Abstract
S-Adenosylmethionine (AdoMet) is an important methyl group donor that plays a central role in many essential biochemical processes. The parasite Leishmania can both synthesize and transport AdoMet. Leishmania cells resistant to the antifolate methotrexate due to a rearrangement in folate biopterin transporter (FBT) genes were cross-resistant to sinefungin, an AdoMet analogue. FBT gene rearrangements were also observed in Leishmania major cells selected for sinefungin resistance. One of the rearranged FBT genes corresponded to the main AdoMet transporter (AdoMetT1) of Leishmania as determined by gene transfection and gene inactivation experiments. AdoMetT1 was determined to be a high affinity plasma membrane transporter expressed constitutively throughout the growth phases of the parasite. Leishmania cells selected for resistance or naturally insensitive to sinefungin had lower expression of AdoMetT1. A new function in one carbon metabolism, also a pathway of interest for chemotherapeutic interventions, is described for a novel class of membrane proteins found in diverse organisms.
Keywords: Drug Resistance, Drug Transport, Folate, Membrane Proteins, S-Adenosylmethionine, Transport Drugs
Introduction
The parasite Leishmania is distributed globally and causes a variety of clinical symptoms ranging from self-healing cutaneous lesions to visceral infections that are usually fatal if left untreated (1, 2). Current first-line chemotherapy against leishmaniasis relies on a rather limited arsenal of drugs, including pentavalent antimonials, liposomal amphotericin B, or miltefosine (2). These drugs are associated with side effects, high costs, and drug resistance, and therefore, the search for new drugs and targets to control this parasite is warranted (3, 4).
S-Adenosylmethionine (AdoMet or SAM)5 is an important biological sulfonium compound recognized as the universal methyl donor for methylation of lipids, proteins, nucleic acids, and xenobiotics in living cells (5). AdoMet is also a donor of propylamine groups for the synthesis of polyamines and participates in the reverse trans-sulfuration pathway where AdoMet is converted into cysteine and glutathione and in Leishmania in the spermidine-glutathione conjugate trypanothione (6, 7). AdoMet is also used as a source of methylene groups, amino groups, and ribosyl groups in the synthesis of fatty acids, biotin, and the modified nucleoside epoxyqueusine of tRNAs (reviewed in Ref. 8).
Most cells are capable of synthesizing AdoMet from l-methionine and ATP, in a process requiring the enzyme methionine adenosyltransferase (MAT). The gene coding for this enzyme is present in most sequenced organisms (reviewed in Ref. 9). However, in some Rickettsia strains, the MAT gene is inactivated by a mutation (10), and in the fungus Pneumocystis carinii, no MAT activity has been detected (11). These organisms have the rare distinction of meeting their AdoMet needs by importing it (12). The Rickettsia transporter is part of the drug/metabolite transporter superfamily (10), and the identity of the Pneumocystis transporter is not known. Transport of AdoMet across the plasma membrane does not occur to a significant extent in mammalian cells, but transport of AdoMet is essential in several organelles. For example, the yeast (13) and human (14) mitochondrial AdoMet transporters and the Arabidopsis AdoMet plastid (and mitochondria) transporter (15, 16) have been functionally characterized. They belong to the mitochondrial carrier protein family (MCF), a class of protein mediating the transport of various substrates (reviewed in Ref. 17).
Other organisms, notably fungus and protozoa, have the ability to both synthesize and transport AdoMet. The only eukaryotic plasma membrane AdoMet transporter characterized to date is the Saccharomyces cerevisiae SAM3 protein (18) belonging to the amino acid permease superfamily. Although SAM3 transports AdoMet, it also transports polyamines with high affinity (19). Sinefungin (SNF) is an analogue of AdoMet with antimicrobial activity, and yeast SNF-resistant mutants were shown to have loss-of function mutations in SAM3 (20). In the protozoan parasites Leishmania (21) and Trypanosoma brucei (22), SNF and AdoMet were shown, on the basis of substrate competition transport experiments, to share a common transporter protein. We report here the cloning and functional characterization of the unique high affinity AdoMet transporter of Leishmania and show that it belongs to the FBT family, a class of proteins present in diverse organisms.
MATERIALS AND METHODS
Cell Lines and Culture Conditions
Leishmania cells (Leishmania infantum MHOM/MA/67/ITMAP-263, L. infantum JPCM5, Leishmania tarentolae, Leishmania major Friedlin, and L. major LV39) were grown in SDM-79 medium supplemented with 10% heat-inactivated fetal bovine serum, 5 μg/ml hemin, and, when required, 10 μm biopterin (Sigma). The L. tarentolae methotrexate (MTX)-resistant mutant TarMTX1000.6 was described previously (23). The L. tarentolae and L. major LV39 SNF-resistant mutants, named TarSNF.8 and LV39SNF4000.4, respectively, were selected with SNF in a step-by-step manner as described in details for other drugs (24). Cell growth was monitored by measuring the absorbance of culture aliquots at 600 nm in a multiwell scanning spectrophotometer. DNA transfection into Leishmania promastigotes has been performed as described previously (25).
AdoMetT1 Gene Transfection and Inactivation
The AdoMetT1 gene of L. infantum (LinJ10_V3.0370) was amplified from genomic DNA using primer pairs 1–2 and 3–4 (see supplemental Table S1). The PCR fragment was first ligated into the pGEM T-easy vector (Invitrogen), digested with the appropriate restriction enzymes, and cloned into the relevant restriction sites within the Leishmania expression vectors pSP72αHYGα (26) and pSPαNEOα-GFP (27).
The L. infantum AdoMetT1 null mutant was obtained by targeted gene replacement. The AdoMetT1 inactivation cassettes were generated using a PCR fusion-based strategy as described previously (28). Primers used to generate the inactivation cassettes are listed in supplemental Table S1. Briefly, the NEO inactivation cassette was generated by using the primer pairs 5–6 and 7–8 to amplify DNA fragments of 730 bp upstream and 680 bp downstream of the AdoMetT1 gene, respectively. The neomycin phosphotransferase (NEO) gene was amplified from the plasmid pSPαNEOα using primers 9 and 10 and then fused to the upstream and downstream DNA fragments of AdoMetT1 using PCR. The same strategy was used to generate the ZEO cassette; primer pairs 5–11 and 12–8 were used to amplify the upstream and downstream fragments of AdoMetT1, whereas primers 13 and 14 were used to amplify the ZEO gene from the pSPαZEOα vector (29).
Cellular Transport Assays
TarMTX1000.6 expressing the AdoMet transporter (AdoMetT1) and the folic acid transporter (FT1) were harvested during the mid-logarithmic phase. 108 cells were washed and resuspended in folate-deficient medium fdDMEL or AdoMet-deficient medium with 115 nm [3H]folic acid (43.2 Ci/mmol) or 60 nm [3H]AdoMet (83 Ci/mmol) (Moravek Biochemicals), respectively. Accumulation was measured as described previously (30). Folic acid uptake was normalized to cell number, and the background transport value was removed by subtracting the accumulation value obtained on ice. The transport kinetic parameters Vmax and apparent Km values for AdoMet were measured while using different [3H]AdoMet concentrations (3–3000 nm) during the linear phase of accumulation (5 min). Transport kinetic parameters were determined by linear regression analysis and Michaelis-Menten analysis.
The transport competition study was performed with 5 × 107 LV39SNF4000.4 cells expressing the AdoMetT1-GFP protein. Briefly, cells were incubated for 10 min in an assay buffer, as described previously (31), containing 50 nm radioactive AdoMet in the presence or absence of various concentrations (100 nm and 1 and 10 μm) of competing molecules. For all transport experiments, the accumulation in cells incubated on ice was subtracted.
Quantitative Real Time Reverse Transcription-PCR
Three independent RNA preparations were used for each real time PCR experiment. RNA extractions, cDNA synthesis, and TaqMan quantification of AdoMetT1 were performed as described previously (27). Primers and Taqman probes are listed in supplemental Table S2. Real time reverse transcription-PCR for the MAT gene was performed as described previously (32) using GAPDH and actin genes as controls (supplemental Table S2).
Western Blot Analysis
Total Leishmania proteins (30 μg) were run on 12% polyacrylamide gels and transferred onto nitrocellulose membranes as described previously (33). The blots were blocked overnight in 5% skimmed milk in 1× phosphate-buffered saline (PBS). A monoclonal anti-α-tubulin antibody (Sigma) directed against a conserved amino-terminal peptide of the bovine α-tubulin or an antibody against the green fluorescent protein (GFP) (Invitrogen) was diluted 1:3000 in PBS containing 0.1% Tween 20 (PBS/Tween) and incubated for 1 h with the membranes. The blots were washed three times for 5 min in PBS/Tween and incubated with horseradish peroxidase-conjugated sheep anti-rabbit IgG for GFP or sheep anti-mouse IgG for α-tubulin (Amersham Biosciences) diluted 1:10,000 in PBS/Tween. The blots were washed as above, incubated with ECL Plus chemiluminescent substrate (Amersham Biosciences), and exposed to x-ray films. The polyclonal rabbit MAT antibody was kindly provided by Prof. Balana-Fouce (University of Leon, Spain) and was used as described previously (34).
Southern Analysis
Leishmania total DNAs were isolated using DNAzol reagent as recommended by the manufacturer (Invitrogen). For Southern blots, genomic DNAs were digested with PvuI to monitor rearrangement events of the FBT family and with PstI for analysis of the AdoMetT1-null mutant. Southern blots, hybridizations, and washes were performed following standard protocols (35), and all probes were obtained by PCR. The FBT gene family was studied with a DNA probe spanning the conserved coding sequences of the FBT genes (27), and AdoMetT1 allele replacement was confirmed by using a probe targeting the 3′-UTR of AdoMetT1.
Fluorescence Microscopy
Live parasites were mounted under poly-l-lysine-coated coverslips. Coverslips were sealed with nail varnish and air-dried for 15 min. Bright field and fluorescence images were taken using a Nikon Eclipse TE300 inverted microscope with a Photometrics coolSNAPfx camera. Visualization of the GFP fluorophore was achieved using a 460/500-nm excitation filter and 510/560-nm emission filter with a 100× objective. The images were processed using the Image-Pro Plus software (version 5.0).
RESULTS
Leishmania Methotrexate-resistant Mutant Is Cross-resistant to Sinefungin
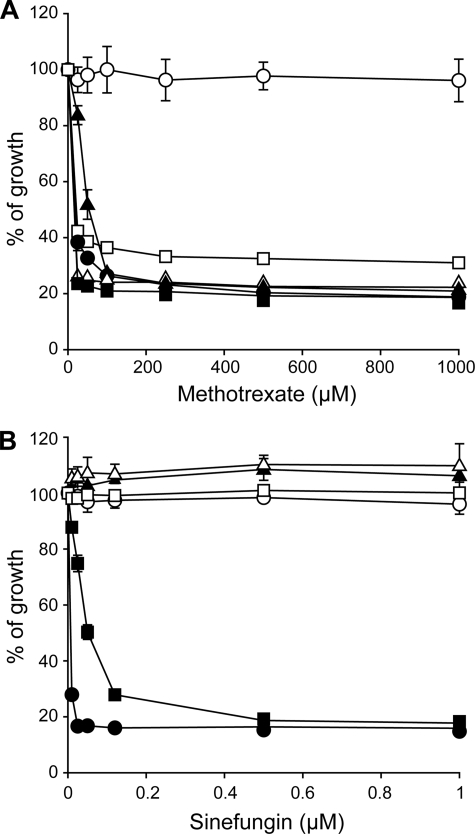
Folic acid (FA) and AdoMet are the one-carbon metabolic donors in cells, and previous reports have demonstrated that AdoMet metabolism is potentially changed in Leishmania cells resistant to the FA analogue MTX (29, 36). Indeed, the L. tarentolae MTX1000.6 mutant is resistant to MTX (Fig. 1A) due to decreased accumulation of MTX or FA (see Fig. 2A and data not shown). This reduced accumulation is due to gene deletion and rearrangements of the FBT transporter FT1 (37). Surprisingly, this mutant was also found to be cross-resistant to the AdoMet analogue SNF (Fig. 1B). This intriguing observation led us to further investigate SNF susceptibility in Leishmania cells. L. tarentolae and L. major wild-type cells were sensitive to MTX (Fig. 1A). However, although L. tarentolae and L. major LV39 were sensitive to SNF, L. major Friedlin was intrinsically resistant to it (Fig. 1B and Table 1). Similarly, whereas the L. infantum 263 strain was highly sensitive to SNF, the genome strain L. infantum JPCM5 was intrinsically resistant (Table 1). L. tarentolae and L. major LV39 cells were selected in a step-by step fashion for resistance to SNF, and these cells were indeed resistant to SNF (Fig. 1B) but not cross-resistant to MTX (Fig. 1A).
FIGURE 1.
Sensitivity of Leishmania cells to methotrexate and sinefungin. Leishmania cells were incubated in SDM medium with varying concentrations of methotrexate (A) and sinefungin (B), and their growth was monitored at 72 h by measuring the absorbance at 600 nm. ●, L. tarentolae wild type; ○, L. tarentolae MTX1000.6; ▴, TarSNF.8; ▵, L. major Friedlin; ■, L. major LV39; □, LV39SNF4000.4. The average of triplicate measurements is shown.
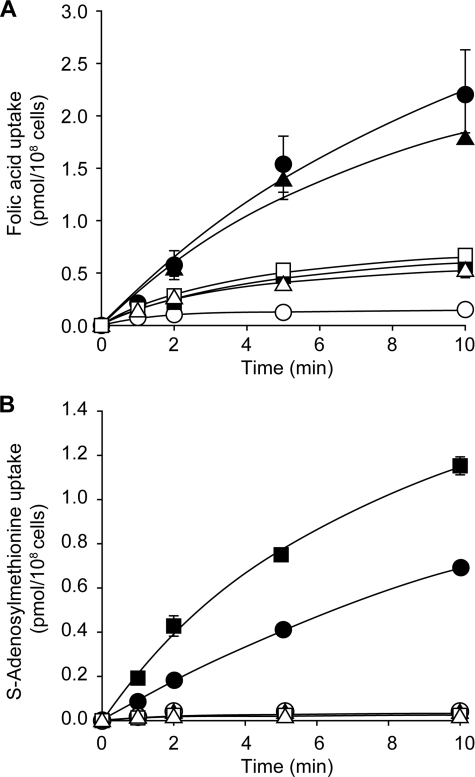
FIGURE 2.
Folic acid and S-adenosylmethionine accumulation in Leishmania cells. The accumulation of 115 nm [3H]folic acid (A) and 60 nm S-[3H]adenosylmethionine (B) was measured in Leishmania cells. ●, L. tarentolae wild type; ○, L. tarentolae MTX1000.6; ▴, TarSNF.8; ▵, L. major Friedlin; ■, L. major LV39; □, LV39SNF4000.4. The average of triplicate measurements is shown.
TABLE 1.
Susceptibility to sinefungin and kinetic properties of S-adenosylmethionine transport in Leishmania spp.
The IC50 values are 50% inhibitory concentrations of sinefungin. The means ± S.D. of at least three independent experiments are given.
| Strains | IC50 value of sinefungin | Km | Vmax |
|---|---|---|---|
| nm | nm | pmol/108 cells/min | |
| L. infantum MHOM/MA/67/ITMAP-263 | 80 ± 18 | 167 ± 27 | 0.56 ± 0.17 |
| L. infantum JPCM5 | 2000 ± 300 | 180 ± 82 | 0.05 ± 0.01 |
| L. major LV39 | 50 ± 4 | 252 ± 10 | 0.57 ± 0.13 |
| L. major Friedlin | >6000 | 320 ± 7 | 0.03 ± 0.01 |
| LV39SNF4000.4 + AdoMetT1-GFP | 10 ± 2 | 284 ± 72 | 0.58 ± 0.24 |
Resistance to MTX in L. tarentolae MTX1000.6 is due to a reduced uptake of the drug (23), which is paralleled by a reduction in the uptake of the analogue FA (Fig. 2A). Accordingly, we tested whether cross-resistance to SNF in L. tarentolae MTX1000.6 correlated with a reduced accumulation of AdoMet and whether cells intrinsically resistant (L. major Friedlin or L. infantum JCPM5) or made resistant to SNF also exhibited a reduced accumulation of AdoMet. Wild-type L. tarentolae and L. major LV39 accumulated AdoMet, but all strains not susceptible to SNF, regardless of whether resistance was intrinsic or acquired, did not (Fig. 2B).
AdoMet Transporter Is a Member of the FBT Family
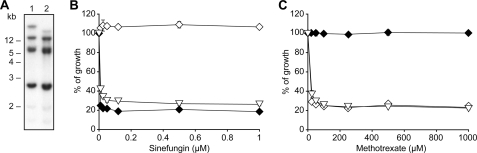
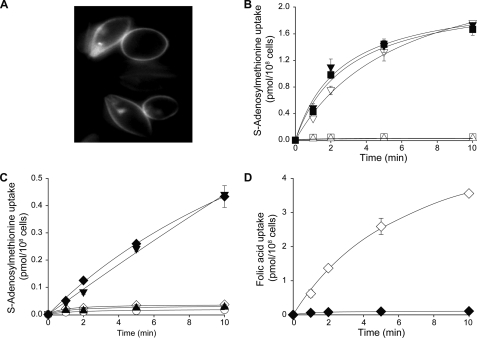
The L. tarentolae MTX1000.6 mutant was resistant to both MTX and SNF (Fig. 1), a phenotype accompanied by a reduced accumulation of FA and AdoMet (Fig. 2). Because there are rearrangements of the FBT gene family in this mutant (30), we hypothesized that the lack of AdoMet transport in this mutant could be correlated with FBT gene rearrangements. This was also investigated in L. tarentolae and L. major cells made resistant to SNF where the genomic DNAs of the mutants were digested with PvuI and hybridized to a FBT probe. There were no clear rearrangements in the L. tarentolae-resistant mutant SNF8 (results not shown). However, there was a clear rearrangement of some of the FBT family members in the L. major-resistant LV39SNF4000.4 mutant compared with its parental wild-type SNF-sensitive isolate (Fig. 3A). According to the published genome sequence of L. major, the high molecular weight (>12 kb) rearranged band most likely encodes three FBT genes (LmjF10.0350, LmjF10.0360, and LmjF10.0370). The corresponding L. infantum orthologues LinJ10_V3.0.0360, LinJ10_V3.0.0370, and LinJ10_V3.0.0380 were cloned into expression vectors and transfected in the SNF-resistant cells L. tarentolae MTX1000.6 and LV39SNF4000.4. Cells transfected with LinJ10_V3.0370 but not the other two constructs became highly sensitive to SNF (Fig. 3B and results not shown). This transporter clearly did not correspond to FT1 (LinJ10_V3.0400). Indeed, L. tarentolae MTX1000.6 transfected with FT1 was still highly resistant to SNF (Fig. 3B) but became highly sensitive to MTX (Fig. 3C). L. tarentolae MTX1000.6 transfected with LinJ10_V3.0370 remained resistant to MTX (Fig. 3C). Because LinJ10_V3.0370 sensitized cells to SNF, we tested whether it could correspond to a plasma membrane AdoMet transporter. Plasma membrane localization was examined by fusing the FBT open reading frame containing the gene of interest to GFP and analyzing the resultant transfectants by fluorescence microscopy (Fig. 4A). The LV39SNF4000.4 mutant and L. major Friedlin did not transport AdoMet (Figs. 2B and 4B). However, when these cells were transfected with LinJ10_V3.0370, we observed an accumulation of AdoMet (Fig. 4B). These results suggested that LinJ10_V3.0370 may catalyze the transport of AdoMet, and it was thus renamed AdoMetT1. L. tarentolae MTX1000.6 or SNF8 cells did not accumulate AdoMet, but the same cells transfected with AdoMetT1 were capable of transporting AdoMet (Fig. 4C). However, L. tarentolae MTX1000.6 cells transfected with AdoMetT1 did not transport FA, whereas the same cells transfected with FT1 did (Fig. 4D).
FIGURE 3.
Identification of the S-adenosylmethionine transporter (AdoMetT1) of Leishmania. A, Southern blot analysis of L. major LV39 and LV39SNF4000.4. Total DNAs of Leishmania cells were digested with PvuI, transferred to a nylon membrane, and hybridized to a 700-bp probe derived from the conserved regions of the FBTs. B, susceptibility of Leishmania cells to sinefungin. C, susceptibility of Leishmania cells to methotrexate. ♦, TarMTX1000.6 transfected with AdoMetT1; ◆, TarMTX1000.6 transfected with FT1; ▿, LV39SNF4000.4 transfected with AdoMetT1. The average of triplicate measurements is shown (for B and C).
FIGURE 4.
Functional characterization of AdoMetT1. A, fluorescence microscopy analysis of LV39SNF4000.4 cells expressing AdoMetT1-GFP. LV39SNF4000.4 cells were transfected with a plasmid where the AdoMetT1 gene and the GFP gene were fused in-frame. Analysis of transfectants by fluorescence microscopy indicated that the fusion protein was localized mainly in the plasma membrane of the parasite. B, transport of 60 nm S-[3H]adenosylmethionine in L. major cells. Δ, L. major Friedlin; ■, L. major Friedlin transfected with AdoMetT1; □, LV39SNF4000.4; ▿, LV39SNF4000.4 transfected with AdoMetT1-GFP; ▾, LV39SNF4000.4 transfected with AdoMetT1. C, transport of 60 nm S-[3H]adenosylmethionine in L. tarentolae cells. ○, TarMTX1000.6 transfected with GFP; ♦, TarMTX1000.6 transfected with AdoMetT1-GFP; ◆, TarMTX1000.6 transfected with FT1; ▴, TarSNF.8; ▾, TarSNF.8 transfected with AdoMetT1-GFP. D, transport of 115 nm [3H]folic acid in L. tarentolae cells. ♦, TarMTX1000.6 transfected with AdoMetT1-GFP; ◆, TarMTX1000.6 transfected with FT1-GFP. The average of triplicate experiments are shown (B–D).
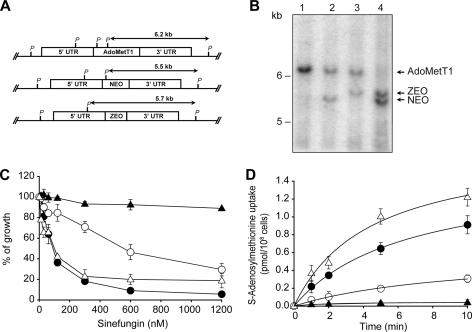
To further link AdoMetT1 to AdoMet transport, we attempted to generate an L. infantum AdoMetT1 null mutant. Two replacement cassettes were made, which allowed AdoMetT1 to be replaced by either the NEO or ZEO genes (Fig. 5A). These constructs were transfected independently in wild-type cells and selected for resistance to either G418 or Zeocin. Leishmania is diploid, and when its DNA is digested with PstI and hybridized to a 3′-UTR AdoMetT1 probe, the resulting hybridized fragment should be 6.2 kb in length (Fig. 5A). Integration of either the NEO or ZEO cassette and digesting with PstI and hybridizing to the same probe should lead to fragments of 5.5 and 5.7 kb, respectively (Fig. 5A). Analysis of wild-type cells and AdoMetT1/NEO or AdoMetT1/ZEO transfectants digested with PstI and hybridized with the 3′-UTR probe were completely consistent with the above scenario (Fig. 5B, lanes 1–3). The ZEO cassette was introduced in an AdoMetT1/NEO clone, and molecular analysis of the resulting transfectant was consistent with the generation of an AdoMetT1 null mutant. Indeed, the 6.2-kb band corresponding to the intact AdoMetT1 gene disappeared, whereas bands corresponding to the integration of NEO and ZEO inactivation cassettes were observed (Fig. 5B, lane 4). The L. infantum-263 wild-type cells were sensitive to SNF (Fig. 5C) and accumulated AdoMet (Fig. 5D). However, the AdoMetT1/NEO cells exhibited intermediate sensitivity to SNF (Fig. 5C) and accumulated less AdoMet (Fig. 5, C and D). The NEO/ZEO AdoMetT1 null mutant was insensitive to SNF and did not accumulate any measurable AdoMet (Fig. 5, C and D). We confirmed that these phenotypes were indeed due to the lack of AdoMetT1, because transfection of an episomal add-back construct encoding AdoMetT1 into the null mutant rescued both the susceptibility to SNF and the transport of AdoMet (Fig. 5, C and D).
FIGURE 5.
Generation and characterization of L. infantum AdoMetT1 null mutant. A, genomic organization of the AdoMetT1 locus. The AdoMetT1 upstream and downstream fragments used for the integration of the NEO and ZEO cassettes are shown. P, PstI. UTR, untranslated region. B, Southern blots of L. infantum genomic DNA digested with PstI and hybridized to a AdoMetT1–3′ UTR probe. Lane 1, L. infantum wild type; lane 2, L. infantum with one AdoMetT1 allele disrupted with the NEO cassette; lane 3, L. infantum with one AdoMetT1 allele disrupted with a ZEO cassette; lane 4, L. infantum AdoMetT1 null mutant with integrated NEO and ZEO cassettes. C, sinefungin susceptibility of Leishmania cells. D, transport of 60 nm S-[3H]adenosylmethionine by Leishmania cells. ●, L. infantum wild type; ○, L. infantum AdoMetT1 with one inactivated allele; ▴, L. infantum AdoMetT1 null mutant; ▵, L. infantum AdoMetT1 null mutant transfected with a add-back AdoMetT1 plasmid. The average of triplicate measurements is shown for C and D.
AdoMetT1 Is a Specific High Affinity AdoMet Transporter Expressed Constitutively
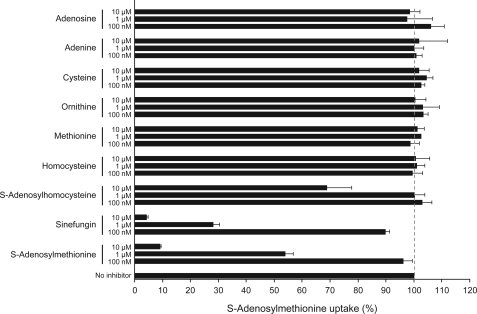
The kinetic properties of AdoMetT1 were studied, and the Km value was determined to be in the 150–300 nm range for both L. major and L. infantum (Table 1). The Vmax values was calculated to be at least 10 times higher in cells sensitive to SNF compared with cells intrinsically less susceptible to SNF (Table 1). We tested whether AdoMetT1 was specific for AdoMet by challenging the accumulation of [3H]AdoMet with related substrates. AdoMet itself and SNF were shown to inhibit the accumulation of the radioactive substrate but neither adenosine nor adenine, cysteine, ornithine, methionine, or homocysteine competed with AdoMet uptake (Fig. 6). Only S-adenosylhomocysteine, at a high concentration, was able to compete with the accumulation of AdoMet (Fig. 6).
FIGURE 6.
Substrate specificity profile of AdoMetT1 in L. major. LV39SNF4000.4 cells expressing AdoMetT1-GFP were co-incubated for 10 min with 50 nm S-[3H]adenosylmethionine and cold substrate competitors at various concentrations (100 nm and 1 and 10 μm). Results represent the percent of S-[3H]adenosylmethionine accumulation in competed samples versus the accumulation of a noncompeted control. The average of triplicate measurements is shown.
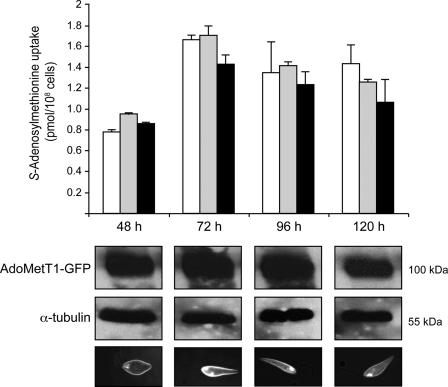
The transport of FA mediated by the FBT member FT1 is stage-regulated in Leishmania, with maximal activity in the logarithmic phase of growth (38). Indeed, during the progression of the parasite toward its stationary phase, there is increased FT1 protein degradation (37). We tested whether AdoMetT1 and AdoMet transport were under similar stage regulation. AdoMet transport increased during log phase, but in contrast to FA, it remained constant during the stationary phase (Fig. 7). Consistent with these results, AdoMetT1-GFP fusions were not degraded in the stationary phase of growth with the fusion protein migrating at the expected 100-kDa size. Moreover, AdoMetT1-GFP was at the level of the plasma membrane in stationary cells as deduced from fluorescence microscopy (Fig. 7). The same lack of growth stage regulation was observed for both L. major and L. infantum (Fig. 7 and results not shown).
FIGURE 7.
Growth stage regulation of S-adenosylmethionine transport activity. Accumulation of 60 nm S-[3H]adenosylmethionine was measured along the growth phases (48, 72, 96, and 120 h) for 10 min in L. infantum wild type (white bars), LV39SNF4000.4 transfected with AdoMetT1-GFP (gray bars), and L. major LV39 wild type (black bars). Average of triplicate measures is shown. Western blot with an anti-GFP antibody or with a control α-tubulin antibody to monitor the amounts of proteins layered was carried out with LV39SNF4000.4 transfected with AdoMetT1-GFP as described under “Materials and Methods.” Cellular localization of AdoMetT1 was deduced from fluorescence microscopy analysis of LV39SNF4000.4 transfected with the AdoMetT1 gene fused in-frame with the GFP gene. The majority of the fluorescence derived from the AdoMetT1-GFP hybrid protein was at the level of the plasma membrane throughout the various growth stages.
Molecular Basis for Intrinsic and Acquired Resistance to Sinefungin
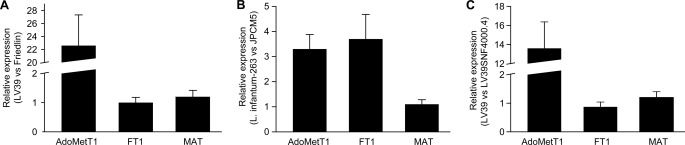
L. major Friedlin and L. infantum JPCM5 were intrinsically resistant to SNF, whereas L. major LV39 and L. infantum-263 were sensitive to it. This correlates with a decrease in the transport of AdoMet (Fig. 2 and Table 1) and most likely SNF (Fig. 6). We sequenced the AdoMetT1 gene in cells incapable of AdoMet transport, and we found that the coding sequence of the gene was identical to the published sequence. We next measured the expression of AdoMetT1 by quantitative reverse transcription-PCR and found that its expression was 20 times higher in L. major LV39 compared with L. major Friedlin (Fig. 8A). The expression of the MAT and FT1 genes was similar in the two L. major strains (Fig. 8A). The expression of AdoMetT1 was also higher (3-fold) in the AdoMet-transporting strain L. infantum-263 compared with L. infantum JPCM5 (Fig. 8B). The expression of the MAT gene was similar in the two L. infantum strains, but the FT1 gene was also expressed at higher level in L. infantum-263 (Fig. 8B). Incidentally, we found that L. infantum-263 was more sensitive to MTX compared with strain JPCM5 (results not shown), an observation that could be attributed to the difference in FT1 expression.
FIGURE 8.
RNA expression of targeted one-carbon metabolism genes in Leishmania cells. The expression of the genes coding for the AdoMet transporter AdoMetT1, of the folate transporter FT1, and of the methionine adenosyltransferase MAT were measured by quantitative real time reverse transcription-PCR as described under “Materials and Methods.” The expression was compared in the following: A, L. major LV39 versus L. major Friedlin, respectively, sensitive and insensitive to SNF; B, L. infantum-263 versus L. infantum JPCM5 (respectively, sensitive and less sensitive to both SNF and MTX); C, L. major LV39 versus the sinefungin-resistant mutant LV39SNF4000.4. For all bars, mean of biological triplicates is presented.
We also sequenced the AdoMetT1 open reading frame in the SNF-resistant mutant LV39SNF4000.4, and we compared that sequence with that of the L. major LV39 strain and found that they were identical (results not shown). However, the expression of AdoMetT1 was down-regulated 11-fold in cells in which resistance was induced compared with the wild-type cells as determined by quantitative reverse transcription-PCR (Fig. 8C). The reduction in AdoMetT1 expression correlated with a decrease of the Vmax value in the transport of AdoMet (Table 1). Unfortunately, this quantitative reverse transcription-PCR assay could not be applied to SNF-sensitive and -resistant L. tarentolae, possibly because the sequence of AdoMetT1 in this species was too divergent.
DISCUSSION
The first indirect evidence for an AdoMet transporter in Leishmania came from the observation that SNF, an AdoMet analogue, had inhibitory effects against the parasite (39). Biochemical evidence was later obtained that demonstrated both the capacity of Leishmania to accumulate AdoMet (40) and that AdoMet and SNF shared the same uptake system (21). Our study now identifies the high affinity AdoMet transporter in Leishmania as AdoMetT1, a membrane protein in the FBT family. The FBT family is a novel class of membrane proteins, which is part of the major facilitator superfamily (41, 42). They were first characterized in Leishmania (30, 43) but are present also in other kinetoplastid parasites (27) or in Apicomplexa parasites such as malaria and Toxoplasma (44, 45), in plants, and in cyanobacteria (46). So far, the only function attributed to these proteins was the transport of unconjugated (47, 48) or conjugated pterins such as FA (30, 37, 46, 49). With 14 members, Leishmania has the largest number of FBTs, but the function of only three members, BT1, FT1, and FT5 is known. The remaining 11 Leishmania proteins in the FBT family are unlikely to correspond to plasma membrane FA transporters (27). The function of a fourth member of the FBT family, namely AdoMetT1, has now been revealed. As such, it would be of interest to investigate the role of AdoMetT1 homologues in other organisms containing FBTs, to determine whether they too can transport AdoMet. This would be particularly important in Trypanosoma brucei, the parasite responsible for sleeping sickness. Indeed, based on the unique biochemical properties of AdoMet transport in T. brucei, it was suggested that the transporter could serve as a novel route for the delivery of drugs (50). T. brucei has seven FBT homologues (27) and determining if any of these function in AdoMet transport would be an important endeavor. In particular, several S-adenosylmethionine decarboxylase inhibitors are being developed against parasites (51–53), and some of these may enter through an AdoMet transporter. Sinefungin, but none of the individual AdoMet building blocks, can inhibit AdoMet uptake in T. brucei (22), and this is consistent with our observations for Leishmania AdoMetT1 (Fig. 6). The Km value of the T. brucei AdoMet transport was reported to be in the millimolar range (54) indicating a lower affinity than Leishmania AdoMetT1, which is in the nanomolar range (Table 1).
FBTs have also been described in plants with nine members in Arabidopsis, one of which transports FA (46). The plastid AdoMet transporter, named SAMT1, is not a member of the FBT family but is part of the MCF (15, 16). MCF proteins are also responsible for AdoMet transport in yeast (13) and human mitochondria (14). Interestingly, Leishmania also encodes MCF proteins, one of which appears to be an orthologue of the mitochondrial AdoMet transporter (55). Disruption of the Arabidopsis SAMT1 gene led to a severe growth defect, but because the plants remained viable, it was suggested that other proteins could be implicated in AdoMet transport (15). It is possible that one of the plant FBTs could be involved as a secondary organellar AdoMet transporter.
The Leishmania AdoMetT1 transports AdoMet specifically in the low nanomolar range, whereas FT1, also a FBT member, exclusively transports FA in the nanomolar range (37, 43). Other FBTs highly homologous to either AdoMetT1 or FT1 transport neither AdoMet nor FA. Because AdoMetT1 and FT1 are 80% identical (27), it should be possible to find key regions and eventually amino acids involved in substrate specificity. Interestingly, AdoMetT1 and FT1 are regulated very differently at the post-translational level with FT1 being degraded in stationary phase (37), whereas AdoMetT1 is expressed constitutively (Fig. 7). Although the signals implicated in the degradation of FT1 are unknown, the production of AdoMetT1/FT1 fusion proteins could aid in identifying motifs involved in protein degradation in Leishmania.
Sinefungin has been found to be active against several Leishmania species (39, 56) but also against other parasites, including malaria (57), and the trypanosomes (58). It was suggested to have the potential to serve as a lead compound for a novel antiparasitic drug (59). However, some Leishmania strains were found to be intrinsically resistant to SNF (60), a finding supported here with the intrinsic SNF resistance observed in L. major Friedlin or L. infantum JPCM5 (Table 1). This phenomenon can now be plausibly explained by the defect in AdoMet transport (Fig. 2B), which is also related to lower AdoMetT1 expression (Fig. 8) and to a reduced Vmax for AdoMet transport (Table 1). The disruption of AdoMetT1 also leads to SNF resistance (Fig. 5). Interestingly, the selection for resistance to SNF in Leishmania was also correlated to a decrease in the expression of AdoMetT1 (Fig. 8). The mechanism by which this happens is not yet clear, but it could be related to the gene rearrangement observed in our study (Fig. 3A). Reduced RNA levels in Leishmania are usually due to a decrease in gene copy number (61, 62), although other mechanisms may also occur (63). Interestingly, in the yeast S. cerevisiae that also transports AdoMet, resistance to SNF was shown to correlate with loss of function mutations in the AdoMet transporter Sam3 (20). Although the overexpression of MAT was shown to confer resistance to SNF in Leishmania (64), it is not involved in the decreased susceptibility observed here because the expression of MAT was not changed in our resistant cells at either the RNA (Fig. 8C) or protein level (result not shown). The expression of MAT was also not increased in cells with reduced expression of AdoMetT1 (Fig. 8) or in cells in which AdoMetT1 was disrupted (results not shown). Leishmania strains can be sensitive to SNF in the low nanomolar range (Fig. 1), but several strains demonstrated no intrinsic susceptibility (Table 1) (60) possibly because of lower AdoMetT1 expression. We could nonetheless envisage using more lipophilic analogues of SNF, which may not require a specific transporter, as novel chemotherapeutic agents against Leishmania.
Pneumocystis appears to depend exclusively on AdoMet uptake to meet its AdoMet needs (11), but Leishmania can either synthesize it or salvage it from its environment. We have inactivated AdoMetT1 in L. infantum (Fig. 5), and the resultant cells were viable, showed no obvious growth defect, and retained the ability to infect macrophages (results not shown). Several organisms use synthesis as their main source of AdoMet, so the presence of proteins implicated in the uptake of exogenous AdoMet in major classes of parasites (kinetoplastids and apicomplexa) is perplexing. Leishmania cycles in various environments (insect vectors and mammalian hosts) with varying concentrations of metabolites that could serve as building blocks for AdoMet synthesis. It may thus be advantageous, under limiting concentrations of metabolic precursors, to import AdoMet directly from the environment. Leishmania is a purine auxotroph with numerous salvage pathways (65), and one could envision that under purine-poor conditions, the uptake of AdoMet could serve as a purine source. Synthesis of AdoMet is energetically expensive because for each molecule of AdoMet synthesized, the three high energy phosphodiester bonds of ATP are hydrolyzed (9). When AdoMet is present in the environment, and under energetic constraint, it may thus be advantageous for the parasite to import AdoMet instead of synthesizing it. The Leishmania MAT activity is maximal during logarithmic phase of growth and negligible in the stationary phase (7). If AdoMet is required during other growth phases of the parasite, it could also be useful for the organism to import it to compensate for a reduced rate of synthesis. Although cells without AdoMetT1 (Fig. 5) or cells with low AdoMetT1 expression (Fig. 8) thrive well under laboratory conditions, it is possible that under specific conditions they may be at a disadvantage or that the expression of the gene is induced. The identification of the AdoMet transporter will now allow the testing of some of these hypotheses in Leishmania and also in other organisms.
Supplementary Material
Acknowledgments
We thank Dr. Jeremy Mottram for the L. infantum JPCM5 strain and Dr. Balana-Fouce for the anti-MAT antibody. We also thank Drs. Danielle Légaré, Angana Mukherjee, Philippe Leprohon, and Jennifer Raven for critical reading of the manuscript.
This work was supported in part by the Canadian Institutes of Health Research group and operating grants (to M. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- AdoMet
- S-adenosylmethionine
- PBS
- phosphate-buffered saline
- GFP
- green fluorescent protein
- FBT
- folate biopterin transporter
- MAT
- methionine adenosyltransferase
- MCF
- mitochondrial carrier protein family
- MTX
- methotrexate
- SNF
- sinefungin
- FA
- folic acid
- UTR
- untranslated region.
REFERENCES
- 1.Herwaldt B. L. (1999) Lancet 354, 1191–1199 [DOI] [PubMed] [Google Scholar]
- 2.Murray H. W., Berman J. D., Davies C. R., Saravia N. G. (2005) Lancet 366, 1561–1577 [DOI] [PubMed] [Google Scholar]
- 3.Ouellette M., Drummelsmith J., Papadopoulou B. (2004) Drug Resist. Updat. 7, 257–266 [DOI] [PubMed] [Google Scholar]
- 4.Croft S. L., Sundar S., Fairlamb A. H. (2006) Clin. Microbiol. Rev. 19, 111–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu S. C., Mato J. M. (2008) J. Gastroenterol. Hepatol. 23, S73–S77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fairlamb A. H., Cerami A. (1992) Annu. Rev. Microbiol. 46, 695–729 [DOI] [PubMed] [Google Scholar]
- 7.Reguera R. M., Redondo C. M., Pérez-Pertejo Y., Balaña-Fouce R. (2007) Mol. Biochem. Parasitol. 152, 1–10 [DOI] [PubMed] [Google Scholar]
- 8.Fontecave M., Atta M., Mulliez E. (2004) Trends Biochem. Sci. 29, 243–249 [DOI] [PubMed] [Google Scholar]
- 9.Mato J. M., Corrales F. J., Lu S. C., Avila M. A. (2002) FASEB J. 16, 15–26 [DOI] [PubMed] [Google Scholar]
- 10.Tucker A. M., Winkler H. H., Driskell L. O., Wood D. O. (2003) J. Bacteriol. 185, 3031–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merali S., Vargas D., Franklin M., Clarkson A. B., Jr. (2000) J. Biol. Chem. 275, 14958–14963 [DOI] [PubMed] [Google Scholar]
- 12.Merali S., Clarkson A. B., Jr. (2004) FEMS Microbiol. Lett. 237, 179–186 [DOI] [PubMed] [Google Scholar]
- 13.Marobbio C. M., Agrimi G., Lasorsa F. M., Palmieri F. (2003) EMBO J. 22, 5975–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agrimi G., Di Noia M. A., Marobbio C. M., Fiermonte G., Lasorsa F. M., Palmieri F. (2004) Biochem. J. 379, 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouvier F., Linka N., Isner J. C., Mutterer J., Weber A. P., Camara B. (2006) Plant Cell 18, 3088–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmieri L., Arrigoni R., Blanco E., Carrari F., Zanor M. I., Studart-Guimaraes C., Fernie A. R., Palmieri F. (2006) Plant Physiol. 142, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haferkamp I. (2007) FEBS Lett. 581, 2375–2379 [DOI] [PubMed] [Google Scholar]
- 18.Rouillon A., Surdin-Kerjan Y., Thomas D. (1999) J. Biol. Chem. 274, 28096–28105 [DOI] [PubMed] [Google Scholar]
- 19.Uemura T., Kashiwagi K., Igarashi K. (2007) J. Biol. Chem. 282, 7733–7741 [DOI] [PubMed] [Google Scholar]
- 20.Zheng S., Shuman S., Schwer B. (2007) Nucleic Acids Res. 35, 6895–6903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phelouzat M. A., Basselin M., Lawrence F., Robert-Gero M. (1995) Biochem. J. 305, 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldberg B., Rattendi D., Lloyd D., Sufrin J. R., Bacchi C. J. (1998) Biochem. Pharmacol. 56, 95–103 [DOI] [PubMed] [Google Scholar]
- 23.Papadopoulou B., Roy G., Ouellette M. (1993) Nucleic Acids Res. 21, 4305–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouellette M., Fase-Fowler F., Borst P. (1990) EMBO J. 9, 1027–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papadopoulou B., Roy G., Ouellette M. (1992) EMBO J. 11, 3601–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El Fadili A., Kündig C., Ouellette M. (2002) Mol. Biochem. Parasitol. 124, 63–71 [DOI] [PubMed] [Google Scholar]
- 27.Ouameur A. A., Girard I., Légaré D., Ouellette M. (2008) Mol. Biochem. Parasitol. 162, 155–164 [DOI] [PubMed] [Google Scholar]
- 28.Moreira W., Leblanc E., Ouellette M. (2009) Free Radic. Biol. Med. 46, 367–375 [DOI] [PubMed] [Google Scholar]
- 29.Drummelsmith J., Girard I., Trudel N., Ouellette M. (2004) J. Biol. Chem. 279, 33273–33280 [DOI] [PubMed] [Google Scholar]
- 30.Richard D., Kündig C., Ouellette M. (2002) J. Biol. Chem. 277, 29460–29467 [DOI] [PubMed] [Google Scholar]
- 31.Al-Salabi M. I., Wallace L. J., De Koning H. P. (2003) Mol. Pharmacol. 63, 814–820 [DOI] [PubMed] [Google Scholar]
- 32.Gagnon D., Foucher A., Girard I., Ouellette M. (2006) Mol. Biochem. Parasitol. 150, 63–71 [DOI] [PubMed] [Google Scholar]
- 33.Foucher A. L., Papadopoulou B., Ouellette M. (2006) J. Proteome Res. 5, 1741–1750 [DOI] [PubMed] [Google Scholar]
- 34.Reguera R. M., Balaña-Fouce R., Pérez-Pertejo Y., Fernández F. J., García-Estrada C., Cubría J. C., Ordóñez C., Ordóñez D. (2002) J. Biol. Chem. 277, 3158–3167 [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, pp. 9.31–9.62, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36.Guimond C., Trudel N., Brochu C., Marquis N., El Fadili A., Peytavi R., Briand G., Richard D., Messier N., Papadopoulou B., Corbeil J., Bergeron M. G., Légaré D., Ouellette M. (2003) Nucleic Acids Res. 31, 5886–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richard D., Leprohon P., Drummelsmith J., Ouellette M. (2004) J. Biol. Chem. 279, 54494–54501 [DOI] [PubMed] [Google Scholar]
- 38.Ellenberger T. E., Beverley S. M. (1987) J. Biol. Chem. 262, 10053–10058 [PubMed] [Google Scholar]
- 39.Bachrach U., Schnur L. F., El-On J., Greenblatt C. L., Pearlman E., Robert-Gero M., Lederer E. (1980) FEBS Lett. 121, 287–291 [DOI] [PubMed] [Google Scholar]
- 40.Avila J. L., Polegre M. A. (1993) Mol. Biochem. Parasitol. 58, 123–134 [DOI] [PubMed] [Google Scholar]
- 41.Saier M. H., Jr., Beatty J. T., Goffeau A., Harley K. T., Heijne W. H., Huang S. C., Jack D. L., Jähn P. S., Lew K., Liu J., Pao S. S., Paulsen I. T., Tseng T. T., Virk P. S. (1999) J. Mol. Microbiol. Biotechnol. 1, 257–279 [PubMed] [Google Scholar]
- 42.Chang A. B., Lin R., Keith Studley W., Tran C. V., Saier M. H., Jr. (2004) Mol. Membr. Biol. 21, 171–181 [DOI] [PubMed] [Google Scholar]
- 43.Dridi L., Haimeur A., Ouellette M. (2010) Biochem. Pharmacol. 79, 30–38 [DOI] [PubMed] [Google Scholar]
- 44.Massimine K. M., Doan L. T., Atreya C. A., Stedman T. T., Anderson K. S., Joiner K. A., Coppens I. (2005) Mol. Biochem. Parasitol. 144, 44–54 [DOI] [PubMed] [Google Scholar]
- 45.Wang P., Wang Q., Sims P. F., Hyde J. E. (2007) Mol. Biochem. Parasitol. 154, 40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klaus S. M., Kunji E. R., Bozzo G. G., Noiriel A., de la Garza R. D., Basset G. J., Ravanel S., Rébeillé F., Gregory J. F., 3rd, Hanson A. D. (2005) J. Biol. Chem. 280, 38457–38463 [DOI] [PubMed] [Google Scholar]
- 47.Kündig C., Haimeur A., Légaré D., Papadopoulou B., Ouellette M. (1999) EMBO J. 18, 2342–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemley C., Yan S., Dole V. S., Madhubala R., Cunningham M. L., Beverley S. M., Myler P. J., Stuart K. D. (1999) Mol. Biochem. Parasitol. 104, 93–105 [DOI] [PubMed] [Google Scholar]
- 49.Cunningham M. L., Beverley S. M. (2001) Mol. Biochem. Parasitol. 113, 199–213 [DOI] [PubMed] [Google Scholar]
- 50.Goldberg B., Yarlett N., Sufrin J., Lloyd D., Bacchi C. J. (1997) FASEB J. 11, 256–260 [DOI] [PubMed] [Google Scholar]
- 51.Bacchi C. J., Barker R. H., Jr., Rodriguez A., Hirth B., Rattendi D., Yarlett N., Hendrick C. L., Sybertz E. (2009) Antimicrob. Agents Chemother. 53, 3269–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirth B., Barker R. H., Jr., Celatka C. A., Klinger J. D., Liu H., Nare B., Nijjar A., Phillips M. A., Sybertz E., Willert E. K., Xiang Y. (2009) Bioorg. Med. Chem. Lett. 19, 2916–2919 [DOI] [PubMed] [Google Scholar]
- 53.Barker R. H., Jr., Liu H., Hirth B., Celatka C. A., Fitzpatrick R., Xiang Y., Willert E. K., Phillips M. A., Kaiser M., Bacchi C. J., Rodriguez A., Yarlett N., Klinger J. D., Sybertz E. (2009) Antimicrob. Agents Chemother. 53, 2052–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goldberg B., Rattendi D., Lloyd D., Yarlett N., Bacchi C. J. (1999) Arch. Biochem. Biophys. 364, 13–18 [DOI] [PubMed] [Google Scholar]
- 55.Colasante C., Peña Diaz P., Clayton C., Voncken F. (2009) Mol. Biochem. Parasitol. 167, 104–117 [DOI] [PubMed] [Google Scholar]
- 56.Paolantonacci P., Lawrence F., Robert-Géro M. (1985) Antimicrob. Agents Chemother. 28, 528–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trager W., Tershakovec M., Chiang P. K., Cantoni G. L. (1980) Exp. Parasitol. 50, 83–89 [DOI] [PubMed] [Google Scholar]
- 58.Dube D. K., Mpimbaza G., Allison A. C., Lederer E., Rovis L. (1983) Am. J. Trop. Med. Hyg. 32, 31–33 [DOI] [PubMed] [Google Scholar]
- 59.Nolan L. L. (1987) Antimicrob. Agents Chemother. 31, 1542–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Avila J. L., Avila A. (1987) Mol. Biochem. Parasitol. 26, 69–75 [DOI] [PubMed] [Google Scholar]
- 61.Ubeda J. M., Légaré D., Raymond F., Ouameur A. A., Boisvert S., Rigault P., Corbeil J., Tremblay M. J., Olivier M., Papadopoulou B., Ouellette M. (2008) Genome Biol. 9, R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leprohon P., Légaré D., Raymond F., Madore E., Hardiman G., Corbeil J., Ouellette M. (2009) Nucleic Acids Res. 37, 1387–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marquis N., Gourbal B., Rosen B. P., Mukhopadhyay R., Ouellette M. (2005) Mol. Microbiol. 57, 1690–1699 [DOI] [PubMed] [Google Scholar]
- 64.Pérez-Pertejo Y., Reguera R. M., Ordóñez D., Balaña-Fouce R. (2006) Biochim. Biophys. Acta 1760, 10–19 [DOI] [PubMed] [Google Scholar]
- 65.Landfear S. M., Ullman B., Carter N. S., Sanchez M. A. (2004) Eukaryot. Cell 3, 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.