Abstract
Members of the transforming growth factor-β superfamily play essential roles in both the pluripotency and differentiation of embryonic stem (ES) cells. Although bone morphogenic proteins (BMPs) maintain pluripotency of undifferentiated mouse ES cells, the role of autocrine Nodal signaling is less clear. Pharmacological, molecular, and genetic methods were used to further understand the roles and potential interactions of these pathways. Treatment of undifferentiated ES cells with SB431542, a pharmacological inhibitor of Smad2 signaling, resulted in a rapid reduction of phosphorylated Smad2 and altered the expression of several putative downstream targets. Unexpectedly, inhibition of the Nodal signaling pathway resulted in enhanced BMP signaling, as assessed by Smad1/5 phosphorylation. SB431542-treated cells also demonstrated significant induction of the Id genes, which are known direct targets of BMP signaling and important factors in ES cell pluripotency. Inhibition of BMP signaling decreased the SB431542-mediated phosphorylation of Smad1/5 and induction of Id genes, suggesting that BMP signaling is necessary for some Smad2-mediated activity. Because Smad7, a known inhibitory factor to both Nodal and BMP signaling, was down-regulated following inhibition of Nodal-Smad2 signaling, the contribution of Smad7 to the cross-talk between the transforming growth factor-β pathways in ES cells was examined. Biochemical manipulation of Smad7 expression, through shRNA knockdown or inducible gene expression, significantly reduced the SB431542-mediated phosphorylation of Smad1/5 and induction of the Id genes. We conclude that autocrine Nodal signaling in undifferentiated mouse ES cells modulates the vital pluripotency pathway of BMP signaling.
Keywords: Cell/Stem, Cytokines/TGF-β, Signal Transduction/Protein Kinases/Serine/Threonine, Stem Cells, Transcription/Development, Transcription/SMAD, Bone Morphogenetic Protein (BMP), Embryonic Stem Cell
Introduction
Mouse embryonic stem (ES)2 cells are derived from the inner cell mass of the early blastocyst. It is from the inner cell mass that the germ layers arise to produce all cell types of the adult. The capacity of ES cells to either self-renew or differentiate into cells of the three germ layers provides an excellent tool for studying early embryonic development, including self-renewal and pluripotency (1). Some of the essential factors controlling ES cell pluripotency have been identified; however, additional work is necessary to identify other functional signaling pathways that impinge upon the ES cell phenotype, to determine genes that are regulated by these pathways, and to discover how the various pathways interact. Applications of this work will yield insights into the promising use of stem cells for regenerative therapies.
ES cells can be maintained as pluripotent cells when grown on mitotically inactivated embryonic fibroblasts or when grown in media supplemented with leukemia inhibitory factor (LIF (2, 3)). Additionally, under serum-free culture conditions, signaling through the bone morphogenetic protein (BMP) pathway is essential for maintaining mouse ES cell pluripotency (4, 5). The combination of LIF and BMP4 inhibits multiple differentiation-inductive signals, a capability that can be replicated without extrinsic stimuli through pharmacological inhibition of fibroblast growth factor 4 and glycogen synthase kinase 3 signaling (6). BMP signaling mediates ES cell pluripotency through induction of the Id genes (Inhibitors of differentiation (5)), which are negative regulators of basic helix-loop-helix transcription factors and play important roles in during early development and in embryonic and somatic stem cells (7, 8). Expression of Id1, a direct target of BMP4, liberates ES cells from BMP or serum dependence (5, 9). The central requirement for active BMP signaling in maintaining ES cells under serum-free conditions is clear, yet its regulation and interactions with other pathways are important questions in ES cell biology.
BMPs are members of the transforming growth factor (TGF)-β superfamily, whose diverse members play vital roles in embryonic development and in ES cell biology (10, 11). The ligands, including TGF-β, Activin, Nodal, and the BMPs, bind to the extracellular domain of the type II receptors. Binding induces activation of type I receptors, including the Activin receptor-like kinases (ALKs) 1–7 (12). Activin and Nodal signal via ALK4, ALK5, and ALK7, whereas the BMPs convey signaling through ALK2, ALK3, and ALK6. In the canonical signaling pathway model, intracellular transduction is mediated by phosphorylation of receptor-regulated Smad proteins via activated type I receptors. Smad2 and Smad3 are activated by Activin and Nodal signaling, whereas Smad1, Smad5, and Smad8 are substrates for BMP-activated receptors (13). Phosphorylated Smads (pSmad) form heteromeric complexes with co-mediator Smad4, enter the nucleus, and interact with co-activators and transcription factors to affect gene transcription. Moreover, inhibitory Smads (Smad6 and Smad7) inhibit activation of receptor-regulated Smad proteins and function as feedback modulators of pathway activity (14).
ES cells have an active Nodal-Smad2 signaling axis (15). Nodal is highly expressed in ES cells, suggesting significant autocrine activity of this pathway. Stimulation of ES cells with recombinant Activin or Nodal enhances Smad2 phosphorylation and increases ES cell proliferation (16). Alternatively, pharmacological inhibition of pSmad2 signaling and inhibition by Smad7 overexpression decrease ES cell proliferation (16). Transcriptional targets of Smad2 in ES cells include many feedback regulatory factors such as Smad7, Lefty, and Bambi (17). However, the actions of downstream target genes of Nodal-Smad2 signaling and interactions with other critical signaling pathways are not known.
In this work, we sought to define the activity of Nodal signaling and its interaction with the BMP pathway in undifferentiated mouse ES cells. Using pharmacological, molecular, and genetic methods, these efforts demonstrated that inhibition of Nodal signaling indirectly enhanced the activation of the BMP substrates Smad1/5 and increased the expression of downstream BMP target genes. Nodal signaling regulated Smad7 expression, which feeds back to inhibit both the Nodal and BMP pathways in ES cells. This work uncovered an interdependence of the Nodal-Smad2 and BMP-Smad1/5 signaling pathways in undifferentiated ES cells and defines potential mechanisms for these pathways in ES cell maintenance.
EXPERIMENTAL PROCEDURES
ES Cell Culture
E14Tg2A (E14) ES cells were maintained on feeder-free, gelatin-coated plates in ES media as described before (18, 19). Experiments were conducted in serum-free, defined media (5, 20), supplemented with 103 units/ml LIF and 10 ng/ml BMP4 (R&D). ES cells were treated with 10 ng/ml BMP4, 10 ng/ml Activin (R&D), 5 μm SB431542 (Sigma), 0.5 μm A-83-01, and 5 μm dorsomorphin (Sigma) at the noted concentration and for 24 h unless specifically noted otherwise. DMSO was applied at the same volume as SB431542, A-83-01, or dorsomorphin as a vehicle control. Analyses of time-course treatments of SB431542 and BMP4 were performed by application of each treatment to sustained cultures; thus, over the length of the time course the media stayed constant and only the treatments were added at each time point.
Standard protocols (19) were used to generate homozygous Smad2m1Mag ES cells from blastocysts. In brief, timed matings were performed by crossing Smad2m1Mag heterozygous females with Smad2m1Mag heterozygous males (21) carrying the Rosa26-lacZ allele (22). Blastocysts were flushed from these matings, and inner cell mass outgrowths were cultured in ES media on gelatin-coated dishes to establish cell lines. DNA was isolated for genotyping of the Smad2m1Mag allele, and X-Gal staining (23) was performed to identify cell lines carrying the Rosa26-lacz allele. Two lines of each genotype were used for the analysis.
RNA Analysis
RNA was isolated using Qiagen columns and initially analyzed with the Mouse Genome 430A Array from Affymetrix. Microarray data were deposited to the Gene Expression Omnibus (GSE17879). For gene expression analysis, cDNA was synthesized (Applied Biosystems), and quantitative real-time PCR analysis was performed using TaqMan primer sets with the 7500 Real Time PCR system (Applied Biosystems). Specific ABI TaqMan Primer/Probe assay identification numbers are available upon request.
Protein Analysis
Cells were lysed using radioimmune precipitation assay buffer with Halt Protease and Phosphatase Inhibitor Cocktails (Pierce) for Western analysis. Primary antibodies for pSmad2 (Millipore), Smad2 (Zymed Laboratories Inc.), pSmad1/5/8 (Cell Signaling), Smad1 (Zymed Laboratories Inc.), glyceraldehyde-3-phosphate dehydrogenase (Santa Cruz Biotechnology), Smad7 (R&D), and FLAG (Sigma) were incubated overnight at 4 °C and followed with incubation of appropriate secondary antibodies.
Luciferase Assays
E14 ES cells were plated 1 day prior to transfection on gelatin-coated 24-well plates and transfected with pGL2-Id1 Luciferase (−1585 to +88 of promoter; Addgene plasmid 16048 (24)), and pGL4.74 [hRluc/TK] (Promega) using Lipofectamine 2000 (Invitrogen). Following transfection, ES media was change to defined media with LIF and BMP4 for 24 h. Cells were then grown in defined media with LIF and treated with BMP4, SB431542, or BMP4 plus SB431542 for an additional 24 h. Firefly luciferase activity in cell lysates was measured and normalized to expression of TK-Renilla luciferase.
The Activin-responsive reporter pGL3-CAGA12-luciferase (25) was co-transfected with pGL4.74 [hRluc/TK] into control and Smad2m1Mag homozygous ES cells. The cells were grown in defined media with LIF and 20 ng/ml BMP4 and treated with Activin or SB431542 for the final 24 h. Lysates were analyzed for luciferase readings.
RNA Knockdown
Two methods were utilized for knockdown of specific target genes. For knockdown of Nodal and TGF-β1, specific siRNAs (Dharmacon) were transfected via lipofection following the manufacturer's instructions. Changes in gene expression were compared with a non-targeting siRNA after 48 h. For knockdown of Smad7, plasmids expressing shRNAs (Open Biosystems) were transfected into ES cells via lipofection and screened for efficient knockdown. The hairpin targeting 5′-CTCCAGATACCCAATGGATTT-3′ (bp 920–940, TRCN0000096053) produced the strongest Smad7 knockdown and was used in subsequent experiments. A non-targeting hairpin (26) was cloned into pLKO.1. The non-targeting and Smad7 shRNAs were transfected into ES cells. One day after transfection, the cells were passaged and plated in defined media with LIF, BMP4, and puromycin to select for cells with transient expression of pLKO.1 constructs. After selection, the ES cells were treated with appropriate media conditions for 24 h after which the cells were harvested for RNA or protein analyses.
Inducible Smad7 Overexpression
CRE-mediated recombination was used to generate an ES cell line with doxycycline-inducible Smad7, as described previously (27). In brief, a cDNA encoding FLAG-Smad7 was cloned into the engineered PALP targeting vector, and the resulting construct was co-electroporated with a Cre-expressing plasmid (pOG231, Addgene Plasmid 17736 (28)) in Ainv15 cells. The Ainv15 cell line, derived from E14 mES cells, contains a reverse tetracycline transactivator for the tet-ON system in the Rosa26 locus and a tet response element, a loxP site, and a neomycin resistance gene lacking an initiation codon in the Hprt locus (27, 29). After selection in media supplemented with G418, clones with appropriate recombination were confirmed via PCR genotyping, tested for doxycycline-inducible expression, and karyotyped. For RNA and protein analyses, cells were treated with doxycycline for 6 h prior to treatment with SB431542 and then co-treated with doxycycline and SB431542 for 24 h.
Data Analysis
Results were graphed to represent means ± S.E. of the mean. RNA analyses were performed in at least triplicate samples. Student t-tests were performed to determine statistical significance of the data with a p value of <0.05 considered statistically significant.
RESULTS
Smad2 Signaling Regulates Transcription of Multiple Feedback and Pluripotency Genes
To determine the target genes of Smad2 signaling in undifferentiated ES cells, changes in gene expression were examined following treatment with recombinant Activin or SB431542 under defined media culture conditions (20). SB431542 specifically inhibits ALK4/5/7 receptor signaling as a competitive inhibitor of ATP binding to the kinase domain and has no effect on BMP-regulated ALKs or other kinase pathways (30, 31). In mouse ES cells, recombinant Activin has been shown to produce stronger Smad2 phosphorylation at lower concentrations than commercially available recombinant Nodal (16); thus, Activin was used to stimulate the Smad2 pathway in this analysis. In non-manipulated ES cells grown in serum-free media, substantial pSmad2 was observed via Western analysis (Fig. 1A), demonstrating an autocrine signaling activity for this pathway. Activin stimulation increased pSmad2 in ES cells after 2 h, while treatment with SB431542 for 24 h virtually eliminated pSmad2. Total Smad2 expression remained unchanged through these manipulations.
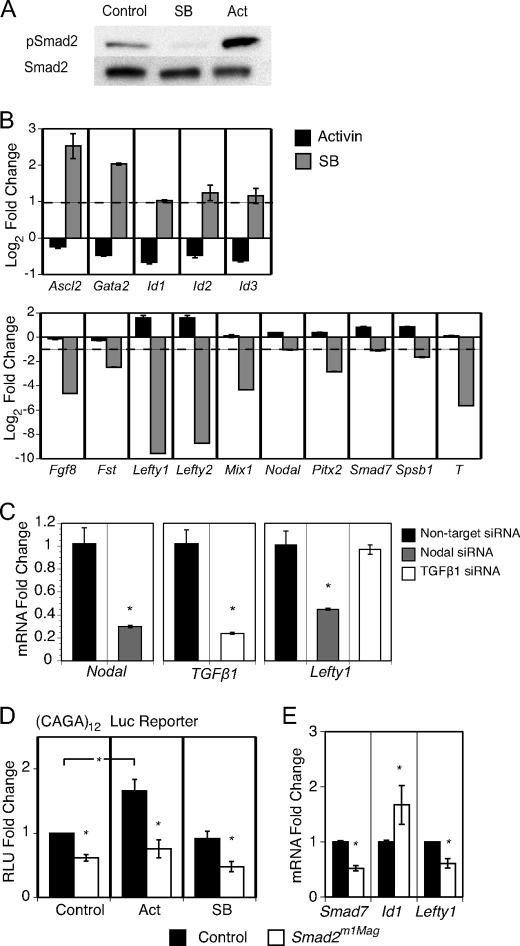
FIGURE 1.
ES cell gene expression analysis following Smad2 stimulation, inhibition, and genetic mutation. ES cells were treated with Activin for 2 h or with SB431542 (SB, pSmad2 inhibitor) for 24 h in defined media with LIF and 20 ng/ml BMP4. A, immunoblot analysis of Smad2 phosphorylation following Activin stimulation or SB treatment. B, gene expression of Activin- and SB-treated ES cells was compared with untreated ES cells using rtPCR. The dotted line represents a 2-fold change in gene expression. C, rtPCR analysis of Lefty1 expression following siRNA knockdown of Nodal and TGF-β1. D, Smad2m1Mag mutant ES cells activated the Smad-binding element (CAGA12) luciferase reporter significantly less than control cells. E, rtPCR analysis revealed that Smad2m1Mag mutant ES cells had significantly decreased Smad7 and Lefty1 expression and increased Id1 expression. *, p < 0.05. Abbreviations: T, Brachyury; Fst, Follistatin.
Changes in gene expression in ES cells treated with Activin or SB431542 was first analyzed through Affymetrix microarrays and then confirmed using real-time quantitative PCR (rtPCR, Fig. 1B). The inhibition of Smad2 signaling resulted in more numerous and larger magnitude changes in gene expression than Smad2 pathway stimulation. This observation is consistent with the idea that Smad2 signaling is active via autocrine mechanisms in mouse ES cells. No changes in the expression of the core pluripotency factors such as Nanog and Pou5f1 (Oct4) were observed in the short timeframe examined in these expression analyses (data not shown). Inhibition of Smad2 signaling acted both to repress and derepress gene expression. Altered in Smad2 activity produced the greatest gene expression changes in the Lefty genes, secreted Nodal antagonists that are highly expressed in ES cells and play vital roles in left/right asymmetry in the developing embryo (32). Loss of pSmad2 reduced the expression of the Lefty genes to an amount greatly reduced from basal expression, whereas stimulation of pSmad2 increased Lefty1 and Lefty2 expression 3-fold, the greatest observed increase in gene expression associated with Activin treatment. Inhibition of Smad2 signaling via SB431542 significantly diminished the expression of multiple other developmentally regulated genes, including Brachyury (T), Follistatin (Fst), Fgf8, Spsb1, Pitx2, and Mixl1, and significantly attenuated the expression of components of the TGF-β family signaling pathway, including Nodal and Smad7.
Interestingly, inhibition of Smad2 signaling enhanced the expression of the Id genes, transcriptional modulators that have been shown to be vital in BMP-mediated ES cell pluripotency (5). Smad2 inhibition induced Id1, Id2, and Id3 expression over 2-fold, whereas Activin treatment had the opposite effect and reduced Id gene expression by almost half (Fig. 1B). Treatment with SB431542 also derepressed the expression of Ascl2 (Mash2) and Gata2, genes that are not expressed in undifferentiated ES cells. Although the developmental expression of Ascl2 and Gata2 is complex, both genes are highly expressed in the developing trophectoderm. These results define a cohort of developmentally important genes that are regulated by Smad2 signaling in undifferentiated mouse ES cells.
The microarray and biochemical results suggest an active autocrine signaling pathway mediated by Smad2 in ES cells. The microarray data were examined to determine which of the TGF-β1-related ligands are expressed in ES cells and could mediate this signaling. The Nodal ligand had by far the strongest mRNA expression in ES cells of all ALK4/5/7 ligands, suggesting the autocrine Smad2 activation is mediated by expression and secretion of Nodal. However, the TGF-β1 ligand demonstrated low expression in ES cells. To confirm that Nodal is the major mediator of pSmad2 activity in ES cells, Nodal and TGF-β1 expression were reduced through siRNA transfection, and known Smad2 gene targets were analyzed. siRNA knockdown of Nodal significantly down-regulated Lefty1, a highly responsive target of Smad2 signaling, whereas TGF-β1 knockdown had no effect of Lefty1 expression (Fig. 1C). Thus, treatment of ES cells with SB431542 primarily inhibits autocrine Nodal signaling.
To confirm the activity of the Nodal-Smad2 axis via non-pharmacological methods, we generated ES cells that carry a homozygous Smad2 mutation, Smad2m1Mag (21). Previous in vivo analysis has shown that the Smad2m1Mag mutation is a severe reduction of the functional allele. The reduced function of this Smad2 mutation was confirmed with the homozygous mutant ES cells by chimeric analysis and via protein interaction analysis (supplemental Fig. 1). The transcriptional activity of the Smad2m1Mag allele was assayed via transient transfection of the CAGA12 luciferase reporter, which is responsive to TGF-β and Activin but not BMP stimulation (25, 33). Smad2m1Mag ES cells activated the CAGA12 reporter significantly less than control cells and failed to enhance reporter activity in response to Activin stimulation (p < 0.05, Fig. 1D). The basal CAGA12 reporter activity in Smad2m1Mag ES cells may be from the low level of Smad3 present in ES cells (33, 34).
To determine if Smad2m1Mag mutant ES cells also exhibited similar gene expression defects as SB431542-treated ES cells, expression of Smad7, Lefty1, and the Id genes were analyzed in the mutant ES cells. Smad2 mutant ES cells expressed significantly lower Smad7 and Lefty1 mRNA than control heterozygous cells (p < 0.05, Fig. 1E). Correspondingly, Id1 expression was significantly up-regulated over 1.5-fold (p < 0.05). Although Id2 and Id3 also had increased expression, the changes did not reach statistical significance. Thus, gene expression changes after pharmacological inhibition of Nodal signaling are consistent with genetic perturbation of Smad2 signaling. Overall, these data support an autocrine signaling pathway in undifferentiated ES cells mediated by Nodal and Smad2 that regulates the expression of a substantial number of genes.
Nodal Inhibition Eliminates pSmad2 with Delayed Induction of pSmad1/5 and Id Gene Expression
Several of the genes identified as being regulated by Smad2 signaling in ES cells include regulators of the Nodal-Smad2 signaling axis itself. Although changes in expression of Lefty1, Lefty2, and Nodal will feedback to affect signaling through the Nodal pathway, modulation of Follistatin and Smad7 have the ability to affect multiple pathways, including BMP-mediated activity (35, 36). Given the importance of the BMP signaling pathway in maintaining ES cell pluripotency (5), we wanted to determine if modulation of Nodal signaling affected the BMP signaling axis in undifferentiated ES cells.
The level of intracellular BMP signaling activity was monitored by examining the phosphorylation of Smad1/5 following inhibition of Nodal signaling in a detailed time course. Treatment with SB431542 significantly decreased pSmad2 levels after 0.5 h (Fig. 2A). Surprisingly, Smad1/5 phosphorylation increased after 3 h of SB431542 treatment. Previous work has indicated that SB431542 is a specific inhibitor of ALK4/5/7 kinase activity and has no affect on the type I BMP receptors that modulate Smad1/5 phosphorylation (30); thus, this effect is not due to direct pharmacological modulation of the BMP receptor kinase activity. The temporal lag of increase in Smad1/5 phosphorylation compared with the decrease in Smad2 phosphorylation suggests that the enhanced phosphorylation of Smad1/5 in response to Nodal inhibition is likely indirect.
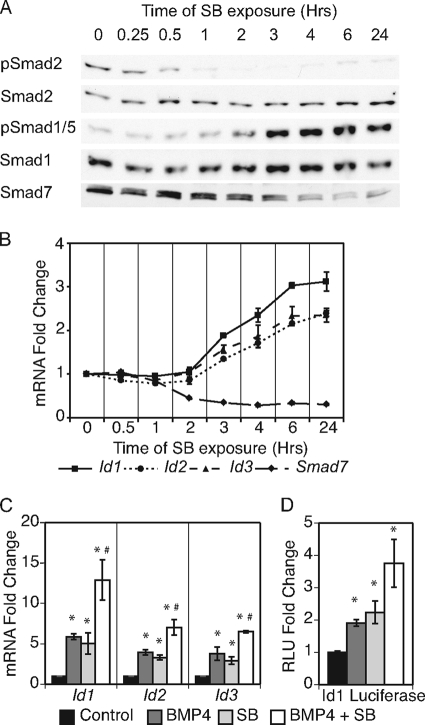
FIGURE 2.
Inhibition of Nodal-Smad2 signaling induced delayed Smad1/5 phosphorylation and Id induction. A, time course of SB431542 (SB) treatment to determine the effects of Nodal-Smad2 inhibition on Smad protein levels and phosphorylation states. B, rtPCR analysis of Id genes and Smad7 during SB time course. Samples are compared with control cells grown in defined media with LIF and BMP4. C, rtPCR analysis of Id genes in response to BMP4, SB, and BMP plus SB. Gene expression compared with cells grown in defined media with LIF. D, dual luciferase reporter assay for Id1 promoter reporter. ES cells were transfected, grown in designated media for 24 h, and assayed for luciferase activity. *, p < 0.05 compared with control; #, p < 0.05 compared with single treatment of BMP4 or SB.
To verify the change in phosphorylation of BMP-regulated Smads in response to inhibition of Nodal signaling, a second small molecule inhibitor of ALK4/5/7, A-83-01, was used. A-83-01 is a more potent inhibitor of the ALK4/5/7 receptors that prevents phosphorylation of Smad2 but has no effect on type I BMP receptors (37). Treatment with either 5 μm SB431542 or 0.5 μm A-83-01 induced similar increases in pSmad1/5 levels after 24 h (supplemental Fig. 2A). The enhancement of pSmad1/5 in response to SB431542 was also observed in cells grown in defined media lacking BMP4 supplementation in the media. Also, stimulation with recombinant Activin, with or without BMP4 supplement, lowered the phosphorylation state of Smad1/5. Thus, multiple modes of Smad2 signal perturbation reveal changes in BMP signaling.
Inhibitory Smad7 is able to antagonize both Nodal and BMP signaling and participate in the cross-talk between the pathways (36). Because Smad7 mRNA expression was reduced, but not eliminated, in response to SB431542 treatment (Fig. 1B), the mRNA and protein expression of Smad7 were analyzed in detail during the time course of SB431542 treatments. Smad7 transcript levels dropped almost 20% after 1 h and declined by more than half within 2 h of SB431542 treatment (Fig. 2B). Smad7 protein levels declined after 2 h of SB431542 treatment and were significantly down-regulated between 3 and 4 h of treatment (Fig. 2A). This result indicates that Smad7 is an immediate early response gene of Nodal signaling. The decline of Smad7 levels in response to SB431542 coincided with the increased phosphorylation of Smad1/5. Correspondingly, the expression of the BMP targets Id1, Id2, and Id3 was significantly enhanced after 3 h of Nodal inhibition and increased 2- to 3-fold by 24 h (Fig. 2B, p < 0.05). The SB431542-mediated Smad7 repression and Id induction was retained during long term culture (supplemental Fig. 2B, p < 0.05). This time-course analysis of the events following inhibition of Smad2 signaling in undifferentiated ES cells suggests that the activation of pSmad1/5 and subsequent induction of BMP-target genes may be controlled indirectly by Nodal-Smad2 signaling.
Active BMP4-Smad1/5 Pathway Is Necessary for Id Induction by Nodal Inhibition
Previous work has indicated that the Id genes are direct targets of BMP signaling in mouse ES cells (5, 9, 38). This was confirmed via a time-course analysis of BMP4 stimulation. ES cells were grown in serum-free conditions lacking BMP4 only for 24 h prior to the time course to retain proper conditions for ES self-renewal. Application of recombinant BMP4 stimulated phosphorylation of Smad1/5 within 45 min (Fig. 3A). Interestingly, the maximum level of Smad1/5 phosphorylation occurred after 1 h of BMP4 stimulation, was reduced at 2 h, and eventually reached steady-state levels by 4 h. The initial spike in BMP-Smad1/5 signaling is likely moderated by multiple feedback mechanisms (36, 39). No significant effect on pSmad2 was observed during the time course of BMP treatment, indicating that BMP activation did not significantly modulate Smad2 signaling.
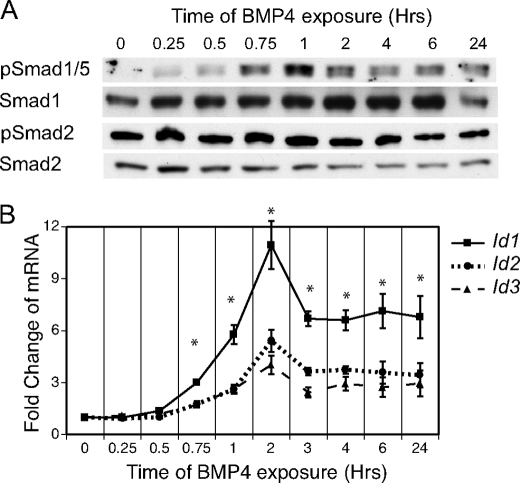
FIGURE 3.
BMP4 time course in ES cells. ES cells were grown in defined media with LIF and BMP4. During the last 24 h, BMP4 was removed from the media and then supplemented at the indicated time points. A, effects of BMP4 stimulation on pSmad1/5 and pSmad2 were assayed through immunoblots with Smad1 and Smad2 expression noting equal loading. B, rtPCR analysis of Id genes and Smad7 during the BMP4 time course. Samples are compared with control cells grown in defined media with LIF. *, p < 0.05.
The timing of Id induction in response to BMP treatment closely followed the phosphorylation and activation of Smad1/5 (Fig. 3B). Expression of Id1, Id2, and Id3 was significantly elevated after 45 min of BMP4 exposure (p < 0.05), a time point that strongly correlates with the phosphorylation of Smad1/5. Id1 expression displayed a peak induction of 11-fold by 2 h after BMP treatment, and a steady-state level of almost 7-fold. Smad7 expression was not significantly affected by BMP4 stimulation (data not shown). Thus, because the Id genes are very early targets of BMP-Smad1/5 signaling, the correlation of the SB431542-mediated Id induction between 3 and 6 h and the phosphorylation of Smad1/5 at 3 h suggests the Id genes may be indirect targets of Nodal inhibition via BMP signaling.
We next determined if co-stimulation with exogenous BMP4 is required for the SB431542-mediated Id gene up-regulation. ES cells were cultured in defined media with LIF and supplemented with BMP4, SB431542, or BMP4 plus SB431542 for 24 h. Treatment with either BMP4 or SB431542 resulted in similar increases in Id expression (Fig. 2C). Concurrent treatment with BMP4 plus SB431542 induced significantly higher Id induction than either treatment alone, suggesting that the transcriptional responses are additive and correlating with higher overall pSmad1/5 levels (supplemental Fig. 2A). Moreover, stimulation with BMP4 and SB431542, alone or in combination, activated an Id1 luciferase reporter (24) that is regulated by the 1.5-kb upstream Id1 promoter sequence (Fig. 2D). Thus, both Smad2 signaling inhibition and BMP4 stimulation enhance Id expression to similar levels in the short term ES cell culture treatments, and this effect is additive with concurrent treatments.
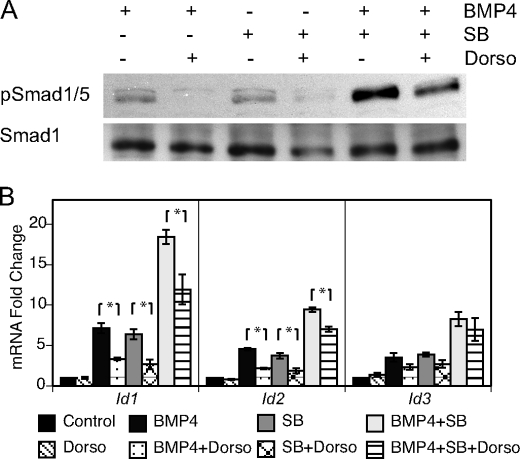
Dorsomorphin, a selective small-molecule inhibitor to the type I BMP-responsive receptors (ALK2/3/6), has recently been identified to block BMP-mediated Smad1/5 phosphorylation (40, 41). To determine if the Id induction elicited by SB431542 treatment is indirectly mediated through BMP signaling, an analysis of cells treated with SB431542 and dorsomorphin was performed. ES cells were cultured for 24 h in BMP4, SB431542, or BMP4 plus SB431542 with and without dorsomorphin. Dorsomorphin treatment resulted in a substantial reduction of pSmad1/5 in response to treatment with either BMP4 or SB431542 (Fig. 4A). Although dorsomorphin also lowered pSmad1/5 levels mediated by simultaneous BMP4 and SB431542 treatments, the amount of activated pSmad1/5 was substantially higher than in singly treated cells. The remaining amount of pSmad1/5 was likely caused by a high number of active ALK2/3/6 receptors stimulated by the dual treatment of BMP4 and SB431542. Thus, inhibition of Nodal signaling primarily induces phosphorylation of Smad1/5 indirectly through BMP-responsive receptors. Furthermore, dorsomorphin treatment of ES cells significantly reduced expression of Id1 and Id2 in response to BMP4, SB431542, or both BMP4 and SB431542 (Fig. 4B, p < 0.05). Id3 expression failed to significantly change in response to ALK2/3/6 inhibition. The protein and transcript analyses indicate that SB431542-mediated Smad1/5 phosphorylation and Id induction require active BMP signaling.
FIGURE 4.
Inhibition of BMP4 signaling reduced SB431542-mediated Smad1/5 phosphorylation and Id induction. ES cells were grown in defined media with LIF and supplemented with combinations of BMP4, SB431542 (SB), and dorsomorphin (Dorso). A, protein lysate was analyzed through immunoblotting to determine the effect of Dorso, an ALK2/3/6 inhibitor, on pSmad1/5 levels induced by BMP4, SB, or BMP plus SB. B, gene expression analysis of Dorso-treated cells by rtPCR demonstrated inhibition of Id1 and Id2 expression induced through BMP4 and SB stimuli. *, p < 0.05.
Manipulation of Smad7 Levels Alters SB431542-mediated Phosphorylation of Smad1/5 and Induction of the Id Genes
The signaling pathways of the two TGF-β subfamilies interact at many levels, including ligand activation, receptor complex formation, and Smad activation (36). To determine the mechanism of interaction between Nodal inhibition and BMP-Smad1/5 activation in ES cells, we analyzed multiple factors that may connect the pathways. First, as the receptor-regulated Smads from the Nodal and BMP pathways both bind to co-Smad4 (39), the Smad4 released by Smad2 inhibition may increase the availability of Smad4 to form heteromeric complexes with pSmad1/5 and therefore activate the BMP pathway. However, when Smad4 is overexpressed in ES cells via transfection of an expression plasmid (42), no effect on BMP signaling is observed, as assessed by phosphorylation of Smad1/5 and Id1 gene expression (supplemental Fig. 3). Also, SB431542 treatment still retained the ability to increase pSmad1/5 and Id1 expression (supplemental Fig. 3). This result indicates that Smad4 levels are not limiting in ES cells.
Second, Follistatin was shown to be down-regulated following Nodal inhibition in ES cells (Fig. 1B). Follistatin is a well characterized antagonist of Activin signaling that can antagonize BMP signaling in some systems (43, 44) but not others (45). To determine if the low level of Follistatin expression in ES cells modulates BMP signaling in mouse ES cells, follistatin levels were reduced via siRNA transfection, and Id1 expression was examined to measure BMP-Smad1/5 activity. siRNAs against follistatin reduced Follistatin expression ∼50%, similar to treatment with SB431542 (supplemental Fig. 4). Decreased Follistatin expression did not affect basal Id1 expression or the SB431542-mediated induction of Id1, suggesting that regulation of follistatin does not contribute to the connectivity between Nodal-Smad2 and BMP-Smad1/5 signaling.
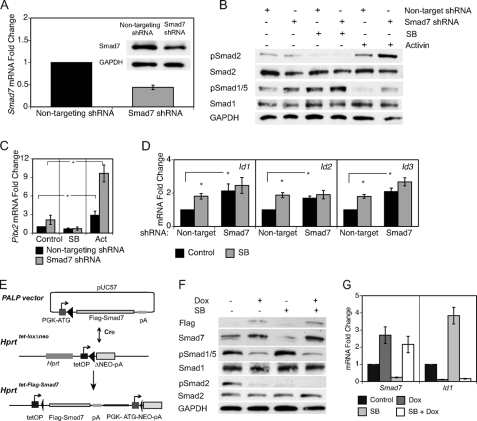
As Smad7 expression was also reduced in response to inhibition of Nodal signaling (Fig. 2A,B), its down-regulation may be responsible for the SB431542-mediated enhancement of the BMP signaling pathway. Smad7 can antagonize both major TGF-β superfamily signaling axes (46, 47) and has been shown to be an important component of the cross-talk between Nodal and BMP signaling (36). Modulation of Smad7 levels was performed to determine the role of Smad7 in interpathway communication in ES cells. To lower Smad7 mRNA levels, ES cells were transiently transfected with plasmids expressing shRNAs against either a non-targeting sequence or Smad7. Cells expressing the shRNA targeting Smad7 produced a reduction of Smad7 mRNA and protein to ∼40% that of control cells (Fig. 5A). This level of knockdown is similar to the decrease of Smad7 mRNA in response to SB431542 treatment. To overexpress Smad7, a doxycycline-inducible system was employed (27, 29). Via CRE-mediated recombination, a FLAG-Smad7 cDNA was inserted into a doxycycline-regulated locus in Ainv15 ES cells (Fig. 5E). Upon treatment with doxycycline, Smad7 is overexpressed 2- to 3-fold (Fig. 5G).
FIGURE 5.
Biochemical manipulation of Smad7 expression inhibits SB431524-induced Smad1/5 phosphorylation and Id expression. Small-hairpin RNAs (shRNA) were utilized to decrease endogenous Smad7 and determine the responses of the TGF-β pathways. ES cells were transfected with pLKO.1-non-targeting shRNA or pLKO.1-Smad7 shRNA, subcultured, and selected with puromycin for 48 h. During the last 24 h, cells were treated with various media conditions. A, the characterized Smad7 shRNA produced about a 60% knockdown of RNA and protein. B, immunoblot analysis of Smad1/5 and Smad2 phosphorylation. C, rtPCR analysis of Pitx2. D, comparison of Id gene expression in cells treated with BMP4 (control) or BMP4 plus SB431542 (SB). FLAG-Smad7 was overexpressed through a doxycycline (dox)-inducible system for expression of transgenes in ES cells (27, 29). E, using CRE-mediated recombination, the FLAG-Smad7 cDNA was inserted into an engineered locus in Ainv15 ES cells. F, immunoblot of Smad7 and phosphorylation levels of Smad1/5 and Smad2. G, rtPCR analysis of Id1 following SB treatment of cells overexpressing Smad7. *, p < 0.05.
The phosphorylation state of receptor-regulated Smad proteins and the expression of downstream genes were analyzed after modulation of Smad7 levels. ES cells demonstrated significantly enhanced Smad1/5 phosphorylation and Id induction following 48-h knockdown of Smad7 (Fig. 5, B and D, p < 0.05). Knockdown of Smad7 also increased the level of pSmad2 in Activin-treated cells and expression of Pitx2, a target of Smad2 signaling identified in the microarray studies (Fig. 5, B and C). Overexpression of Smad7 greatly diminished phosphorylation of Smad2 and Smad1/5 and dramatically reduced expression of Id1 (Fig. 5, F and G). Thus, Smad7 actively represses and regulates the endogenous signaling activity of both the Nodal-Smad2 and BMP-Smad1/5 pathways in undifferentiated ES cells.
To analyze the role of Smad7 in mediating TGF-β interpathway communication in ES cells, Smad7 expression was knocked down or overexpressed in SB431542-treated cells. SB431542 treatment of ES cells containing the Smad7 shRNA had lower Smad7 expression than the Smad7 knockdown alone (supplemental Fig. 5). Correspondingly, the SB431542-treated Smad7 shRNA cells also demonstrated enhanced Smad1/5 phosphorylation (Fig. 5B), likely due to the additional reduction of inhibitory Smad7. ES cells overexpressing Smad7 demonstrated no enhancement of pSmad1/5 to Nodal-Smad2 inhibition (Fig. 5F). Furthermore, while SB431542 induced significantly higher expression of the Id genes in cells transfected with the non-targeting shRNA, ES cells containing the Smad7 shRNA failed to significantly induce Id1, Id2, or Id3 expression in response to SB431542 treatment (Fig. 5D, p < 0.05). Moreover, ES cells overexpressing Smad7 failed to induce Id1 following SB431542 application (Fig. 5G). These results clearly demonstrate that reduction of Smad7 expression is the basis for enhanced BMP activity in response to inhibition of Nodal signaling. These results define a link between Nodal and BMP signaling pathways through the inhibitory Smad7 in undifferentiated ES cells.
DISCUSSION
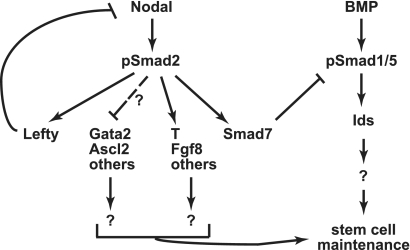
Pharmacological, molecular, and genetic methods were used to demonstrate a previously uncharacterized function for Nodal-Smad2 signaling in modulating the activity of BMP-Smad1/5 signaling in undifferentiated mouse ES cells. Given that the BMP signaling axis is an essential pathway in maintaining mouse ES cell pluripotency, these data suggest that Nodal signaling has the capacity to modulate BMP-mediated ES cell pluripotency. Via decreased Smad7 expression, Nodal inhibition induces expression of the Id genes through modulation of BMP signaling. Thus, the Nodal pathway modulates a signaling pathway important for ES cell pluripotency through regulation of the Id factors (Fig. 6), which help to maintain ES cell pluripotency and inhibit the differentiation of multiple cell types (4, 5, 7).
FIGURE 6.
Model of Nodal-Smad2 signaling regulation of the BMP pluripotency pathway in mouse ES cells. Nodal-Smad2 signaling regulates a significant number of genes in undifferentiated ES cells. Positively regulated targets include genes characteristic of the mesendoderm developmental lineage. Genes negatively regulated by Nodal signaling include some trophoblast markers; the mechanism of this regulation remains to be determined. The regulation of Id gene expression by Nodal-Smad2 is indirect, via the BMP signaling pathway. The transcriptional regulation of Smad7 by Nodal signaling is the major contributor to the modulation of the BMP signaling pathway. BMP pathway activation and Id gene expression are essential components to maintain ES cell pluripotency.
Smad7 Is the Critical Component of Nodal Regulation of BMP Activity in ES Cells
Although Nodal signaling regulates the expression of numerous genes, our work shows that the down-regulation of Smad7 is the major contributor to the interconnectivity of the Nodal and BMP signaling pathways. Smad7, which is a direct transcriptional target of Smad2 signaling (17), had enhanced expression following pSmad2 stimulation and decreased expression after Nodal inhibition. Increased expression of Smad7 slowed the proliferation of ES cells (Ref. 16 and data not shown) and inhibited the phosphorylation of Smad2 and Smad1/5. The inhibitory activity of Smad7 impinges upon both signaling pathways, and Smad7 has been a well characterized factor in the cross-talk between the TGF-β subfamilies (36). Inhibition via Smad7 works through multiple potential mechanisms: 1) compete with Smads for binding to activated type I receptor, 2) engage E3-ubiquitin ligases to degrade activated type 1 receptors, and 3) recruit phosphatases to activated type 1 receptor causing dephosphorylation and deactivation (48). Our work does not distinguish from these potential models of Smad7 function in ES cells; thus, further work is necessary to define the mechanism of BMP signaling modulation by Smad7 in ES cells.
Little work has been done to understand the complex transcriptional regulation of Smad7 in mouse ES cells. Our results clearly show a role for Nodal signaling in the transcriptional regulation of Smad7 in ES cells. Smad7 levels are reduced 0.5-fold upon inhibition of Nodal signaling by SB431542. In contrast, alterations of BMP signaling have very little effect on Smad7 transcript levels, suggesting BMP signaling is not a major regulator of Smad7 in ES cells. Although our work clearly shows that Nodal signaling can attenuate BMP signaling, our work does not address the converse question of whether regulation of Nodal signaling is regulated by the BMP pathway. Because Smad7 is not substantially regulated by BMP signaling, any potential regulation of Nodal signaling by the BMP pathway would likely be via a Smad7-independent mode. Interestingly, upon inhibition of Nodal signaling by SB431542, ES cells still retain a significant amount of Smad7 expression (Fig. 2A), indicating an additional Nodal-independent regulatory pathway that also controls some Smad7 expression. Genome-wide localization studies have shown that both Oct4 and Nanog occupy distal upstream sites upstream of the Smad7 transcriptional start site (49), the functions of which have not yet been determined. Because Smad7 regulates both the BMP and Nodal signaling pathways, a better understanding of Smad7 regulation will provide valuable information about the control of these pathways.
Nodal Functions in Pluripotency and Prodifferentiation Pathways
The microarray analysis of mouse ES cells with altered Nodal signaling identified a cohort of Smad2-regulated genes, which confirms and expands on previously published studies examining pSmad2 positively regulated genes via expression of an activated type I receptor (17). Although it remains to be shown, many of these genes are likely directly regulated by Nodal through transcriptional activation via Smad2. Many of the genes such as Brachyury, Pitx2, and Mixl1 also have significant developmental roles, with a bias toward genes with roles in gastrulation and mesoderm differentiation. However, the expression of many of these genes is complex and not restricted to the mesoderm lineage and include expression in the inner cell mass of blastocyst stage embryos (50–52). Thus, the expression of many of the developmentally regulated genes in ES cells likely does not represent overtly differentiated cell types.
Notably, Nodal inhibition also caused a striking derepression of genes not expressed in ES cells, including those characteristic of the trophectoderm lineage such as Ascl2 and Gata2. Nodal signaling has complex roles in regulating trophoblast differentiation (53). Our findings suggest that Nodal signaling may function to suppress elements of this cell fate in undifferentiated in ES cells. More work needs to be done to determine if this derepression of trophectoderm genes is BMP-dependent. Mouse ES cells do not readily contribute to the trophectoderm lineages when introduced to a blastocyst; however, some differentiation conditions have been described to allow such differentiation in vitro (54, 55). Thus, further work is needed to ascertain whether autocrine Nodal signaling inhibits the propensity of ES cells to differentiate to a trophectoderm pathway, and if this function is achieved by attenuating BMP activity. Nodal activity in ES cells is thus complex and modulates both pluripotency and prodifferentiation pathways.
Our data point to a role for Nodal signaling in attenuating, but not abolishing, BMP signaling in undifferentiated ES cells. Given that BMP signaling is essential for maintaining ES cells under serum-free conditions, an important question remains as to why ES cells utilize autocrine Nodal signaling to attenuate BMP signaling. Both ES cells and the early epiblast are thought to represent a “poised state,” in which a variety of prodifferentiation pathways are present but inhibited by extrinsic signals such as BMP and LIF to maintain ES cell identity and pluripotency (6, 56). Although BMP signaling is essential for maintaining ES cells in an undifferentiated state, numerous studies have shown that under differentiating conditions BMP signaling also strongly promotes differentiation of ES cells toward certain mesodermal lineages (57), in many ways mimicking the developmental progression of mesoderm differentiation in the gastrulating embryo. Thus, attenuated BMP activity in undifferentiated ES cells may be important to repress the differentiation of ES cells to some mesodermal, and perhaps trophectodermal pathways. In addition to functioning to modulate BMP signaling, the active Nodal signaling pathway in ES cells may also represent a poised signaling pathway, be invoked upon BMP withdrawal, and be further enhanced during differentiation to certain early developmental lineages. Multiple studies have shown that Activin and Nodal signaling promote the differentiation of definitive endoderm and mesoderm fate choices (58, 59). Thus, the activities of both the Nodal signaling and BMP signaling are multifaceted in ES cells, and appear to be essential to both maintain ES cell homeostasis while also required for certain differentiation pathways. The multifaceted functions of these pathways in both pluripotency and differentiation would thus require a complex means to regulating the activity of these pathways to control the ES cell phenotype.
Interactions between Nodal and BMP Signaling in Other Experimental Systems
Our data point to a regulation of the BMP pathway in ES cells via Nodal signaling through an intracellular regulation of Smad7. Activin/Nodal signaling can modulate BMP signaling activity in a variety of systems, including dorsal-ventral patterning of the mesoderm during gastrulation (60) and induction of trophoblast differentiation from human ES cells (61). In many contexts, Activin/Nodal signaling has a negative effect on BMP signaling via regulation of secreted BMP ligand antagonists such as noggin, follistatin, and chordin (35). These extracellular antagonists permit localized activity of both Activin/Nodal and BMP signaling in the prospective dorsal and ventral sides of the embryo, respectively. The connectivity between the Nodal-Smad2 pathway with the pro-pluripotency program of BMP-Smad1/5 suggests that Nodal signaling modulates the pluripotent state of ES cells. Pharmacological inhibition of Smad2 signaling demonstrated that Smad2 signaling is required for the efficient outgrowth of the inner cell mast from blastocysts yet is not necessary for maintenance of Oct4 expression in mouse ES cells (62). Inhibition of Nodal-Smad2 signaling reduces proliferation of ES cells grown in media with serum or knock-out serum replacement (16). We confirmed that long term Nodal-Smad2 inhibition slows the proliferation of ES cells in a defined media supplemented with LIF and BMP4 and, of interest, the SB431542 treatment appears to reduce the spontaneous differentiation on the edge of ES cell colonies (data not shown). However, long term treatment of ES cells with SB431542 in the absence of BMP supplement resulted in a decline of the pluripotent marker Nanog expression by over 50% (supplemental Fig. 2B). Thus, although the cross-talk between the Nodal and BMP pathways is long lasting, inhibition of Smad2 signaling alone is not sufficient to support ES cell self-renewal.
The culture conditions required to induce and sustain the pluripotent state of mouse and human ES cells greatly differ. LIF maintains undifferentiated mouse ES cells (2, 3), whereas human ES cells rely on Activin/Nodal signaling in combination with fibroblast growth factors (63–65). Activated BMP signaling stimulates differentiation in human ES cells (66) but maintains pluripotency in mouse ES cells (5). The disparities between extrinsic pluripotent signals of the mouse and human cells may in part arise from these cells representing slightly different stages of embryonic development (67). In common with our analyses, human ES cells induce Smad1 phosphorylation following inhibition of Smad2 signaling and reduce pSmad1 levels after Activin stimulation (61, 62). The molecular mechanisms of Nodal and BMP interconnectivity, including Smad7 modulation, may be shared in ES cells of different species even though the functionalities of these pathways differ.
Functions of Nodal-related Signaling in iPSCs
Induced pluripotent stem cells (iPSCs) were first generated through viral transduction of specific transcription factors (Oct4, Sox2, c-Myc, and Klf4) into somatic cells to reprogram them to the embryonic state (68, 69). Current work is focusing on potential small molecules to replace the oncogenic factors and increase reprogramming efficiency (70). In combination with inhibition of the MEK pathway, SB431542 increased efficiency of fully reprogrammed human iPSCs from fibroblasts over 100-fold (71). Chemical inhibition of Smad2 reduced spontaneous differentiation and increased clonal expansion efficiency of iPSCs (72). Further analysis revealed that inhibition of Smad2 signaling replaces the need for transduction of Sox2 by inducing expression of Nanog in iPSCs (73). The biochemical mechanism for the enhanced reprogramming of somatic cells by Smad2 inhibition is unknown. SB431542 treatment of mouse ES cells grown in ES media increases the percentage of Nanog-positive cells and expression of Oct4, Nanog, Stella, and Id genes (data not shown). Thus, the linkage between Nodal-Smad2 inhibition and BMP-Smad1/5 activation found in mouse ES cells may also apply to iPSCs. Future work will be needed to determine if the molecular mechanisms by which inhibition of Nodal signaling enhances the pluripotency of ES cells and iPSCs have a common molecular basis.
Supplementary Material
Acknowledgments
We gratefully acknowledge the technical expertise of Lucy Liu. We thank the University of Kansas Medical Center-Microarray Facility for generating array data sets and Aris Moustakas for providing the CAGA12 plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grant RR022888 and COBRE P20RR024214 (to J. L. V.) and by NIH postdoctoral training grant T32HD007455 (to K. E. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, references, and Figs. 1–5.
- ES
- embryonic stem
- TGF-β
- transforming growth factor-β
- LIF
- leukemia inhibitory factor
- T
- Brachyury
- Fst
- Follistatin
- BMP
- bone morphogenetic protein
- ALK
- Activin receptor-like kinase
- pSmad
- phosphorylated Smad
- Id
- inhibitors of differentiation
- SB
- SB431542
- shRNA
- small hairpin RNA
- siRNA
- small interference RNA
- rtPCR
- real-time quantitative PCR
- iPSC
- induced pluripotent stem cell.
REFERENCES
- 1.Evans M. J., Kaufman M. H. (1981) Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- 2.Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M., Rogers D. (1988) Nature 336, 688–690 [DOI] [PubMed] [Google Scholar]
- 3.Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., Wagner E. F., Metcalf D., Nicola N. A., Gough N. M. (1988) Nature 336, 684–687 [DOI] [PubMed] [Google Scholar]
- 4.Qi X., Li T. G., Hao J., Hu J., Wang J., Simmons H., Miura S., Mishina Y., Zhao G. Q. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6027–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ying Q. L., Nichols J., Chambers I., Smith A. (2003) Cell 115, 281–292 [DOI] [PubMed] [Google Scholar]
- 6.Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008) Nature 453, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lasorella A., Uo T., Iavarone A. (2001) Oncogene 20, 8326–8333 [DOI] [PubMed] [Google Scholar]
- 8.Sikder H. A., Devlin M. K., Dunlap S., Ryu B., Alani R. M. (2003) Cancer Cell 3, 525–530 [DOI] [PubMed] [Google Scholar]
- 9.Hollnagel A., Oehlmann V., Heymer J., Rüther U., Nordheim A. (1999) J. Biol. Chem. 274, 19838–19845 [DOI] [PubMed] [Google Scholar]
- 10.Pucéat M. (2007) Cardiovasc. Res. 74, 256–261 [DOI] [PubMed] [Google Scholar]
- 11.Kitisin K., Saha T., Blake T., Golestaneh N., Deng M., Kim C., Tang Y., Shetty K., Mishra B., Mishra L. (2007) Sci. STKE 2007, cm1. [DOI] [PubMed] [Google Scholar]
- 12.Miyazawa K., Shinozaki M., Hara T., Furuya T., Miyazono K. (2002) Genes Cells 7, 1191–1204 [DOI] [PubMed] [Google Scholar]
- 13.Massagué J., Seoane J., Wotton D. (2005) Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 14.Nakao A., Afrakhte M., Morén A., Nakayama T., Christian J. L., Heuchel R., Itoh S., Kawabata M., Heldin N. E., Heldin C. H., ten Dijke P. (1997) Nature 389, 631–635 [DOI] [PubMed] [Google Scholar]
- 15.Watabe T., Miyazono K. (2009) Cell Res. 19, 103–115 [DOI] [PubMed] [Google Scholar]
- 16.Ogawa K., Saito A., Matsui H., Suzuki H., Ohtsuka S., Shimosato D., Morishita Y., Watabe T., Niwa H., Miyazono K. (2007) J. Cell Sci. 120, 55–65 [DOI] [PubMed] [Google Scholar]
- 17.Guzman-Ayala M., Lee K. L., Mavrakis K. J., Goggolidou P., Norris D. P., Episkopou V. (2009) PLoS One 4, e4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skarnes W. C. (2000) Methods Enzymol. 328, 592–615 [DOI] [PubMed] [Google Scholar]
- 19.Nagy A., Gertsenstein M., Vintersten K., Behringer R. (2003) Manipulating the Mouse Embryo, 3rd Ed., pp. 359–397, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20.Ying Q. L., Smith A. G. (2003) Methods Enzymol. 365, 327–341 [DOI] [PubMed] [Google Scholar]
- 21.Vivian J. L., Chen Y., Yee D., Schneider E., Magnuson T. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15542–15547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedrich G., Soriano P. (1991) Genes Dev. 5, 1513–1523 [DOI] [PubMed] [Google Scholar]
- 23.Vivian J. L., Klein W. H., Hasty P. (1999) BioTechniques 27, 154–162 [DOI] [PubMed] [Google Scholar]
- 24.Tournay O., Benezra R. (1996) Mol. Cell Biol. 16, 2418–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pardali K., Kurisaki A., Morén A., ten Dijke P., Kardassis D., Moustakas A. (2000) J. Biol. Chem. 275, 29244–29256 [DOI] [PubMed] [Google Scholar]
- 26.Ro S., Hwang S. J., Ordög T., Sanders K. M. (2005) BioTechniques 38, 625–627 [DOI] [PubMed] [Google Scholar]
- 27.Kyba M., Perlingeiro R. C., Daley G. Q. (2002) Cell 109, 29–37 [DOI] [PubMed] [Google Scholar]
- 28.O'Gorman S., Fox D. T., Wahl G. M. (1991) Science 251, 1351–1355 [DOI] [PubMed] [Google Scholar]
- 29.Hooper M., Hardy K., Handyside A., Hunter S., Monk M. (1987) Nature 326, 292–295 [DOI] [PubMed] [Google Scholar]
- 30.Inman G. J., Nicolás F. J., Callahan J. F., Harling J. D., Gaster L. M., Reith A. D., Laping N. J., Hill C. S. (2002) Mol. Pharmacol. 62, 65–74 [DOI] [PubMed] [Google Scholar]
- 31.Laping N. J., Grygielko E., Mathur A., Butter S., Bomberger J., Tweed C., Martin W., Fornwald J., Lehr R., Harling J., Gaster L., Callahan J. F., Olson B. A. (2002) Mol. Pharmacol. 62, 58–64 [DOI] [PubMed] [Google Scholar]
- 32.Tabibzadeh S., Hemmati-Brivanlou A. (2006) Stem Cells 24, 1998–2006 [DOI] [PubMed] [Google Scholar]
- 33.Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. (1998) EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao X., Nicholls J. M., Chen Y. G. (2008) J. Biol. Chem. 283, 3272–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balemans W., Van Hul W. (2002) Dev. Biol. 250, 231–250 [PubMed] [Google Scholar]
- 36.Yan X., Liu Z., Chen Y. (2009) Acta Biochim. Biophys. Sin 41, 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tojo M., Hamashima Y., Hanyu A., Kajimoto T., Saitoh M., Miyazono K., Node M., Imamura T. (2005) Cancer Sci. 96, 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyazono K., Miyazawa K. (2002) Sci. STKE 2002, pe40. [DOI] [PubMed] [Google Scholar]
- 39.Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 40.Hao J., Daleo M. A., Murphy C. K., Yu P. B., Ho J. N., Hu J., Peterson R. T., Hatzopoulos A. K., Hong C. C. (2008) PLoS One 3, e2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu P. B., Hong C. C., Sachidanandan C., Babitt J. L., Deng D. Y., Hoyng S. A., Lin H. Y., Bloch K. D., Peterson R. T. (2008) Nat. Chem. Biol. 4, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagna G., Hata A., Hemmati-Brivanlou A., Massagué J. (1996) Nature 383, 832–836 [DOI] [PubMed] [Google Scholar]
- 43.Amthor H., Christ B., Rashid-Doubell F., Kemp C. F., Lang E., Patel K. (2002) Dev. Biol. 243, 115–127 [DOI] [PubMed] [Google Scholar]
- 44.Iemura S., Yamamoto T. S., Takagi C., Uchiyama H., Natsume T., Shimasaki S., Sugino H., Ueno N. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9337–9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pierre A., Pisselet C., Monget P., Monniaux D., Fabre S. (2005) Reprod. Nutr. Dev. 45, 419–425 [DOI] [PubMed] [Google Scholar]
- 46.Hanyu A., Ishidou Y., Ebisawa T., Shimanuki T., Imamura T., Miyazono K. (2001) J. Cell Biol. 155, 1017–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishisaki A., Yamato K., Hashimoto S., Nakao A., Tamaki K., Nonaka K., ten Dijke P., Sugino H., Nishihara T. (1999) J. Biol. Chem. 274, 13637–13642 [DOI] [PubMed] [Google Scholar]
- 48.ten Dijke P., Hill C. S. (2004) Trends Biochem. Sci. 29, 265–273 [DOI] [PubMed] [Google Scholar]
- 49.Loh Y. H., Wu Q., Chew J. L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wong K. Y., Sung K. W., Lee C. W., Zhao X. D., Chiu K. P., Lipovich L., Kuznetsov V. A., Robson P., Stanton L. W., Wei C. L., Ruan Y., Lim B., Ng H. H. (2006) Nat. Genet. 38, 431–440 [DOI] [PubMed] [Google Scholar]
- 50.Takaoka K., Yamamoto M., Shiratori H., Meno C., Rossant J., Saijoh Y., Hamada H. (2006) Dev. Cell 10, 451–459 [DOI] [PubMed] [Google Scholar]
- 51.Yoshikawa T., Piao Y., Zhong J., Matoba R., Carter M. G., Wang Y., Goldberg I., Ko M. S. (2006) Gene Expr. Patterns 6, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong W., Wang Q. T., Sun T., Wang F., Liu J., Leach R., Johnson A., Puscheck E. E., Rappolee D. A. (2006) Mol. Reprod. Dev. 73, 540–550 [DOI] [PubMed] [Google Scholar]
- 53.Ma G. T., Soloveva V., Tzeng S. J., Lowe L. A., Pfendler K. C., Iannaccone P. M., Kuehn M. R., Linzer D. I. (2001) Dev. Biol. 236, 124–135 [DOI] [PubMed] [Google Scholar]
- 54.He S., Pant D., Schiffmacher A., Meece A., Keefer C. L. (2008) Stem Cells 26, 842–849 [DOI] [PubMed] [Google Scholar]
- 55.Hayashi Y., Furue M. K., Tanaka S., Hirose M., Wakisaka N., Danno H., Ohnuma K., Oeda S., Aihara Y., Shiota K., Ogura A., Ishiura S., Asashima M. (2010) In Vitro Cell Dev. Biol. Anim. 46, 416–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva J., Smith A. (2008) Cell 132, 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park C., Afrikanova I., Chung Y. S., Zhang W. J., Arentson E., Fong Gh G., Rosendahl A., Choi K. (2004) Development 131, 2749–2762 [DOI] [PubMed] [Google Scholar]
- 58.Takenaga M., Fukumoto M., Hori Y. (2007) J. Cell Sci. 120, 2078–2090 [DOI] [PubMed] [Google Scholar]
- 59.Kubo A., Shinozaki K., Shannon J. M., Kouskoff V., Kennedy M., Woo S., Fehling H. J., Keller G. (2004) Development 131, 1651–1662 [DOI] [PubMed] [Google Scholar]
- 60.Niehrs C. (2004) Nat. Rev. Genet. 5, 425–434 [DOI] [PubMed] [Google Scholar]
- 61.Wu Z., Zhang W., Chen G., Cheng L., Liao J., Jia N., Gao Y., Dai H., Yuan J., Cheng L., Xiao L. (2008) J. Biol. Chem. 283, 24991–25002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.James D., Levine A. J., Besser D., Hemmati-Brivanlou A. (2005) Development 132, 1273–1282 [DOI] [PubMed] [Google Scholar]
- 63.Beattie G. M., Lopez A. D., Bucay N., Hinton A., Firpo M. T., King C. C., Hayek A. (2005) Stem Cells 23, 489–495 [DOI] [PubMed] [Google Scholar]
- 64.Xiao L., Yuan X., Sharkis S. J. (2006) Stem Cells 24, 1476–1486 [DOI] [PubMed] [Google Scholar]
- 65.Vallier L., Alexander M., Pedersen R. A. (2005) J. Cell Sci. 118, 4495–4509 [DOI] [PubMed] [Google Scholar]
- 66.Xu R. H., Chen X., Li D. S., Li R., Addicks G. C., Glennon C., Zwaka T. P., Thomson J. A. (2002) Nat. Biotechnol. 20, 1261–1264 [DOI] [PubMed] [Google Scholar]
- 67.Ginis I., Luo Y., Miura T., Thies S., Brandenberger R., Gerecht-Nir S., Amit M., Hoke A., Carpenter M. K., Itskovitz-Eldor J., Rao M. S. (2004) Dev. Biol. 269, 360–380 [DOI] [PubMed] [Google Scholar]
- 68.Park I. H., Zhao R., West J. A., Yabuuchi A., Huo H., Ince T. A., Lerou P. H., Lensch M. W., Daley G. Q. (2008) Nature 451, 141–146 [DOI] [PubMed] [Google Scholar]
- 69.Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. (2007) Nature 448, 318–324 [DOI] [PubMed] [Google Scholar]
- 70.Seifinejad A., Tabebordbar M., Baharvand H., Boyer L. A., Hosseini Salekdeh G. (2010) Stem Cell Rev., in press [DOI] [PubMed] [Google Scholar]
- 71.Lin T., Ambasudhan R., Yuan X., Li W., Hilcove S., Abujarour R., Lin X., Hahm H. S., Hao E., Hayek A., Ding S. (2009) Nat. Methods 6, 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li W., Wei W., Zhu S., Zhu J., Shi Y., Lin T., Hao E., Hayek A., Deng H., Ding S. (2009) Cell Stem Cell 4, 16–19 [DOI] [PubMed] [Google Scholar]
- 73.Ichida J. K., Blanchard J., Lam K., Son E. Y., Chung J. E., Egli D., Loh K. M., Carter A. C., Di Giorgio F. P., Koszka K., Huangfu D., Akutsu H., Liu D. R., Rubin L. L., Eggan K. (2009) Cell Stem Cell 5, 491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.