Abstract
MTA1 (metastasis-associated protein 1), an integral component of the nucleosome remodeling and deacetylase complex, has recently been implicated in the ionizing radiation-induced DNA damage response. However, whether MTA1 also participates in the UV-induced DNA damage checkpoint pathway remains unknown. In response to UV radiation, ATR (ataxia teleangiectasia- and Rad3-related) is the major kinase activated that orchestrates cell cycle progression with DNA repair machinery by phosphorylating and activating a number of downstream substrates, such as Chk1 (checkpoint kinase 1) and H2AX (histone 2A variant X). Here, we report that UV radiation stabilizes MTA1 in an ATR-dependent manner and increases MTA1 binding to ATR. On the other hand, depletion of MTA1 compromises the ATR-mediated Chk1 activation following UV treatment, accompanied by a marked down-regulation of Chk1 and its interacting partner Claspin, an adaptor protein that is required for the phosphorylation and activation of Chk1 by ATR. Furthermore, MTA1 deficiency decreases the induction of phosphorylated H2AX (referred to as γ-H2AX) and γ-H2AX focus formation after UV treatment. Consequently, depletion of MTA1 results in a defect in the G2-M checkpoint and increases cellular sensitivity to UV-induced DNA damage. Thus, MTA1 is required for the activation of the ATR-Claspin-Chk1 and ATR-H2AX pathways following UV treatment, and the noted abrogation of the DNA damage checkpoint in the MTA1-depleted cells may be, at least in part, a consequence of dysregulation of the expression of these two pathways. These findings suggest that, in addition to its role in the repair of double strand breaks caused by ionizing radiation, MTA1 also participates in the UV-induced ATR-mediated DNA damage checkpoint pathway.
Keywords: Checkpoint Control, Chromatin Remodeling, DNA Damage, DNA Repair, PI 3-Kinase, Protein-Protein Interactions
Introduction
MTA1 (metastasis-associated protein 1), the founding member of the MTA2 family, is widely up-regulated in human cancers and plays an important role in tumorigenesis, tumor invasion, and metastasis (1–3). As a dual function coregulator (3, 4), MTA1 functions not only as a transcriptional repressor of estrogen receptor-α (5), BRCA1 (breast cancer type 1 susceptibility protein) (6), Six3 (7), and p21WAF11 (8) genes but also as a transcriptional activator via interacting with RNA polymerase II on the BCAS3 (breast cancer-amplified sequence 3) (9) and Pax5 (paired box gene 5) (10) promoters. The co-repressor versus co-activator activity of MTA1 might be influenced by its binding partners on the promoter region of various genes. In addition to its paramount role in cancer and coregulator biology, emerging evidence suggests that MTA1 is a DNA damage-responsive protein and facilitates DNA double strand break (DSB) repair following ionizing radiation (IR) treatment (11, 12). In support of these findings, recent studies have demonstrated that MTA1 regulates p53-dependent and -independent DNA repair processes following IR treatment by modulating p53-p53R2 and p21WAF1-proliferating cell nuclear antigen pathways, respectively (8, 13). However, the new functions and related signaling transduction pathways of MTA1 remain to be further explored.
Interestingly, an earlier study revealed that MTA1 could co-immunoprecipitate with ATR (ataxia teleangiectasia- and Rad3-related protein) (14), one of the key regulators for transducing DNA damage signals to checkpoint control proteins (15–17). It is well established that ATR is activated by stalled replication forks and agents that produce bulky adducts, such as UV radiation, whereas ATM (ataxia teleangiectasia mutated) is activated in response to DSB-causing agents, such as IR (15, 17, 18). Indeed, ATR exhibited higher affinity to UV-damaged DNA than undamaged DNA (19). Once activated, ATR coordinates cell cycle progression with DNA repair machinery by phosphorylating and activating a number of downstream substrates (15, 17, 18, 20).
Although the list of ATR substrates is rapidly expanding due to large scale proteomic profiling methodologies (21–24), the best studied is the serine/threonine kinase Chk1 (checkpoint kinase 1), an evolutionarily conserved protein kinase that amplifies ATR signaling and directs it to the desired cell cycle and DNA repair effectors (25–29). Chk1 is phosphorylated at Ser345 and Ser317 by ATR kinase in response to stalled replication and genotoxic stresses, and these phosphorylations are critical for Chk1 activation (17, 25, 30–32). In addition, evidence has been presented that a number of factors act in concert with ATR to facilitate Chk1 phosphorylation, including ATR-interacting protein (ATRIP), Rad17-replication factor C, Rad9-Hus1-Rad1 (9-1-1) complex, TopBP1 (topoisomerase II-binding protein 1), and Claspin (15). Once activated, Chk1 phosphorylates Cdc25 and blocks its ability to activate Cdc2, thereby preventing cell cycle transition through the G2-M boundary (33–35). Such a rapid and reversible cell cycle arrest is critical to allow successful completion of DNA repair (17, 36). In fact, loss or inhibition of Chk1 leads to inability to shut down the cell cycle progression and, accordingly, allows cells to progress into mitosis before completion of DNA repair (37). This, in turn, results in high accidence of mitotic catastrophe and increased sensitivity of cancer cells to various genotoxic drugs (38, 39).
In addition to Chk1, another key substrate of ATR is H2AX. H2AX is a core histone variant distributed throughout chromosomes and is required for genomic stability and cellular survival after DNA damage (40, 41). It is phosphorylated on its C terminus (referred to as γ-H2AX) at sites of DNA damage and surrounding megabase regions (42), which is associated with the recruitment of repair factors to damaged DNA. Phosphorylation of H2AX is dependent on ATM after IR (43) and on ATR during replication stress induced by UV (44, 45). In support of this notion, overexpression of kinase-inactive ATR inhibits the phosphorylation and formation of foci of H2AX upon treatment with UV (44).
The association of MTA1 with ATR (14) and the key role of ATR in the cellular response to UV radiation (17, 18) indicate that MTA1 may play a role in the UV-induced DNA damage response in mammalian cells and contribute to the regulation of DNA damage checkpoints. Here, we report that MTA1 is stabilized in response to UV radiation in an ATR-dependent manner. On the other hand, UV-induced activation of ATR kinase requires MTA1. As a result, depletion of MTA1 compromises the ATR-mediated Chk1 activation following UV treatment, accompanied by a marked down-regulation of Chk1 and its interacting partner Claspin, an adaptor protein that mediates the phosphorylation and activation of Chk1 by ATR (46–48). Furthermore, we found that deficiency of MTA1 results in a defect in G2-M checkpoints, decreased γ-H2AX induction, and increased cellular sensitivity to UV. Thus, our findings suggest that, in addition to its role in the repair of DSBs caused by IR, MTA1 also participates in UV-induced ATR-mediated DNA damage checkpoint pathway.
EXPERIMENTAL PROCEDURES
Cell Culture, Mice, and UV Radiation
Human osteosarcoma cell line U2OS, lung carcinoma cell line A549, cervix adenocarcinoma cell line HeLa, keratinocyte cell line 1102, and mouse epidermal cell line JB6 were obtained from the ATCC (Manassas, VA). The spontaneously transformed human keratinocyte cell line HaCaT was maintained in our laboratory, and the in vitro characteristics and cytogenetic patterns of this cell line have been described in detail elsewhere (49). Doxycycline-inducible U2OS cells expressing wild-type (ATRWT) (clone GK33) or kinase-dead (ATRKD) (clone GK41) ATR (50, 51) were provided by Dr. Paul Nghiem (University of Washington, Seattle, WA). Wild-type (MTA1+/+) and MTA1 knock-out (MTA1−/−) mouse embryonic fibroblasts (MEFs) were generated in our laboratory from embryos at day 9 of development by using a standard protocol (7).
Keratinocyte cell line 1102 was maintained in the keratinocyte serum-free medium (Invitrogen) supplemented with 50 μg/ml bovine pituitary extract and 5 ng/ml epidermal growth factor (Invitrogen). ATRWT or ATRKD cells were grown in Dulbecco's modified Eagle's medium/F-12 medium containing 10% fetal bovine serum (Hyclone, Logan, UT), 200 μg/ml Geneticin (Invitrogen), and 50 μg/ml hygromycin (Calbiochem) for selective pressure of the transfected constructs (50). To induce ATR expression, doxycycline (Sigma) was added to a final concentration of 1 μg/ml for 48 h prior to assay. All of the other cell lines were maintained in the recommended media by the providers in a humidified 5% CO2 at 37 °C. Cell culture medium and additives were obtained from Invitrogen if not otherwise stated.
For UV radiation, cells were washed twice with prewarmed phosphate-buffered saline and then exposed to a 254-nm wavelength UV source using a Stratagene Stratalinker (Stratagene, La Jolla, CA). The culture dishes were replenished with fresh culture medium immediately after irradiation and incubated for the indicated periods of time. Control cells were subjected to the identical procedure without being UV-exposed. For cycloheximide chase assays, cells were treated with 100 μg/ml cycloheximide (Sigma) and harvested at the indicated time points for Western blot analysis. For in vivo experiments, 1 day prior to irradiation, the dorsal hair of mice was shaved, and the age- and sex-matched MTA1+/+ and MTA1−/− mice were subjected to whole body irradiation using a Stratalinker UV cross-linker (Stratagene). Mice were killed at different time points after irradiation, and tissue samples were harvested for further experiments. All of the mice were maintained under standard conditions, and all animal protocols followed the guidelines of the Institutional Animal Care and Use Committee.
siRNA and Transfections
ON-TARGETplus SMARTpool siRNAs targeting human MTA1 (catalog no. M-004127-00-0005), human ATR (catalog no. L-003202-00-0005), ON-TARGETplus non-targeting control siRNA (catalog no. D-001810-10-05), siGENOME SMARTpool siRNA targeting MTA1 (catalog no. M-004127-02-0005), and siGENOME non-targeting control siRNA (catalog no. D-001210-01-05) were obtained from Dharmacon (Lafayette, CO). The transfection of siRNA was performed twice at 24-h intervals with Oligofectamine reagent (Invitrogen) according to the manufacturer's protocol. Cells were collected after 48–72 h of transfection for further analyses.
Antibodies and Chemicals
Rabbit polyclonal anti-phospho-ATR Ser428, anti-phospho-Chk1 Ser345 (133D3), anti-phosphohistone H3 (Ser10), anti-ATR, anti-ATRIP, anti-Claspin, and mouse monoclonal anti-ubiquitin (P4D1) were obtained from Cell Signaling Technology (Danvers, MA). Goat polyclonal anti-ATR (N-19), anti-TopBP1, anti-MTA1 (C17), anti-Rad9 (C-20), mouse monoclonal anti-Chk1 (G-4), anti-Hus1 (G-3), rabbit polyclonal anti-Mi-2 (H-242), anti-Rad17 (H-300), and anti-ATRIP (H-300) were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Rabbit polyclonal anti-MTA1, anti-MTA2, and anti-MTA3 were obtained from Bethyl Laboratories (Montgomery, TX). Mouse monoclonal anti-phosphohistone H2AX (Ser139) and rabbit polyclonal anti-H2AX were obtained from Upstate Biotechnology, Inc. (Lake Placid, NY). Mouse monoclonal anti-FLAG (M2) and anti-vinculin (clone hVIN-1) antibodies were purchased from Sigma. Horseradish peroxidase-conjugated secondary antibodies and ECL reagents were obtained from Amersham Biosciences. All of the primary antibodies were used following the manufacturer's instructions. Micrococcal nuclease (or S7 nuclease) was obtained from Roche Applied Science. All reagents were obtained from Sigma unless otherwise stated. Cycloheximide was resuspended in DMSO, and doxycycline was resuspended at 1 mg/ml in an ethanol/water mix (1:1).
Clonogenic Survival Assay
Cells were irradiated at doses of 50, 100, and 250 mJ/cm2 from a 254-nm wavelength UV source (Stratagene). After UV exposure, cells were plated in quadruplicate at five different cell concentrations, 200, 300, 400, 800, and 1200 cells/60-mm2 dish, to obtain a significant number of colonies at all doses. After 14–21 days, the formed colonies were stained with 0.5% crystal violet in 70% ethanol and quantified. Survival was calculated as a percentage of colonies formed relative to untreated controls.
Cell Cycle Analysis and Phosphohistone H3 Staining
MTA1+/+ or MTA1−/− MEFs were treated with or without UV, harvested at the indicated time points by trypsinization, and fixed with 70% ethanol. After centrifugation, the cell pellet was suspended in 100 μl of PBS containing 0.5% bovine serum albumin and 1:50 diluted phosphohistone H3 (Ser10) antibody, followed by staining with fluorescein isothiocyanate-labeled secondary antibody as described previously (52). After 1 h of incubation at room temperature, the cells were stained with 50 μg/ml propidium iodide and 10 μg/ml ribonuclease A. Cellular fluorescence was measured by a BD Biosciences FACScan, and the data were analyzed using Cell Quest software.
Quantitative Real-time PCR
Total RNA was isolated by using TRIzol reagent (Invitrogen), and a 2-μg aliquot of the total RNA was reverse-transcribed by using the SuperScript III first strand synthesis system for reverse transcription-PCR (Invitrogen). Quantitative real-time PCR was done by using iQTM SYBR Green Supermix (Bio-Rad) on an iCycler iQ real-time PCR detection system (Bio-Rad). The values for specific genes were normalized to actin housekeeping controls. Primers for human MTA1 were 5′-TGCTCAACGGGAAGTCCTACC-3′ (forward) and 5′-GGGCATGTAGAACACGTCACC-3′ (reverse); for human actin, primers were 5′-TCCCTGGAGAAGAGCTACGA-3′ (forward) and 5′-GTACTTGCGCTCAGGAGGAG-3′ (reverse).
Western blot analysis, immunoprecipitation (IP), indirect immunofluorescent staining, and pulse-chase experiments using metabolic labeling of cells with [35S]methionine (11, 13), EtBr, and micrococcal nuclease treatment of cleared lysates (54) have been described previously in detail.
RESULTS AND DISCUSSION
Induction of MTA1 Protein in Vitro and in Vivo following UV Radiation
Exposure of mammalian cells to UV radiation leads to activation of a large number of proteins, such as p53 and NF-κB by signaling cascades, generally known as UV response (55, 56). The interaction between MTA1 and ATR (14) and the key role of ATR in the cellular response to UV irradiation (17, 18) promoted us to investigate the possible role of MTA1 protein in the UV-induced DNA damage response.
Because UV radiation has a variety of effects on the skin associated with carcinogenesis, including DNA damage and effects on signal transduction (57), we initially characterized the kinetics and dose dependence of UV induction of MTA1 in human keratinocyte cell lines HaCaT (49) and 1102 and murine epidermal cell line JB6 by Western blot analysis. We found that exposure of various cell lines with increasing doses of UV resulted in a marked increase in the protein levels of MTA1, but not MTA2 and MTA3 (Fig. 1A). In agreement with previous report (58), UV radiation also increases the protein expression of Mi-2, another core subunit of the Mi-2·NuRD complex (3, 59, 60). We further demonstrated that this effect is not cell type-specific, because the increase of MTA1 protein following UV treatment was also observed in the MEFs (Fig. 1B), human U2OS osteosarcoma cells, and A549 lung adenocarcinoma cells (Fig. 1C). Interestingly, we found that MTA1 protein levels were increased, whereas MTA3 protein levels were decreased following UV treatment in MEFs (Fig. 1B). Previous study has demonstrated that transient overexpression of MTA1 in the MCF7 and T47D breast cancer cell lines with MTA1-expressing adenovirus leads to dramatic decreases in MTA3 protein as well as MTA2 protein (61). It was proposed that transient overexpression of MTA1 leads to a shift in the composition of the cellular pool of MTA family members from predominantly MTA2 and MTA3 to predominantly MTA1 (61). In our study, we also found that knock-out of MTA1 led to an increase in MTA3 protein levels in the MTA1−/− MEFs as compared with its wild-type controls (see Fig. 4D). One possible explanation is that there is cross-talk between MTA family members, and the underlying mechanism for this observation remains to be determined.
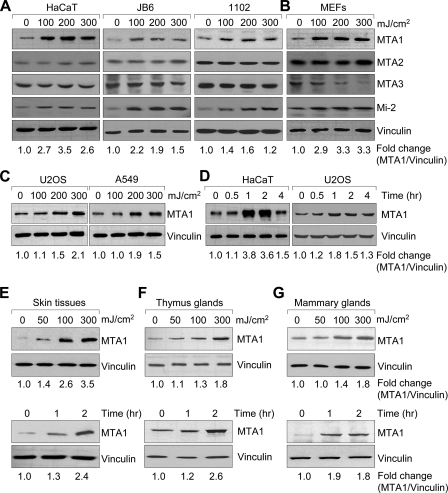
FIGURE 1.
MTA1 is induced following exposure to UV. A–C, cells were irradiated or mock-irradiated with UV at the indicated doses and harvested after 2 h of exposure for Western blot analysis with the indicated antibodies. The expression of vinculin was used as the loading control. D, HaCaT and U2OS cells were treated with 200 mJ/cm2 UV and harvested at the indicated time points for Western blot analysis using the indicated antibodies. E–G, whole-body mice were irradiated or mock-irradiated with UV at the indicated doses, and skin tissues (E), thymus glands (F), and mammary glands (G) were collected at the indicated time points for Western blot analysis with the antibody against MTA1 or vinculin. These experiments were repeated three times, and a representative blot is shown. The fold change (MTA1/vinculin) is shown at the bottom.
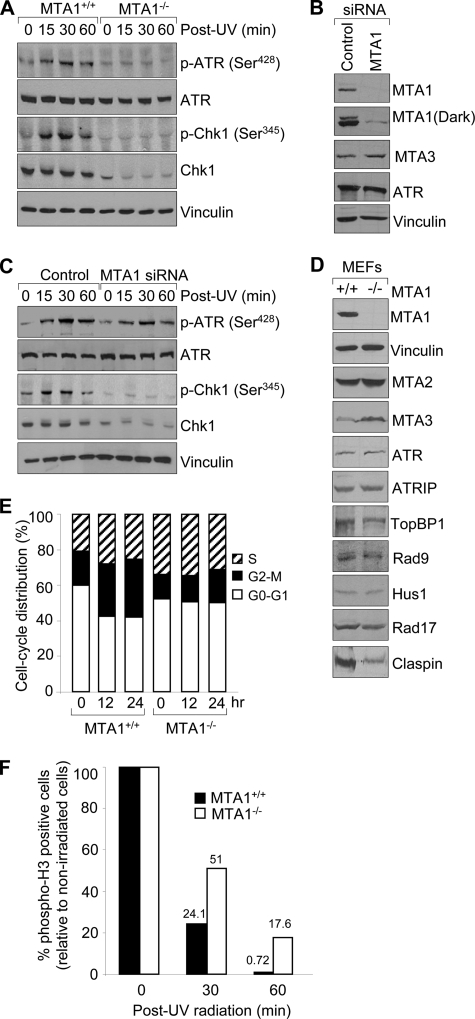
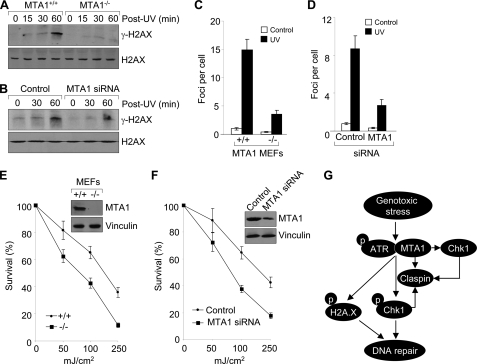
FIGURE 4.
Depletion of MTA1 results in decreased activation of ATR-Chk1 pathway and a defect of G2/M checkpoint. A, MTA1+/+ or MTA1−/− MEFs were treated with 200 mJ/cm2 UV and harvested at the indicated time points for Western blot analysis with the indicated antibodies. B, HaCaT cells were transfected with control siRNAs or specific siRNAs targeting MTA1 and then subjected to Western blot analysis with the indicated antibodies. C, control siRNA- or MTA1 siRNA-transfected HaCaT cells were exposed to 200 mJ/cm2 UV and harvested at the indicated time points for Western blot as described above. D, Western blot analysis of MTA1+/+ and MTA1−/− MEFs using the corresponding specific antibodies. E, MTA1+/+ or MTA1−/− MEFs were treated with 200 mJ/cm2 UV and harvested at the indicated time points for fluorescence-activated cell sorting analysis. The graph data represent the average values of cell cycle distribution from two independent experiments. F, MTA1+/+ or MTA1−/− MEFs were treated with or without 200 mJ/cm2 UV and harvested at the indicated time points. Cells were stained with anti-phosphohistone H3 (Ser10) antibody and then subjected to fluorescence-activated cell sorting analysis. The percentage of phospho-H3-positive cells in UV-treated cells was normalized to that in non-irradiated cells, and the average values of the results of two independent experiments are shown. p-ATR, phospho-ATR; p-Chk1, phospho-Chk1.
Following these observations, we next performed time course analysis of MTA1 induction in the HaCaT and U2OS cells and found that MTA1 protein levels were increased within 30 min and maximized at 1–2 h after UV treatment (Fig. 1D). We next examined whether MTA1 protein levels were affected by UV radiation in a whole animal setting. To conduct these studies, whole-body mice were exposed to increasing doses of UV, and tissue samples were harvested at the indicated time points for Western blot analysis of MTA1 protein. Consistent with in vitro observations, we found that MTA1 protein was also dramatically increased after UV treatment in all tissues that were examined, including skin tissues (Fig. 1E), thymus glands (Fig. 1F), and mammary glands (Fig. 1G), in a dose- and time-dependent manner. Taken together, these results suggest that MTA1 protein is induced following UV radiation in vitro and in vivo.
UV Radiation Stabilizes MTA1 by Inhibiting Its Ubiquitination
To determine the mechanism by which UV induces MTA1 protein expression, the levels of MTA1 transcripts after UV exposure were measured using a quantitative real-time PCR approach. To do this, HaCaT cells were exposed to UV and harvested at the indicated time points for quantitative real-time PCR analysis. As shown in supplemental Fig. S1, we found no discernible difference in the expression levels of MTA1 mRNA in the HaCaT cells with or without treatment of increasing doses of UV at all time points, indicating that induction of MTA1 protein might be through a posttranslational mechanism. To test this notion, we monitored the effect of cycloheximide, an inhibitor of protein biosynthesis in eukaryotic organisms (62), on the rate of decline of the endogenous MTA1 in UV-treated cells versus controls. We found that UV radiation led to a marked increase in the half-life of MTA1 in UV-treated cells in comparison with unirradiated controls (Fig. 2A). To further confirm the above findings, we performed the classic pulse-chase experiments using metabolic labeling of cells with [35S]methionine. In agreement with the above observations, we found that UV radiation increases the half-life of endogenous MTA1 protein in the HaCaT cells (Fig. 2B). These findings suggest that UV radiation up-regulates MTA1 through its posttranslational modifications.
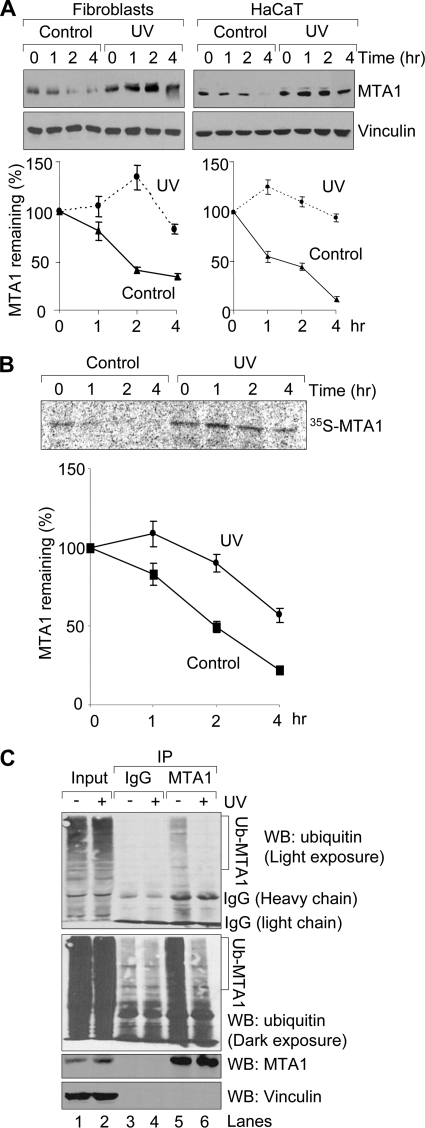
FIGURE 2.
UV stabilizes MTA1 by inhibiting its ubiquitination. A, HaCaT cells or fibroblasts were untreated or treated with 200 mJ/cm2 UV. After 1 h of UV treatment, cells were incubated with 100 μg/ml cycloheximide and harvested at the indicated time points for Western blotting analysis using the indicated antibodies. Western blots were subjected to densitometric analysis, and results were normalized based on vinculin expression levels and reported in a graph (bottom). The mean values from three independent experiments are shown. B, HaCaT cells were treated with 200 mJ/cm2 UV for 1 h prior to pulse-chase analysis using [35S]methionine labeling. Cells were harvested at various time points during the chase period and immunoprecipitated using an anti-MTA1 antibody. Complexes were resolved by SDS-PAGE and exposed to storage phosphor screens. The intensity of the labeled MTA1 band was quantified by PhosphorImager analysis using ImageQuant software (Amersham Biosciences), and the percentage of MTA1 remaining was calculated relative to that at the beginning of the chase period (time 0). The mean values from three independent experiments are shown. C, HaCaT cells were treated with 20 μm MG-132 for 1 h and then exposed to 200 mJ/cm2 UV. Protein extracts were prepared after 2 h of UV treatment and then subjected to sequential IP/Western blot (WB) analysis with the indicated antibodies.
Recent studies have demonstrated that MTA1 is an ubiquitinated protein and targeted by the RING-finger E3 ubiquitin ligase COP1 (constitutive photomorphogenesis protein 1) for degradation via the ubiquitin-proteasome pathway (11). To test the possibility that UV radiation stabilizes MTA1 by inhibiting its ubiquitination, HaCaT cells were pretreated with 20 μm proteasome inhibitor MG-132 for 1 h to enhance the signal of the ubiquitination of endogenous MTA1 and then subjected to UV radiation. Total cellular lysates were prepared after 2 h of UV radiation and then subjected to IP analysis with an anti-MTA1 antibody or IgG control, followed by Western blot analysis with the indicated antibodies. Results showed that treatment of HaCaT cells with UV resulted in a marked decrease in the ubiquitination of endogenous MTA1 (Fig. 2C, compare lane 6 with lane 5). Collectively, these results suggest that UV radiation increases the protein stability of MTA1 by, at least in part, inhibiting its ubiquitination.
Induction of MTA1 by UV Radiation Is Dependent on ATR
Because DNA damage produced by UV light activates the ATR checkpoint, which is a major part of the cell response to UV radiation from yeast to humans (17, 18, 63), we next determined whether the noted increase of MTA1 in response to UV requires ATR protein. Because ATR−/− cells are nonviable, we used human osteosarcoma U2OS cells expressing either ATRWT or ATRKD through a tetracycline-inducible system to determine the role of ATR in MTA1 protein stability following UV treatment (50, 51). The expression of ATRKD resulted in dominant negative effects, including inhibition of ATR downstream signaling and disruption of the cell cycle checkpoints (50, 51). As shown in Fig. 3A, expression of ATR was confirmed by Western blotting using an anti-FLAG antibody 48 h after the addition of doxycycline. Following UV radiation, the protein levels of MTA1 were increased in the ATRWT-expressing cells (Fig. 3A); however, cells expressing ATRKD showed little to no increase in MTA1 levels in response to UV radiation (Fig. 3A).
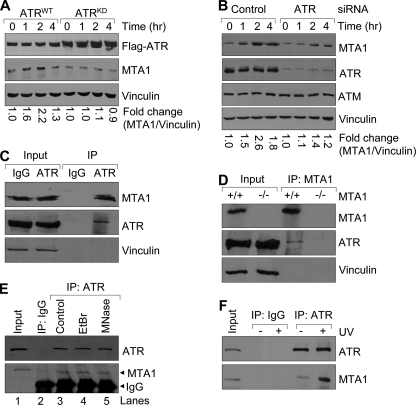
FIGURE 3.
Induction of MTA1 following exposure to UV is dependent on ATR. A, ATRWT- or ATRKD-expressing U2OS cells were incubated with 1 μg/ml doxycycline for 48 h and then exposed to 200 mJ/cm2 UV. Total cellular lysates were prepared at the indicated time points for Western blot analysis with the indicated antibodies. The density of bands was measured using the ImageQuest program and normalized to that of vinculin. The relative -fold change (MTA1/vinculin) is shown at the bottom. B, U2OS cells were transfected with specific siRNAs targeting human ATR or non-targeting control siRNAs. After 48 h of transfection, cells were treated with or without 200 mJ/cm2 of UV and then subjected to Western blot analysis with the indicated antibodies. The relative -fold change (MTA1/vinculin) is shown at the bottom. C, protein extracts from HeLa cells were subjected to IP analysis with an anti-ATR antibody or IgG control, followed by immunoblotting with the antibodies against ATR and MTA1. The immunoprecipitates were immunoblotted with vinculin, an abundant protein not expected to be part of this complex, as a negative control. D, protein extracts from MTA1+/+ or MTA1−/− (negative control) MEFs were subjected to IP analysis with an anti-MTA1 antibody, followed by immunoblotting with the indicated antibodies. E, HeLa cellular lysates were incubated with 100 μg/ml EtBr on ice for 30 min. Precipitates were removed by centrifugation for 5 min at 4 °C in a microcentrifuge, and the supernatant was transferred to a fresh tube. The resulting lysate was then ready for the IP assay. For micrococcal nuclease treatment, the immunoprecipitates bound to Protein G-agarose beads were incubated with micrococcal nuclease at 37 °C for 1 h, and the samples were washed twice with 1 ml of digestion buffer prior to SDS-PAGE. F, HeLa cells were irradiated with 200 mJ/cm2 UV and harvested after 2 h of UV treatment for sequential IP/Western blot analysis with the indicated antibodies.
To further confirm the above observations, we next knocked down the endogenous ATR in the U2OS cells by transfecting cells with specific siRNAs targeting human ATR or non-targeting control siRNAs. After 48 h of transfection, cells were treated with or without 200 mJ/cm2 of UV and then subjected to Western blot analysis with the indicated antibodies. Results showed that ATR siRNA effectively knocked down the endogenous ATR in the U2OS cells but did not alter the expression of ATM, another DNA damage-responsive protein (63), or of vinculin as an internal control (Fig. 3B). Consistent with our above observation, we found that knockdown of ATR by specific siRNAs compromised the induction of MTA1 following UV radiation (Fig. 3B). Taken together, these results consistently demonstrate that MTA1 induction after UV radiation is dependent on ATR.
Because MTA1 has been shown to associate with ATR under normal conditions (14), we next sought to determine whether the association between both proteins is affected by UV radiation. Consistent with a previous study (14), we found an interaction between MTA1 and ATR proteins at the endogenous levels in the HeLa cells by IP analysis using an anti-ATR antibody followed by Western blot analysis with an anti-MTA1 antibody (Fig. 3C). Furthermore, using MTA1−/− MEFs (7) as negative controls, we also confirmed the interaction between endogenous MTA1 and endogenous ATR in the MTA1+/+ but not MTA1−/− MEFs (Fig. 3D). When the immunoprecipitates were immunoblotted for vinculin, an abundant protein not expected to be part of this complex, no bands were seen in these precipitates even after long exposure (Fig. 3, C and D, bottom). By contrast, the expression of vinculin was clearly seen in the total cellular lysates (input) (Fig. 3, C and D, bottom).
To examine the possibility that the observed interaction between MTA1 and ATR is dependent on DNA, we next carried out the IP analysis with an anti-ATR antibody using the HeLa cellular lysates in the presence or absence of the DNA intercalator EtBr, which interferes generally with protein-DNA interaction (64, 65) according to the published method (54), followed by Western blot analysis with an anti-MTA1 antibody. Results showed that the association of ATR with MTA1 was not disrupted by the addition of EtBr (Fig. 3E, compare lane 4 with lane 3), suggesting that the interaction between ATR and MTA1 is DNA-independent. To further confirm these observations, we next treated the ATR immunoprecipitates with micrococcal nuclease (S7 nuclease or micrococcal nuclease) after IP from HeLa cellular extracts according to the method described previously (54). Micrococcal nuclease is an endo-exonuclease that preferentially digests single-stranded nucleic acids but will also cleave double-stranded DNA or RNA. Therefore, the enzyme is widely implicated for removing nucleic acids from protein preparation. As did the addition of EtBr, micrococcal nuclease treatment had no effect on association of ATR with MTA1 (Fig. 3E, compare lane 5 with lane 3). These results suggest that the interaction between MTA1 and ATR is independent of DNA.
To further determine whether the association between both proteins is affected by UV radiation, HeLa cells were irradiated with 200 mJ/cm2 UV, and protein extracts were prepared after 2 h of UV treatment for IP analysis with an anti-ATR antibody or IgG control, followed by Western blot analysis with the indicated antibodies. As shown in Fig. 3F, although the cellular amounts of the ATR protein remained unchanged following UV radiation, the relative amount of MTA1 bound to ATR increased significantly following radiation. Taken together, these results suggest that induction of MTA1 following UV radiation is dependent on ATR, and UV radiation increases the binding of MTA1 to ATR.
MTA1 Controls ATR-dependent Phosphorylation of Chk1
ATR is activated in response to UV radiation and initiates a checkpoint signaling cascade by phosphorylating a number of downstream substrates, including Chk1 (15, 25, 32). Mammalian Chk1 has recently been shown to be phosphorylated on Ser317 and Ser345 by ATR following UV treatment (25, 32). In support of this notion, overexpression of kinase-inactive ATR completely abolishes Chk1 phosphorylation in 293T cells (25). In addition, immunodepletion of ATR from a Xenopus cell-free system not only abolished UV-induced Chk1 phosphorylation but also severely compromised checkpoint responses to replication blocks and UV-induced DNA damage (28, 29). Consistently, Chk1 containing non-phosphorylatable residues at conserved ATR phosphorylation sites was deficient in the DNA replication checkpoint (28). These studies suggest that ATR and Chk1 are required for checkpoint responses to UV-induced DNA damage and incomplete DNA synthesis.
Because MTA1 interacts with ATR (14), we next examined whether MTA1 plays a role in ATR activation after UV treatment. It is worth pointing out that, despite the essential role of ATR in cell cycle signaling and DNA repair processes (15), little is known about its activation. To explore the possible role of MTA1 in ATR activation following UV treatment, MTA1+/+ or MTA1−/− MEFs were irradiated with 200 mJ/cm2 UV and harvested at the indicated time points, and activation of the ATR kinase was assayed by immunoblotting of cellular lysates with a specific antibody against activated forms of ATR (phospho-ATR Ser428). As shown in Fig. 4A, we found a marked decrease in ATR phosphorylation on Ser428 in the MTA1−/− MEFs relative to the MTA1+/+ controls following UV treatment. Interestingly, levels of total ATR protein were not affected by MTA1 deficiency or altered by UV exposure, suggesting that MTA1 regulates ATR activation rather than its expression.
Chk1 is a downstream effector of ATR that has been shown to be phosphorylated on Ser345 and Ser317 by ATR following exposure to UV (25, 32), and these phosphorylations are thought to be necessary for Chk1 activation (32). Thus, we next addressed whether activation of Chk1 is regulated by knock-out of MTA1. To do this, we assayed the phosphorylation status of Chk1 on Ser345 in the MTA1+/+ or MTA1−/− MEFs following UV treatment by Western blotting analysis with an anti-phospho-Chk1 (Ser345) antibody. Consistent with the above observations, Chk1 phosphorylation on Ser345 was almost completely abrogated in the MTA1−/− MEFs compared with the MTA1+/+ controls (Fig. 4A). Strikingly, we found that total Chk1 levels are lower in the MTA1−/− cells but not low enough to account for the complete absence of Chk1 phosphorylation. Thus, these results suggest that MTA1 is required for phosphorylation of Chk1 following UV-induced DNA damage as well as for the expression of total Chk1 protein levels.
To further confirm that the ATR/Chk1 phosphorylation defects observed in the MTA1−/− MEFs after UV exposure (Fig. 4A) were a general phenomenon and not restricted to MEFs, we next performed similar experiments in the HaCaT cells in which endogenous MTA1 was knocked down by using specific siRNAs targeting MTA1. As shown in Fig. 4B, the effects of MTA1 siRNA knockdown were specific, because MTA1 siRNA did not alter the expression of ATR, another DNA damage checkpoint protein (15), or of vinculin as an internal control. In addition, we found that knockdown of MTA1 resulted in a slight increase in MTA3 protein levels. This result is consistent with a previous study (61), in which the authors reported that transient overexpression of MTA1 in the MCF7 and T47D breast cancer cell lines with MTA1-expressing adenovirus led to dramatic decreases in MTA3 protein. Consistent with our above observations, we found that treatment of control siRNA-transfected HaCaT cells with UV resulted in a significant increase in phosphorylation of ATR at Ser428 as well as Chk1 at Ser345 (Fig. 4C). In contrast, in cells where MTA1 expression was knocked down, phosphorylation of ATR and Chk1 was compromised after UV treatment. Consistently, the expression levels of Chk1 were also reduced in MTA1 knockdown cells (Fig. 4C).
To further rule out the potential off-target effects of MTA1 siRNA, we used two additional specific siRNAs targeting human MTA1 (Qiagen) and repeated the above experiments. The efficiency and specificity of MTA1 siRNAs were demonstrated by Western blot analysis (supplemental Fig. S2A). Both MTA1 siRNAs effectively knocked down MTA1 but not another MTA family member MTA2 as well as internal control vinculin (supplemental Fig. S2A). Consistent with our above findings (Fig. 4C), we demonstrate that knockdown of MTA1 by siRNAs compromised the induction of phospho-ATR Ser428 and phospho-Chk1 Ser345 in response to UV radiation (supplemental Fig. S2B). Taken together, these results indicate that MTA1 is important for the proper phosphorylation of ATR/Chk1 in the UV-induced DNA damage checkpoint signaling cascade.
It has been shown that a number of factors act in concert with ATR to facilitate Chk1 phosphorylation, including ATRIP, Rad17-replication factor C, Rad9·Rad1·Hus1 (9-1-1) complex, TopBP1, and Claspin (15). We next examined the expression levels of these known regulators of the ATR/Chk1 pathway in the MTA1+/+ and MTA1−/− MEFs by Western blot analyses using the corresponding specific antibodies. Results showed that there are no changes in the protein expression levels of ATR, ATRIP, TopBP1, Rad17, Rad9, and Hus1 between the MTA1+/+ and MTA1−/− cells (Fig. 4D). Interestingly, we found that knock-out of MTA1 led to a marked down-regulation of the Chk1-interacting partner Claspin in the MTA1−/− MEFs relative to its wild-type controls. All of these data are consistent with our hypothesis that MTA1 is required for the ATR-mediated phosphorylation and activation of Chk1.
Claspin is an “adaptor” or “mediator” protein that is crucial for the ATR-mediated phosphorylation and activation of Chk1, functioning to bring ATR and Chk1 together (46), and its degradation plays a critical role in DNA damage checkpoint recovery (66). Genetic data with human cells and in vitro data with Xenopus egg extracts have demonstrated that the kinase activity of ATR toward Chk1 kinase depends on the mediator protein Claspin (46). A most recent study using an in vitro system with purified human proteins also demonstrated that the ATR-dependent phosphorylation of Chk1, but not p53, is strongly stimulated by Claspin (47). Consistently, the RNA interference-mediated ablation of Claspin selectively abrogated the ability of ATR to phosphorylate Chk1 but not other ATR targets (48). In the present study, we found that knock-out of MTA1 compromised the ATR-mediated Chk1 activation following UV radiation, accompanied by a marked down-regulation of Claspin in the MTA1−/− cells. Thus, we propose that MTA1 is required for activation of the ATR-Chk1 checkpoint pathway through, at least in part, regulation of Claspin.
In addition, evidence has been presented that Chk1 is required to maintain Claspin stability (67). Down-regulation of Chk1 expression by siRNA or inhibition of Chk1 activity by UCN01 decreases Claspin levels in cells. Conversely, overexpression of Chk1 increases Claspin levels (67). In the present work, we found that knock-out of MTA1 resulted in a marked decrease in expression levels of both Chk1 and Claspin, but whether regulation of Claspin by MTA1 is Chk1-dependent remains to be examined. Based on these findings, we propose that UV radiation stabilizes MTA1, which in turn regulates the expression of Claspin, resulting in increased Chk1 activation by ATR. Once Chk1 is activated, it stabilizes Claspin, which in turn helps to maintain the Chk1 activation by ATR (67). On the other hand, MTA1 might directly regulate the expression of Chk1, which stabilizes Claspin, resulting in further signal amplification of the ATR-Claspin-Chk1 pathway (Fig. 5G). Thus, these proteins, including MTA1, ATR, Chk1, and Claspin, regulate each other and thus ensure the proper cell cycle progression and replication checkpoint control.
FIGURE 5.
Depletion of MTA1 results in decreased γ-H2AX induction in response to UV and increased cellular sensitivity to UV irradiation. A, MTA1+/+ or MTA1−/− MEFs were treated with 200 mJ/cm2 UV and harvested at the indicated time points for Western blot analysis with the indicated antibodies. B, HaCaT cells were transfected with control siRNAs or specific siRNAs targeting MTA1. After 48 h of transfection, cells were exposed to 200 mJ/cm2 UV and harvested at the indicated time points for Western blot as described above. C, MTA1+/+ or MTA1−/− MEFs were treated with 200 mJ/cm2 UV and fixed after 1 h for immunofluorescent staining with an anti-phospho-H2AX (Ser139) antibody. The nuclei were visualized by 4′,6-diamidino-2-phenylindole staining. Representative confocal images of γ-H2AX foci are shown in supplemental Fig. S4. The graph shows the average number of γ-H2AX foci determined by counting about 50 nuclei/sample. The error bars indicate mean ± S.D. from three independent experiments. D, HaCaT cells were transfected with control siRNA or specific siRNA targeting MTA1. After 48 h of transfection, cells were irradiated with UV and then subjected to γ-H2AX staining as described above. Representative confocal images of γ-H2AX foci were shown in supplemental Fig. S5. The graph shows the average number of γ-H2AX foci determined by counting about 50 nuclei per sample. The error bars indicate mean ± S.D. from three independent experiments. E, clonogenic survival assay of MTA1+/+ or MTA1−/− MEFs untreated or treated with increasing doses of UV. Cells were counted, plated, and subjected to the indicated doses of radiation and colonies formed over 14–21 days. Surviving colonies were plotted as a function of cells plated and normalized by the plating efficiency for each condition. Knock-out of MTA1 was confirmed by Western blot analysis (upper right). F, HaCaT cells were transfected with control siRNA or specific siRNA targeting MTA1. After 36 h of transfection, cells were subjected to a clonogenic survival assay as described above. Depletion of MTA1 was confirmed by Western blot analysis (upper right). G, working model summarizing the findings presented here. MTA1 regulates the expression of Claspin, which facilitates ATR-dependent Chk1 activation. Once Chk1 is activated, it stabilizes Claspin, which in turn helps to maintain the Chk1 activation by ATR. On the other hand, MTA1 also regulates the expression of Chk1, which stabilizes Claspin, resulting in further signal amplification of the ATR-Claspin-Chk1 pathway. Furthermore, MTA1 deficiency decreases the induction of γ-H2AX and compromises γ-H2AX formation of foci in response to UV. Consequently, MTA1 deficiency results in a defective G2/M DNA damage checkpoint and increased cellular sensitivity to UV radiation.
Depletion of MTA1 Results in a Defect of G2/M Checkpoint
Chk1 is a major checkpoint kinase and has been shown to be mainly responsible for the G2-M checkpoint (25). Conditional Chk1−/− embryonic stem cells have a defective G2-M DNA damage checkpoint (25, 26, 68). Similarly, disruption of the IR-induced G2-M DNA damage checkpoint was obtained by elimination of Chk1 by short interfering RNA (52, 69, 70). We next sought to address whether the failure of ATR to phosphorylate and activate Chk1 in response to UV in MTA1-depleted cells would also result in a defect in the G2-M checkpoint. To this end, MTA1+/+ or MTA1−/− MEFs were treated with 200 mJ/cm2 of UV and harvested at the indicated time points for the DNA profile analysis by propidium iodide staining. As shown in Fig. 4E and supplemental Fig. S3, MTA1+/+ MEFs were arrested with a G2-M DNA content, indicating the presence of an intact DNA damage checkpoint in this cell line. In contrast, G2-M checkpoint activation was compromised in the MTA1−/− MEFs upon UV treatment, thus reiterating the important role of MTA1 in the G2-M checkpoint, and these activities of MTA1 might result from its regulation of the expression or activation of the checkpoint regulator Chk1.
To further demonstrate these observations, we next carried out the G2/M checkpoint assay as described previously (52). Briefly, MTA1+/+ and MTA1−/− MEFs were treated with 200 mJ/cm2 UV and harvested at the indicated time points, followed by flow cytometric assessment of anti-phosphohistone H3 antibody to distinguish mitotic cells from G2 cells (52). As shown in Fig. 4F, MTA1−/− cells exhibited a higher level of phospho-H3 staining than did the MTA1+/+ cells at the corresponding time points after UV radiation, suggesting a failure of the MTA1−/− cells to fully arrest or to maintain an arrest in G2/M. These results suggest that MTA1 is required for a normal G2/M cell cycle delay after UV radiation. Activation of the DNA damage checkpoint and the transient block to mitotic entry correlate with improved survival following DNA damage (71). Cells lacking Chk1 fail to arrest the cell cycle in response to DNA damage, enter mitosis with damaged DNA, and die (72, 73). These results further indicate that MTA1 status could affect cell survival in response to UV-induced DNA damage (see below).
MTA1 Controls ATR-dependent Phosphorylation of H2AX
H2AX phosphorylation on Ser139 in response to DNA damage is required for the concentration of DNA repair proteins to the damaged chromatin (42). Phosphorylation of H2AX is dependent on ATM after IR (43) and on ATR during replication stress induced by UV (44, 45). We next examined the effect of MTA1 deficiency on the levels of γ-H2AX by Western blot analysis using an anti-phospho-H2AX (Ser139) antibody. Results showed that in response to UV, the levels of γ-H2AX were greatly impaired in the MTA1−/− MEFs as compared with those in the MTA1+/+ controls (Fig. 5A). Interestingly, there was no change in total H2AX protein levels between MTA1+/+ and MTA1−/− MEFs with or without UV treatment, indicating that MTA1 is critical for the efficient induction of H2AX phosphorylation in response to UV radiation. To further confirm these findings, we depleted endogenous MTA1 in HaCaT cells using specific siRNAs targeting MTA1. As shown in Fig. 5B, γ-H2AX was significantly induced after 1 h of UV treatment in the control siRNA-treated cells. In contrast, UV-induced γ-H2AX was substantially reduced in the MTA1 siRNA-transfected cells.
Intensive DNA damage can induce the formation of thousands of molecules of γ-H2AX, which tend to accumulate into microscopically observable nuclear foci (74). Once formed, γ-H2AX may further recruit other proteins involved in DNA repair, such as Rad50, Rad51, BRCA1, Nbs1, p53BP1, and MDC1. We next examined whether MTA1 deficiency affects the formation of γ-H2AX foci by immunofluorescent staining using an anti-phospho-H2AX (Ser139) antibody and found that depletion of MTA1 decreased the average number of the γ-H2AX-containing foci per cell as compared with the MTA1+/+ controls (Fig. 5C and supplemental Fig. S4), suggesting that MTA1 is critical for the formation of γ-H2AX foci after DNA damage. Furthermore, these effects of MTA1 depletion on γ-H2AX formation of foci were also observed in HaCaT cells (Fig. 5D and supplemental Fig. S5). We found that knockdown of MTA1 in HaCaT cells using specific siRNAs targeting MTA1 results in a significant decrease in γ-H2AX formation as compared with control siRNA-transfected cells following UV exposure.
H2AX is critical for facilitating the assembly of specific DNA repair complexes on damaged DNA. Therefore, H2AX−/− mice are radiation-sensitive, and immortalized MEFs exhibit defective DNA repair (41). Our above observations that depletion of MTA1 results in decreased γ-H2AX induction after UV exposure (Fig. 5, A–D) led us to further examine whether MTA1 has a role in the repair of DNA damage produced by UV. One of the hallmarks of defective DNA repair is increased radiation sensitivity. We first examined the effect of MTA1 deficiency on cell survival in response to UV exposure by clonogenic survival assay (75). We found that MTA1−/− MEFs were hypersensitive to UV exposure and exhibited a decreased clonogenic survival compared with its wild-type controls (Fig. 5E), suggesting a defect in DNA repair in MTA1-deficient cells. In support of this finding, we further demonstrated that knockdown of MTA1 in HaCaT cells results in increased cellular sensitivity to UV-induced DNA damage (Fig. 5F). In all of the MTA1 knockdown experiments, MTA1 depletion was confirmed by Western blot analysis with a specific anti-MTA1 antibody (Fig. 5F). In brief, these results suggest that MTA1 deficiency or knockdown compromises the ability of cells to respond to DNA damage and increases cell sensitivity to UV.
In summary, here we demonstrate a vital role for MTA1 in the UV-induced ATR-mediated DNA checkpoint pathway (Fig. 5G). An earlier study has shown that ATR is associated with the components of the Mi-2·NuRD complex, including MTA1 and Mi-2, suggesting that there may be a linkage between the role of ATR in mediating checkpoints induced by DNA damage and chromatin modulation via remodeling and deacetylation (14). However, the biochemical function of ATR-MTA1 interaction was not explored in that study. Interestingly, a recent study demonstrated that UV radiation induces the protein expression of Mi-2, another core subunit of the Mi-2·NuRD complex (3, 59, 60), by regulating protein translation and stability (58).
In support of the earlier finding of the MTA1-ATR interaction (14), here we found that UV radiation induces MTA1 protein expression in an ATR-dependent manner. More interestingly, MTA1 is required for activation of ATR following UV radiation because depletion or knockdown of MTA1 severely impaired ATR-dependent phosphorylation of Chk1 and H2AX in response to UV damage. As a result, depletion of MTA1 results in the abrogation of the G2-M checkpoint and increased cellular sensitivity to UV-induced DNA damage. Thus, MTA1 is required for the activation of ATR-Claspin-Chk1 and ATR-H2AX pathways following exposure to UV, and the abrogation of the DNA damage checkpoints in the MTA1-depleted cells may be, at least in part, a consequence of dysregulation of the expression of these two checkpoint pathways (Fig. 5G).
The molecular mechanism for the requirement of MTA1 in the activation of both checkpoint pathways is currently being investigated in our laboratory. Accumulating evidence shows that chromatin remodeling complexes, such as SWI/SNF and INO80, play an important role not only in transcription, but also in DSB repair via interaction with γ-H2AX (76–78). As a part of the Mi-2·NuRD chromatin remodeling complex (53), it is well established that MTA1 modulates transcription by influencing chromatin remodeling (3, 59). Furthermore, recent studies from our laboratory have demonstrated that MTA1, like other chromatin remodeling factors, is also implicated in DSB repair following IR treatment (11–13). Depletion of MTA1 results in increased DNA damage sensitivity and decreased DSB repair (11, 13). It is possible that MTA1 as a chromatin modifier could alter chromatin structure in an unknown way in response to DNA damage, resulting in increased accessibility of damaged DNA to repair factors (12). Taken together, these findings suggest that, in addition to its role in the repair of DSBs caused by IR, MTA1 also participates in the UV-induced ATR-mediated DNA damage checkpoint pathway.
Supplementary Material
Acknowledgments
We thank Paul Nghiem for providing ATR-inducible U2OS cell lines. We also thank Vasudha S. Nair and Sujit S. Nair for immunofluorescence staining and the checkpoint assays.
This work was supported, in whole or in part, by National Institutes of Health Grants CA98823 and CA98823-S1 (to R. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- MTA
- metastasis-associated protein
- IR
- ionizing radiation
- IP
- immunoprecipitation
- MEF
- mouse embryonic fibroblast
- DSB
- double strand break
- ATRIP
- ATR-interacting protein
- siRNA
- small interfering RNA
- NuRD
- nucleosome remodeling and deacetylase.
REFERENCES
- 1.Toh Y., Pencil S. D., Nicolson G. L. (1994) J. Biol. Chem. 269, 22958–22963 [PubMed] [Google Scholar]
- 2.Kumar R., Wang R. A., Bagheri-Yarmand R. (2003) Semin. Oncol. 30, 30–37 [DOI] [PubMed] [Google Scholar]
- 3.Manavathi B., Kumar R. (2007) J. Biol. Chem. 282, 1529–1533 [DOI] [PubMed] [Google Scholar]
- 4.O'Malley B. W., Kumar R. (2009) Cancer Res. 69, 8217–8222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazumdar A., Wang R. A., Mishra S. K., Adam L., Bagheri-Yarmand R., Mandal M., Vadlamudi R. K., Kumar R. (2001) Nat. Cell Biol. 3, 30–37 [DOI] [PubMed] [Google Scholar]
- 6.Molli P. R., Singh R. R., Lee S. W., Kumar R. (2008) Oncogene 27, 1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manavathi B., Peng S., Rayala S. K., Talukder A. H., Wang M. H., Wang R. A., Balasenthil S., Agarwal N., Frishman L. J., Kumar R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13128–13133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D. Q., Pakala S. B., Reddy S. D., Ohshiro K., Peng S. H., Lian Y., Fu S. W., Kumar R. (2010) J. Biol. Chem. 285, 10044–10052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gururaj A. E., Singh R. R., Rayala S. K., Holm C., den Hollander P., Zhang H., Balasenthil S., Talukder A. H., Landberg G., Kumar R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6670–6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balasenthil S., Gururaj A. E., Talukder A. H., Bagheri-Yarmand R., Arrington T., Haas B. J., Braisted J. C., Kim I., Lee N. H., Kumar R. (2007) Cancer Res. 67, 7132–7138 [DOI] [PubMed] [Google Scholar]
- 11.Li D. Q., Ohshiro K., Reddy S. D., Pakala S. B., Lee M. H., Zhang Y., Rayala S. K., Kumar R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17493–17498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li D. Q., Kumar R. (2010) Cell Cycle 9, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li D. Q., Divijendra Natha Reddy S., Pakala S. B., Wu X., Zhang Y., Rayala S. K., Kumar R. (2009) J. Biol. Chem. 284, 34545–34552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt D. R., Schreiber S. L. (1999) Biochemistry 38, 14711–14717 [DOI] [PubMed] [Google Scholar]
- 15.Cimprich K. A., Cortez D. (2008) Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakkenist C. J., Kastan M. B. (2004) Cell 118, 9–17 [DOI] [PubMed] [Google Scholar]
- 17.Zhou B. B., Elledge S. J. (2000) Nature 408, 433–439 [DOI] [PubMed] [Google Scholar]
- 18.Abraham R. T. (2001) Genes Dev. 15, 2177–2196 [DOI] [PubMed] [Google Scholar]
- 19.Unsal-Kaçmaz K., Makhov A. M., Griffith J. D., Sancar A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6673–6678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiloh Y. (2001) Curr. Opin. Genet. Dev. 11, 71–77 [DOI] [PubMed] [Google Scholar]
- 21.Mu J. J., Wang Y., Luo H., Leng M., Zhang J., Yang T., Besusso D., Jung S. Y., Qin J. (2007) J. Biol. Chem. 282, 17330–17334 [DOI] [PubMed] [Google Scholar]
- 22.Smolka M. B., Albuquerque C. P., Chen S. H., Zhou H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10364–10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stokes M. P., Rush J., Macneill J., Ren J. M., Sprott K., Nardone J., Yang V., Beausoleil S. A., Gygi S. P., Livingstone M., Zhang H., Polakiewicz R. D., Comb M. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19855–19860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 25.Liu Q., Guntuku S., Cui X. S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Donehower L. A., Elledge S. J. (2000) Genes Dev. 14, 1448–1459 [PMC free article] [PubMed] [Google Scholar]
- 26.Takai H., Tominaga K., Motoyama N., Minamishima Y. A., Nagahama H., Tsukiyama T., Ikeda K., Nakayama K., Nakanishi M., Nakayama K. (2000) Genes Dev. 14, 1439–1447 [PMC free article] [PubMed] [Google Scholar]
- 27.Bartek J., Lukas J. (2003) Cancer Cell 3, 421–429 [DOI] [PubMed] [Google Scholar]
- 28.Guo Z., Kumagai A., Wang S. X., Dunphy W. G. (2000) Genes Dev. 14, 2745–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hekmat-Nejad M., You Z., Yee M. C., Newport J. W., Cimprich K. A. (2000) Curr. Biol. 10, 1565–1573 [DOI] [PubMed] [Google Scholar]
- 30.Kastan M. B., Bartek J. (2004) Nature 432, 316–323 [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Girona A., Tanaka K., Chen X. B., Baber B. A., McGowan C. H., Russell P. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11289–11294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao H., Piwnica-Worms H. (2001) Mol. Cell Biol. 21, 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furnari B., Rhind N., Russell P. (1997) Science 277, 1495–1497 [DOI] [PubMed] [Google Scholar]
- 34.Peng C. Y., Graves P. R., Thoma R. S., Wu Z., Shaw A. S., Piwnica-Worms H. (1997) Science 277, 1501–1505 [DOI] [PubMed] [Google Scholar]
- 35.Sanchez Y., Wong C., Thoma R. S., Richman R., Wu Z., Piwnica-Worms H., Elledge S. J. (1997) Science 277, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 36.Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 37.Chen Y., Sanchez Y. (2004) DNA Repair. 3, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 38.Chen Z., Xiao Z., Gu W. Z., Xue J., Bui M. H., Kovar P., Li G., Wang G., Tao Z. F., Tong Y., Lin N. H., Sham H. L., Wang J. Y., Sowin T. J., Rosenberg S. H., Zhang H. (2006) Int. J. Cancer 119, 2784–2794 [DOI] [PubMed] [Google Scholar]
- 39.Tse A. N., Schwartz G. K. (2004) Cancer Res. 64, 6635–6644 [DOI] [PubMed] [Google Scholar]
- 40.Bassing C. H., Chua K. F., Sekiguchi J., Suh H., Whitlow S. R., Fleming J. C., Monroe B. C., Ciccone D. N., Yan C., Vlasakova K., Livingston D. M., Ferguson D. O., Scully R., Alt F. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8173–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Celeste A., Petersen S., Romanienko P. J., Fernandez-Capetillo O., Chen H. T., Sedelnikova O. A., Reina-San-Martin B., Coppola V., Meffre E., Difilippantonio M. J., Redon C., Pilch D. R., Olaru A., Eckhaus M., Camerini-Otero R. D., Tessarollo L., Livak F., Manova K., Bonner W. M., Nussenzweig M. C., Nussenzweig A. (2002) Science 296, 922–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Redon C., Pilch D., Rogakou E., Sedelnikova O., Newrock K., Bonner W. (2002) Curr. Opin. Genet. Dev. 12, 162–169 [DOI] [PubMed] [Google Scholar]
- 43.Burma S., Chen B. P., Murphy M., Kurimasa A., Chen D. J. (2001) J. Biol. Chem. 276, 42462–42467 [DOI] [PubMed] [Google Scholar]
- 44.Ward I. M., Chen J. (2001) J. Biol. Chem. 276, 47759–47762 [DOI] [PubMed] [Google Scholar]
- 45.O'Driscoll M., Ruiz-Perez V. L., Woods C. G., Jeggo P. A., Goodship J. A. (2003) Nat. Genet. 33, 497–501 [DOI] [PubMed] [Google Scholar]
- 46.Kumagai A., Dunphy W. G. (2000) Mol. Cell 6, 839–849 [DOI] [PubMed] [Google Scholar]
- 47.Lindsey-Boltz L. A., Serçin O., Choi J. H., Sancar A. (2009) J. Biol. Chem. 284, 33107–33114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu S., Bekker-Jensen S., Mailand N., Lukas C., Bartek J., Lukas J. (2006) Mol. Cell Biol. 26, 6056–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boukamp P., Petrussevska R. T., Breitkreutz D., Hornung J., Markham A., Fusenig N. E. (1988) J. Cell Biol. 106, 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nghiem P., Park P. K., Kim Y., Vaziri C., Schreiber S. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9092–9097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nghiem P., Park P. K., Kim Y., Desai B. N., Schreiber S. L. (2002) J. Biol. Chem. 277, 4428–4434 [DOI] [PubMed] [Google Scholar]
- 52.Gatei M., Sloper K., Sorensen C., Syljuäsen R., Falck J., Hobson K., Savage K., Lukas J., Zhou B. B., Bartek J., Khanna K. K. (2003) J. Biol. Chem. 278, 14806–14811 [DOI] [PubMed] [Google Scholar]
- 53.Bao Y., Shen X. (2007) Cell 129, 632. [DOI] [PubMed] [Google Scholar]
- 54.Lai J. S., Herr W. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 6958–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herrlich P., Sachsenmaier C., Radler-Pohl A., Gebel S., Blattner C., Rahmsdorf H. J. (1994) Adv. Enzyme Regul. 34, 381–395 [DOI] [PubMed] [Google Scholar]
- 56.Cooper S. J., Bowden G. T. (2007) Curr. Cancer Drug Targets 7, 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Gruijl F. R. (1999) Eur. J. Cancer 35, 2003–2009 [DOI] [PubMed] [Google Scholar]
- 58.Burd C. J., Kinyamu H. K., Miller F. W., Archer T. K. (2008) J. Biol. Chem. 283, 34976–34982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Denslow S. A., Wade P. A. (2007) Oncogene 26, 5433–5438 [DOI] [PubMed] [Google Scholar]
- 60.Feng Q., Zhang Y. (2003) Curr. Top. Microbiol. Immunol. 274, 269–290 [DOI] [PubMed] [Google Scholar]
- 61.Fujita N., Kajita M., Taysavang P., Wade P. A. (2004) Mol. Endocrinol. 18, 2937–2949 [DOI] [PubMed] [Google Scholar]
- 62.Baliga B. S., Pronczuk A. W., Munro H. N. (1969) J. Biol. Chem. 244, 4480–4489 [PubMed] [Google Scholar]
- 63.Shiloh Y. (2003) Nat. Rev. Cancer 3, 155–168 [DOI] [PubMed] [Google Scholar]
- 64.Parker R. C., Watson R. M., Vinograd J. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schröter H., Maier G., Ponstingl H., Nordheim A. (1985) EMBO J. 4, 3867–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Freire R., van Vugt M. A., Mamely I., Medema R. H. (2006) Cell Cycle 5, 2831–2834 [DOI] [PubMed] [Google Scholar]
- 67.Chini C. C., Wood J., Chen J. (2006) Oncogene 25, 4165–4171 [DOI] [PubMed] [Google Scholar]
- 68.Zachos G., Rainey M. D., Gillespie D. A. (2003) EMBO J. 22, 713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X., Khadpe J., Hu B., Iliakis G., Wang Y. (2003) J. Biol. Chem. 278, 30869–30874 [DOI] [PubMed] [Google Scholar]
- 70.Zhao H., Watkins J. L., Piwnica-Worms H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14795–14800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.al-Khodairy F., Carr A. M. (1992) EMBO J. 11, 1343–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walworth N., Davey S., Beach D. (1993) Nature 363, 368–371 [DOI] [PubMed] [Google Scholar]
- 73.al-Khodairy F., Fotou E., Sheldrick K. S., Griffiths D. J., Lehmann A. R., Carr A. M. (1994) Mol. Biol. Cell 5, 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rogakou E. P., Boon C., Redon C., Bonner W. M. (1999) J. Cell Biol. 146, 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Franken N. A., Rodermond H. M., Stap J., Haveman J., van Bree C. (2006) Nat. Protoc. 1, 2315–2319 [DOI] [PubMed] [Google Scholar]
- 76.Park J. H., Park E. J., Lee H. S., Kim S. J., Hur S. K., Imbalzano A. N., Kwon J. (2006) EMBO J. 25, 3986–3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morrison A. J., Highland J., Krogan N. J., Arbel-Eden A., Greenblatt J. F., Haber J. E., Shen X. (2004) Cell 119, 767–775 [DOI] [PubMed] [Google Scholar]
- 78.Bao Y., Shen X. (2007) Curr. Opin. Genet. Dev. 17, 126–131 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.