Abstract
The cytoplasmic tyrosine phosphatase SHP1 has been shown to inhibit the oncogenic fusion protein nucleophosmin (NPM)-anaplastic lymphoma kinase (ALK), and loss of SHP1 contributes to NPM-ALK-mediated tumorigenesis. In this study, we aimed to further understand how SHP1 interacts and regulates NPM-ALK. We employed an in vitro model in which GP293 cells were transfected with various combinations of NPM-ALK (or mutants) and SHP1 (or mutants) expression vectors. We found that SHP1 co-immunoprecipitated with NPM-ALK, but not the enzymatically inactive NPM-ALKK210R mutant, or the mutant in which all three functionally important tyrosine residues (namely, Tyr338, Tyr342, and Tyr343) in the kinase activation loop (KAL) of ALK were mutated. Interestingly, whereas mutation of Tyr338 or Tyr342 did not result in any substantial change in the NPM-ALK/SHP1 binding (assessed by co-immunoprecipitation), mutation of Tyr343 abrogated this interaction. Furthermore, the NPM-ALK/SHP1 binding was readily detectable when each of the remaining 8 tyrosine residues known to be phosphorylated were mutated. Although the expression of SHP1 effectively reduced the level of tyrosine phosphorylation of NPM-ALK, it did not affect that of the NPM-ALKY343F mutant. In soft agar clonogenic assay, SHP1 expression significantly reduced the tumorigenicity of NPM-ALK but not that of NPM-ALKY343F. In conclusion, we identified Tyr343 of NPM-ALK as the crucial site for mediating the NPM-ALK/SHP1 interaction. Our results also support the notion that the tumor suppressor effects of SHP1 on NPM-ALK are dependent on its ability to bind to this oncogenic protein.
Keywords: Oncogene, Protein Phosphorylation, Signal Transduction, Tyrosine-Protein Kinase (Tyrosine Kinase), Tyrosine-Protein Phosphatase (Tyrosine Phosphatase), NPM-ALK, SHP1
Introduction
ALK2-positive anaplastic large cell lymphoma (ALK+ALCL) is a distinct type of aggressive lymphoma of T/null cell immunophenotypes (1). Approximately 80% of these neoplasms have the chromosomal translocation t(2;5)(p23;q35), which brings the nucleophosmin (NPM) gene at 5q35 in juxtaposition with the anaplastic lymphoma kinase (ALK) gene at 2p23, leading to the formation of NPM-ALK (2–5). ALK is normally a receptor-tyrosine kinase expressed exclusively on the cell surface of embryonic neuronal cells (6). It has been shown that the oligomerization domain of the NPM portion in this fusion gene protein induces dimerization of ALK, which results in autophosphorylation and constitutive activation of the ALK tyrosine kinase (7, 8). By virtue of its constitutively active tyrosine kinase, it is believed that NPM-ALK promotes tumorigenesis by aberrantly phosphorylating various tyrosine residues of a host of cellular signaling proteins, thereby deregulating a large number of cellular signaling pathways (9).
In addition to the expression of NPM-ALK, recent studies also revealed the existence of multiple coexisting biochemical defects in NPM-ALK-expressing lymphomas (10–15). The loss of SHP1, a cytoplasmic tyrosine phosphatase, serves as an example (14). SHP1 is normally expressed at the highest level in hematopoietic cells and it is known to act as a negative regulator of various cell signaling pathways, such as the JAK/STAT pathway and that of the T-/B-cell receptor (16). The biological importance of SHP1 is highlighted by the phenotype of the so-called “moth-eaten” mice; specifically, homozygous mutations of the SHP1 gene, which results in a complete loss of SHP1 expression, are associated with severe dysregulation of the leukocyte development and systemic autoimmunity (17, 18). SHP1 also has been shown to have tumor suppressor functions. Specifically, loss of SHP1 has been demonstrated in a proportion of T-cell lymphomas, including the majority of the ALK+ALCL cases (10, 12, 19). Importantly, restoration of SHP1 expression has been shown to decrease the growth of ALK+ALCL cells in vitro. In another study, decreased expression of SHP1 has been shown to be associated with the progression of chronic myeloid leukemia (20). Previous studies have shown that SHP1 is physically associated with NPM-ALK, and SHP1 dephosphorylates NPM-ALK (14, 21). However, the molecular details underlying the physical and functional interaction between SHP-1 and NPM-ALK are incompletely understood.
Because NPM-ALK is known to interact with its binding partners via the phospho-Tyr/SH2 motif (7, 22–24), one of the main objectives of this study was to identify the exact tyrosine residue of NPM-ALK involved in mediating its interaction with SHP1. We focused our search on the 11 tyrosine residues of NPM-ALK that have been previously found to be phosphorylated (25). Similarly, because SHP1 is known to interact with its substrates via one or both of its two SH2 domains, commonly termed N-SH2 (i.e. close to the N terminus) and C-SH2 (i.e. close to the C terminus) (26), we focused our studies on the roles of these two domains of SHP1 in the NPM-ALK/SHP1 binding. After we identified the exact tyrosine residue of NPM-ALK that is important for the physical interaction between NPM-ALK and SHP1, we examined the functional importance of this physical interaction in SHP1-mediated suppression on NPM-ALK.
MATERIALS AND METHODS
Cell Lines and Tissue Culture
An ALK+ALCL cell line, Karpas 299, was used. These cells were from the American Type Culture Collection (Manassas, VA) and cultured in RPMI 1640 (Invitrogen) containing 2 mm l-glutamine supplemented with 10% fetal bovine serum (FBS)(Invitrogen). GP293, a human embryonic kidney cell line, was cultured in Dulbecco's modified Eagle's medium (Invitrogen) containing 4 g/liter of glucose supplemented with 10% FBS.
Vectors and Plasmids
The wild-type SHP1 plasmid has been previously described (14). SHP1 mutants with Arg → Lys mutation of one or both of the two functionally important arginine residues (Arg32 and Arg138) located in the N-terminal and C-terminal SH2 domains, respectively, were kind gifts from Dr. F. Böhmer (Friedrich-Schiller-University, Jena, Germany) (27). Phosphatase inactive SHP1 (SHP1C445S) that has a point mutation (Cys → Ser) at the critical cysteine residue in the phosphatase domain was a kind gift from Dr. Zhenbao Yu (National Research Council of Canada, Montreal, Canada) (28). The NPM-ALK expression vector, a kind gift from Dr. S. Morris (St. Jude's Children Research Hospital, Memphis, TN), was cloned into the pcDNA3.1(+) vector (Invitrogen). The use of the “kinase-dead” NPM-ALK (NPM-ALKK210R) has been previously described (29). The NPM-ALKFFF mutant, which was generated by mutating all three tyrosine residues in the kinase activation loop of ALK (Tyr338, Tyr342, and Tyr343) in a pcDNA3.1(+)/His-tagged NPM-ALK backbone, has also been described (25). Similarly, the single Tyr → Phe mutants of NPM-ALK (i.e. site-directed mutagenesis of Tyr138, Tyr152, Tyr156, Tyr191, Tyr419, Tyr567, Tyr644, and Tyr664) were constructed on a pcDNA3.1(+)/NPM-ALK backbone. The coding sequences of NPM-ALK and all mutants were confirmed to ensure that no artificial mutations were acquired.
Gene Transfection
GP293 cells were plated in a 100-mm cell culture dish 24 h before gene transfection to obtain cell confluence of ∼70%. Gene transfection of NPM-ALK alone or in combination with SHP1 were done using TurboFectTM in vitro Transfection Reagent (Fermentas, Burlington, Ontario, Canada) according to the manufacturer's protocol. Transient transfection of Karpas 299 cells (4 × 106 cells) with 10 μg of DNA of wild-type SHP1 or different SHP1 mutants were performed using the Electro square electroporator, BTX ECM 800 (San Diego, CA) at 225 V (8.5 ms, 03 pulses).
Co-immunoprecipitation, Antibodies, and Western Blot Analysis
Cell lysates were prepared 24 h after gene transfection. For immunoprecipitation, a standard protocol was used as previously described (30). Briefly, cells were washed with cold phosphate-buffered saline and lysed using Cell Lytic Buffer M (Sigma) supplemented with 0.1 mm phenylmethylsulfonyl fluoride (Sigma), a protease inhibitor mixture (Nacalai Inc., San Diego, CA), and phosphatase inhibitor mixture (Calbiochem, EMD Biosciences, Darmstadt, Germany). After incubating the lysate on ice for 30 min, it was centrifuged at 15,000 × g for 15 min. Two micrograms of the primary antibody was added to 500 μg of protein lysate and rotated overnight at 4 °C. Negative control samples with the primary antibody omitted were included. 50 μl of protein (A/G Plus-agarose) beads (Santa Cruz Biotechnology, Santa Cruz, CA) was added to both the test and control lysates and rocked for 2 h at 4 °C. The beads were then washed 3 times with cold phosphate-buffered saline. For co-immunoprecipitation experiments, the final wash was done using cold cell lysis buffer. For immunoprecipitation experiments, the final wash was done using RIPA buffer. Proteins were then eluted from the beads in 20 μl of SDS protein loading buffer by boiling for 5 min at 100 °C. The complex was then subjected to SDS-polyacrylamide gel electrophoresis and Western blotting, and the proteins were visualized using enzyme chemiluminescence (Pierce ECL Western Blotting Substrate, Thermo Scientific, Rockford, IL). The following antibodies were used for immunoprecipitation and immunoblotting: goat polyclonal anti-ALK, rabbit polyclonal anti-SHP1 (both were from Santa Cruz Biotechnology), monoclonal anti-ALK (Dako, Glostrup, Denmark), mouse anti-SHP1 monoclonal antibody (BD Transduction Laboratories, Mississauga, Ontario, Canada), rabbit polyclonal anti-phospho-ALK (Tyr1604), anti-phosphotyrosine (both were from Cell Signaling, Danvers, MA), and anti-β-actin (Sigma). Western blot analysis is described as follows: cells were washed with cold phosphate-buffered saline, and cellular proteins were precipitated using RIPA buffer containing 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS, 50 mm Tris (pH 8), which was supplemented with 40 μg/ml of leupeptin, 1 μm pepstatin, 1 mm 4-(2-aminoethyl)-benzenesulfonyl fluoride, and 0.1 mm phenylmethylsulfonyl fluoride. The protein concentration of the samples was determined using the BCA Protein assay kit (Pierce, Thermo Fisher Scientific Inc.). Cell lysates were then electrophoresed on 8 or 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad). Subsequently, the membranes were blocked with 5% milk in Tris-buffered saline, 0.1% Tween buffer for 1 h (20 mm Tris-HCl, pH 7.6, 150 mm NaCl) and then incubated with the primary antibodies overnight at 4 °C. After 3 washes with Tris-buffered saline, 0.1% Tween, the membranes were incubated with the isotype-specific secondary antibody conjugated with the horseradish peroxidase (Cedarlane Laboratories, Burlington, Ontario, Canada) for 1 h at room temperature. This was followed by 3 washes with Tris-buffered saline, 0.1% Tween, and the protein was detected using the chemiluminescence detection kit (Pierce).
Colony Formation in Soft Agar
The soft agar consisted of two layers, both of which were prepared from a stock 1.2% Bacto-agar (Difco, Detroit, MI) dissolved in distilled water and autoclaved. For the bottom layer, Dulbecco's modified Eagle's medium supplemented with 10% FBS was added to the stock agar to achieve a 0.6% agar concentration. For the top layer, cell suspension (20,000 cells in 2 ml of Dulbecco's modified Eagle's medium supplemented with 10% FBS) was mixed with the stock agar to achieve a final agar concentration of 0.3%. Cells in the agar were fed with 200 μl of Dulbecco's modified Eagle's medium, 10% FBS every 2 days. Colonies were stained and visualized with 0.05% crystal violet after 4 weeks of culture.
Statistical Analysis
The association between SHP1 and the number of colonies formed in soft agar was evaluated using Student's t test. A p value ≤ 0.05 is considered statistically significant.
RESULTS
SHP1 Interacts with NPM-ALK but Not the NPM-ALKK210R Kinase-dead Mutant
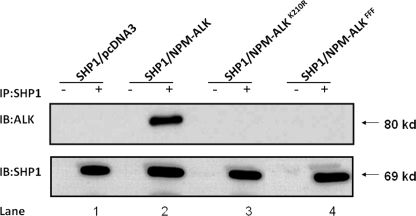
Based on a number of previously published studies (23, 29, 31, 32), the binding between NPM-ALK and partners is typically dependent on the autophosphorylation and activation status of NPM-ALK. Thus, we asked if the activation status of NPM-ALK is important to the physical interaction between SHP1 and NPM-ALK. To address this question, we co-transfected GP293 cells with SHP1 and NPM-ALK or the enzymatically inactive NPM-ALKK210R mutant. As shown in Fig. 1, SHP1 co-immunoprecipitated with NPM-ALK (lane 2); it did not bind the NPM-ALKK210R mutant (lane 3). We performed similar experiments using another NPM-ALK mutant, in which all three functionally important tyrosine residues in the kinase activation loop (KAL) of ALK have been mutated (labeled as FFF) (25, 33). As shown in lane 4, SHP1 also did not associate with the FFF mutant.
FIGURE 1.
The activation status of NPM-ALK is important for its binding to SHP1. Co-immunoprecipitation (IP) experiments using GP293 cells co-transfected with NPM-ALK (or its mutants) and SHP1 revealed binding of SHP1 to NPM-ALK (lane 2), but not the enzymatically inactive NPM-ALKK210R mutant (lane 3) or the NPM-ALKFFF mutant (lane 4). Immunoblotting (IB) with anti-SHP1 revealed a relatively equal amount of immunoprecipitated SHP1 proteins. Negative control reactions (−) were performed by omitting the use of anti-SHP1 antibody. Cells co-transfected with SHP1 and an empty vector (i.e. pcDNA3) were used as a negative control. Results shown are representative of three independent experiments.
The Importance of the KAL of ALK for the NPM-ALK/SHP1 Binding
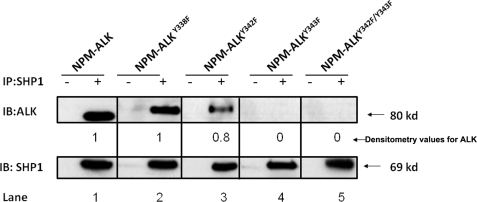
We have previously shown that mutations of one or more of the three tyrosine residues in the KAL of ALK result in a dramatic decrease in the number of phosphorylated tyrosine residues on NPM-ALK, its ability to phosphorylate substrates and its tumorigenicity (25). Our previously published data showed that Tyr338 is the first tyrosine residue in the KAL to be phosphorylated and it appears to be functionally more important than Tyr342 and Tyr343 (25). With this background, we assessed how mutation of each one of these three tyrosine residues in the KAL affects the NPM-ALK/SHP1 binding. As shown in Fig. 2, mutation of Tyr338, which was previously shown to dramatically decrease the overall level of tyrosine phosphorylation of NPM-ALK and the ability of NPM-ALK to phosphorylate various downstream targets, did not result in any substantial change in the binding between NPM-ALK and SHP1 (lane 2). Similarly, a relatively strong SHP1 binding was evident with the Tyr342 mutant (lane 3). In contrast, mutation of Tyr343 resulted in a complete loss of SHP1 binding detectable by our co-immunoprecipitation experiments (lane 4). Double mutation of Tyr342 and Tyr343 also abrogated the binding (lane 5), highlighting the importance of the Tyr343 residue in mediating the NPM-ALK/SHP1 interaction.
FIGURE 2.
The importance of the three tyrosine residues in the KAL of ALK for the binding between NPM-ALK and SHP1. Single mutation of the Tyr338 of the KAL showed no detectable change in its binding to SHP1 (lane 2) as compared with NPM-ALK (lane 1). A readily detectable SHP1 binding was also evident for the Tyr342 mutant (lane 3). In contrast, the Tyr343 mutant did not show detectable SHP1 binding (lane 4). In addition, double mutant NPM-ALKY342F/Y343F also showed a complete loss of SHP1 binding. The degree of SHP1 binding seen in each lane was assessed by densitometry and the results were normalized to the SHP1 protein level and determined relative to the SHP1 binding to unaltered NPM-ALK (lane 1). Results are representative of three independent experiments. Of note, the NPM-ALK vectors used for lanes 2–5 contained a His tag, which explains the slightly slower migration pattern seen in these lanes. IB, immunoblot.
Tyr343 Falls into a Consensus Sequence That Is Recognized by SHP1
To explain why mutation of Tyr343 results in the dramatic loss of SHP1 binding, we performed peptide sequence analysis, and assessed if Tyr343 falls into any of the known consensus sequences that can be recognized by SHP1. In this regard, we found a consensus sequence, (XXpY(Y/F)K/R) (34), present in a segment spanning Tyr343 (namely ASY342Y343R). In contrast, Tyr338 of NPM-ALK was not found to fall into any specific consensus sequences recognizable by SHP1. This finding further supports that Tyr343 represents the crucial binding site between NPM-ALK and SHP1, and explains why mutation of Tyr343, but not Tyr338 or Tyr342, results in a dramatic change in the binding between NPM-ALK and SHP1.
The Loss of SHP1 Binding Is Specific for the NPM-ALKTyr-343 Mutant
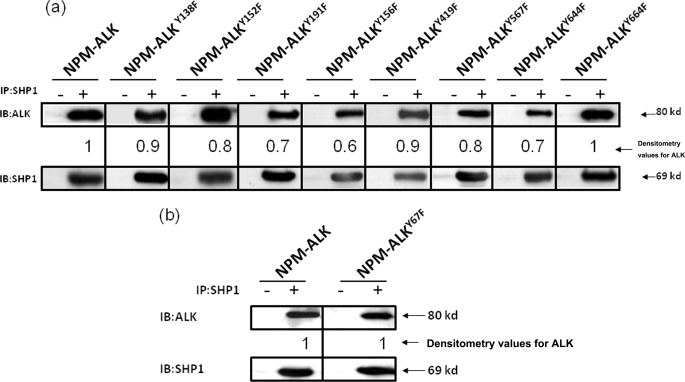
We then asked if the loss of SHP1 binding can be seen in mutations outside the KAL of ALK. Because our recent mass spectrometry studies revealed only 11 tyrosine residues (including the three in the KAL of ALK) that are phosphorylated (25), we performed site-directed mutagenesis of each of the remaining 8 tyrosine residues outside the KAL, including Tyr138, Tyr152, Tyr156, Tyr191, Tyr419, Tyr567, Tyr644, and Tyr664. Using co-immunoprecipitation experiments, we found that mutations of all of these 8 tyrosine residues resulted in readily detectable binding between NPM-ALK and SHP1 (Fig. 3a), and these findings are in sharp contrast with that for the Tyr343 mutant (Fig. 2). We also examined one randomly chosen tyrosine residue of NPM-ALK that is not involved in the phosphorylation, namely Tyr67; no loss of SHP1 binding was detected (Fig. 3b).
FIGURE 3.
The loss of SHP1 binding is specific for the NPM-ALKTyr-343 mutant. a, co-immunoprecipitation (IP) experiments using GP293 cells co-expressing SHP1 and 8 different NPM-ALK mutants (each of which contained a single mutation of the 8 remaining tyrosine residues known to be phosphorylated) showed no substantial or relatively minimal loss of SHP1 binding. b, a randomly selected mutant, Tyr67, which has been shown previously not to be phosphorylated, showed no loss of binding to SHP1. Results shown are representative of three independent experiments. The degree of SHP1 binding seen in each lane was assessed by densitometry, and the results were normalized to the SHP1 protein level and determined relative to the SHP1 binding to unaltered NPM-ALK. IB, immunoblot.
Tyrosine Dephosphorylation of NPM-ALK by SHP1 Is Dependent on Their Physical Interaction
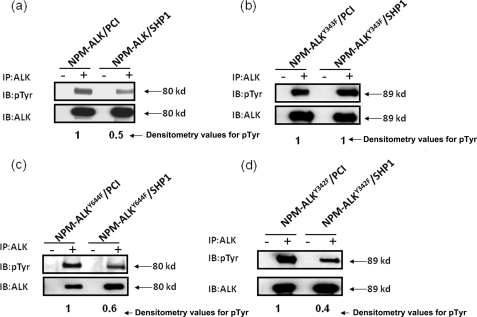
Because we identified Tyr343 of NPM-ALK as the crucial binding site for SHP1, we then asked if the SHP1-mediated tyrosine dephosphorylation of NPM-ALK is dependent on their physical interaction. A dependence on the physical interaction suggests that SHP1 directly dephosphorylates NPM-ALK. On the other hand, an independence could suggest that SHP1 dephosphorylates NPM-ALK indirectly (i.e. involves another mediator). As shown in Fig. 4a, the expression of SHP1 and NPM-ALK resulted in a 50% reduction of the tyrosine phosphorylation level of NPM-ALK. This level of tyrosine dephosphorylation induced by SHP1 is similar to that seen previously (14, 35). In contrast, SHP1 did not result in any detectable change in the tyrosine phosphorylation level of the Tyr343 mutant (Fig. 4b). We repeated similar experiments using other NPM-ALK mutants, including (i) the Tyr644 mutant, which is known to be phosphorylated and resides outside the KAL; and (ii) the Tyr342 mutant, which is known to be phosphorylated and resides in the KAL. As shown in Fig. 4, c and d, SHP1 dephosphorylated these mutants as effectively as it did for NPM-ALK.
FIGURE 4.
Tyrosine dephosphorylation of NPM-ALK by SHP1 is dependent on their physical interaction. GP293 cells co-transfected with NPM-ALK (or its mutants) and SHP1 (or the empty vector, PCI) were used for immunoprecipitation (IP) of NPM-ALK, followed by detection of the phosphorylation level of NPM-ALK using phosphotyrosine antibody. There is almost a 50% reduction of NPM-ALK tyrosine phosphorylation when SHP1 was coexpressed (a). In contrast, there was no detectable difference in the tyrosine phosphorylation of NPM-ALKY343F when it was coexpressed with SHP1 or an empty vector (b). The NPM-ALKY644F and NPM-ALKY342F showed similar results as NPM-ALK (c and d). Results shown are representative of three independent experiments. IB, immunoblot.
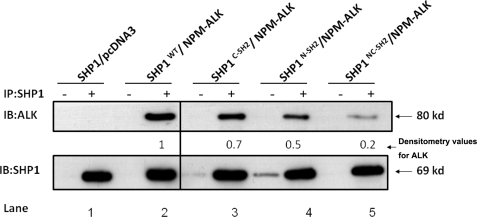
The SHP1/NPM-ALK Binding Is Dependent on Both SH2 Domains of SHP1, but Not Its Tyrosine Phosphatase Activity
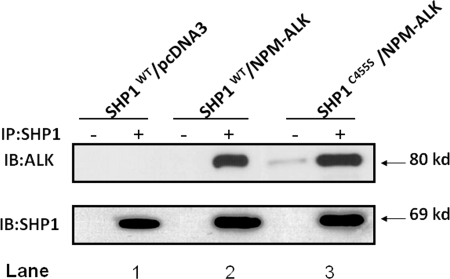
As shown in Fig. 5, in GP293 cells co-transfected with SHP1 and NPM-ALK, SHP1 co-immunoprecipitated with NPM-ALK (lane 2). In contrast, as shown in lanes 3-5, all three SHP1 mutants (i.e. with mutations in the N-SH2 domain, the C-SH2 domains, or both the N-SH2 and C-SH2 domains) had reduced binding to NPM-ALK. Thus, it is evident that both the N-SH2 and C-SH2 domains of SHP1 are important in mediating its binding to NPM-ALK. Reciprocal experiments using anti-ALK antibody for immunoprecipitation yielded similar conclusions regarding the importance of N-SH2 and C-SH2 domains of SHP1 in mediating the interaction between these two proteins (data not shown). Using the same experimental approach, we assessed whether the tyrosine phosphatase activity of SHP1 is important for the SHP1/NPM-ALK binding. As shown in Fig. 6, NPM-ALK co-immunoprecipitated with the SHP1C445S mutant (lane 3), previously reported to be “phosphatase-dead” (28).
FIGURE 5.
The SHP1/NPM-ALK binding is dependent on both of the SH2 domains of SHP1. Co-immunoprecipitation (IP) experiments using GP293 cells co-expressing NPM-ALK and SHP1 or its SH2 mutants showed reduced NPM-ALK/SHP1 interaction in all three mutants (lanes 3–5). Results shown are representative of three independent experiments. IB, immunoblot.
FIGURE 6.
The SHP1/NPM-ALK binding is not dependent on the phosphatase activity of SHP1. Co-immunoprecipitation (IP) experiments using GP293 cells showed that NPM-ALK/SHP1 binding was the same between NPM-ALK/SHP1 (lane 2) and NPM-ALK/SHP1C455S (lane 3). Results shown are representative of three independent experiments. IB, immunoblot.
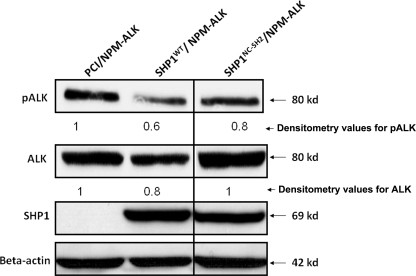
Mutation of the SH2 Domains of SHP1 Results in a Partial Loss of Its Inhibitory Effects on NPM-ALK
We then asked if mutations of the SH2 domain of SHP1 also affect its ability to dephosphorylate NPM-ALK. To address this question, we transfected SHP1 or its double SH2 mutant into Karpas 299 cells, a SHP1-negative ALK+ALCL cell line (14). As shown in Fig. 7, mutations of both N-SH2 and C-SH2 domains of SHP1 resulted in a partial decrease in the level of tyrosine phosphorylation of NPM-ALK, as compared with the parental SHP1 construct. This partial decrease was expected, as mutation of the SH2 domain of SHP1 did not completely abrogate the SHP1/NPM-ALK binding (Fig. 5).
FIGURE 7.
Mutation of the SH2 domains of SHP1 results in a partial loss of its inhibitory effects on NPM-ALK. Transfection of SHP1 in ALK+ALCL cell line Karpas 299 resulted in a dramatic down-regulation of phosphorylated NPM-ALK as well as the total ALK protein level. In contrast, the SHP1 mutant (mutation in both the N-SH2 and C-SH2 domains, SHP1NC-SH2) resulted in a partial decrease in the level of tyrosine phosphorylation of NPM-ALK, as compared with SHP1. The densitometry values were determined after normalization to the β-actin band. Results shown are representative of three independent experiments.
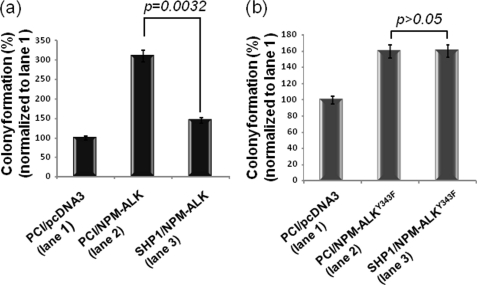
Soft Agar Clonogenic Assay
Using a soft agar clonogenic assay, we assessed how the physical interaction between SHP1 and NPM-ALK affects NPM-ALK-driven tumorigenicity. As shown in Fig. 8, the tumorigenicity of NPM-ALK in cells co-expressing SHP1 was significantly less than that seen in cells co-expressing an empty vector (p value = 0.0032, Student's t test). In contrast, there was no significant difference between tumorigenicity of the Y343F mutant in cells coexpressing either empty vector or SHP1 (p > 0.05, Student's t test).
FIGURE 8.
The tumorigenicity of NPM-ALK, but not that of NPM-ALKY343F, was suppressed by SHP1, as assessed by soft agar clonogenic assay. The colony formation induced by NPM-ALK in GP293 cells was significantly reduced by SHP1 (p = 0.0032). In contrast, the colony formation induced by NPM-ALKY343F was not significantly affected by SHP1 (p > 0.05). These experiments were performed in triplicates.
DISCUSSION
In normal cells, the tyrosine phosphorylation status of various proteins is tightly regulated by interaction of a host of kinases and phosphatases (36, 37). SHP1, a cytoplasmic tyrosine phosphatase largely expressed in hematopoietic cells, is known to dephosphorylate and inhibit various proteins including cytokine receptors (38–42) as well as cell-surface receptors involved in immune response (43–47). By virtue of the two SH2 domains, SHP1 physically interacts with specific phosphotyrosine residues in its substrates. Upon binding to these substrates, SHP1 undergoes intramolecular changes that set its tyrosine phosphatase domain free from the hindering effects of its N-SH2 domain (48). The “exposed” tyrosine phosphatase domain is able to dephosphorylate the substrates and thereby down-regulate their activities.
Reduction or loss of SHP1 expression has been found in a number of hematopoietic neoplasms (12, 20, 49–51). This loss of expression can be attributed to gene methylation, and restoration of SHP1 expression by 5-azathioprine has been shown in a number of hematopoietic cell lines (35, 50). In ALK+ALCL, the loss of SHP1 expression can be identified in up to 80% of these tumors, and this finding correlates with gene methylation of SHP1 (12). Its tumor suppressor function and inhibitory effects on NPM-ALK in these tumors have also been shown previously. Specifically, gene transfection of SHP1 down-regulated the activation/phosphorylation level of NPM-ALK and inhibits its cell growth-promoting effects (14, 21, 35).
Although the molecular events underlying the interactions between SHP1 and various substrates in normal cells have been extensively studied, how SHP1 binds and regulates oncogenic tyrosine kinases is incompletely understood. To our knowledge, there are only 3 published studies in which the physical and/or functional interactions between SHP1 and different oncogenic tyrosine kinases were examined (52–54). In the first study, Lim et al. (52) examined the effect of SHP1 on BCR-ABL and concluded that the physical binding between SHP1 and BCR-ABL is important for down-regulating the phosphorylation level and tumorigenicity of BCR-ABL. In contrast with our study, this study employed an artificially created fusion protein containing the catalytic domain of SHP1 and the ABL binding domain of RIN1 (a known binding partner of c-ABL). Thus, whether the SH2 domains of SHP1 are required for this physical interaction could not be determined. In addition, the tyrosine residue(s) of BCR-ABL involved in this interaction was not determined. In the second study, Hennige et al. (53) examined the inhibitory effect of SHP1 on Ret, an oncogenic tyrosine kinase expressed in medullary thyroid carcinoma. Although it is nicely demonstrated that SHP1 reduces the phosphorylation status of Ret and its oncogenic activity, details of the physical interaction of these two proteins and the importance of this interaction were not examined. Finally, Roccato et al. (54) identified the site of interaction between SHP1 and TRK-T3 (an oncogenic tyrosine kinase characteristic for papillary thyroid tumors) although the functional significance of the physical interaction between these two proteins was not addressed.
The requirement for both SH2 domains of SHP1 for its optimal interaction with NPM-ALK is in contrast with the previous finding that only the C-SH2 domain is important for mediating the physical interaction between SHP1 and FcγRIIb1 (43). Nevertheless, the requirement for both SH2 domains of SHP1 has been described for the SHP1-epidermal growth factor receptor interaction (27). Thus, the relative roles of N-SH2 and C-SH2 of SHP1 appear to vary among different substrates. Because mutations of both SH2 domains of SHP1 did not completely abrogate its binding to NPM-ALK, it is likely that other portions of SHP1 may contribute to the binding. In this regard, it has been previously reported that SHP1 binds the insulin receptor via a unique sequence located in the C-terminal tail of SHP1 (55). Because ALK is a member of the insulin receptor subfamily, this portion of SHP1 may also contribute to the NPM-ALK/SHP1 binding.
A recent study from our laboratory revealed that only 11 tyrosine residues in NPM-ALK are phosphorylated (25). Three of these 11 tyrosine residues are found in the KAL (Tyr338, Tyr342, and Tyr343). The other eight tyrosine residues were found to be confined to the ALK portion of the fusion protein, and they include Tyr138, Tyr152, Tyr191, Tyr156, Tyr419, Tyr567, Tyr644, and Tyr664. In the same study, we found that mutation of any of the three tyrosine residues in the KAL did not affect the phosphorylation status of the remaining two tyrosine residues. Furthermore, the tyrosine phosphorylation pattern outside the KAL is identical between the Tyr338 mutant and the Tyr343 mutant. These findings are relevant to this study, because we can conclude that, as compared with the Tyr338 mutant, the dramatic loss of SHP1 binding seen in the Tyr343 mutant is not because of a loss of phosphorylation in any tyrosine residue other than Tyr343 itself. Because Tyr343 is located in the KAL of ALK, one may have to consider the possibility that this loss of SHP1/NPM-ALK binding is related to the loss of the overall activity of NPM-ALK. We consider this possibility to be unlikely, because the Tyr338 or Tyr342 mutants, both of which have been shown to have significantly lower tumorigenicity than the Tyr343 mutant (25), did not show a substantial loss of SHP1 binding. To further show that Tyr343 is the specific binding site of SHP1, we found no substantial loss of SHP1 binding when we introduced mutations of each of the remaining 8 different tyrosine residues known to be involved in the phosphorylation of NPM-ALK. Of note, because the enzymatically inactive NPM-ALKK210R mutant resulted in abrogation of the SHP1/NPM-ALK binding, some degree of (but not full) phosphorylation and activation of NPM-ALK is required for its physical interaction with SHP1. The requirement for at least some degree of tyrosine phosphorylation is in keeping with the observation that SHP1 generally binds only the phosphorylated substrates (16).
Based on the presented data, we have developed a hypothetical model to describe the SHP1/NPM-ALK interaction. We speculate that one of the two SH2 domains of SHP1 directly binds Tyr343 of NPM-ALK, whereas the second SH2 domain of SHP1 is responsible for providing the optimal three-dimensional conformation for this interaction. This second SH2 domain may also interact with another specific tyrosine residue of NPM-ALK, although this interaction by itself is so weak that mutation of this specific tyrosine residue alone did not result in a dramatic effect in our co-immunoprecipitation experiments. This model explains why mutation of either SH2 domain of SHP1 resulted in a reduction in the NPM-ALK/SHP1 binding.
In conclusion, we identified Tyr343 of NPM-ALK as the crucial binding site for SHP1. Furthermore, the inhibitory effects of SHP1 on NPM-ALK require some degree of (but not full) activation of NPM-ALK. Their physical interaction is also partly dependent on the two SH2 domains of SHP1. Our results support the model that SHP1 exerts its inhibitory effects directly on NPM-ALK. To our knowledge, this is one of the most comprehensive studies of how SHP1 physically and functionally interacts with an oncogenic tyrosine kinase.
Acknowledgments
We thank Dr. Peter Kaiser for providing the HB-vector, Dr. Stephen Morris for providing the NPM-ALK cDNA plasmid, Dr. Frank Bohmer for providing the SHP1 vectors, and Dr. Zhenbao Yu for providing the SHP1C445S vector.
This work was supported by grants from the Canadian Institutes of Health Research and the Alberta Cancer Research Institute (to R. L.).
- ALK
- anaplastic lymphoma kinase
- NPM
- nucleophosmin
- ALCL
- anaplastic large cell lymphoma
- KAL
- kinase activation loop
- FBS
- fetal bovine serum
- SH2
- Src homology domain 2.
REFERENCES
- 1.Delsol G. (2008) Ann. Pathol. 28, S20–24 [DOI] [PubMed] [Google Scholar]
- 2.Morgan R., Hecht B. K., Sandberg A. A., Hecht F., Smith S. D. (1986) N. Engl. J. Med. 314, 1322. [DOI] [PubMed] [Google Scholar]
- 3.Rimokh R., Magaud J. P., Berger F., Samarut J., Coiffier B., Germain D., Mason D. Y. (1989) Br. J. Haematol. 71, 31–36 [DOI] [PubMed] [Google Scholar]
- 4.Le Beau M. M., Bitter M. A., Larson R. A., Doane L. A., Ellis E. D., Franklin W. A., Rubin C. M., Kadin M. E., Vardiman J. W. (1989) Leukemia 3, 866–870 [PubMed] [Google Scholar]
- 5.Mathew P., Sanger W. G., Weisenburger D. D., Valentine M., Valentine V., Pickering D., Higgins C., Hess M., Cui X., Srivastava D. K., Morris S. W. (1997) Blood 89, 1678–1685 [PubMed] [Google Scholar]
- 6.Morris S. W., Naeve C., Mathew P., James P. L., Kirstein M. N., Cui X., Witte D. P. (1997) Oncogene 14, 2175–2188 [DOI] [PubMed] [Google Scholar]
- 7.Shiota M., Fujimoto J., Semba T., Satoh H., Yamamoto T., Mori S. (1994) Oncogene 9, 1567–1574 [PubMed] [Google Scholar]
- 8.Bischof D., Pulford K., Mason D. Y., Morris S. W. (1997) Mol. Cell. Biol. 17, 2312–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amin H. M., Lai R. (2007) Blood 110, 2259–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Q., Raghunath P. N., Vonderheid E., Odum N., Wasik M. A. (2000) Am. J. Pathol. 157, 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonvini P., Gastaldi T., Falini B., Rosolen A. (2002) Cancer Res. 62, 1559–1566 [PubMed] [Google Scholar]
- 12.Khoury J. D., Rassidakis G. Z., Medeiros L. J., Amin H. M., Lai R. (2004) Blood 104, 1580–1581 [DOI] [PubMed] [Google Scholar]
- 13.Qiu L., Lai R., Lin Q., Lau E., Thomazy D. M., Calame D., Ford R. J., Kwak L. W., Kirken R. A., Amin H. M. (2006) Blood 108, 2407–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Y., Amin H. M., Franko B., Frantz C., Shi X., Lai R. (2006) Blood 108, 2796–2803 [DOI] [PubMed] [Google Scholar]
- 15.Dien Bard J., Gelebart P., Anand M., Zak Z., Hegazy S. A., Amin H. M., Lai R. (2009) Am. J. Pathol. 175, 825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J., Somani A. K., Siminovitch K. A. (2000) Semin. Immunol. 12, 361–378 [DOI] [PubMed] [Google Scholar]
- 17.Kozlowski M., Mlinaric-Rascan I., Feng G. S., Shen R., Pawson T., Siminovitch K. A. (1993) J. Exp. Med. 178, 2157–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenz U., Bergemann A. D., Steinberg H. N., Flanagan J. G., Li X., Galli S. J., Neel B. G. (1996) J. Exp. Med. 184, 1111–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witkiewicz A., Raghunath P., Wasik A., Junkins-Hopkins J. M., Jones D., Zhang Q., Odum N., Wasik M. A. (2007) Hum. Pathol. 38, 462–467 [DOI] [PubMed] [Google Scholar]
- 20.Amin H. M., Hoshino K., Yang H., Lin Q., Lai R., Garcia-Manero G. (2007) J. Pathol. 212, 402–410 [DOI] [PubMed] [Google Scholar]
- 21.Honorat J. F., Ragab A., Lamant L., Delsol G., Ragab-Thomas J. (2006) Blood 107, 4130–4138 [DOI] [PubMed] [Google Scholar]
- 22.Bai R. Y., Dieter P., Peschel C., Morris S. W., Duyster J. (1998) Mol. Cell. Biol. 18, 6951–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai R. Y., Ouyang T., Miething C., Morris S. W., Peschel C., Duyster J. (2000) Blood 96, 4319–4327 [PubMed] [Google Scholar]
- 24.Cussac D., Greenland C., Roche S., Bai R. Y., Duyster J., Morris S. W., Delsol G., Allouche M., Payrastre B. (2004) Blood 103, 1464–1471 [DOI] [PubMed] [Google Scholar]
- 25.Wang P., Wu F., Ma Y., Li L., Lai R., Young L. C. (2010) J. Biol. Chem. 285, 95–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plutzky J., Neel B. G., Rosenberg R. D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 1123–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenev T., Keilhack H., Tomic S., Stoyanov B., Stein-Gerlach M., Lammers R., Krivtsov A. V., Ullrich A., Böhmer F. D. (1997) J. Biol. Chem. 272, 5966–5973 [DOI] [PubMed] [Google Scholar]
- 28.Yu Z., Su L., Hoglinger O., Jaramillo M. L., Banville D., Shen S. H. (1998) J. Biol. Chem. 273, 3687–3694 [DOI] [PubMed] [Google Scholar]
- 29.Ambrogio C., Voena C., Manazza A. D., Piva R., Riera L., Barberis L., Costa C., Tarone G., Defilippi P., Hirsch E., Boeri Erba E., Mohammed S., Jensen O. N., Palestro G., Inghirami G., Chiarle R. (2005) Blood 106, 3907–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amin H. M., Medeiros L. J., Ma Y., Feretzaki M., Das P., Leventaki V., Rassidakis G. Z., O'Connor S. L., McDonnell T. J., Lai R. (2003) Oncogene 22, 5399–5407 [DOI] [PubMed] [Google Scholar]
- 31.Zamo A., Chiarle R., Piva R., Howes J., Fan Y., Chilosi M., Levy D. E., Inghirami G. (2002) Oncogene 21, 1038–1047 [DOI] [PubMed] [Google Scholar]
- 32.Voena C., Conte C., Ambrogio C., Boeri Erba E., Boccalatte F., Mohammed S., Jensen O. N., Palestro G., Inghirami G., Chiarle R. (2007) Cancer Res. 67, 4278–4286 [DOI] [PubMed] [Google Scholar]
- 33.Tartari C. J., Gunby R. H., Coluccia A. M., Sottocornola R., Cimbro B., Scapozza L., Donella-Deana A., Pinna L. A., Gambacorti-Passerini C. (2008) J. Biol. Chem. 283, 3743–3750 [DOI] [PubMed] [Google Scholar]
- 34.Hampel K., Kaufhold I., Zacharias M., Böhmer F. D., Imhof D. (2006) ChemMedChem 1, 869–877 [DOI] [PubMed] [Google Scholar]
- 35.Han Y., Amin H. M., Frantz C., Franko B., Lee J., Lin Q., Lai R. (2006) Leukemia 20, 1602–1609 [DOI] [PubMed] [Google Scholar]
- 36.Wu C., Sun M., Liu L., Zhou G. W. (2003) Gene 306, 1–12 [DOI] [PubMed] [Google Scholar]
- 37.Tonks N. K. (2006) Nat. Rev. Mol. Cell Biol. 7, 833–846 [DOI] [PubMed] [Google Scholar]
- 38.Yi T., Zhang J., Miura O., Ihle J. N. (1995) Blood 85, 87–95 [PubMed] [Google Scholar]
- 39.Klingmüller U., Lorenz U., Cantley L. C., Neel B. G., Lodish H. F. (1995) Cell 80, 729–738 [DOI] [PubMed] [Google Scholar]
- 40.David M., Chen H. E., Goelz S., Larner A. C., Neel B. G. (1995) Mol. Cell. Biol. 15, 7050–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi T., Mui A. L., Krystal G., Ihle J. N. (1993) Mol. Cell. Biol. 13, 7577–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Migone T. S., Cacalano N. A., Taylor N., Yi T., Waldmann T. A., Johnston J. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3845–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D'Ambrosio D., Hippen K. L., Minskoff S. A., Mellman I., Pani G., Siminovitch K. A., Cambier J. C. (1995) Science 268, 293–297 [DOI] [PubMed] [Google Scholar]
- 44.Otipoby K. L., Draves K. E., Clark E. A. (2001) J. Biol. Chem. 276, 44315–44322 [DOI] [PubMed] [Google Scholar]
- 45.Wu Y., Nadler M. J., Brennan L. A., Gish G. D., Timms J. F., Fusaki N., Jongstra-Bilen J., Tada N., Pawson T., Wither J., Neel B. G., Hozumi N. (1998) Curr. Biol. 8, 1009–1017 [DOI] [PubMed] [Google Scholar]
- 46.Adachi T., Flaswinkel H., Yakura H., Reth M., Tsubata T. (1998) J. Immunol. 160, 4662–4665 [PubMed] [Google Scholar]
- 47.Kosugi A., Sakakura J., Yasuda K., Ogata M., Hamaoka T. (2001) Immunity 14, 669–680 [DOI] [PubMed] [Google Scholar]
- 48.Yang J., Liu L., He D., Song X., Liang X., Zhao Z. J., Zhou G. W. (2003) J. Biol. Chem. 278, 6516–6520 [DOI] [PubMed] [Google Scholar]
- 49.Oka T., Ouchida M., Koyama M., Ogama Y., Takada S., Nakatani Y., Tanaka T., Yoshino T., Hayashi K., Ohara N., Kondo E., Takahashi K., Tsuchiyama J., Tanimoto M., Shimizu K., Akagi T. (2002) Cancer Res. 62, 6390–6394 [PubMed] [Google Scholar]
- 50.Koyama M., Oka T., Ouchida M., Nakatani Y., Nishiuchi R., Yoshino T., Hayashi K., Akagi T., Seino Y. (2003) Lab. Invest. 83, 1849–1858 [DOI] [PubMed] [Google Scholar]
- 51.Chim C. S., Wong K. Y., Loong F., Srivastava G. (2004) Leukemia 18, 356–358 [DOI] [PubMed] [Google Scholar]
- 52.Lim Y. M., Wong S., Lau G., Witte O. N., Colicelli J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12233–12238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hennige A. M., Lammers R., Höppner W., Arlt D., Strack V., Teichmann R., Machicao F., Ullrich A., Häring H. U., Kellerer M. (2001) Endocrinology 142, 4441–4447 [DOI] [PubMed] [Google Scholar]
- 54.Roccato E., Miranda C., Raho G., Pagliardini S., Pierotti M. A., Greco A. (2005) J. Biol. Chem. 280, 3382–3389 [DOI] [PubMed] [Google Scholar]
- 55.Uchida T., Matozaki T., Noguchi T., Yamao T., Horita K., Suzuki T., Fujioka Y., Sakamoto C., Kasuga M. (1994) J. Biol. Chem. 269, 12220–12228 [PubMed] [Google Scholar]