Abstract
Agrobacterium tumefaciens is a plant pathogen that utilizes a type IV secretion system (T4SS) to transfer DNA and effector proteins into host cells. In this study we discovered that an α-crystallin type small heat-shock protein (α-Hsp), HspL, is a molecular chaperone for VirB8, a T4SS assembly factor. HspL is a typical α-Hsp capable of protecting the heat-labile model substrate citrate synthase from thermal aggregation. It forms oligomers in a concentration-dependent manner in vitro. Biochemical fractionation revealed that HspL is mainly localized in the inner membrane and formed large complexes with certain VirB protein subassemblies. Protein-protein interaction studies indicated that HspL interacts with VirB8, a bitopic integral inner membrane protein that is essential for T4SS assembly. Most importantly, HspL is able to prevent the aggregation of VirB8 fused with glutathione S-transferase in vitro, suggesting that it plays a role as VirB8 chaperone. The chaperone activity of two HspL variants with amino acid substitutions (F98A and G118A) for both citrate synthase and glutathione S-transferase-VirB8 was reduced and correlated with HspL functions in T4SS-mediated DNA transfer and virulence. This study directly links in vitro and in vivo functions of an α-Hsp and reveals a novel α-Hsp function in T4SS stability and bacterial virulence.
Keywords: Bacteria, Chaperone Chaperonin, Heat Shock Protein, Membrane Proteins, Protein Secretion, Agrobacterium tumefaciens, DNA Transfer, Type IV Secretion System, Small Heat-shock Protein, Virulence
Introduction
α-Crystallin-type small heat-shock proteins (α-Hsps)2 are ubiquitous ATP-independent low molecular mass chaperones consisting of a conserved α-crystallin domain of 80–100 amino acid residues flanked by a variable N-terminal region and a C-terminal extension (1). A number of in vitro and in vivo studies demonstrated that α-Hsps bind partially denatured proteins to prevent irreversible protein aggregation resulting from stresses or diseases (2). In contrast, little is known about the actual physiological substrates or clients of α-Hsps and the molecular mechanisms underlying their in vivo functions. Studies with α-Hsp variants demonstrated that the chaperone activity determined in vitro is critical for their function in vivo, that is very important for organisms. Point mutations in human αA-crystallin are associated with hereditary cataracts (3–5). The αA-crystallin R49C protein causes both loss of chaperone activity in vitro (6) and increases protein insolubility in vivo (7, 8), suggesting the importance of its chaperone activity for eye lens transparency. Mutation studies of Hsp16.6 from the cyanobacterium Synechocystis indicated a link between in vitro chaperone activity and cellular thermotolerance (9–11). Almost all characterized α-Hsps have been studied using thermal-labile artificial model substrates such as citrate synthase (CS), firefly luciferase, or malate dehydrogenase to determine their ability in preventing aggregation of these substrates in vitro (12, 13). Occasionally, different model substrates provided conflicting results emphasizing an urgent need for the identification of the actual physiological α-Hsp substrates or clients.
In general, the expression of bacterial α-Hsp genes is low or undetectable under normal growth conditions but is massively induced under heat shock or as consequence of other non-heat-shock stresses (1). The induction of bacterial α-Hsp genes during the process of bacterial infection or under virulence-induced conditions was previously reported in the human pathogens Mycobacterium tuberculosis (14–16), Mycobacterium leprae (17), and the plant pathogen Agrobacterium tumefaciens (18). Although the α-Hsp genes in M. tuberculosis and A. tumefaciens are required for full virulence, the molecular mechanisms underlying their role in bacterial virulence are not clear. Multiple α-Hsp genes are present in rhizobial bacteria (19–21) including A. tumefaciens containing at least four members of α-Hsp genes (hspC; hspL; hspAT1; hspAT2) (22). The latter three were induced by heat shock (22), whereas only hspL was up-regulated by the virulence gene inducer acetosyringone (AS) (18, 23).
A. tumefaciens is a Gram-negative plant pathogenic bacterium with the unique ability to conduct interkingdom DNA transfer to plant genomes. This horizontal gene transfer is mediated by an evolutionarily conserved type IV secretion system (T4SS) comprising the VirD4-coupling protein and 11 VirB proteins (VirB1 to VirB11) that also assembles T-pili (24, 25). Accumulating biochemical and genetic data suggest that VirB6 and VirB8 are inner membrane proteins functioning as an inner membrane base complex to connect the VirB7/VirB9/VirB10 core channel across the double membrane (26–28). Crystallography and protein-protein interaction studies also suggest that VirB8 dimerization and interactions with VirB4 and VirB10 are important for its role in T4SS assembly (29, 30). As monitored by transfer DNA immunoprecipitation assays, the T-complex (a covalent complex of single-stranded DNA and VirD2) is translocated across the cell envelope via four discrete steps of sequential interactions with VirD4, VirB11, VirB6/VirB8, and VirB2/VirB9 (31). In addition to transporting the T-complex and effector proteins from bacteria into plant cells, the T4SS can translocate the Q group incompatibility plasmid RSF1010 from A. tumefaciens into plant cells (32) or between agrobacteria (33).
We previously discovered that HspL is a VirB-induced α-Hsp involved in VirB protein accumulation, T4SS-mediated DNA transfer, that is required for full virulence of A. tumefaciens (18). Here, we demonstrate that HspL is a typical molecular chaperone interacting not only with the model substrate CS but also with VirB8. HspL is able to protect both target proteins from aggregation in vitro. The altered chaperone activity of two HspL variants correlated well with reduced HspL functions in promoting T4SS-mediated DNA transfer and virulence. The provided evidence strongly suggests that HspL functions as a VirB8 chaperone and that this chaperone activity is responsible for its role in DNA transfer and virulence of A. tumefaciens.
EXPERIMENTAL PROCEDURES
Plasmids, Bacterial Strains, and Growth Conditions
The bacterial strains and plasmids constructed or used in this study were summarized in supplemental Table 1. The sequences of primers used for plasmid construction were listed in supplemental Table 2. A. tumefaciens strains were routinely grown at 28 °C in 523 medium (34), and Escherichia coli strains were grown at 37 °C in Luria-Bertani medium (35). Antibiotics were added at the following final concentrations: 100 μg/ml ampicillin, 20 μg/ml of tetracyclin (Tc), and 10 μg/ml of gentamicin (Gm) for E. coli; 50 μg/ml erythromycin, 50 μg/ml of rifampicin, 250 μg/ml of spectinomycin, 20 μg/ml of Tc, and 50 μg/ml of Gm for A. tumefaciens. The A. tumefaciens virulence gene induction was performed as described (18) by growing the agrobacterial cells in I-medium (AB-MES, pH 5.5) (36) at 25 °C for 16 h in the presence of 200 μm AS (Sigma) without the addition of any antibiotics.
SDS-PAGE and Western Blot Analysis
Proteins were resolved by either glycine-SDS-PAGE (35) or Tricine-SDS-PAGE (37). Western blot analysis was performed as described previously (36) using primary polyclonal antibodies against HspL (18), VirB (38, 39), or GroEL (40) followed by secondary antibody using horseradish peroxidase-conjugated goat anti-rabbit IgG (Chemicon International, Inc.) and detection using the Western Lightning System (PerkinElmer Life Sciences). Chemiluminescent signals were visualized on an x-ray film (Eastman Kodak Co.).
Protein Expression and Purification
The expression vectors pET22b(+) and pET42b(+) were used to construct the plasmids for overexpression of His-tagged HspL and glutathione S-transferase (GST)-fused VirB8, respectively. The overexpression of HspL-His (∼19 kDa) and GST-VirB8 (∼52 kDa) in the E. coli BL21(DE3) strain was performed according to the instructions of the pET user manual (Novagen, EMD Biosciences, Inc.) by isopropyl 1-thio-β-d-galactopyranoside (0.4 mm) induction at 37 °C for 1.5 h (HspL-His) or 28 °C for 2 h (GST-VirB8). His-tagged HspL and variants were purified via Ni-NTA His Bind Resins, and GST-VirB8 was purified via glutathione-agarose (Sigma).
Thermal Aggregation Protection Assay
The artificial model substrate CS (Sigma) or native client VirB8 fused with GST were used as substrates for thermal aggregation protection assays (41). Thermal aggregation of CS and GST-VirB8 were induced at 43 and 50 °C, respectively, and they were co-incubated with His-tagged HspL or two inactive variants (HspLF98A and HspLG118A) in 50 mm sodium phosphate, pH 6.8, to assess the HspL chaperone activity in protecting substrates from aggregation. CS was dialyzed against TE buffer (10 mm Tris-HCl, 1 mm EDTA, pH 8.0) and stored at −20 °C before use.
Size Exclusion Chromatography (SEC)
Native sizes of His-tagged HspL and its variants were determined by analytical SEC on a Superdex 200 pg Hiload 16/60 column (GE Healthcare) in the cold room (4–8 °C) after equilibration with mobile buffer (1 m NaCl, 50 mm sodium phosphate pH 8.0, 10% glycerol). Purified proteins were separated on an ÄKTATM explorer system (GE Healthcare) at a flow rate of 0.7 ml/min. Absorbance was recorded at a wavelength of 280 nm. Blue dextran (2000 kDa) was used to determine the void volume that we routinely obtained at 43.3-ml elution with the following calibration proteins: thyroglobulin (669 kDa), apoferritin (443 kDa), amylase (200 kDa), alcohol dehydrogenase (150 kDa), albumin (66 kDa), carbonic anhydrase (29 kDa), and vitamin B12 (1.3 kDa).
Biochemical Fractionations, Membrane Protein Solubilization, and Spheroplast Protease Accessibility Assay
Cellular fractions were isolated as described (42) with the exception of lysozyme (Sigma) treatment that was conducted on ice for 1 h after cell lysis. Protein solubilization experiments with detergents were performed as described (43) with minor modifications. Insoluble membrane fractions (1 mg/ml protein concentration) were resuspended in 200 μl of 50 mm Tris-HCl, pH 7.5 (buffer only), or in the presence of 1 m NaCl, 0.1% Na2CO3, 1% Triton X-100, 6 m urea, or 1% SDS in the buffer. After incubation on ice for 30 min, the samples were centrifuged at 150,000 × g for 1 h to sediment insoluble proteins. The resulting supernatant and the pellets resuspended in 200 μl of 50 mm Tris-HCl, pH 7.5, were analyzed by Western blotting. Spheroplasts were prepared as described (44) with minor modifications. A. tumefaciens cell culture was harvested by centrifugation at 10,000 × g at 4 °C for 10 min and resuspended in buffer (50 mm Tris-HCl, pH 7.5, 20% sucrose, 2 mm EDTA, 0.2 mm dithiothreitol, 0.25 mm phenylmethylsulfonyl fluoride) to A600 = 10. The cell suspension was treated with lysozyme (0.5 mg/ml) and incubated on ice for 1 h. Spheroplasts were collected by centrifugation at 5000 × g for 5 min, gently washed once, and resuspended in the same volume of buffer (50 mm Tris-HCl, pH 7.5, 20% sucrose, 10 mm MgSO4) followed by treatment with Streptomyces griseus protease (Sigma) at the final concentration of 100, 50, or 25 μg/ml for 10 min on ice. The reaction was stopped by adding 2× SDS sample buffer (35), and the samples were then boiled immediately for 15 min.
Dodecyl-β-d-maltopyranoside (DDM)-extracted HspL Complex and Its Interacting Protein(s)
The membrane isolation, extraction of membrane-associated proteins with DDM and SEC for analysis of HspL and VirB protein complexes, were conducted as described (45) with minor modifications. Membrane fractions isolated from A. tumefaciens strains NT1RE(pJK270) or ΔhspL(pHspL-His) grown in AS-induced I-medium at 25 °C for 16 h were resuspended at 0.2 mg/ml protein concentration in lysis buffer (50 mm sodium phosphate, 250 mm NaCl, pH 8.0) containing 2% DDM and incubated with rocking at 4 °C overnight to extract membrane protein complexes. The DDM-extracted soluble fractions were collected by centrifugation at 150,000 × g at 4 °C for 1 h, which sedimented the insoluble fraction. 1 ml of DDM-extracted proteins was analyzed by SEC on a Superdex 200 HiLoad 16/60 column as described above. When applicable, the samples were 4-fold-diluted in lysis buffer and used to pull down potential HspL-interacting protein(s) purified via a Ni-NTA His Bind Resin column (Novagen). Purified HspL-His and its interacting protein(s) were resolved by glycine-SDS-PAGE and detected by Western blotting with specific antibodies.
Yeast Two-hybrid Analysis
MatchmakerTM yeast two-hybrid analysis was performed according to the instructions of the user manual (Clontech, Mountain View, CA). The PCR-amplified full-length virB8 (VirB8-Y2H, digested by NdeI/EcoRI) and hspL (HspL-Y2H, digested by NdeI/BamHI) were cloned into pGADT7 to create an N-terminal fusion to the activation domain and into pGBKT7 to create an N-terminal fusion to the DNA binding domain (Clontech) at the same restriction enzyme sites, respectively. Each pair of plasmids was transformed into Saccharomyces cerevisiae strain AH109 and selected on synthetic dextrose minimal medium in the absence of leucine and tryptophan grown at 28 °C for 3 days. The positive clones were selected twice by their growth on synthetic dextrose medium in the absence of Ade, His, Leu, and Trp grown at 28 °C for 3 days.
Co-purification of HspL-His and VirB8 from E. coli
The proteins encoded by pETHspL and pTrcB8 plasmids were co-expressed in E. coli BL21(DE3) by isopropyl 1-thio-β-d-galactopyranoside induction at 37 °C (0.4 mm) for 2 h. Soluble HspL-His and its interacting protein(s) were bound to a Ni-NTA His Bind Resin column (Novagen) and washed with washing buffer (50 mm NaH2PO4, 300 mm NaCl, 50 mm imidazole, pH 8.0) following the manufacturer's instructions. The bound proteins were eluted with elution buffer (50 mm sodium phosphate, 300 mm NaCl, 250 mm imidazole, pH 8.0), resolved by glycine-SDS-PAGE, and detected by Western blotting with VirB8- and HspL-specific antibodies.
Conjugal Transfer Analysis of Q Group Incompatibility Plasmid RSF1010
The conjugation assays were carried out using NT1RE(pJK270), and its derivatives as donor strains and NT1RE-Sp as the recipient strain is as described (18).
Tumor Assay on Potato Tuber Discs
Quantitative tumorigenesis assays using potato tuber discs were performed following the method described (18) with minor modifications. The potato tuber discs were placed on water agar, infected with 10 μl of bacterial culture, and incubated at 24 °C for 24 h. To prevent bacterial growth, infected potato discs were dipped into phosphate-buffered saline containing Timentin (25 μg/ml) for a few seconds before further incubation at 24 °C on water agar supplemented with Timentin (100 μg/ml). Tumors were scored after 3 weeks.
RESULTS
HspL Possesses Concentration-dependent Chaperone Activity and Oligomerizes in Vitro
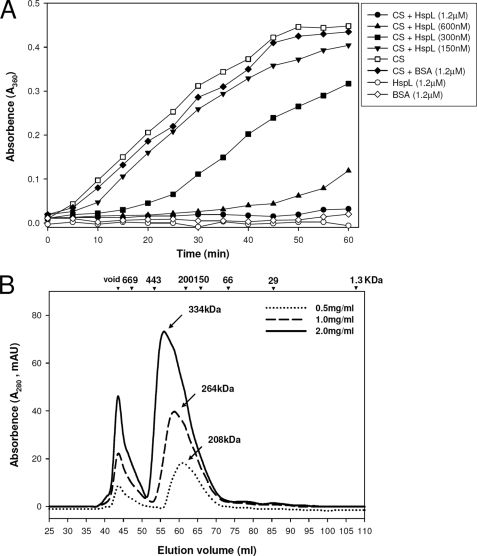
To understand the molecular mechanism underlying the contribution of HspL to T4SS-mediated DNA transfer and virulence of A. tumefaciens, we first investigated whether HspL possesses typical chaperone activity in preventing a model substrate from thermal aggregation. His-tagged HspL was purified and co-incubated with the standard heat-labile substrate CS at 43 °C. Although CS alone or in the presence of bovine serum albumin as a control aggregated rapidly as measured by light scattering (absorbance at 360 nm), thermal aggregation was inhibited in the presence of HspL (Fig. 1A). CS aggregation was reduced with increasing HspL concentrations, reaching full protection of CS from aggregation when HspL/CS co-incubation was set at a molar ratio of 2:1 (Fig. 1A). This result indicated that HspL is a typical α-Hsp possessing chaperone activity in protecting CS from thermal aggregation in a concentration-dependent manner.
FIGURE 1.
The chaperone activity and oligomerization property of HspL-His6. A, thermal aggregation protection assays using 600 nm CS as substrate were performed at 43 °C in the absence (□) and presence of HspL-His6 with concentrations at 150 nm (▾), 300 nm (■), 600 nm (▴), and 1.2 μm (●) or of 1.2 μm bovine serum albumin (BSA; (♦). 1.2 μm HspL-His6 (○) or BSA (◆) alone were also treated at the same condition as controls. Aggregation was monitored in absorbance at 360 nm and presented as a function of time. B, 0.5 ml of purified HspL-His at concentrations of 2.0 mg/ml (continuous line), 1.0 mg/ml (broken line), and 0.5 mg/ml (dotted line) were analyzed via SEC on Superdex 200 HiLoad 16/60. Blue dextran (2000 kDa) was used to determine the void volume at ∼44-ml elution volume and calibration markers are indicated. Protein elution was monitored by absorbance at 280 nm. mAU, milliabsorbance units.
Another property of α-Hsp is the formation of large oligomers, which is a prerequisite for chaperone activity (2). SEC analysis of purified soluble HspL resulted in two major peaks, the first corresponding to the void volume. Both peaks increased with increasing protein concentrations, and the second peaks corresponded to molecular masses of 208, 264, and 330 kDa at 0.5, 1.0, and 2.0 mg/ml protein concentration, respectively (Fig. 1B). Based on SDS-PAGE analysis, the majority of HspL eluted in the second peaks, but we also detected low amounts eluted in the void volume (∼44 ml of elution volume) (supplemental Fig. S1). The data suggest that native HspL predominantly forms large complexes containing appropriately 12–18 subunits.
Two HspL Variants Causing Reduced HspL Chaperone Activity and Altered Oligomerization Patterns
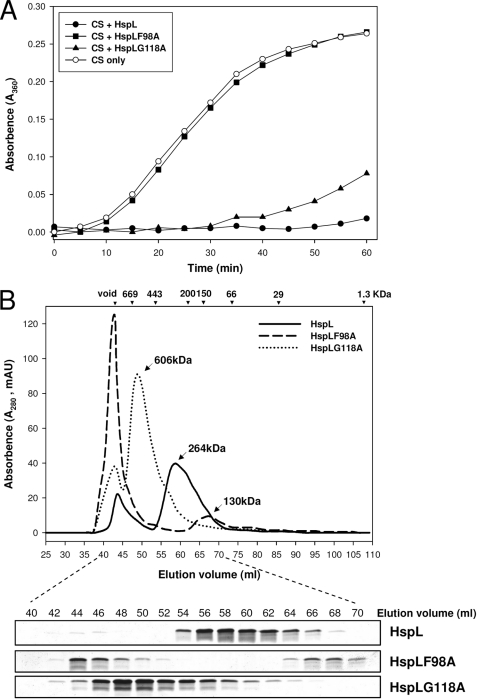
The α-crystallin domain of α-Hsps plays a critical role in its oligomerization and chaperone activity (9, 41). Multiple amino acid sequence alignments of HspL and selected homologs encoded by Rhizobiaceae and Homo sapiens α-crystallin A and B revealed the presence of a typical α-crystallin domain with conserved amino acid residues in HspL (supplemental Fig. S2). Several amino acid residues are strikingly conserved in all α-Hsps, strongly suggesting that these conserved amino acids are critical for α-Hsp function. Here, we generated two HspL variants in which the conserved residues Phe-98 or Gly-118 were substituted by Ala (F98A and G118A, supplemental Fig. S2). The importance of these two conserved amino acids has been documented previously in assays for both in vitro chaperone activity and in vivo function (9, 41). Although CS was fully protected from thermal aggregation by wild type HspL, HspLF98A completely lost the ability to prevent CS aggregation, and HspLG118A possessed reduced chaperone activity (Fig. 2A). The defect in chaperone activity was not due to precipitation of the HspL variants that remained mostly soluble after heat treatment (supplemental Fig. S3).
FIGURE 2.
HspL protein variants (HspLF98A and HspLG118A) were impaired in chaperone activity with altered oligomerization patterns. A, thermal aggregation protection assays using model substrate CS (600 nm) was carried out at 43 °C in the absence (○) or presence of HspL-His6 (●), HspLF98A-His6 (■), and HspLG118A-His6 (▴) at concentration of 1.2 μm. Aggregation was monitored in absorbance at 360 nm and presented as a function of time. B, 0.5 ml (1.0 mg/ml) of HspL-His6 (continuous line), HspLF98A-His6 (broken line), and HspLG118A-His6 (dotted line) were analyzed via SEC on Superdex 200 HiLoad 16/60. Blue dextran (2000 kDa) was used to determine the void volume at ∼44-ml elution volume, and calibration markers are indicated. Protein elution was monitored by absorbance at 280 nm. 10 μl of each eluted 2-ml interval was resolved in 15% glycine SDS-PAGE followed by Coomassie Blue staining.
To determine whether the altered chaperone activity correlated with changes in oligomerization, we examined HspLF98A and HspLG118A by SEC and found that oligomerization was different from the wild type in both cases (Fig. 2B and supplemental Fig. S1). In comparison to the 264-kDa oligomer of wild type HspL, HspLF98A formed two different complexes, one eluting with the void volume and the other at 130 kDa. HspLG118A formed a large 600-kDa oligomer. These results suggest that formation of a stable and defined oligomer is important for HspL chaperone activity.
HspL Chaperone Activity Correlates with Its Function in Promoting T4SS-mediated DNA Transfer and Virulence
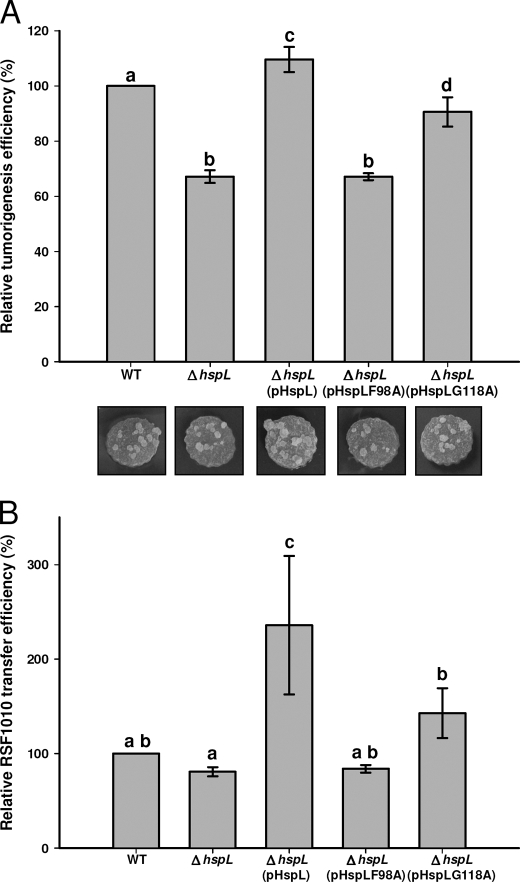
We next examined whether the altered in vitro chaperone activity of HspLF98A and HspLG118A correlated with a change in HspL function in vivo. The expression of wild type hspL driven by its native promoter on an IncP plasmid rescued the defect of the hspL deletion mutant (ΔhspL) in tumorigenesis efficiency on potato tuber discs (Ref. 18, Fig. 3A, and supplemental Table 3). In contrast, the expression of pHspLF98A in ΔhspL did not restore wild type tumor formation, and expression of pHspLG118A gave an intermediate phenotype. Similar to our previous findings (18), the deletion of hspL had only modest effects on T4SS-mediated plasmid transfer, but the expression of HspL stimulated transfer to levels higher than wild type (Fig. 3B and supplemental Table 4). This effect may be explained by the modest overexpression of HspL from the IncP plasmid (18), and the stimulation underlines the positive contribution of HspL to T4SS function. The expression of pHspLF98A in ΔhspL did not rescue the reduced transfer efficiency caused by the absence of HspL (Fig. 3B, supplemental Table 4). Consistent with its residual in vitro activity, pHspLG118A caused the increased T4SS-mediated transfer efficiency in ΔhspL to a level between ΔhspL and ΔhspL expressing wild type HspL. The positive correlation between chaperone activity, T4SS-mediated DNA transfer, and virulence strongly suggested that the chaperone activity of HspL is critical for its in vivo functions participation in T4SS.
FIGURE 3.
HspL protein variants (HspLF98A and HspLG118A) were impaired in complementing the tumorigenesis efficiency (A) and T4SS-mediated DNA transfer in ΔhspL mutant (B). For virulence assay, A. tumefaciens wild type (WT) strain NT1RE(pJK270), ΔhspL mutant, ΔhspL(pHspL), ΔhspL(pHspLF98A), and ΔhspL(pHspLG118A) were analyzed for their tumorigenesis efficiency (number of tumors per disc) on potato tuber discs by inoculation with 10 μl (108 cfu/ml) of bacterial cells. For T4SS-mediated RSF1010 transfer, all A. tumefaciens strains harboring the Q group incompatibility plasmid RSF1010 served as the donor strains and co-incubated with the recipient strain NT1RE-Sp (18) on the I-medium agar in the presence of 200 μm AS at 25 °C for 3 days. The transfer efficiency was evaluated as the number of transconjugants per input donor. Average values for relative tumorigenesis or RSF1010 transfer efficiency from three independent experiments are shown with S.E., in which the efficiency of wild type strain NT1RE(pJK270) was set at 100% and that of other strains is shown relative to that of NT1RE(pJK270). Means annotated with the same letter (a–d) are not significantly different; those with different letters are significantly different (p < 0.05) according to Duncan's multiple range test. Representative potato tuber discs with tumors were shown. The absolute tumorigenesis and RSF1010 transfer efficiency obtained from three independent experiments were shown in supplemental Table 3 and 4, respectively.
HspL Co-fractionates with Inner and Outer Membranes
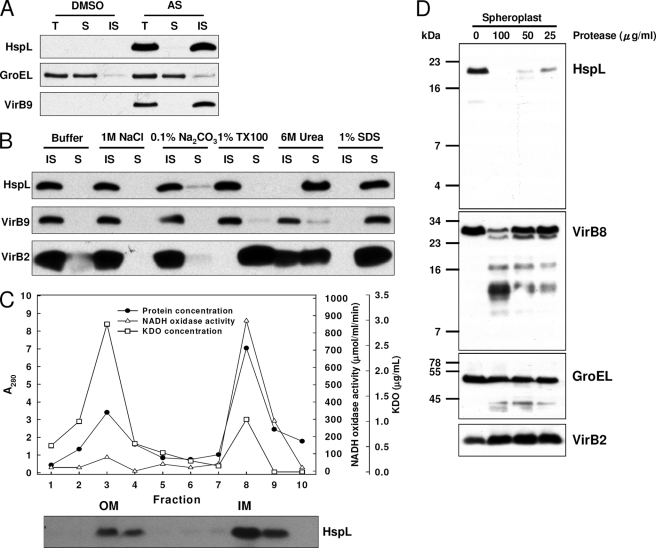
We showed the importance of HspL chaperone activity in promoting DNA transfer, and this raises the question of how HspL impacts T4SS functions. Our previous study revealed that HspL protein accumulation was triggered in response to certain VirB proteins, and optimal VirB protein accumulation required HspL (18). Thus, we hypothesize that HspL may function as a VirB chaperone to prevent VirB proteins from aggregation and/or degradation. To assess this possibility, we first examined the subcellular localization of HspL to determine whether it co-fractionates with VirB proteins in the membrane. Biochemical fractionations revealed that HspL was present in the insoluble membrane fractions, whereas little signal was detected in the soluble cytoplasmic and periplasmic fractions (Fig. 4A). HspL protein was tightly associated with membranes, because it was not solubilized by high salt concentrations (1 m NaCl or 0.1% Na2CO3) or mild nonionic detergent (1% Triton X-100). Only strong denaturant (6 m urea) or ionic detergent (1% SDS) succeeded in solubilizing the chaperone (Fig. 4B). Further analysis of the membrane fractions by sucrose gradient separation revealed that HspL localized primarily in the inner membrane but also associated with the outer membrane (Fig. 4C). The presence of HspL in both inner and outer membranes provoked us to determine whether HspL is exposed across the inner membrane. We converted agrobacterial cells to spheroplasts and performed protease sensitivity assays. We observed the complete digestion of HspL without traces of truncated products, and this suggested that membrane-associated HspL is exposed toward the periplasm (Fig. 4D). As controls, cytoplasmic GroEL and the integral inner membrane VirB2 proteins were analyzed and shown to be resistant to protease digestion, whereas the bitopic inner membrane VirB8 was amenable to proteolytic cleavage, resulting in truncated products.
FIGURE 4.
Biochemical fractionations of HspL. A, equal volumes of total proteins (T), soluble fraction (S), and insoluble membrane fraction (IS) of wild type strain NT1RE(pJK270) grown in I-medium at 25 °C for 16 h with the addition of DMSO or AS were subjected to Western blotting. The quality of fractionation was assessed by monitoring the distribution of GroEL (cytosolic marker) and VirB9 (membrane marker). B, the insoluble membrane fraction was treated in 50 mm Tris-HCl, pH 7.5 (buffer only), in the presence of salts or detergents as indicated. Soluble (S) and insoluble (IS) fractions were separated by ultracentrifugation and subjected to Western blotting. C, the insoluble membrane fractions separated by sucrose density gradient centrifugation at 120,000 × g at 4 °C for 18 h were collected from the top of the gradient and analyzed. The fractions were detected by Western blotting. The fractions containing the outer membranes (OM) and inner membranes (IM) were identified on the basis of the activity of the inner membrane specific enzyme NADH oxidase (open triangle) and the outer membrane specific lipopolysaccharide 2-keto-3-deoxyoctonate (KDO, open square) (57). D, spheroplasts were treated with protease at different concentrations (0, 25, 50, or 100 μg/ml) and subjected to Western blotting. The resistance of cytosolic GroEL to protease digestion indicates the quality of spheroplast preparation.
HspL Co-fractionates with VirB Complexes and Interacts with VirB8, a Key T4SS Assembly Factor
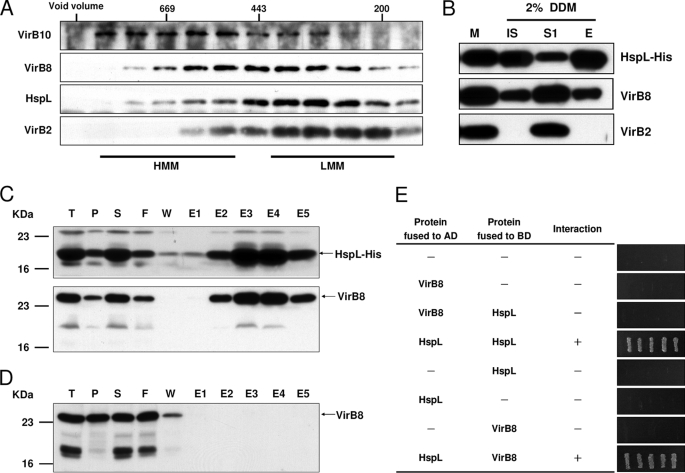
HspL is predicted to be a soluble protein without signal peptide or transmembrane domain (23), and therefore, its co-fractionation with membranes may be due to its association with membrane-associated VirB protein complexes. To assess this possibility, we extracted membrane proteins with the mild detergent DDM and then size-fractionated the DDM-extracted proteins by SEC (Fig. 5A). The eluted fractions were analyzed by Western blot with VirB10 as a marker of high molecular mass T4SS core components, VirB2 as a marker of low molecular mass pilus assembly complex components, and VirB8 as a marker present in both subassemblies (45, 46). HspL was detected in fractions across both T4SS subassemblies (Fig. 5A), raising the possibility that HspL may associate with certain VirB proteins such as VirB8 and VirB2 based on their largely overlapped eluted fractions. To test this possibility, we expressed HspL-His driven by its native promoter in the hspL deletion mutant under AS-inducing conditions, collected the DDM-extracted membrane proteins, and purified HspL-His and its interacting proteins via a Ni-NTA column. Western blot analysis showed that VirB8 co-eluted with HspL-His but not with VirB2 (Fig. 5B). This result suggested that HspL forms a protein complex with VirB8 either by a direct or by an indirect interaction via other proteins.
FIGURE 5.
HspL co-fractionated with VirB complexes and physically interacted with VirB8. A, membrane fractions of A. tumefaciens wild type strain NT1RE(pJK270) grown in I-medium in the presence of AS at 25 °C for 16 h were subjected for 2% DDM extraction followed by ultracentrifugation. The DDM-solubilized membrane proteins were further fractionated by SEC on Superdex 200 HiLoad 16/60, and the elution of HspL and of the T4SS components VirB10 (marker of high molecular mass core complex (HMM)), VirB2 (marker of low molecular mass pilus assembly complex (LMM)), and VirB8 (marker present in both subassemblies) was analyzed by SDS-PAGE and Western blotting with specific antisera as indicated. B, the membrane fraction of AS-induced A. tumefaciens ΔhspL(pHspL-His) strain was extracted with 2% DDM followed by ultracentrifugation to separate DDM-extracted soluble proteins (S1) from insoluble pellets (IS). The soluble fraction was incubated with Ni-NTA and eluted by elution buffer (E) and then analyzed by Western blotting detected with specific antisera as indicated. E. coli BL21(DE3) strain containing both pETHspL and pTrcB8 (C) or pTrcB8 only (D) was induced by 0.4 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 2 h. The soluble protein extracts were run through Ni-NTA His Bind Resins to purify HspL-His and its interacting proteins. The total protein (T), insoluble pellet (P), soluble supernatant (S), flow-through (F), wash fraction (W), and eluted fractions (E1∼E5) were analyzed by Western blot with HspL and VirB8 antisera. E, yeast two-hybrid protein-protein interaction results using combinations of various full-length HspL and VirB8 are indicated. For each transformant, five independent colonies were streaked out on synthetic dextrose medium in the absence of Ade, His, Leu, and Trp grown at 30 °C for 3 days to select the transformant expressing the interacting proteins. AD, activation domain; BD, binding domain.
To confirm the HspL-VirB8 interaction, we performed co-purification experiments after co-expressing VirB8 and HspL-His in E. coli. VirB8 co-eluted with HspL-His from a Ni-NTA column, and its amount correlated with that of purified HspL-His in each elution fraction (Fig. 5C). In contrast, VirB8 alone was not retained by the nickel column in the absence of HspL (Fig. 5D), suggesting that VirB8 co-purified with HspL-His via a direct physical interaction. The notion that HspL and VirB8 interact was further supported by yeast two-hybrid analysis. No interaction was detected when each of the fusion proteins was expressed alone with the paired empty vector (data not shown). In agreement with the biochemical data, we detected self-interaction of HspL and interaction between HspL fused to the GAL4 activation domain and VirB8 fused to the DNA binding domain (Fig. 5E). These protein-protein interaction studies in A. tumefaciens and in the heterologous E. coli and yeast backgrounds demonstrated that HspL physically interacts with VirB8.
HspL Is a VirB8 Chaperone in Protecting GST-VirB8 from Aggregation in Vitro
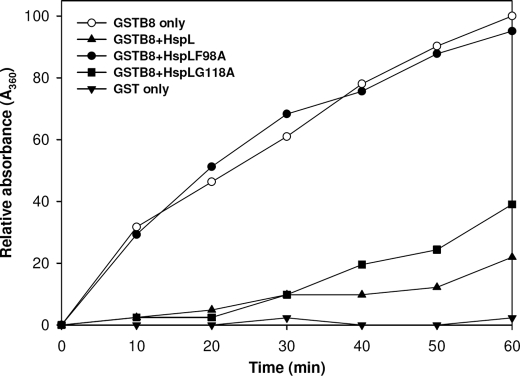
VirB8 is a bitopic inner membrane protein with a short N terminus facing the cytoplasm and a large C-terminal domain localized in the periplasmic space (26). Its association with both T4SS core and pilus subassemblies and direct interaction with several VirB proteins localized in both inner and outer membranes provided strong evidence that VirB8 plays a key role in T4SS assembly (26). Based on the results presented above, it is possible that HspL impacts T4SS function via functioning as VirB8 chaperone. To test this hypothesis, we expressed full-length VirB8 fused with GST at its N terminus and purified the soluble GST-VirB8 fusion protein (supplemental Fig. S4) for chaperone activity assay. Soluble GST-VirB8 but not GST alone gradually aggregated during heat treatment (Fig. 6). When the experiment was conducted in the presence of HspL, GST-VirB8 was protected from thermal aggregation (Fig. 6). As in the CS assay (Fig. 2A), HspLF98A did not protect GST-VirB8 from aggregation, and HspLG118A possessed reduced chaperone activity (Fig. 6). The chaperone activity of HspL for both the model substrate CS and VirB8 fused to GST suggests that HspL is a chaperone and that VirB8 is a native substrate in the T4SS complex.
FIGURE 6.
Chaperone activity assays of HspL and its variants (HspLF98A and HspLG118A) using GST-VirB8 as substrate. Thermal aggregation protection assays using GST-VirB8 (600 nm) was carried out at 50 °C in the absence (○) or presence of HspL-His6 (▴), HspLF98A-His6 (●), and HspLG118A-His6 (■) at concentrations of 1.2 μm, respectively. GST (▾) alone incubated under the same conditions was used as a negative control. Aggregation was monitored in absorbance at 360 nm and presented as a function of time.
DISCUSSION
The chaperone activity of α-Hsps is usually analyzed by testing their ability to prevent aggregation of thermally denatured artificial model substrates in vitro. In most cases it remains an open question of whether the in vitro chaperone activity reflects the respective biological function. In this study we provided strong evidence that the virulence-induced A. tumefaciens small heat-shock protein HspL is a VirB8 chaperone. The correlation between reduced chaperone activity of two HspL variants and the effects on T4SS-mediated DNA transfer and virulence of A. tumefaciens provided a direct link between in vitro and in vivo functions. Our study also revealed the first example of a small heat-shock protein as a molecular chaperone involved in T4SS functions and bacterial virulence.
Similar to many α-Hsps including HspH of Bradyrhizobium japonicum (41), HspL oligomerizes in a concentration-dependent manner ranging from appropriately 12 to 18 subunits in vitro (Fig. 1B). Two available α-Hsp crystal structures, MjHsp16.5 from the archaeon Methanococcus jannaschii (47) and TaHsp16.9 from wheat (48), both revealed a hollow, football-like 24-mer structure of native α-Hsp. Thus, it is possible that HspL may form a spherical structure yet to be determined.
Two HspL protein variants with amino acid substitutions at specific amino acids in the conserved α-crystallin domain (HspLF98A and HspLG118A) had reduced chaperone activity (Figs. 2A and 6), and the oligomeric states were altered (Fig. 2B). We noticed that all SEC analysis resulted in two major peaks, the first corresponding to the void volume, and both peaks contained HspL proteins (Fig. 2B and supplemental Fig. S1), but those of HspL and HspLG118A in the void volume represent only small amounts and do not likely represent a significant fraction. Although the majority of wild type HspL forms the 264-kDa oligomer, HspLF98A eluted as a smaller 130-kDa oligomer and in the void volume (Fig. 2B). HspL and its variants were purified as soluble proteins, and the majority remained soluble after heat treatment (supplemental Fig. S3). Therefore, the significant amounts of HspLF98A eluted in the void volume, and earlier fractions likely represent a soluble complex larger than 669 kDa; the largest protein marker could be resolved by Superdex 200 (GE Healthcare).
The absence of chaperone activity of the HspLF98A is consistent with respective HspH F94A (or F94D) protein of B. japonicum (41) and Hsp16.6 F102A protein of Synechocystis (9). Although HspH G114A in B. japonicum caused a complete loss of chaperone activity, the HspLG118A still retained partial chaperone activity, similar with that observed in the respective Hsp16.6 G124A protein in Synechocystis (9). The importance of G118 for proper oligomerization is consistent with critical roles of equivalent Synechocystis Hsp16.6 G124 and B. japonicum HspH G114 in maintaining oligomers (9, 41). The HspL F98A protein formed both larger and smaller oligomers (Fig. 2B) but did not render the protein monomeric (∼19-kDa) like its counterparts, HspH F94A or F84D in B. japonicum (41), or form dimers (∼38 Da), like the equivalent Hsp16.6 F102A in Synechocystis (9). The somewhat distinct functions of conserved amino acids in different α-Hsps in dimerization or oligomerization imply that different α-Hsps may not fold into oligomers in the same manner. Nevertheless, our results clearly indicate that the amino acid residues Phe-98 and Gly-118 of HspL are critical for maintaining proper oligomerization (Fig. 2B).
The lost or reduced chaperone activity observed in case of the two HspL variants (F98A and G118) (Figs. 2A and 6) correlated with the loss or reduction of their ability in promoting T4SS-mediated RSF1010 transfer between agrobacteria and tumorigenesis (Fig. 3). The link between in vitro and in vivo functions has also been observed for the equivalent Hsp16.6 F102A and G124A that resulted in loss and severe reduction in thermotolerance of Synechocystis, respectively (9). Basha et al. (49) showed that Hsp16.6 interacts with several heat-labile proteins during heat stress in vivo, and these substrates could be released by ATP-dependent chaperones in vitro. Importantly, this work also demonstrated that Hsp16.6 interacts with serine esterase, one of the identified substrates, and protects it from aggregation in a heat-dependent manner in vitro. Consistent with its induction by the virulence gene inducer AS (and not heat), we here demonstrate that HspL interacts with its substrate VirB8 in a large complex in A. tumefaciens (Fig. 5, A and B). Moreover, we provided direct evidence that HspL is a VirB8 chaperone in protecting it from aggregation in vitro (Fig. 6). This argument is further supported by the observation that more soluble VirB8 was detected when VirB8 was co-expressed with HspL-His than when it was expressed alone in E. coli (Fig. 5, C and D), suggesting that HspL protects VirB8 from aggregation in vivo.
The identification of HspL tightly associated with the membrane and exposed to the periplasm (Fig. 4) suggested that HspL may interact with membrane lipids and/or with membrane proteins such as VirB8. Despite the lack of any obvious transmembrane domain or Sec-dependent signal peptide, α-Hsp associations with membranes have been well documented (2). E. coli IbpA/B localized in the outer membrane and functioned in protecting cells from heat and oxidative stress (51–53). M. tuberculosis Hsp16.3, a major immunodominant protein, was found to be a major membrane protein (54). In addition, Synechocystis Hsp17 binds to specific membrane lipids and is important for thermostability of thylakoid membranes (55, 56). It would be interesting to understand how different α-Hsps are translocated into and/or across the bacterial inner membrane.
VirB8 is a key T4SS assembly factor interacting with many VirB proteins (25, 26, 50). Our results suggest that HspL may function as a specific VirB8 chaperone to maintain VirB8 in a folding-competent state. This may be critical for the stability of VirB8, and as a consequence, it also affects the stability of its interacting proteins (18). Nevertheless, HspL may also interact directly with other T4SS components, and this possibility will be tested in the future. To dissect the contribution of HspL, it would be of particular interest to identify HspL variants that uncouple their chaperone activity and interaction with VirB8, and this may shed light on the contribution of HspL to T4SS function and Agrobacterium virulence.
Supplementary Material
Acknowledgments
We are grateful to Clarence Kado for providing antibodies of VirB2 and VirB9 and Ban-Yang Chang for GroEL antibody. We also thank Chan Gao and Sonja Klüsener for assistance at the initial stage of this work and the Lai laboratory members for discussion and reading the manuscript.
This work was supported in part by Taiwan National Science Council (NSC) Grant NSC 95-2320-B-001-009 (to E.-M. L.), an NSC postdoctoral fellowship (to Y.-L. T.), and a joint grant from the German Academic Exchange Service (DAAD) and Taiwan National Science Council PPP Grant 0970029248P (to F. N. and E.-M. L.). This work was also supported by Canadian Institutes of Health Research Grant MOP-84239, the Canada Foundation for Innovation, and the Fonds de la Recherche en Santé du Québec.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1–S4.
- α-Hsp
- α-crystallin-type small heat-shock protein
- CS
- citrate synthase
- AS
- acetosyringone
- α-GST
- glutathione S-transferase
- AS
- acetosyringone
- T4SS
- type IV secretion system
- SEC
- size exclusion chromatography
- DDM
- dodecyl-β-d-maltopyranoside
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1.Narberhaus F. (2002) Microbiol. Mol. Biol. Rev. 66, 64–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamoto H., Vígh L. (2007) Cell. Mol. Life Sci. 64, 294–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Litt M., Kramer P., LaMorticella D. M., Murphey W., Lovrien E. W., Weleber R. G. (1998) Hum. Mol. Genet. 7, 471–474 [DOI] [PubMed] [Google Scholar]
- 4.Mackay D. S., Andley U. P., Shiels A. (2003) Eur. J. Hum. Genet 11, 784–793 [DOI] [PubMed] [Google Scholar]
- 5.Graw J. (2009) Exp. Eye Res. 88, 173–189 [DOI] [PubMed] [Google Scholar]
- 6.Biswas A., Miller A., Oya-Ito T., Santhoshkumar P., Bhat M., Nagaraj R. H. (2006) Biochemistry 45, 4569–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andley U. P., Hamilton P. D., Ravi N. (2008) Biochemistry 47, 9697–9706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xi J. H., Bai F., Gross J., Townsend R. R., Menko A. S., Andley U. P. (2008) J. Biol. Chem. 283, 5801–5814 [DOI] [PubMed] [Google Scholar]
- 9.Giese K. C., Basha E., Catague B. Y., Vierling E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18896–18901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giese K. C., Vierling E. (2002) J. Biol. Chem. 277, 46310–46318 [DOI] [PubMed] [Google Scholar]
- 11.Giese K. C., Vierling E. (2004) J. Biol. Chem. 279, 32674–32683 [DOI] [PubMed] [Google Scholar]
- 12.Basha E., Friedrich K. L., Vierling E. (2006) J. Biol. Chem. 281, 39943–39952 [DOI] [PubMed] [Google Scholar]
- 13.Jaya N., Garcia V., Vierling E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15604–15609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart G. R., Newton S. M., Wilkinson K. A., Humphreys I. R., Murphy H. N., Robertson B. D., Wilkinson R. J., Young D. B. (2005) Mol. Microbiol. 55, 1127–1137 [DOI] [PubMed] [Google Scholar]
- 15.Stewart J. N., Rivera H. N., Karls R., Quinn F. D., Roman J., Rivera-Marrero C. A. (2006) Microbiology 152, 233–244 [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson K. A., Stewart G. R., Newton S. M., Vordermeier H. M., Wain J. R., Murphy H. N., Horner K., Young D. B., Wilkinson R. J. (2005) J. Immunol. 174, 4237–4243 [DOI] [PubMed] [Google Scholar]
- 17.Dellagostin O. A., Esposito G., Eales L. J., Dale J. W., McFadden J. (1995) Microbiology 141, 1785–1792 [DOI] [PubMed] [Google Scholar]
- 18.Tsai Y. L., Wang M. H., Gao C., Klüsener S., Baron C., Narberhaus F., Lai E. M. (2009) Microbiology 155, 3270–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Münchbach M., Dainese P., Staudenmann W., Narberhaus F., James P. (1999) Eur. J. Biochem. 264, 39–48 [DOI] [PubMed] [Google Scholar]
- 20.Münchbach M., Nocker A., Narberhaus F. (1999) J. Bacteriol. 181, 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosen R., Büttner K., Becher D., Nakahigashi K., Yura T., Hecker M., Ron E. Z. (2002) J. Bacteriol. 184, 1772–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balsiger S., Ragaz C., Baron C., Narberhaus F. (2004) J. Bacteriol. 186, 6824–6829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai E. M., Shih H. W., Wen S. R., Cheng M. W., Hwang H. H., Chiu S. H. (2006) Proteomics 6, 4130–4136 [DOI] [PubMed] [Google Scholar]
- 24.Lai E. M., Kado C. I. (2000) Trends Microbiol. 8, 361–369 [DOI] [PubMed] [Google Scholar]
- 25.Christie P. J., Atmakuri K., Krishnamoorthy V., Jakubowski S., Cascales E. (2005) Annu. Rev. Microbiol. 59, 451–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baron C. (2006) Biochem. Cell Biol. 84, 890–899 [DOI] [PubMed] [Google Scholar]
- 27.Fronzes R., Christie P. J., Waksman G. (2009) Nat. Rev. Microbiol. 7, 703–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alvarez-Martinez C. E., Christie P. J. (2009) Microbiol. Mol. Biol. Rev. 73, 775–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey S., Ward D., Middleton R., Grossmann J. G., Zambryski P. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2582–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paschos A., Patey G., Sivanesan D., Gao C., Bayliss R., Waksman G., O'callaghan D., Baron C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7252–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cascales E., Christie P. J. (2004) Science 304, 1170–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchanan-Wollaston V., Passiatore J. E., Cannon F. (1987) Nature 328, 172–175 [Google Scholar]
- 33.Beijersbergen A., Dulk-Ras A. D., Schilperoort R. A., Hooykaas P. J. (1992) Science 256, 1324–1327 [DOI] [PubMed] [Google Scholar]
- 34.Kado C. I., Heskett M. G. (1970) Phytopathology 60, 969–976 [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J., Russell D. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 36.Lai E. M., Kado C. I. (1998) J. Bacteriol. 180, 2711–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schägger H., von Jagow G. (1987) Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 38.Baron C., Domke N., Beinhofer M., Hapfelmeier S. (2001) J. Bacteriol. 183, 6852–6861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shirasu K., Kado C. I. (1993) FEMS Microbiol. Lett. 111, 287–294 [DOI] [PubMed] [Google Scholar]
- 40.Chang B. Y., Chen K. Y., Wen Y. D., Liao C. T. (1994) J. Bacteriol. 176, 3102–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lentze N., Studer S., Narberhaus F. (2003) J. Mol. Biol. 328, 927–937 [DOI] [PubMed] [Google Scholar]
- 42.Ma L. S., Lin J. S., Lai E. M. (2009) J. Bacteriol. 191, 4316–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rashkova S., Spudich G. M., Christie P. J. (1997) J. Bacteriol. 179, 583–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cascales E., Christie P. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17228–17233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan Q., Carle A., Gao C., Sivanesan D., Aly K. A., Höppner C., Krall L., Domke N., Baron C. (2005) J. Biol. Chem. 280, 26349–26359 [DOI] [PubMed] [Google Scholar]
- 46.Krall L., Wiedemann U., Unsin G., Weiss S., Domke N., Baron C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11405–11410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim K. K., Kim R., Kim S. H. (1998) Nature 394, 595–599 [DOI] [PubMed] [Google Scholar]
- 48.van Montfort R. L., Basha E., Friedrich K. L., Slingsby C., Vierling E. (2001) Nat. Struct. Biol. 8, 1025–1030 [DOI] [PubMed] [Google Scholar]
- 49.Basha E., Lee G. J., Breci L. A., Hausrath A. C., Buan N. R., Giese K. C., Vierling E. (2004) J. Biol. Chem. 279, 7566–7575 [DOI] [PubMed] [Google Scholar]
- 50.Ward D. V., Draper O., Zupan J. R., Zambryski P. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11493–11500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitagawa M., Miyakawa M., Matsumura Y., Tsuchido T. (2002) Eur. J. Biochem. 269, 2907–2917 [DOI] [PubMed] [Google Scholar]
- 52.Kitagawa M., Matsumura Y., Tsuchido T. (2000) FEMS Microbiol. Lett. 184, 165–171 [DOI] [PubMed] [Google Scholar]
- 53.Laskowska E., Bohdanowicz J., Kuczyńska-Wiśnik D., Matuszewska E., Kedzierska S., Taylor A. (2004) Microbiology 150, 247–259 [DOI] [PubMed] [Google Scholar]
- 54.Lee B. Y., Hefta S. A., Brennan P. J. (1992) Infect. Immun. 60, 2066–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horváth I., Glatz A., Varvasovszki V., Török Z., Páli T., Balogh G., Kovács E., Nádasdi L., Benkö S., Joó F., Vígh L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3513–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakamoto H., Suzuki N., Roy S. K. (2000) FEBS Lett. 483, 169–174 [DOI] [PubMed] [Google Scholar]
- 57.de Maagd R. A., Lugtenberg B. (1986) J. Bacteriol. 167, 1083–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.