swap PS1 Mutants
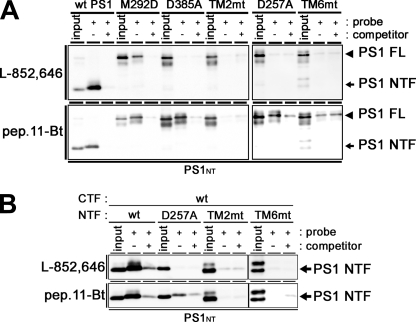
The biochemical characters of the HMW complex containing the TM2mt or TM6mt PS1 were almost similar to that with WT PS1. To gain further insights into the molecular mechanisms underlying the enzymatic defects of these mutants, we took the photoaffinity labeling approach. The helical peptide-type GSI-based photoprobe pep.11-Bt and the transition state analogue-type GSI-based photoprobe L-852,646 are directly targeted to the initial substrate-binding site and the catalytic site, respectively (5, 6, 21). These probes specifically labeled PS1 NTF as well as PS1/M292D holoprotein, whereas WT PS1 holoprotein was never positively labeled, supporting the notion that the PS1 fragments forming the stabilized HMW complex are the proteolytically active form and that these two distinct functional sites are formed independently of the endoproteolysis (Fig. 7A). Moreover, pep.11-Bt bound to the catalytic site mutants (i.e. PS1/D257A and PS1/D385A), whereas L-852,646 did not specifically label these catalytic site mutants. Thus, formation of the initial substrate-binding site of γ-secretase appears to take place independently of that of the catalytic site, in agreement with the previous prediction (4). In contrast, TM2mt and TM6mt PS1 were labeled neither by the pep.11-Bt-binding site nor by the L-852,646 catalytic site photoprobes. Photoaffinity labeling experiments combined with intramolecular complementation (i.e. co-expression of TM2mt or TM6mt NTF and WT CTF) also resulted in no specific binding of the probes (Fig. 7B). These results suggest that TMD2 and the luminal side of TMD6 have critical role(s) in the formation of the initial substrate-binding site and the catalytic site.
FIGURE 7.
Photoaffinity labeling experiments using TMD-swap PS1 mutants. A, CHAPSO-solubilized DKO cell membranes (DKO cells expressing PS1 holoproteins) were photoactivated in the absence (−) or presence (+) of the parent compound as a competitor and analyzed by immunoblotting with anti-PS1 NTF antibody (PS1NT). PS1 FL, full-length PS1. B, CHAPSO-solubilized DKO cell membranes (DKO cells co-expressing PS1 NTF and PS1 CTF individually from separate vectors) were photoactivated in the absence (−) or presence (+) of the parent compound as a competitor and analyzed by immunoblotting with anti-PS1NT antibody. Probes used are indicated at left of the panel.
TMD2 and -6 Locate in Proximity to TMD9
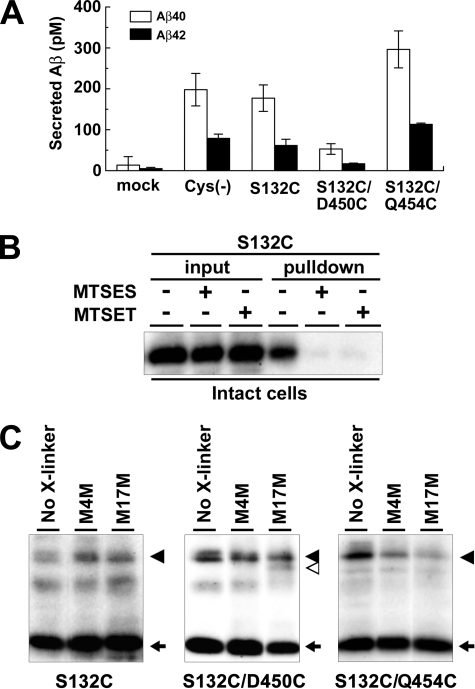
We have previously reported the functional role of the extracellular/luminal side of PS1 TMD9 in the initial substrate binding of the γ-secretase by SCAM using MTS reagents (9, 11). Of note, a sulfhydryl-based cross-linking experiment revealed that TMD9 locates in proximity to the residue Leu250 in TMD6. To test whether TMD2 also locates in proximity to TMD9, we generated mutant PS1 that harbors a single cysteine substituted at residue Ser132, which is placed at the juxtamembrane region of TMD2 on a Cys-less PS1 basis (PS1Cys(−)/S132C). Amino acid substitution of Ser132 to cysteine did not affect the γ-secretase activity (Fig. 8A). SCAM labeling using N-biotinylaminoethyl methanethiosulfonate and competition experiments by charged MTS reagents (i.e. negatively charged 2-sulfonatoethyl methanethiosulfonate (MTSES) and the bulkier, positively charged 2-(trimethylammonium)-ethyl methanethiosulfonate (MTSET) (9, 11)) revealed that Ser132 faces an open hydrophilic environment at the extracellular/luminal side (Fig. 8B). Next we tested whether S132C was cross-linked with residues Asp450 and Gln454 replaced with Cys, which are located at TMD9 and are the most C terminal, respectively (Fig. 8C). We utilized MTS cross-linkers MTS-4-MTS (M4M) and MTS-14-MTS (M14M) with spacer moieties of 7.8 and 20.8 Å long, respectively. We found that the N- and C-terminal fragments of PS1Cys(−)/S132C/D450C double mutant are cross-linked. In contrast, no cross-linked product of the fragments of PS1Cys(−)/S132C/Q454C was detected. These data imply that TMD2 is also located in proximity to TMD9 similarly to TMD6. Taken together, we concluded that TMD2, -6, and -9 of PS1 collaborate to form the initial substrate-binding site of the γ-secretase complex.
FIGURE 8.
Cross-linking experiment using MTS cross-linkers. A, sandwich ELISA analysis of secreted Aβ from APPNL-stable DKO cells transiently expressing cysteine PS1 mutants. Error bars indicate S.E. B, SCAM analysis of S132C mutant PS1 using intact cells. MTSES, 2-sulfonatoethyl methanethiosulfonate; MTSET, 2-(trimethylammonium)-ethyl methanethiosulfonate. C, cross-linking experiments of single- (S132C) or double-Cys (S132C/D450C or S132C/Q454C) mutant PS1 in microsomes and immunoblot analysis using anti-PS1NT antibody. Locations of cysteine mutations are shown below the panel. PS1 FL, NTF, and cross-linked product (NTF-CTF heterodimer) are shown by black arrowheads, black arrows, and white arrowhead, respectively. No X-linker, no cross-linker; M4M, MTS-4-MTS; M17M, MTS-17-MTS.
DISCUSSION
A number of studies have revealed the seminal roles of the TMDs of the γ-secretase components in the assembly of the active complex and acquisition of its proteolytic activity (12, 15, 17, 18, 24, 25, 32, 33). Previously we and others found the requirement of TMD4 in the binding with Pen-2 (12, 33). Here we furthered the extensive analysis of a series of TMD-swap PS1 proteins and examined their ability to form functional γ-secretase complex in PS null background. The replacement of TMD1, -2, -5, -6, -8, or -9 with irrelevant polypeptide abolished the ability to generate Aβ or NICD, whereas these mutants formed the HMW complex with other γ-secretase components (i.e. Nct, Aph-1, and Pen-2). Among the mutants, swap of TMD1, -5, -8, or -9 affected the stability of HMW complex, suggesting that the acquisition of stability of the complex is required for the proteolytic activity. Notably, using the photoaffinity labeling approach, we found that the TMD2 and luminal side of TMD6 of PS1 contribute to the formation of the initial substrate-binding site and the catalytic site. Moreover, an intramolecular complementation assay suggested the role of TMD3 in the acquisition of proper γ-secretase activity after endoproteolysis. The predicted scheme of the relationship between the assembly of the γ-secretase and the function of PS1 TMDs is shown in Fig. 9.
FIGURE 9.
Malfunction of TMD-swap PS1 mutants in the maturation process of the γ-secretase complex. The maturation process of the γ-secretase complex is indicated as boxes with arrows. PS1 mutants are indicated at right of the process that is defective in the mutant.
Active γ-secretase is known to form a highly stable protein complex (34). Systematic mutational studies of PS support our previous view that the formation of stabilized HMW complex is tightly related to its proteolytic activity (15, 18, 34–36). Nct-Aph-1 subcomplex has been considered as the stabilization factor for the γ-secretase complex (7, 8). In fact, we have recently shown that single chain variable fragment against Nct diminished the stability and activity of the γ-secretase complex (31). However, TM1mt, TM5mt, TM8mt, and TM9mt PS1 failed to be stabilized despite their capability to bind Nct-Aph-1 subcomplex and form the HMW complex. These PS1 mutants recovered complex-type N-glycosylated Nct polypeptides that acquired trypsin resistance, indicating that conformational maturation of Nct and the trafficking of the γ-secretase complex occur independently of the stability of PS1. Thus, it is reasonable to conclude that the interaction of PS1 with the Nct-Aph-1 subcomplex is not sufficient for the stability of the γ-secretase. Brunkan et al. (36) reported on similar results about TMD1 of PS1. Lack of significant difference in the band patterns of trypsin-digested PS1 mutant polypeptides suggests that the topology of TMDs in the lipid bilayer is not affected by mutagenesis. Instead, these TMDs might contribute to the stabilization as well as structural integrity of PS1 within the functional γ-secretase architecture, which is critical to the enzymatic activity.
Both D257A/PS1 and D385A/PS1 were labeled by pep.11-Bt, but not L-852,646, in the photoaffinity labeling experiments. This is consistent with the previous result that transition state analogue-type and helical peptide-type GSIs target spatially different regions in PS1 and indicates that the formation of the initial substrate-binding site occurs independently of the catalytically active conformation of the γ-secretase complex (4–6). Thus, the result that TM2mt and TM6mt failed to interact with both types of photoprobes suggests that TMD2 and the luminal side of TMD6 are responsible for the formation of the initial substrate-binding site after or during stabilization. In addition, we have found that both TMD2 (this study) and TMD6 (9, 11) are located in proximity to TMD9 within the PS1 CTF. Thus, the former two TMDs might be responsible for the formation of initial substrate-binding site in PS1. However, we cannot fully exclude the possibility that other portions of PS1 are also involved in the substrate binding, and the integrity of these portions was disrupted by conformational changes caused by TMD-swap mutagenesis.
Molecular mechanisms whereby the intramembrane-cleaving enzymes hydrolyze membrane-embedded substrates still remain enigmatic. However, x-ray crystallographic and subsequent biochemical analyses of rhomboid and site-2 proteases, the serine-type and metalloprotease-type intramembrane-cleaving enzymes, respectively, provided some clue to the atypical proteolytic process; initial substrate-enzyme interaction should occur via helix-helix interface in a hydrophobic environment (37). Then the substrate is transferred into the hydrophilic catalytic pore through the lateral gate. TMD9 of PS1 is implicated in the substrate binding and the gating mechanism, whereas structural information regarding PS1 at higher resolution has been lacking. Of note, using recombinant protein-based binding assay, a region containing PS1 TMD2 is implicated in the direct substrate binding (38). In regard to TMD6, we have shown that the luminal side of TMD6 is exposed to a hydrophilic environment along with the catalytic aspartate residue, Asp257, by SCAM (9, 10). Pharmacological and chemical biological analyses have indicated that the initial substrate binding and the catalytic sites are located in proximity (5, 6). Thus, the luminal side of TMD6 might be one of the structural constituents not only of the catalytic site but also of the initial substrate-binding site. The luminal half of TMD6 has been shown to comprise an α-helical structure, of which one plane formed by residues Ala246, Leu250, and Asp257 is exposed to the hydrophilic milieu of the catalytic pore (9). We propose that the reverse side of this segment that faces the lipid bilayer might comprise the initial substrate-binding interface. Binding of the substrate to the initial binding site composed of TMD2, -6, and -9 may induce a conformational change of PS1 and facilitate the access of the substrate to the catalytic site. A similar substrate-binding and catalytic site relationship has been reported in the rhomboid (39).
In sum, we have revealed that PS1 TMD1, -5, -8, and -9 are required for the stabilization of the HMW complex and that TMD2 and the luminal side of TMD6 contribute to the formation of the initial substrate-binding site of the γ-secretase. However, we cannot fully exclude the possibility that the TMD swapping might alter the overall conformation of the presenilin polypeptide in a way that could alter its function. Additional mutagenesis studies will be needed to further identify the critical domains in PS1 for the substrate recognition. X-ray crystallographic analyses would eventually reveal the mechanism by which the γ-secretase complex recognizes the substrate and the substrate enters the active site.
Acknowledgments
We thank Drs. B. De Strooper (Katholieke Universiteit Leuven, Leuven, Belgium), R. Kopan (Washington University in St. Louis, St. Louis, MO), T. Kitamura (The University of Tokyo, Tokyo, Japan), Y. M. Li (Sloan Kettering Cancer Center, New York, NY), G. Thinakaran (The University of Chicago, Chicago, IL), A. Takashima (RIKEN, Saitama, Japan), and T. E. Golde (Mayo Clinic, Jacksonville, FL) for valuable reagents, Takeda Pharmaceutical Company (Osaka, Japan) for Aβ ELISA, and our current/previous laboratory members for helpful discussions and technical assistance.
This work was supported in part by grants-in-aid for young scientists (S) from the Japan Society for the Promotion of Science (JSPS), Scientific Research on Priority Areas “Research on Pathomechanisms of Brain Disorders” from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), by the Targeted Proteins Research Program of the Japan Science and Technology Corporation (JST), and by the Core Research for Evolutional Science and Technology of JST, Japan.
N. Watanabe, T. Iwatsubo, and T. Tomita, unpublished data.
- APP
- β-amyloid precursor protein
- Aβ
- amyloid β peptide
- HMW
- high molecular weight
- CHAPSO
- 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate
- CTF
- carboxyl-terminal fragment
- NTF
- amino-terminal fragment
- DKO
- PS1/PS2 double knockout fibroblasts
- ELISA
- enzyme-linked immunosorbent assay
- Nct
- nicastrin
- mt
- mutant
- PS
- presenilin
- TMD
- transmembrane domain
- WT
- wild-type
- BN-PAGE
- Blue Native-PAGE
- SCAM
- substituted cysteine-scanning method
- NICD
- Notch intracellular domain
- SPP
- signal peptide peptidase
- MTS
- methanethiosulfonate
- GSI
- γ-secretase inhibitor.
REFERENCES
- 1.Tomita T. (2009) Expert Rev. Neurother. 9, 661–679 [DOI] [PubMed] [Google Scholar]
- 2.Wolfe M. S. (2009) Semin. Cell Dev. Biol. 20, 219–224 [DOI] [PubMed] [Google Scholar]
- 3.Takasugi N., Tomita T., Hayashi I., Tsuruoka M., Niimura M., Takahashi Y., Thinakaran G., Iwatsubo T. (2003) Nature 422, 438–441 [DOI] [PubMed] [Google Scholar]
- 4.Esler W. P., Kimberly W. T., Ostaszewski B. L., Ye W., Diehl T. S., Selkoe D. J., Wolfe M. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2720–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kornilova A. Y., Bihel F., Das C., Wolfe M. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3230–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imamura Y., Watanabe N., Umezawa N., Iwatsubo T., Kato N., Tomita T., Higuchi T. (2009) J. Am. Chem. Soc. 131, 7353–7359 [DOI] [PubMed] [Google Scholar]
- 7.LaVoie M. J., Fraering P. C., Ostaszewski B. L., Ye W., Kimberly W. T., Wolfe M. S., Selkoe D. J. (2003) J. Biol. Chem. 278, 37213–37222 [DOI] [PubMed] [Google Scholar]
- 8.Niimura M., Isoo N., Takasugi N., Tsuruoka M., Ui-Tei K., Saigo K., Morohashi Y., Tomita T., Iwatsubo T. (2005) J. Biol. Chem. 280, 12967–12975 [DOI] [PubMed] [Google Scholar]
- 9.Sato C., Morohashi Y., Tomita T., Iwatsubo T. (2006) J. Neurosci. 26, 12081–12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tolia A., Chávez-Gutiérrez L., De Strooper B. (2006) J. Biol. Chem. 281, 27633–27642 [DOI] [PubMed] [Google Scholar]
- 11.Sato C., Takagi S., Tomita T., Iwatsubo T. (2008) J. Neurosci. 28, 6264–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe N., Tomita T., Sato C., Kitamura T., Morohashi Y., Iwatsubo T. (2005) J. Biol. Chem. 280, 41967–41975 [DOI] [PubMed] [Google Scholar]
- 13.Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. (2003) Exp. Hematol. 31, 1007–1014 [PubMed] [Google Scholar]
- 14.Morohashi Y., Kan T., Tominari Y., Fuwa H., Okamura Y., Watanabe N., Sato C., Natsugari H., Fukuyama T., Iwatsubo T., Tomita T. (2006) J. Biol. Chem. 281, 14670–14676 [DOI] [PubMed] [Google Scholar]
- 15.Tomita T., Takikawa R., Koyama A., Morohashi Y., Takasugi N., Saido T. C., Maruyama K., Iwatsubo T. (1999) J. Neurosci. 19, 10627–10634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morohashi Y., Hatano N., Ohya S., Takikawa R., Watabiki T., Takasugi N., Imaizumi Y., Tomita T., Iwatsubo T. (2002) J. Biol. Chem. 277, 14965–14975 [DOI] [PubMed] [Google Scholar]
- 17.Leem J. Y., Saura C. A., Pietrzik C., Christianson J., Wanamaker C., King L. T., Veselits M. L., Tomita T., Gasparini L., Iwatsubo T., Xu H., Green W. N., Koo E. H., Thinakaran G. (2002) Neurobiol. Dis. 11, 64–82 [DOI] [PubMed] [Google Scholar]
- 18.Saura C. A., Tomita T., Soriano S., Takahashi M., Leem J. Y., Honda T., Koo E. H., Iwatsubo T., Thinakaran G. (2000) J. Biol. Chem. 275, 17136–17142 [DOI] [PubMed] [Google Scholar]
- 19.Nyborg A. C., Kornilova A. Y., Jansen K., Ladd T. B., Wolfe M. S., Golde T. E. (2004) J. Biol. Chem. 279, 15153–15160 [DOI] [PubMed] [Google Scholar]
- 20.Tomita T., Maruyama K., Saido T. C., Kume H., Shinozaki K., Tokuhiro S., Capell A., Walter J., Grünberg J., Haass C., Iwatsubo T., Obata K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y. M., Xu M., Lai M. T., Huang Q., Castro J. L., DiMuzio-Mower J., Harrison T., Lellis C., Nadin A., Neduvelil J. G., Register R. B., Sardana M. K., Shearman M. S., Smith A. L., Shi X. P., Yin K. C., Shafer J. A., Gardell S. J. (2000) Nature 405, 689–694 [DOI] [PubMed] [Google Scholar]
- 22.Fuwa H., Takahashi Y., Konno Y., Watanabe N., Miyashita H., Sasaki M., Natsugari H., Kan T., Fukuyama T., Tomita T., Iwatsubo T. (2007) ACS Chem. Biol. 2, 408–418 [DOI] [PubMed] [Google Scholar]
- 23.Isoo N., Sato C., Miyashita H., Shinohara M., Takasugi N., Morohashi Y., Tsuji S., Tomita T., Iwatsubo T. (2007) J. Biol. Chem. 282, 12388–12396 [DOI] [PubMed] [Google Scholar]
- 24.Bergman A., Laudon H., Winblad B., Lundkvist J., Näslund J. (2004) J. Biol. Chem. 279, 45564–45572 [DOI] [PubMed] [Google Scholar]
- 25.Laudon H., Mathews P. M., Karlström H., Bergman A., Farmery M. R., Nixon R. A., Winblad B., Gandy S. E., Lendahl U., Lundkvist J., Näslund J. (2004) J. Neurochem. 89, 44–53 [DOI] [PubMed] [Google Scholar]
- 26.Zhao B., Yu M., Neitzel M., Marugg J., Jagodzinski J., Lee M., Hu K., Schenk D., Yednock T., Basi G. (2008) J. Biol. Chem. 283, 2927–2938 [DOI] [PubMed] [Google Scholar]
- 27.Shirotani K., Edbauer D., Capell A., Schmitz J., Steiner H., Haass C. (2003) J. Biol. Chem. 278, 16474–16477 [DOI] [PubMed] [Google Scholar]
- 28.Leem J. Y., Vijayan S., Han P., Cai D., Machura M., Lopes K. O., Veselits M. L., Xu H., Thinakaran G. (2002) J. Biol. Chem. 277, 19236–19240 [DOI] [PubMed] [Google Scholar]
- 29.Kaether C., Lammich S., Edbauer D., Ertl M., Rietdorf J., Capell A., Steiner H., Haass C. (2002) J. Cell Biol. 158, 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herreman A., Van Gassen G., Bentahir M., Nyabi O., Craessaerts K., Mueller U., Annaert W., De Strooper B. (2003) J. Cell Sci. 116, 1127–1136 [DOI] [PubMed] [Google Scholar]
- 31.Hayashi I., Takatori S., Urano Y., Iwanari H., Isoo N., Osawa S., Fukuda M. A., Kodama T., Hamakubo T., Li T., Wong P. C., Tomita T., Iwatsubo T. (2009) J. Biol. Chem. 284, 27838–27847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saura C. A., Tomita T., Davenport F., Harris C. L., Iwatsubo T., Thinakaran G. (1999) J. Biol. Chem. 274, 13818–13823 [DOI] [PubMed] [Google Scholar]
- 33.Kim S. H., Sisodia S. S. (2005) J. Biol. Chem. 280, 41953–41966 [DOI] [PubMed] [Google Scholar]
- 34.Ratovitski T., Slunt H. H., Thinakaran G., Price D. L., Sisodia S. S., Borchelt D. R. (1997) J. Biol. Chem. 272, 24536–24541 [DOI] [PubMed] [Google Scholar]
- 35.Tomita T., Watabiki T., Takikawa R., Morohashi Y., Takasugi N., Kopan R., De Strooper B., Iwatsubo T. (2001) J. Biol. Chem. 276, 33273–33281 [DOI] [PubMed] [Google Scholar]
- 36.Brunkan A. L., Martinez M., Wang J., Walker E. S., Beher D., Shearman M. S., Goate A. M. (2005) J. Neurochem. 94, 1315–1328 [DOI] [PubMed] [Google Scholar]
- 37.Erez E., Fass D., Bibi E. (2009) Nature 459, 371–378 [DOI] [PubMed] [Google Scholar]
- 38.Annaert W. G., Esselens C., Baert V., Boeve C., Snellings G., Cupers P., Craessaerts K., De Strooper B. (2001) Neuron 32, 579–589 [DOI] [PubMed] [Google Scholar]
- 39.Baker R. P., Young K., Feng L., Shi Y., Urban S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8257–8262 [DOI] [PMC free article] [PubMed] [Google Scholar]