Abstract
Neuronal nicotinic acetylcholine receptors (nAChR) composed of α4 + β2 subunits, the high affinity nicotine-binding site in the mammalian brain, up-regulate in response to chronic nicotine exposure. The identities of endogenous mediators of this process are unknown. We find that choline also up-regulates α4 + β2 nAChRs stably expressed by HEK293 cells as measured by increased [3H]epibatidine density. Choline-mediated up-regulation is dose-dependent and corresponds with an increase in β2 subunit protein expression. The choline kinase inhibitor hemicholinium-3 inhibits ∼60% of choline-mediated up-regulation revealing both an HC3-dependent and -independent pathway. Furthermore, choline-mediated up-regulation is not additive with up-regulation agents such as nicotine, but it is additive with weaker promoters of the up-regulation process. When co-applied with the pro-inflammatory cytokine tumor necrosis factor α, choline-mediated up-regulation is increased further through a mechanism that includes an increase in both α4 and β2 protein expression, and this is inhibited by the p38 MAPK inhibitor SB202190. These findings extend the view that up-regulation of α4 + β2 nAChRs is a normal physiological response to altered metabolic and inflammatory conditions.
Keywords: Inflammation, Nicotinic Acetylcholine Receptors, p38 MAPK, Receptor Regulation, Tumor Necrosis Factor (TNF), Choline
Introduction
One response by neuronal nicotinic acetylcholine receptors (nAChR)2 composed of α4 + β2 subunits (α4β2), the high affinity nicotine-binding site in the mammalian brain, is to up-regulate in response to chronic nicotine exposure (1–4). This process, as measured by an increase in the density of high affinity ligand-binding sites, is associated with multiple physiological processes ranging from nicotine addiction to a therapeutic target for intervention in age-related neurological disorders, including Alzheimer or Parkinson disease where α4β2 nAChR expression becomes dysregulated (for recent discussion see Ref. 3). The mechanism(s) leading to receptor up-regulation are complex and may vary among different cell systems. For example, up-regulation has been attributed to enhanced receptor assembly, increased efficiency of receptor trafficking (possibly through the interaction with chaperones or ligands that stabilize subunit interactions), changes in degradation rates, altered subunit stoichiometry, or changes in conformational states (5–7). Another feature of up-regulation is that compounds that bind to the receptor ligand-binding site (both agonists and antagonists) are sufficient to promote this process even when receptors are expressed in heterologous systems such as Xenopus oocytes or transfected mammalian cultured cells (3, 4). Up-regulation to nicotine is the most common context in which this receptor property is examined. However, this process can also be responsive to certain pro-inflammatory cytokines (8, 9). In particular, the simultaneous exposure of α4β2 to receptor ligands such as nicotine and the pro-inflammatory cytokine tumor necrosis factor α (TNFα) strongly enhances up-regulation of α4β2 high affinity binding sites in an almost synergistic manner (9). Therefore, while up-regulation is a key feature of nAChR biology and clearly important to many of the alterations in physiological responses leading to addiction (3, 4), the normal physiological context of this receptor response and the identity of natural endogenous mediators that impart it are unknown. Choline is another endogenous agent of importance to nAChR function. Many examples exist where oral choline supplementation to a normal diet is correlated to increased brain acetylcholine concentrations and enhanced performance on memory tasks that in part are related to increased nAChR function (e.g. see Refs. 10–12). In addition to its obvious role as a metabolic precursor to the principal endogenous cholinergic neurotransmitter, acetylcholine, choline can also act to modify receptor activation. For example, choline is a full and potent agonist of α7 or α9 containing nAChRs, and it can modify the function of certain other nAChRs such as those composed of α3β4 subunits (13–15). The direct interaction of choline with the high affinity nicotine-binding site of α4β2 is at most weak and is probably unlikely to occur normally (14, 16). Thus, despite the actions of choline on other nAChRs, it has not been examined in detail for its impact on the expression of high affinity α4β2 nAChRs nor within a pro-inflammatory context where the impact of choline is documented (e.g. Refs. 10, 17, 18).
While examining how nAChRs participate in inflammatory interactions (8, 9, 19), we noted that choline alone can promote up-regulation of α4β2 nAChRs expressed by 293 cells stably expressing this receptor. Choline-mediated up-regulation, as measured by an increase in the density of the high affinity α4β2 ligand, [3H]epibatidine ([3H]EB), is dose-dependent and corresponds with increased expression of the β2 subunit. Approximately 60% of choline-mediated up-regulation was inhibited by the choline kinase inhibitor hemicholinium-3 (HC3) indicating that choline acts through a choline kinase-dependent and -independent pathway (21). Choline-mediated up-regulation was additive with weaker inducers of ligand-mediated up-regulation but not when stronger mediators of this process (such as nicotine) were co-applied. In the presence of TNFα and choline, up-regulation exhibits a more additive effect that is distinctly different from the dramatic impact this cytokine has on nicotine-mediated up-regulation (9). Our findings extend the view that α4β2 up-regulation is responsive to endogenous mediators, including this important nutrient as well as pro-inflammatory mediators such as TNFα. These findings also suggest that up-regulation could serve as an important component of the normal role of α4β2 and the nAChR-inflammatory interaction, especially in the absence of potent exogenous agents such as nicotine.
EXPERIMENTAL PROCEDURES
Cells Lines and Culture Conditions
The stably co-transfected 293 cell lines expressing nAChR subunits α4 + β2, α4 + β4, or α3 + β4 were provided by Drs. Ken Kellar and Yingxian Xiao (Department of Pharmacology, Georgetown, University) and were cultured as described previously (9, 14). Because choline is an essential nutrient, it is present in cultured cell media, and it is rapidly depleted by cells when added. Consequently, care must be exercised to control concentrations. For this study, HEK293 (293) cells were passed into 100-mm culture dishes containing 10 ml of Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum and geneticin (1.0 g/liter) as described previously (9, 14). Cells were cultured at 37 °C and 95% air, 5% CO2 atmosphere for 48 h before treatment as indicated. Up-regulation of nAChRs was measured 18–24 h thereafter.
Radioligand Binding
The binding of [3H]EB (PerkinElmer Life Sciences) to cell membrane preparations was done as described previously (9, 14). Briefly, 18–24 h after treatment, cells were gently washed with room temperature phosphate-buffered saline, scraped into ice-cold 50 mm Tris buffer, pH 7.4, pelleted, resuspended, gently homogenized, and centrifuged again at low speed (100 × g; 5 min) before removing the supernatant, which was again centrifuged at 20,000 × g for 10 min. Binding assays, done in triplicate, used 5 μg of resuspended crude membranes that were incubated with 5 nm [3H]EB for 2–4 h at room temperature. Addition of 300 μm nicotine hydrogen tartrate (Sigma) for 30 min to parallel tubes before adding [3H]EB served to measure nonspecific binding. Samples were then vacuum filtrated through Whatman GF/C filters to collect bound ligand and then quantitated using standard scintillation counting. The specific binding was calculated by averaging the total binding minus the nonspecific (nicotine-blocked) binding. Data were analyzed using Prism 4 (GraphPad Software Inc., San Diego, version 4.03 (9, 14)).
Western Blot Analysis
Western blots were as described previously (8). Briefly, cells were harvested, and crude membranes were prepared before mixing a fixed amount (20 μg) in gel-loading buffer containing dithiothreitol and heated to 95 °C for 10–15 min. The samples were then fractionated by SDS-PAGE fractionation followed by semi-dry transfer to polyvinylidene difluoride membrane, blocked with 5% dry milk and 0.05% Tween 20 (PBS-T), and rocked overnight at 4 °C in primary antibody. Antibodies were rabbit polyclonal 5009 or 4964 (α4) and 4842 or 305 (β2) as described elsewhere (9, 14). Blots were washed, incubated at room temperature for 1 h in blocking solution containing peroxidase-conjugated secondary antibody, washed again, and the bands detected on film after developing with the enhanced chemiluminescence system (PerkinElmer Life Sciences). Gels were recorded on a computer scanner, and the images were assembled using either Photoshop version 5.5 or Image Pro-plus (version 4.5, Media Cybernetics) software.
Sucrose Gradients
Detailed descriptions of sucrose gradient analysis were described previously (8). Briefly, transfected 293 cells were solubilized (Triton X-100), and the cleared supernatant was layered onto a 5-ml sucrose gradient (continuous 5–20% (w/w); 20 mm Tris, pH 7.5). Gradients were centrifuged at 54,000 rpm for 2 h (SW55Ti rotor, Beckman Instruments, Fullerton, CA) at 4 °C. Fractions were collected at various volumes to ensure desired resolution with an Auto Densiflow and FC203B fraction collector (Labconco, Kansas City, MO; Gilson, Middleton, WI). Sucrose gradient markers were the 20 S proteosome subunit (20 S, from Dr. M. Rechsteiner, University of Utah, Salt Lake City), catalase (11.2 S, Sigma), β-amylase (8.9 S, Sigma), and bovine serum albumin (4.2 S, Pierce).
RESULTS
A well known feature of nAChRs is that subunit composition determines their pharmacological and functional characteristics, including the magnitude of the up-regulation response (3, 4). The nAChR receptor consisting of α4 + β2 (α4β2) subunits is the receptor in the mammalian central nervous system that provides both the majority of high affinity nicotine binding and significant up-regulation (3–4-fold) in response to chronic agonist exposure (1, 2). The up-regulation response is recapitulated in cultured cells stably transfected with cDNAs encoding α4 and β2 subunits (2, 4, 16, 22–25). As noted above, during the course of experiments designed to evaluate the impact of nicotine and pro-inflammatory cytokines on the process of up-regulation, we noted that certain conditions of culture impacted upon this process. One candidate for this effect was choline, an essential nutrient that is closely associated with multiple nAChR functions and is rapidly depleted from the media by cells undergoing cell growth.
Choline Modifies nAChR Expression Consistent with Up-regulation
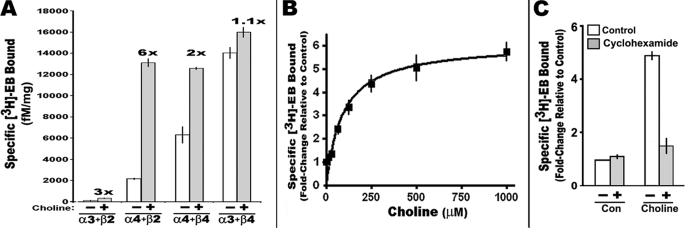
Because receptor subunit composition corresponds with the magnitude of up-regulation as measured by an increase in [3H]EB-binding site density, the impact of choline treatment on this process for four different nAChR receptor subtypes expressed stably by independent 293 cell lines was measured after exposure to 500 μm choline for 24 h (see “Experimental Procedures”). The nAChRs examined were of the α4β2, α4β4, α3β2, or α3β4 subunit subtypes, all of which exhibit characteristic levels of up-regulation following nicotine exposure (4, 9, 16, 26). The results are shown in Fig. 1A. The most dramatic enhancement of ligand binding was for α4β2-expressing cells where 6-fold increases can occur. To a lesser extent, but still significant, choline induced up-regulation of both α3β2 (∼3-fold) and α4β4 (∼2-fold), although native α3β2 receptor density was very low and could be difficult to detect in nontreated cells (16). Cells expressing α3β4 exhibited no significant choline-mediated enhancement of [3H]EB-binding sites which is similar to the response by this receptor subtype to nicotine (9, 16). This result is essentially identical to the results for up-regulation to nicotine reported previously for these cell lines (9, 14).
FIGURE 1.
Influence of choline on [3H]EB-binding site density in 293 cells stably expressing nAChRs of different subunit composition. A, cells expressing the indicated nicotinic receptor subunits were grown in the presence of 500 μm choline chloride for 24 h before measuring specific [3H]EB binding to the crude cell membrane fraction (see under “Experimental Procedures”). Results are expressed as the density of nAChR [3H]EB-binding sites per fm/mg protein. The calculated fold-change of choline versus control binding is shown above each cell group. B, dose response for choline-mediated up-regulation of [3H]EB-binding sites of α4β2 receptors. The Kd value for up-regulation by choline was 92.5 μm (nonlinear least square regression and one-site saturation binding model of the Prism 4.03 software). C, choline up-regulation of [3H]EB sites was inhibited by 10 μm cycloheximide. All values are normalized to the control (con) (saline and saline plus cycloheximide-only treatments). Error bars reflect mean ± S.E. calculated from three to six independent measurements for each experiment.
Choline-mediated increased [3H]EB binding by α4β2 is dose-dependent (Fig. 1B). In these experiments, 250 μm choline produced effectively the maximal up-regulation. Because choline impacts many cellular processes, this concentration was selected for subsequent experiments. Of note is that maintaining choline in the media at the amounts indicated is difficult due to its cellular uptake and metabolism. Therefore, doses are defined in terms of the concentration of choline added to the media 2 days after plating.
The process of up-regulation examined by us previously using this transfected 293 cell line also required protein synthesis (9). This is also the case for choline-mediated up-regulation as shown in Fig. 1C. For this experiment, an inhibitor of protein synthesis (cycloheximide) was added to cultures together with choline (250 μm). Although choline-mediated up-regulation in the absence of cycloheximide was ∼5-fold, inhibition of protein synthesis completely abolished all up-regulation relative to matched control cultures.
HC3 Inhibition of Choline-mediated Up-regulation Reveals Two Intracellular Pathways
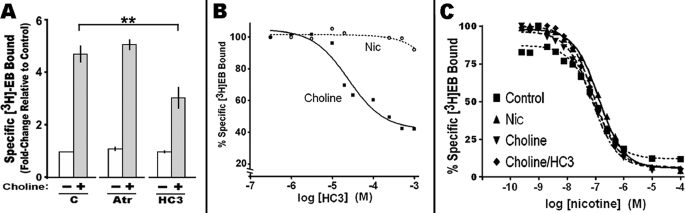
How choline elicits up-regulation was examined. It was first determined if an interaction of choline with muscarinic receptors contributed to the up-regulation effect. As shown in Fig. 2A, the muscarinic antagonist, atropine at 10 μm, failed to inhibit choline-mediated up-regulation of α4β2 [3H]EB binding. Similar results were obtained for scopolamine (10 μm; data not shown). Thus, muscarinic receptors are not necessary for choline-mediated up-regulation. Choline is actively transported into the cell via three choline transporter activities (27). These include the sodium-dependent high affinity uptake transporter, which is predominantly expressed by adult neurons, the choline-like transporter, which is sodium-independent and low affinity and expressed by cells throughout the body, and the blood-brain barrier transporter. Once in the cell, a major fate of choline is phosphorylation by choline kinase activity (28). An effective inhibitor of both the high affinity choline transporter and choline kinase activity is HC3 (21, 29–32). When this compound is added to cells prior to choline, it partially inhibits choline-mediated up-regulation (Fig. 2A). To determine which HC3 target dominates this response, the expression of choline transporter transcripts was measured by real time-PCR using commercially available primers (ABI) specific to each transporter transcript. Transcripts for the choline-like transporter were detected in these 293 cells (27 cycle threshold compared with 17 for β-actin; data not shown), but no evidence for expression of the high affinity transporter and only a weak, if any, signal for the BBB transporter was found (not shown). Consistent with the absence of high affinity choline transporters in these 293 α4β2 cells, no specific high affinity [3H]HC3 binding to cell membrane preparations was detected (data not shown and see Ref. 29). Thus, because the high affinity transporter was absent in these cells, the impact of HC3 in this system should be dominated by inhibition of the choline kinase activity.
FIGURE 2.
Choline-mediated up-regulation is through an atropine-insensitive, HC3 partially sensitive mechanism. A, cells expressing α4β2 were treated with atropine (Atr) (10 μm) or the choline kinase inhibitor HC3 before the addition of choline. Choline up-regulation relative to the control (C) was partially inhibited by HC3. B, dose response of the impact by HC3 on [3H]EB binding measured in the presence of either choline or nicotine (Nic). The plot curves were generated using the Prism 4.03 software. C, nicotine competition of [3H]EB binding to receptors from cells treated with nicotine, choline, or choline + HC3 versus the control (no treatment). The Ki for nicotine was ∼13.5 nm in all samples regardless of the treatment used to produce up-regulation. Error bars reflect mean ± S.E. from three independent measurements. **, p < 0.01.
To determine the dose of HC3 that achieves optimal inhibition of up-regulation, HC3 at different concentrations ranging from 6.6 × 10−9 to 1 × 10−3 m was added 15 min prior to adding 250 μm choline. Each dish was harvested; membranes were prepared, and [3H]EB was measured/μg of membrane protein. These values were then normalized to control cells receiving choline but no HC3. The results of this experiment are shown in Fig. 2B. The maximum inhibition of choline-mediated up-regulation achieved using HC3 was only 60%. At greater concentrations of HC3 (1 mm), indications of cell stress and possible toxicity were present. However, the dose-response curve in Fig. 2B suggests the effect of HC3 is saturating prior to reaching the maximum dose tested. The HC3 inhibition curve was best fit by a single regression (assuming saturation by HC3 at 500 μm) for a calculated Ki of ∼25 μm. This is in agreement with the IC50 values of 20–57 μm reported for human choline kinase (20, 21, 34). In contrast, the choline-like low affinity transporter is relatively poorly inhibited by HC3 (Ki of ∼ 200 μm) (32) suggesting choline kinase as the likely target of HC3 in this system.
The interpretation of HC3 inhibition is complicated by reports that choline acts on cells through both HC3-dependent and -independent mechanisms that are tissue-specific and developmentally regulated (33). To determine whether the HC3-insensitive component of up-regulation reflects altered α4β2 ligand binding affinity under these measurement conditions (Triton-solubilized membranes), competition for [3H]EB binding by nicotine was measured on membranes prepared from cells treated with nicotine, choline, or choline + HC3. Regardless of whether nicotine, choline or choline in the presence of HC3 was used to promote α4β2 receptor up-regulation (Fig. 2C), the Ki for [3H]EB was ∼13 nm. Thus, HC3-sensitive and -insensitive pathways leading to up-regulation that function independently of the high affinity choline transporter are present in these 293 cells. Of note is that choline acting through both HC3-dependent and -independent mechanisms that are developmentally regulated and tissue-specific have been described previously for other cell systems (21, 33).
Choline-mediated Up-regulation Corresponds with Increased β2 Subunit Expression
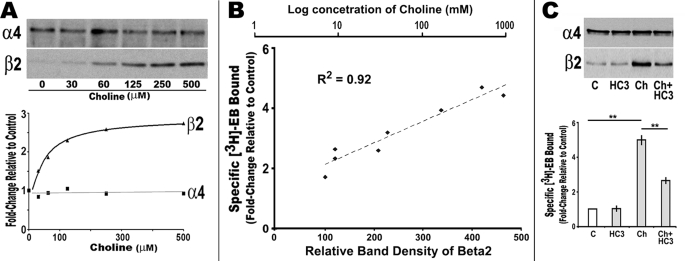
Because both α4 and β2 transcriptions in these stably transfected cells are from heterologous promoters (16, 26), and choline-mediated up-regulations are sensitive to protein synthesis, post-translational mechanisms of up-regulation were examined (22). To begin, the relative expression of α4 or β2 protein was examined using Western blot analysis of cells subjected to increasing choline concentrations. As shown in Fig. 3A, no change in α4 expression was observed, but there was a striking increase in the amount of β2 subunit protein as the choline concentration is increased. The maximum increase in β2 was again at 250 μm choline. The relationship between the β2 subunit increase and ligand binding was also very strong. When ligand binding data are collected on the same membranes used for protein analysis, there was a very highly significant correlation (R2 = 0.92) between the choline dose, the relative Western blot band intensity of β2, and [3H]EB ligand binding.
FIGURE 3.
Choline-mediated α4β2 up-regulation corresponds with increased β2 protein expression. A, cells expressing α4β2 were treated with varying amounts of choline (30–500 μm) for 18 h before collecting crude membranes for Western blot analysis of α4 and β2 expression (see under “Experimental Procedures”). The relative band density for each subunit as a fold-change relative to the control is shown below the gels, and a best fit line for each subunit is superimposed (GraphPad version 4.03). B, comparison of the relative band density of β2 to the specific fold-change in [3H]EB and choline treatment concentration as labeled. There is a highly significant correlation between the amount of β2 subunit protein measured and the relative increase in ligand binding induced by choline. C, HC3 decreases choline-mediated β2 subunit protein and up-regulation of [3H]EB. The relative expression of α4 and β2 protein and [3H]EB binding was measured in crude membrane fractions prepared from cells treated with HC3, choline (Ch) or choline + HC3 as indicated. The results of Western blot analysis and ligand binding are shown in the respective panels. Error bars reflect mean ± S.E. determined from three to six measurements. **, p < 0.01.
Whether the choline-induced increase in β2 was sensitive to HC3 was also determined using Western blot analysis. For these measurements, crude membrane fractions prepared from cultures treated with choline, HC3, or both agents were compared with controls for both α4 and β2 subunit expression. The dramatic increase of β2 after choline treatment was partially inhibited by HC3 revealing that both the HC3-sensitive and -insensitive processes contribute to this effect (Fig. 3C). Collectively, the results support the conclusion that choline acts to promote up-regulation through enhancing β2 expression, presumably through favoring assembly with already established protein pools of α4.
Choline Complements Other Up-regulation Processes
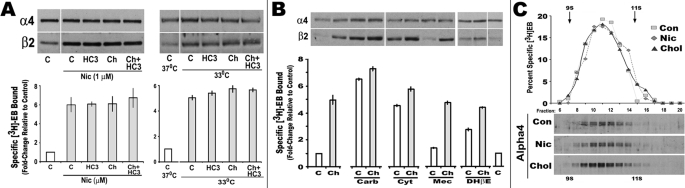
The relationship between up-regulation and increased β2 expression is documented in many systems examining up-regulation (22, 24, 35, 36). A possibility raised by these results is that HC3 could inhibit other up-regulation processes where there is an increase in β2 such as nicotine or low temperature. This was tested by co-application of choline, HC3, and choline + HC3 with either nicotine (ligand-dependent up-regulation) or by reduced culture temperatures (33 °C, ligand-independent (22)). Both nicotine and reduced temperatures increased β2 subunit expression and [3H]EB ligand binding (Fig. 4A). Similar to nicotine, but not as robust, low temperatures also increased ligand binding over 4-fold (from 2500 fm/mg (37 °C control) to 10,500 fm/mg (33 °C for 24 h). When the values were normalized to 1.0 (control) and the relative impact of choline and/or HC3 on up-regulation was measured, neither choline nor HC3 had any additional effect on up-regulation produced by either nicotine or 33 °C (Fig. 4A). No effect was observed on α4 expression in any of these treatment groups. This result indicates that the mechanism of up-regulation by choline was overlapping with these other up-regulation processes or that these other processes were saturating, and no further up-regulation was possible.
FIGURE 4.
Choline-mediated shifts in β2 expression complement up-regulation processes without altering receptor shape. A, cells were then treated with choline (Ch), HC3, and/or nicotine (Nic) as indicated. Cells placed at 33 °C were stabilized at this temperature for 1 h before treating them as labeled and then continuing culture at 33 °C for at least an additional 18 h. Western blots for α4 or β2 were prepared as shown. [3H]EB binding was measured in crude membrane fractions prepared from cells treated in parallel. The results are for the average fold-increase from three experiments where all values were normalized to the control (1.0) and then summed. Error bars reflect mean ± S.E. B, cells were treated with other drugs that interact with nicotinic receptors either at the ligand-binding site, including carbacholamine (Carb), cytosine (Cyt), dihydro-β-erythroidine (DHβE), or as an open channel blocker as for mecamylamine (Mec). Cells were then treated with either vehicle (C, saline) or choline (Ch, 250 μm) and harvested for either as indicated. Parallel cells membrane preparations measured [3H]EB binding as in A. C, sedimentation profiles on sucrose gradients were performed on cells treated with either nicotine (Nic) or choline (Chol). Crude membranes from cells treated with choline or nicotine were prepared for this analysis as described under “Experimental Procedures.” Fractions were collected across the gradient, and the specific [3H]EB binding profiles are shown. Below is the Western blot analysis for α4 protein measured in fractions in the vicinity of 9 S to 11 S as indicated. The majority of ligand binding and α4 protein was localized to the 10 S to 10.5 S fractions. Con, control.
The same experiments were repeated, but only choline (250 μm) was co-applied to cultures with other agents that impact upon up-regulation (both agonists and antagonists). This includes the open channel blocker mecamylamine that in our system produces weak (if any) up-regulation (9). In all cases, the increased β2 expression (but not α4 expression) corresponded with an increase in [3H]EB ligand binding (Fig. 4B). Addition of choline did increase up-regulation by the weaker agents, cytisine and dihydro-β-erythroidine. Similar to nicotine, choline did not increase up-regulation by carbacholamine. In the case of mecamylamine (9), which alone failed to increase either β2 protein or [3H]EB binding, choline-mediated up-regulation was similar to nontreated cells. Thus, for weaker inducers of up-regulation, choline enhances the process consistent with an increase in β2 protein production.
The possibility that choline acts differently from other agents, such as nicotine, to increase up-regulation by a mechanism that impacts upon physical parameters of the receptor, such as shape, was examined using sucrose density gradient centrifugation. Fractions were collected across the sedimentation profile, and either [3H]EB binding or Western blot analysis for α4 expression was measured and compared with similar profiles from untreated cells. As shown in Fig. 4C, the majority of immunoreactivity to α4 and corresponding [3H]EB sediment to between 9 S and 11 S (peak at ∼10 S to 10.5 S). Thus, no difference between controls, nicotine-treated or choline-treated α4β2 sedimentation profiles, occurred suggesting the complexes generated under these different treatments, despite differences in subunit expression and ligand binding density, do not vary significantly. Collectively, choline-mediated up-regulation is additive with other agents that induce this process, at least to saturation, and this does not involve an obvious change in receptor shape.
Choline-mediated Up-regulation Is Increased by TNFα
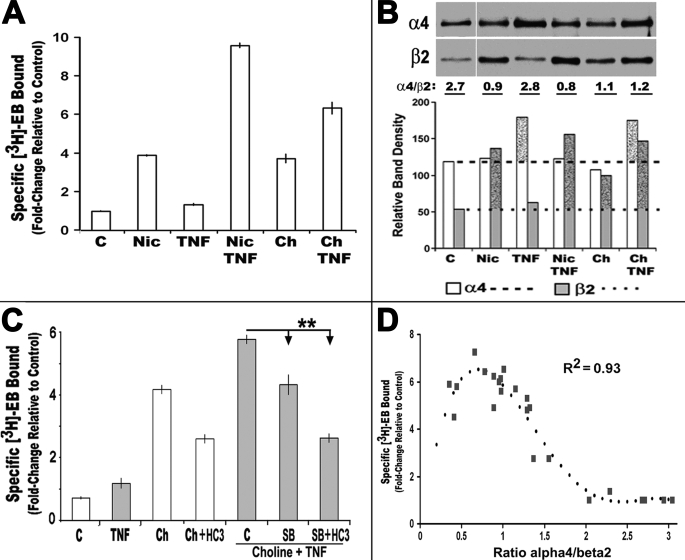
Previously, we reported that the pro-inflammatory cytokine TNFα promotes assembly of α4 + β2 receptors, and it enhances nicotine-mediated up-regulation dramatically (9). Thus, TNFα should also impact upon α4β2 up-regulation in the presence of choline. To test this possibility, we added TNFα, choline, or both agents to 293 cells expressing α4β2 nAChRs for 18 h. Choline increased [3H]EB density by ∼5-fold compared with an average 6-fold increase produced by nicotine. In both cases, combining TNFα with either nicotine or choline resulted in an enhancement of up-regulation of an additional 1.6-fold (Fig. 5A). TNFα alone had only a small impact on ligand binding of ∼1.2-fold over control values that varied among experiments. However, the overall increase to up-regulation imparted by co-application of choline and TNFα reached ∼8-fold over the control. Western blot analyses of these treatment groups (Fig. 5B) revealed that in cells treated with TNFα there was a corresponding increase in α4 rather than β2 expression. Notably, the increase in α4 protein could be observed as early as 3 h after cytokine application, and it was insensitive to HC3 (data not shown). This reveals a route to promote up-regulation where choline + TNFα simultaneously increase both α4 and β2 and consequently the subunit pool available for receptor assembly. However, the shift in α4 is not observed when nicotine is substituted for choline (Fig. 5B) in the presence of TNFα. This suggests that TNFα enhancement of up-regulation by nicotine versus choline differs because the α4 increase is best observed when TNFα is co-applied with choline. The reason for this difference has not been established, although possible explanations are discussed below.
FIGURE 5.
TNFα alone does not increase up-regulation of [3H]EB sites but enhances choline-mediated up-regulation. A, average fold-change in specific [3H]EB ligand binding by crude cell membranes from 293 cells treated with choline, nicotine, and/or TNFα, as indicated. (C, control; Nic, nicotine (1 μm); TNF, TNFα (25 nm); Ch, choline (250 μm)). B, Western blot analysis of α4 or β2 expression from crude membranes of cells treated as indicated. Cells exposed to choline increase β2 expression, and those exposed to TNFα exhibit increased protein for α4. The ratio of protein bands that is typical of multiple experiments is indicated below the gel as is the quantitative measure of band density for each subunit in each treatment group in this experiment. Dotted lines mark the respective control level of expression, and stippled bars indicate increase over control. C, TNFα enhancement of choline-mediated up-regulation is significantly inhibited by the highly specific p38 MAPK inhibitor, SB2020190 (10 μm). Choline (Ch) or choline plus TNFα (TNF)-treated cells were also treated with a combination of SB202190 (SB) and/or HC3 (**, p < 0.01). Each panel reflects results from three to eight independent experiments. D, up-regulation of α4β2 in stably transfected 293 cells is related to an optimal ratio of α4/β2 protein as measured on Western blots. Comparing this ratio with [3H]EB density collected from all treatments described in this study results in a best fit (optimal R2) for these data with the following equation: Y = −0.47 + 23.9x − 26.1x2 + 9.7x3 − 1.2x4 as calculated by the Prism version 4.03 software. Error bars are removed for clarity.
In a previous report (9), we found that the influence of TNFα on up-regulation was sensitive to inhibition of the p38 MAPK pathway. In particular, the highly specific inhibitor SB202190 essentially inhibited all TNFα enhancements of nicotine-mediated up-regulation. This same experiment was performed using cells treated with variations of choline and HC3 treatment in the presence of TNFα. For these experiments, the inhibitor SB202190 (10 μm) and/or HC3 was applied to cells 15 min in advance of choline + TNFα. The control received only SB2020190. After 18 h, the cells were harvested, and [3H]EB binding was determined. Choline-mediated up-regulation was enhanced by co-application of TNFα. The results in Fig. 5C show that SB202190 inhibited the TNFα-enhanced component of choline + TNFα up-regulation. Furthermore, co-application of SB202190 with HC3 diminished choline + TNFα up-regulation to that observed in cells treated with choline + HC3 (the HC3-independent level). This is consistent with the previous finding that TNFα impacts upon the α4β2 up-regulation mechanism through a p38 MAPK pathway. Collectively, these results reveal a unique mode of up-regulation relative to other agents because choline + TNFα has been the only condition we have examined thus far to increase both α4 and β2 simultaneously. Consequently, these results are consistent with early studies showing that TNFα promotes increased nAChR expression. Thus, combining the influence of TNFα on α4 expression with the impact of choline on β2 imparts an additive effect that would increase the total subunit pool for assembly into high affinity ligand-binding sites. However, the much stronger enhancement of up-regulation by TNFα when nicotine is present might reflect the addition of yet other processes attributed to up-regulation by this exogenous ligand (3, 22).
Subunit Protein Expression Ratio of α4 to β2 Is an Important Predictor of [3H]EB Binding in Transfected 293 Cells
As defined above, in this system up-regulation by choline and/or TNFα corresponds well to the starting subunit protein amount. For example, a strong increase in β2 subunit expression by choline and the proportional impact by HC3 appears closely related to the increase in [3H]EB up-regulation. Such a finding is reminiscent of stochastic models that describe receptor assembly from starting pools to receptor assembly to be best fit by a 4th order polynomial (36, 37). We compared the α4/β2 ratios and [3H]EB-binding site density from all of the experiments conducted in this study, and the results are shown in Fig. 5D. For this system, the best fit of the data was also a 4th order polynomial consistent with other models for nAChRs and other receptors that are assembled from heterologous starting subunit pools (36, 37). In terms of the optimal α4/β2 ratio to achieve maximum up-regulation, the relative ratio for nicotine and TNFα would be the best candidate which is 0.8. Of note is that the ratio for choline plus TNFα is consistently 1.2, and thus slightly off this optimal ratio. Also, the increase in α4 by TNFα alone could account for the slight increase observed in up-regulation, although if this is the case receptors dominated by α4 over β2 would be expected. This possibility has not been pursued, but it might be predicted to alter channel or ligand binding properties of the mature receptors under these different conditions, as proposed by others for receptors of varied stoichiometry, or leave them differentially vulnerable to shifts in affinity or other post-translational modifications.
DISCUSSION
Up-regulation of high affinity nicotine-binding sites in response to receptor ligands such as nicotine was reported over 20 years ago in studies of rodents receiving chronic nicotine (38, 39). The same process is observed in the human brain where it is a correlate of nicotine addiction (40, 41). Up-regulation of nAChRs containing α4β2 subunits contributes the majority of this response to nicotine in the mammalian brain (1–3). In this study, we demonstrate that choline is sufficient to up-regulate nAChRs consisting of α4β2 subunits. However, the mechanism(s) involved in the choline-induced expression of these receptors is distinct from other agents in that the choline kinase activity is, in part, responsible.
Most agents enhance processes of up-regulation through occupying the receptor ligand-binding site, and this may also include a corresponding increase in the β2 protein expression (3, 4, 24, 42). However, some forms of up-regulation vary dramatically from this generalization such as up-regulation by reduced temperature (22, 43). This process proceeds independently of ligand binding, although β2 protein is increased (e.g. Fig. 4). In terms of ligand binding, although choline can act as an agonist or antagonist of other nAChRs (3), we are unaware of any data suggesting choline binding to nAChRs with sufficient affinities to produce the effects reported here. In terms of function, choline can inhibit native hippocampal α4β2 (type II) nAChRs very weakly (IC50 of 370 μm), but it has no effect when these receptors are expressed by cultured hippocampal neurons (44). Also, in terms of competition assays against [3H]EB conducted on the 293 cell lines used here (16, 26), choline exhibits an apparent Ki of 35 μm, which is 3 to 4 orders of magnitude less than acetylcholine or nicotine, respectively. Our results do show that choline-induced up-regulation of α4β2 receptors does involve an increase in β2 protein expression (Fig. 3). This result is further supported by the observation that the increase in β2 protein is directly related to up-regulation of ligand binding density (Fig. 3) and that this relationship includes the ratio of α4 and β2 subunits (Fig. 5D). The data in Fig. 5D fit well with other studies that have proposed an equilibrium model of assembly where the preferred assembly of nAChRs into pentamers is through an orderly process determined by subunit availability and preference of subunit interactions that optimize the stoichiometry (3, 4, 45). In this scenario, at least two α subunits (i.e. α4) are incorporated at intervals that separate them by at least two β2 subunits before the fifth subunit (in this case it could be either α4 or β2) is added to close the pentamer. This could lead to nAChRs of different stoichiometries as well as structural differences that would lend themselves to altered ligand affinity under different measurement conditions (4, 22). These possibilities were not examined in this study. What is suggested is that although a rather simple subunit ratio appears to correspond to the optimal outcome for formation of nAChRs that bind ligand with sufficient affinity to be measured in this cell system, it is highly probable that this parameter is only one of several that contribute to optimal nAChR assembly and expression (3, 4).
The expression of different nAChRs is also likely to contribute to cell-specific responses to choline and the inflammatory environment. For example, when other subunits such as α3 or β4 are present, an altered up-regulation response to choline is expected. As shown in Fig. 1 and reported elsewhere (8, 9), both α3 + β4 receptors tend to exhibit a dampened up-regulation response. Also, the β subunit does impact this process because co-expression of β4 with α4 and β2 significantly alters the impact cytokines, such as TNFα or interleukin-1β, have on modifying the final subunit composition of assembled receptors (8, 9). Because both α3 and β4 are prominently expressed in peripheral systems such as autonomic ganglia, this would suggest that they would buffer these cells against the fluctuations in ligand and cytokines common to this environment. Another place of interest in terms choline impact on up-regulation would be the basal ganglia. In these structures the high affinity nicotine-binding sites include co-assembly among α4, β2, α6, and β3 subunits (4, 22). These receptors undergo up-regulation responses that are very different from those described for α4β2 and in some cases may actually include decreased ligand binding density. Thus, how choline and/or TNFα would impact upon this system is unknown. A final speculation regarding the influence of other nAChRs is the role of α7 receptors. In addition to modulating neurotransmission, this nAChR also reduces TNFα signaling (19, 46–48). In a more complete cell system, this leaves the possibility open that α7 (activated by choline), TNFα, and choline-mediated up-regulation of α4β2 are interactive under normal physiological conditions. In any case, the nAChR-inflammatory interaction is likely to be more involved than previously thought.
A significant issue remains regarding whether this mechanism is relevant to the in vivo system. Unfortunately, these measurements are complicated by the inability to control or be certain of local choline concentrations. Although reports of dietary choline inducing up-regulation of α4β2-type receptors in the adult rodent brain exist (49), as well as other nAChRs (50), and choline concentrations in cerebral spinal fluid reach 7 mm (51), the availability of free choline concentrations near receptors is likely to be far less. Adult animals express high affinity choline transporters whose relative spatio-temporal distributions vary considerably among different brain regions as do the enzymes important for choline production and catabolism (21, 52–54). In addition to this, nAChR subtype expression can vary considerably among adjacent cells or differing brain regions and tissues. Thus, the local response in terms of nicotinic receptor function would be anticipated to be highly customized. In fact, where free choline levels may be most relevant to nAChR expression could be during pre-natal development when the high affinity choline transporters are not widely expressed (54), and the ratio of the HC3-sensitive to -insensitive components of choline kinase activity differs considerably from that measured in the adult (21). The impact of pre-natal dietary choline supplementation and its ability to impart long term changes in brain function, and cholinergic and neurotransmitter “tone,” may not appear until much later in the life (for examples see Refs. 55–58). Whether long term modifications of α4β2 expression contribute to these mechanisms is unknown. However, in this context an immature choline uptake system coupled with the expanding role of pro-inflammatory cytokines such as TNFα could have a notable impact on regulating this system during development (59, 60). Therefore, in addition to the many extended roles in physiology that choline and the pro-inflammatory environment have, a modulatory role on nAChRs is suggested. Such effects would differ in magnitude and would impact on this system throughout life.
This work was supported, in whole or in part, by National Institutes of Health Grants AG017517 (to S. W. R.), AG029838 (to L. C. G.), and DA025057 (to L. C. G. and S. W. R.). This work was also supported by a Veterans Affairs Merit award (to L. C. G.).
- nAChR
- nicotinic acetylcholine receptor
- HC3
- hemicholinium-3
- EB
- epibatidine
- TNF
- tumor necrosis factor.
REFERENCES
- 1.Flores C. M., Rogers S. W., Pabreza L. A., Wolfe B. B., Kellar K. J. (1992) Mol. Pharmacol. 41, 31–37 [PubMed] [Google Scholar]
- 2.Mao D., Perry D. C., Yasuda R. P., Wolfe B. B., Kellar K. J. (2008) J. Neurochem. 104, 446–456 [DOI] [PubMed] [Google Scholar]
- 3.Albuquerque E. X., Pereira E. F., Alkondon M., Rogers S. W. (2009) Physiol. Rev. 89, 73–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Govind A. P., Vezina P., Green W. N. (2009) Biochem. Pharmacol. 78, 756–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buisson B., Bertrand D. (2001) J. Neurosci. 21, 1819–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vallejo Y. F., Buisson B., Bertrand D., Green W. N. (2005) J. Neurosci. 25, 5563–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alkondon M., Albuquerque E. X. (2005) J. Pharmacol. Exp. Ther. 313, 740–750 [DOI] [PubMed] [Google Scholar]
- 8.Gahring L. C., Days E. L., Kaasch T., González de Mendoza M., Owen L., Persiyanov K., Rogers S. W. (2005) J. Neuroimmunol. 166, 88–101 [DOI] [PubMed] [Google Scholar]
- 9.Gahring L. C., Osborne-Hereford A. V., Vasquez-Opazo G. A., Rogers S. W. (2008) J. Biol. Chem. 283, 693–699 [DOI] [PubMed] [Google Scholar]
- 10.Guseva M. V., Hopkins D. M., Scheff S. W., Pauly J. R. (2008) J. Neurotrauma 25, 975–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanders L. M., Zeisel S. H. (2007) Nutr. Today 42, 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeisel S. H. (2000) J. Am. Coll. Nutr. 19, 528S–531S [DOI] [PubMed] [Google Scholar]
- 13.Mandelzys A., De Koninck P., Cooper E. (1995) J. Neurophysiol. 74, 1212–1221 [DOI] [PubMed] [Google Scholar]
- 14.Alkondon M., Pereira E. F., Cortes W. S., Maelicke A., Albuquerque E. X. (1997) Eur. J. Neurosci. 9, 2734–2742 [DOI] [PubMed] [Google Scholar]
- 15.Alkondon M., Pereira E. F., Eisenberg H. M., Albuquerque E. X. (1999) J. Neurosci. 19, 2693–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao Y., Kellar K. J. (2004) J. Pharmacol. Exp. Ther. 310, 98–107 [DOI] [PubMed] [Google Scholar]
- 17.Detopoulou P., Panagiotakos D. B., Antonopoulou S., Pitsavos C., Stefanadis C. (2008) Am. J. Clin. Nutr. 87, 424–430 [DOI] [PubMed] [Google Scholar]
- 18.Rivera C. A., Wheeler M. D., Enomoto N., Thurman R. G. (1998) Am. J. Physiol. 275, G862–G867 [DOI] [PubMed] [Google Scholar]
- 19.Carlson N. G., Bacchi A., Rogers S. W., Gahring L. C. (1998) J. Neurobiol. 35, 29–36 [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez-González A., Ramírez de Molina A., Benítez-Rajal J., Lacal J. C. (2003) Prog. Cell Cycle Res. 5, 191–201 [PubMed] [Google Scholar]
- 21.Burt A. M. (1977) J. Neurochem. 28, 961–966 [DOI] [PubMed] [Google Scholar]
- 22.Walsh H., Govind A. P., Mastro R., Hoda J. C., Bertrand D., Vallejo Y., Green W. N. (2008) J. Biol. Chem. 283, 6022–6032 [DOI] [PubMed] [Google Scholar]
- 23.Nelson M. E., Kuryatov A., Choi C. H., Zhou Y., Lindstrom J. (2003) Mol. Pharmacol. 63, 332–341 [DOI] [PubMed] [Google Scholar]
- 24.Corringer P. J., Sallette J., Changeux J. P. (2006) J. Physiol. 99, 162–171 [DOI] [PubMed] [Google Scholar]
- 25.Sallette J., Pons S., Devillers-Thiery A., Soudant M., Prado de Carvalho L., Changeux J. P., Corringer P. J. (2005) Neuron 46, 595–607 [DOI] [PubMed] [Google Scholar]
- 26.Xiao Y., Baydyuk M., Wang H. P., Davis H. E., Kellar K. J. (2004) Bioorg. Med. Chem. Lett. 14, 1845–1848 [DOI] [PubMed] [Google Scholar]
- 27.Lockman P. R., Allen D. D. (2002) Drug. Dev. Ind. Pharm. 28, 749–771 [DOI] [PubMed] [Google Scholar]
- 28.Aoyama C., Liao H., Ishidate K. (2004) Prog. Lipid Res. 43, 266–281 [DOI] [PubMed] [Google Scholar]
- 29.Apparsundaram S., Ferguson S. M., George A. L., Jr., Blakely R. D. (2000) Biochem. Biophys. Res. Commun. 276, 862–867 [DOI] [PubMed] [Google Scholar]
- 30.Ribeiro F. M., Alves-Silva J., Volknandt W., Martins-Silva C., Mahmud H., Wilhelm A., Gomez M. V., Rylett R. J., Ferguson S. S., Prado V. F., Prado M. A. (2003) J. Neurochem. 87, 136–146 [DOI] [PubMed] [Google Scholar]
- 31.Campos J., Núñez M. C., Conejo-García A., Sánchez-Martín R. M., Hernández-Alcoceba R., Rodríguez-González A., Lacal J. C., Gallo M. A., Espinosa A. (2003) Curr. Med. Chem. 10, 1095–1112 [DOI] [PubMed] [Google Scholar]
- 32.Michel V., Yuan Z., Ramsubir S., Bakovic M. (2006) Exp. Biol. Med. 231, 490–504 [DOI] [PubMed] [Google Scholar]
- 33.Abreu-Villaca Y., Filgueiras C. C., Manhaes A. C. (2010) Behav. Brain Res., [DOI] [PubMed] [Google Scholar]
- 34.Janardhan S., Srivani P., Sastry G. N. (2006) Curr. Med. Chem. 13, 1169–1186 [DOI] [PubMed] [Google Scholar]
- 35.Harkness P. C., Millar N. S. (2002) J. Neurosci. 22, 10172–10181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.López-Hernández G. Y., Sánchez-Padilla J., Ortiz-Acevedo A., Lizardi-Ortiz J., Salas-Vincenty J., Rojas L. V., Lasalde-Dominicci J. A. (2004) J. Biol. Chem. 279, 38007–38015 [DOI] [PubMed] [Google Scholar]
- 37.Brorson J. R., Li D., Suzuki T. (2004) J. Neurosci. 24, 3461–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz R. D., Kellar K. J. (1985) J. Neurochem. 45, 427–433 [DOI] [PubMed] [Google Scholar]
- 39.Marks M. J., Collins A. C. (1985) Pharmacol. Biochem. Behav. 22, 283–291 [DOI] [PubMed] [Google Scholar]
- 40.Benwell M. E., Balfour D. J., Anderson J. M. (1988) J. Neurochem. 50, 1243–1247 [DOI] [PubMed] [Google Scholar]
- 41.Perry D. C., Dávila-García M. I., Stockmeier C. A., Kellar K. J. (1999) J. Pharmacol. Exp. Ther. 289, 1545–1552 [PubMed] [Google Scholar]
- 42.Sallette J., Bohler S., Benoit P., Soudant M., Pons S., Le Novère N., Changeux J. P., Corringer P. J. (2004) J. Biol. Chem. 279, 18767–18775 [DOI] [PubMed] [Google Scholar]
- 43.Cooper S. T., Harkness P. C., Baker E. R., Millar N. S. (1999) J. Biol. Chem. 274, 27145–27152 [DOI] [PubMed] [Google Scholar]
- 44.Alkondon M., Albuquerque E. X. (2006) J. Pharmacol. Exp. Ther. 318, 268–275 [DOI] [PubMed] [Google Scholar]
- 45.Tapia L., Kuryatov A., Lindstrom J. (2007) Mol. Pharmacol. 71, 769–776 [DOI] [PubMed] [Google Scholar]
- 46.Giebelen I. A., van Westerloo D. J., LaRosa G. J., de Vos A. F., van der Poll T. (2007) Shock 6, 700–703 [DOI] [PubMed] [Google Scholar]
- 47.Wang H., Yu M., Ochani M., Amella C. A., Tanovic M., Susarla S., Li J. H., Wang H., Yang H., Ulloa L., Al-Abed Y., Czura C. J., Tracey K. J. (2003) Nature 421, 384–388 [DOI] [PubMed] [Google Scholar]
- 48.Gahring L. C., Rogers S. W. (2005) AAPS Journal 7, E885–E894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coutcher J. B., Cawley G., Wecker L. (1992) J. Pharmacol. Exp. Ther. 262, 1128–1132 [PubMed] [Google Scholar]
- 50.Guseva M. V., Hopkins D. M., Pauly J. R. (2006) Pharmacol. Biochem. Behav. 84, 26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein J., Gonzalez R., Köppen A., Löffelholz K. (1993) Neurochem. Int. 22, 293–300 [DOI] [PubMed] [Google Scholar]
- 52.Zahalka E. A., Seidler F. J., Lappi S. E., Yanai J., Slotkin T. A. (1993) Brain Res. 601, 221–229 [DOI] [PubMed] [Google Scholar]
- 53.Happe H. K., Murrin L. C. (1993) J. Neurochem. 60, 1191–1201 [DOI] [PubMed] [Google Scholar]
- 54.Klein J., Weichel O., Ruhr J., Dvorak C., Löffelholz K. (2002) J. Neurochem. 80, 843–849 [DOI] [PubMed] [Google Scholar]
- 55.Meck W. H., Smith R. A., Williams C. L. (1989) Behav. Neurosci. 103, 1234–1241 [DOI] [PubMed] [Google Scholar]
- 56.Cermak J. M., Holler T., Jackson D. A., Blusztajn J. K. (1998) FASEB J. 12, 349–357 [DOI] [PubMed] [Google Scholar]
- 57.Montoya D. A., White A. M., Williams C. L., Blusztajn J. K., Meck W. H., Swartzwelder H. S. (2000) Brain Res. Dev. Brain Res. 123, 25–32 [DOI] [PubMed] [Google Scholar]
- 58.Slotkin T. A., Seidler F. J., Qiao D., Aldridge J. E., Tate C. A., Cousins M. M., Proskocil B. J., Sekhon H. S., Clark J. A., Lupo S. L., Spindel E. R. (2005) Neuropsychopharmacology 30, 129–144 [DOI] [PubMed] [Google Scholar]
- 59.Matsumoto M. (1999) J. Med. Invest. 46, 141–150 [PubMed] [Google Scholar]
- 60.Melnick M., Chen H., Zhou Y., Jaskoll T. (2001) Anat. Rec. 262, 318–330 [DOI] [PubMed] [Google Scholar]