Abstract
Type 1 diabetes is an autoimmune disorder characterized by chronic inflammation and pancreatic β-cell loss. Here, we demonstrate that the proinflammatory cytokine interleukin-1β, combined with interferon-γ, induces the expression of the Bcl-2 homology 3 (BH3)-only activator PUMA (p53 up-regulated modulator of apoptosis) in β-cells. Transcriptional activation of PUMA is regulated by nuclear factor-κB and endoplasmic reticulum stress but is independent of p53. PUMA activation leads to mitochondrial Bax translocation, cytochrome c release, and caspase-3 cleavage resulting in β-cell demise. The antiapoptotic Bcl-XL protein is localized mainly at the mitochondria of the β-cells and antagonizes PUMA action, but Bcl-XL is inactivated by the BH3-only sensitizer DP5/Hrk in cytokine-exposed β-cells. Moreover, a pharmacological mimic of the BH3-only sensitizer Bad, which inhibits Bcl-XL and Bcl-2, induces PUMA-dependent β-cell death and potentiates cytokine-induced apoptosis. Our data support a hierarchical activation of BH3-only proteins controlling the intrinsic pathway of β-cell apoptosis in the context of inflammation and type 1 diabetes.
Keywords: Apoptosis, Cell Death, Diabetes, Endoplasmic Reticulum stress, Signal Transduction, PUMA, Pancreatic β-Cells
Introduction
Diabetes affects the quality of life of millions of individuals worldwide (1). The two main forms of diabetes, type 1 (T1D)4 and type 2, are characterized by progressive pancreatic β-cell loss (2). In T1D, β-cells are selectively targeted by the immune system and die through apoptosis (3). Previous work has demonstrated that the proinflammatory cytokines interleukin (IL)-1β and interferon (IFN)-γ contribute to β-cell apoptosis and the build-up of insulitis in the early stages of T1D (2–4). In vitro exposure of β-cells to IL-1β + IFN-γ causes functional changes similar to those observed in pre-T1D patients: (i) elevated proinsulin/insulin ratio (5), (ii) a preferential loss of the first-phase insulin response to glucose (6), and (iii) β-cell death (2, 7). Proinflammatory cytokines modulate the activity of several target genes and proteins in β-cells (2). The final outcome is depletion of endoplasmic reticulum (ER) Ca2+ and ER stress (8), mitochondrial Bax translocation, cytochrome c release, and activation of caspases (9, 10). The mechanism(s) responsible for cytokine-induced β-cell death, however, remains to be clarified.
The Bcl-2 homology 3 (BH3)-only proteins participate in several, if not all, apoptotic pathways (11). These Bcl-2 members interact through their BH3 domain with other pro- and antiapoptotic Bcl-2 proteins modulating cell death or survival. The mechanisms and outcome depend on the cell type and apoptotic stimulus. Recent evidence suggests that BH3-only proteins can be divided in two subgroups: the sensitizers (DP5/Hrk, Bik, Bad, and Noxa) and the activators (Bid, Bim, and PUMA) (12). According to this model, the first subgroup inactivates prosurvival Bcl-2 proteins (Bcl-2, Bcl-XL, Mcl-1, Bcl-W, and A1), whereas the second activates the proapoptotic members Bax and/or Bak. Once activated, Bax translocates from the cytosol to the mitochondria and forms pores in the membrane, releasing proapoptotic proteins, such as cytochrome c, to the cytoplasm (13). After a specific stimulus, the activator BH3-only proteins directly bind and activate Bax and Bak (14); this process is antagonized by prosurvival Bcl-2 proteins (15). In this context, the proposed role for the BH3-only sensitizers (first subgroup) is to displace activators from prosurvival proteins, allowing them to bind and activate Bax and Bak. Recent data from our group demonstrated that the sensitizer DP5, also known as Hrk, is central for β-cell demise after exposure to proinflammatory cytokines or chemical ER stressors (16). The nature of the putative activator molecule downstream of DP5 remains, however, to be clarified.
In the present work, we provide evidence that the BH3-only activator p53 up-regulated modulator of apoptosis (PUMA) is induced in β-cells after cytokine or chemical ER stressor exposure, leading to mitochondrial Bax translocation and cell death. Knockdown of PUMA prevents Bax activation, mitochondrial cytochrome c release, and caspase-3 cleavage, protecting β-cells from apoptosis. These results suggest that PUMA is an interesting target for prevention of β-cell demise and inhibition of the amplification of the autoimmune response in T1D.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments
Human islets were isolated from nondiabetic organ donors in Pisa, Italy, with the approval of the local Ethics Committee. Islets were isolated by enzymatic digestion and density-gradient purification, as described previously (17), placed in M199 culture medium containing 5.5 mm glucose and cultured in a CO2 incubator, and then shipped to Brussels for study within 1–5 days. After overnight recovery in Ham's F-10 medium containing 6.1 mm glucose, 2 mm GlutaMAX, 50 μm 3-isobutyl-1-methylxanthine, 1% bovine serum albumin, 50 units/ml penicillin, 50 μg/ml streptomycin, and 10% fetal calf serum, whole (for real time RT-PCR experiments) or dispersed (for viability experiments) islets were exposed to cytokines (human recombinant IL-1β (50 units/ml) + human recombinant IFN-γ (1000 units/ml; R&D systems)) as described (18). Of note, exposure of human or rat β-cells to the IL-1β + IFN-γ combination, but not to either cytokine alone, triggers apoptosis (2, 3). The percentage of β-cells in the human preparations (37–70%) was assessed in dispersed islets following staining with mouse monoclonal anti-insulin antibody (1:1000; Sigma) and donkey anti-mouse IgG rhodamine (1:200; Jackson ImmunoResearch Europe, Soham, Cambridgeshire, UK) (18).
Pancreatic islets were isolated from adult Wistar rats (Charles River Laboratories Belgium, Brussels, Belgium), housed, and used according to the guidelines of the Belgian Regulations for Animal Care. All experimental protocols utilized were approved by the Ethical Committee for Animal Experiments of the Université Libre de Bruxelles. Islets were isolated by collagenase digestion, and β-cells were purified by FACS (fluorescence-activated cell sorting, FACStar; Becton-Dickinson and Co.) and precultured for 48 h in Ham's F-10 medium before subsequent experimental procedures (8, 19, 20). Insulin-producing INS-1E cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 5% fetal calf serum (21). Recombinant rat IFN-γ (R&D Systems) and human recombinant IL-1β (a kind gift from Dr. C. W. Reinolds, NCI, National Institutes of Health) were used at the following concentrations: IFN-γ, 100 units/ml and 500 units/ml and IL-1β, 10 units/ml and 50 units/ml, for INS-1E cells and primary β-cells, respectively. The nitric oxide (NO) blocker l-NMA (NG-methyl-l-arginine; Sigma) was used at 2.5 mm. Culture medium was collected for nitrite determination (nitrite is a stable product of NO oxidation) by the Griess method (21). Cyclopiazonic acid (CPA; 20 μm), tunicamycin (5 μm), and thapsigargin (1 μm) were purchased from Sigma and dissolved in dimethyl sulfoxide. The Bcl-XL/Bcl-2 inhibitor ABT-737 was purchased from Selleck Chemicals LLC (Houston, TX) and used according to the manufacturer's instructions. Cytokines, l-NMA, CPA, tunicamycin, thapsigargin, and ABT-737 concentrations were selected based on time course and dose-response studies (data not shown and Refs. 3, 21, 22).
Immunofluorescence and Microscopy Confocal Analysis
Cells were plated on polylysine-coated coverslips, treated with cytokines, fixed with 4% paraformaldehyde, and permeabilized with 0.3% Triton X-100 (Sigma). The cells were then incubated overnight with the primary antibodies rabbit anti-Bax (1:200; Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-ATP synthase β (1:1000; Sigma), and mouse anti-cytochrome c (1:200; BD Biosciences). Fluorescein isothiocyanate- or rhodamine-conjugated secondary antibodies (1:200; Jackson ImmunoResearch, Westgrove, PA) were used for visualization by inverted fluorescence microscopy (Zeiss Axiovert 200, Oberkochen, Germany).
For confocal microscopy, cells were fixed and treated as described above using rabbit anti-Bcl-XL (1:200; Cell Signaling, Danvers, MA), rabbit anti-Bax (1:200; Santa Cruz Biotechnology), and mouse anti-cytochrome c (1:200; BD Biosciences) antibodies. The cells were analyzed with an LSM510 NLO multiphoton confocal microscope fitted on an Axiovert M200 inverted microscope equipped with C-Apochromat 40×/1.2 N.A. and 63×/1.2 N.A. water immersion objectives (Zeiss, Iena, Germany). The datasets generated were merged and displayed with the Zeiss LSM510 software. DNA staining was performed with Hoechst 33342 (10 μg/ml).
Real Time RT-PCR
mRNA expression was determined by real time RT-PCR using SYBR Green fluorescence on a LightCycler instrument (Roche Applied Science). Primer sequences for rat and human PUMA, Bcl-XL, and Bim are provided in supplemental Table S1. Expression of the gene of interest was divided by the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and presented as fold induction of control. GAPDH expression is not modified under the present experimental conditions (data not shown) (18, 21).
Western Blotting
Equal amounts of proteins were resolved by 10% SDS-PAGE. Immunoblot analysis was performed with mouse anti-PUMA (1:400, ProSci, Inc.; Poway, CA), rabbit anti-cleaved caspase-3, rabbit anti-Bcl-XL, rabbit anti-Bim (1:1000; Cell Signaling), and mouse anti-α-tubulin antibodies (1:5000; Sigma). The proteins were detected using horseradish peroxidase-conjugated secondary antibody (1:5000; Santa Cruz Biotechnology) and chemiluminescence Supersignal (Pierce). Protein loading was normalized for α-tubulin signal. Band intensity was quantified versus a control sample (considered as 1), using Aida1D analysis software (Fujifilm, London, UK).
Small Interfering RNA (siRNA) Treatment
Cells were transfected overnight with 30 nm PUMA smart Pool (Dharmacon; Chicago, IL), siRNA PUMA (ACGAGCGGCGGAGACAAGAAGAGCA), siRNA Bim 1 (GAGUUCAAUGAGACUUACACGAGGA), siRNA Bim 2 (CGAGGAGGGCGUUUGCAAACGAUUA), siRNA Bcl-XL 1 (GCGUAGACAAGGAGAUGCAGGUAUU), siRNA Bcl-XL 2 (AGAGAAAGUCAACCACCAGCUCCCG), siRNA DP5 (UCACAGUUUCUUGGUGCUAAGUGUA), or medium GC content inactive control siRNA (Invitrogen) using 1 μl of DharmaFECTTM lipid reagent (Dharmacon) to 150 nm siRNA in Opti-MemR (Invitrogen), with an efficiency of transfection >90% (18, 20, 23). After overnight incubation, transfection medium was replaced by regular culture medium for cell recovery.
Cell Viability
The percentage cell death was determined in at least 600 cells/experimental condition by inverted fluorescence microscopy after staining with the DNA dyes Hoechst 33342 (10 μg/ml) and propidium iodide (5 μg/ml) (HO/PI) (8, 16, 20, 21). Viability was evaluated by at least two observers, one of whom was unaware of sample identity. Apoptosis was also confirmed by alternative methods (e.g. caspase-3 activation, Bax translocation, and cytochrome c release) as suggested previously (24).
PUMA Reporter Assay
Previously described PUMA promoter constructs driving the luciferase reporter gene were used (25). INS-1E cells were transfected for 6 h using Lipofectamine (Invitrogen) with 250 ng of different luciferase reporter constructs and the pRL-CMV plasmid (50 ng, with Renilla luciferase used as internal control for transfection efficiency). 24 h after transfection, the cells were exposed to IL-1β + IFN-γ for an additional 24 h. Luciferase activity of cell lysates was then determined and represented as firefly/Renilla (relative luciferase activity).
Infection with Recombinant Adenoviruses
After 48 h preculture, cells were infected either with AdLUC (control luciferase expressing virus) or AdIκB(SA)2 (encoding a previously described NF-κB superrepressor protein) (26). INS-1E cells were infected for 2 h at 37 °C with a multiplicity of infection of 10.
Immunoprecipitation
Immunoprecipitation was performed using immunoprecipitation buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1% Nonidet P-40, protease inhibitor mixture (Roche Applied Science)). Cells were lysed with immunoprecipitation buffer for 30 min on ice, followed by centrifugation at 10,000 × g for 15 min. Total protein was used as the starting material for immunoprecipitations. Rabbit anti-Bcl-XL antibody (Cell Signaling) was added to a final concentration of 5 mg/ml and incubated overnight at 4 °C. Antibody-protein complexes were collected with the 50% protein A-agarose slurry (Santa Cruz Biotechnology), washed, and then boiled in sample buffer to remove the antibody-protein complex from the protein A slurry. Samples were then subjected to SDS-PAGE and immunoblotted with rabbit anti-PUMA or anti-Bcl-XL antibody (Cell Signaling).
Statistical Analysis
Data are means ± S.E. of three to six independent experiments. Comparisons between groups are made by paired t test or by analysis of variance followed by t test with the Bonferroni correction.
RESULTS
BH3-only Activator Protein PUMA Is Induced by Cytokines in β-Cells
The proinflammatory cytokines IL-1β + IFN-γ induced mitochondrial Bax translocation, cytochrome c release. and caspase-9, -7, and -3 activation in β-cells (Fig. 1A and supplemental Fig. S1, A–C), events that characterize the intrinsic pathway of apoptosis (11). Of note, caspase-8 cleavage was not detected (supplemental Fig. S1C). Previous studies in other cell types demonstrated that PUMA is an efficient activator of the intrinsic apoptotic pathway (27, 28). Importantly, proinflammatory cytokines induced PUMA expression in four human islet preparations (Fig. 1B). PUMA induction was also observed in FACS-purified primary rat β-cells and insulin-producing INS-1E cells by real time RT-PCR and Western blotting (Fig. 1, C–E and supplemental Fig. S1D). There was a relative dissociation between cytokine-induced PUMA mRNA (Fig. 1D) and protein expression (Fig. 1E), which may be due to delayed protein translation and/or longer protein half-life. IL-1β or IFN-γ alone did not induce PUMA mRNA transcription (supplemental Fig. S1E), in keeping with the fact that neither of these cytokines alone induced β-cell apoptosis (3, 4), but the cytokine combination increased 8-fold PUMA expression and caused cell death. On the other hand, IL-1β + IFN-γ did not significantly modify the different isoforms generated by alternative mRNA splicing of the BH3-only activator Bim (BimEL, BimL, and BimS; Fig. 1, F and G, and supplemental Fig. S1D).
FIGURE 1.
IL-1β + IFN-γ induce mitochondrial Bax translocation and PUMA activation in β-cells. A, representative immunofluorescence images of cytochrome c and Bax localization in FACS-purified primary β-cells treated for 24 h with IL-1β + IFN-γ. Arrows indicate mitochondrial Bax translocation and cytochrome c release, n = 3. Scale bar, 20 μm. B, expression of PUMA in human islets isolated from four organ donors, under control condition or following treatment with IL-1β + IFN-γ. PUMA expression was determined by real time RT-PCR and is represented as fold induction compared with control (nontreated cells). C, PUMA mRNA expression in primary β-cells exposed to cytokines. Real time RT-PCR for PUMA was performed at the indicated time points after cytokine addition, n = 4–5. *, p < 0.05 versus control. D, time course of PUMA mRNA expression after cytokine exposure of INS-1E cells, as evaluated by real-time RT-PCR. Data are means ± S.E., n = 3. *, p < 0.05 versus control (time 0 h). E, Western blot demonstrating increased expression of PUMA protein in cytokine-treated INS-1E cells. Densitometric quantification of bands is shown on the right as fold induction of control after correction for α-tubulin, n = 5. *, p < 0.05 versus control. F, time course of Bim mRNA expression in INS-1E cells after cytokine exposure, evaluated by real time RT-PCR. Data are means ± S.E., n = 3. G, time course of Bim protein expression after cytokine treatment. Cell lysates were subjected to Western blotting with antibodies detecting BimEL, BimL, BimS, or α-tubulin as loading control (left side of the figure). Quantification of bands is shown on the right as fold induction of control after correction for α-tubulin, n = 3.
PUMA Knockdown by siRNAs Prevents the Triggering of the Intrinsic Apoptotic Pathway
The role of PUMA in IL-1β + IFN-γ-induced β-cell apoptosis was evaluated by using siRNA technology. Both smart Pool and single-sequence PUMA siRNAs were used to knock down PUMA, achieving more than 60% silencing compared with an inactive control siRNA in INS-1E cells (Fig. 2, A and B, and supplemental Fig. S2A). PUMA knockdown did not affect viability under basal conditions, but it decreased cytokine-induced apoptosis by 2-fold at 24 h (Fig. 2C). Knockdown of Bim by two different siRNAs, on the other hand, increased basal cell death and did not affect apoptosis mediated by cytokines (Fig. 2, A–C). Importantly, primary β-cells were also partially protected against IL-1β + IFN-γ-induced apoptosis by PUMA knockdown (Fig. 2D and supplemental Fig. S2B). In line with these findings, knockdown of PUMA diminished cytokine-induced mitochondrial Bax translocation, cytochrome c release, and caspase-3 activation in INS-1E cells (Fig. 2, E–G).
FIGURE 2.
PUMA knockdown protects β-cells from cytokine-induced apoptosis. A, INS-1E cells were transfected with inactive siRNA (Control), a single-sequence siRNA against PUMA (siRNA PUMA), PUMA smart Pool siRNAs (smart Pool), or two different siRNAs against Bim (siRNA Bim 1 and siRNA Bim 2) as indicated. Real time RT-PCR for PUMA or Bim and GAPDH mRNA expression was performed 72 h after siRNA transfection, n = 3. *, p < 0.05; **, p < 0.01 versus control. B, Western blot analysis of PUMA and Bim expression in INS-1E cells was performed after siRNA transfection and subsequent 16-h cytokine treatment. Densitometric quantification of the PUMA and BimEL bands (with correction for the housekeeping protein α-tubulin) is shown in supplemental Fig. S2A, n = 5. C, INS-1E cells were transfected with PUMA, Bim, or control siRNAs and then exposed for 24 h to IL-1β + IFN-γ. Cell death was measured by HO/PI, n = 5. *, p < 0.05; **, p < 0.01; ***, p < 0.001. D, PUMA knockdown partially prevents primary β-cell death after 48-h cytokine treatment, n = 4. **, p < 0.01. E and F, untreated (control) or cytokine-treated INS-1E cells transfected with siRNA control or siRNA PUMA were stained with antibodies against Bax or cytochrome c and Hoechst for nuclear morphology evaluation, followed by immunofluorescence analysis. The percentage of cells exhibiting Bax translocation (E; arrows) or diffuse or no cytochrome c staining (F; arrows) was determined. Scale bar, 20 μm; n = 3. *, p < 0.05. G, INS-1E cells were transfected with control or PUMA siRNAs, and Western blotting for cleaved caspase-3 was performed 16 h after cytokine addition, n = 3. *, p < 0.05.
NF-κB and ER Stress Induce PUMA Expression in β-Cells
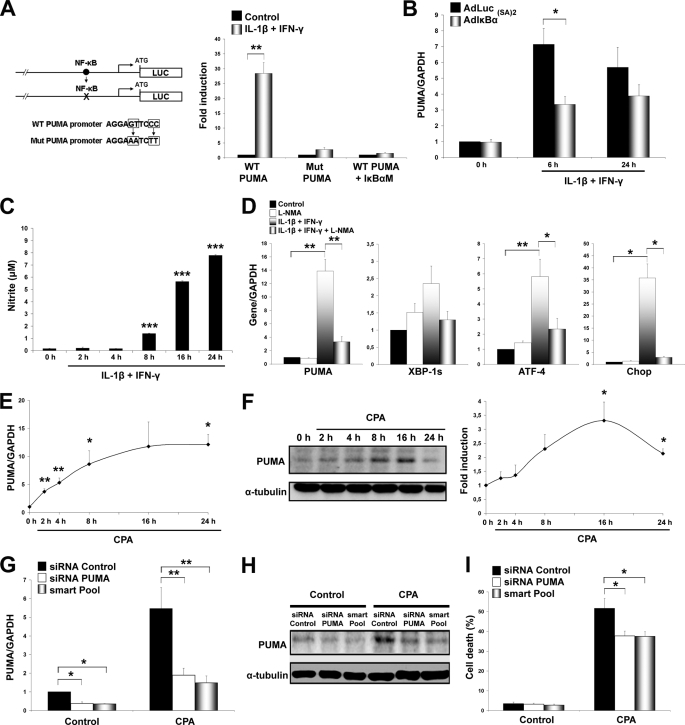
The transcription factor p53 was the first identified modulator of PUMA (27, 28). Cytokine exposure did not modify p53 expression, nor did it induce its nuclear translocation in β-cells (supplemental Fig. S3). Regulation of PUMA transcription also involves p53-independent mechanisms, including the mitogen-activated protein kinase pathway leading to JNK and c-Jun phosphorylation (29). Inactivation of JNK, however, did not affect cytokine-mediated PUMA expression in β-cells (supplemental Fig. S4). We have shown previously that the concentrations of SP600125 and peptide JNK inhibitor utilized here prevent cytokine-induced JNK activation in β-cells (16). Recent evidence indicates that the transcription factor NF-κB induces PUMA up-regulation following tumor necrosis factor α exposure (25). NF-κB is induced early after cytokine exposure (16), and we thus analyzed luciferase reporter constructs containing the NF-κB binding site in the PUMA promoter (Fig. 3A). IL-1β + IFN-γ treatment markedly activated the PUMA reporter. Mutations in the NF-κB binding site of the PUMA promoter abolished the responsiveness of the reporter to cytokines, indicating that this NF-κB site is crucial for PUMA gene transcription. Cotransfection of PUMA promoter constructs with plasmids expressing an IκB superrepressor (25), but not the control vector, also abolished the PUMA promoter activation by cytokines (Fig. 3A). Consistently, infection with a NF-κB-inactivating adenovirus AdIκB(SA)2 (26) prevented cytokine-induced PUMA up-regulation in INS-1E cells (Fig. 3B). The inactivation was only partial, suggesting that NF-κB likely synergizes with another mechanism(s).
FIGURE 3.
PUMA induction is regulated by NF-κB and ER stress. A, mutation of the NF-κB site abolished activation of the PUMA promoter. The wild type NF-κB site was mutated as is represented (left panel). INS-1E cells were cotransfected with PUMA luciferase reporter constructs containing the wild type (WT PUMA) or NF-κB mutated site (Mut PUMA), in combination or not with plasmids expressing an IκB superrepressor (IκBαM) or a pcDNA control plasmid and then treated for 24 h with IL-1β + IFN-γ. Luciferase activity was expressed as fold induction of respective controls, n = 6. **, p < 0.01. B, AdIκB(SA)2 prevents cytokine-induced PUMA expression. INS-1E cells were infected with AdLUC or AdIκB(SA)2 and then exposed to IL-1β + IFN-γ for 16 h. PUMA expression was determined by real time RT-PCR, n = 4. *, p < 0.05. C, nitrite accumulated in the medium of cytokine-treated INS-1E cells over time, n = 3. ***, p < 0.001 versus control (time 0 h). D, INS-1E cells were treated for 16 h with IL-1β + IFN-γ, l-NMA or combination, as indicated. Expression of mRNAs encoding PUMA and ER stress markers was measured by real time RT-PCR, n = 3. *, p < 0.05; **, p < 0.01. E, time course of CPA-induced PUMA mRNA in INS-1E cells, as evaluated by real-time RT-PCR. Data are means ± S.E., n = 3. *, p < 0.05; **, p < 0.01 versus 0 h. F, Western blot demonstrating increased expression of PUMA protein in CPA-treated INS-1E cells. Densitometric quantification of bands is shown on the right as fold induction of control after correction for α-tubulin, n = 3. *, p < 0.05 versus control (time 0 h). G, INS-1E cells were transfected with inactive (control), single, or smart Pool PUMA siRNAs and PUMA mRNA expression was evaluated by real-time RT-PCR after 8 h CPA exposure, n = 4. *, p < 0.05; **, p < 0.01. H, INS-1E cells were transfected with inactive (control), PUMA or smart Pool siRNAs and exposed to CPA for 8 h and Western blotting for PUMA was performed. Densitometric quantification of the bands is shown in supplemental Fig. S6E, n = 4. I, PUMA knockdown decreased INS-1E cell death following a 24-h exposure to CPA, n = 6. *, p < 0.05.
NO, generated by the inducible form of NO synthase (iNOS), contributes to cytokine-induced gene expression, ER stress, and apoptosis in β-cells (3, 8, 21). Cytokines induced iNOS expression (supplemental Figs. S1, C and D, and S5A) and NO formation (Fig. 3C) as previously reported (3, 8, 26). Inhibition of NO formation by l-NMA (supplemental Fig. S5B) significantly decreased PUMA expression in β-cells exposed to cytokines (Fig. 3D). Cytokine-induced NO down-regulates the SERCA2 pump, depleting ER Ca2+ stores and triggering ER stress (8). Thus, inhibition of iNOS decreased cytokine induction of the ER stress markers ATF-4, Chop, and to a lesser extent XBP-1s (Fig. 3D). We next examined whether ER stress contributes to PUMA activation using two different chemical ER stressors, namely CPA and tunicamycin. Chemical ER stress up-regulated PUMA mRNA and protein levels in INS-1E cells (Fig. 3, E and F, and supplemental Fig. S6A). Increased PUMA mRNA expression (6-fold, n = 4) was also observed following exposure to the ER stressor thapsigargin at 8, 16, and 24 h (p < 0.05). Knockdown of PUMA by siRNA significantly attenuated CPA- and tunicamycin-mediated cell death in INS-1E cells and CPA-induced apoptosis in primary β-cells (Fig. 3, G–I and supplemental Fig. S6, B–E). Together, these results suggest that PUMA is an important downstream effector of ER stress-induced β-cell death and that ER stress may synergize with NF-κB to transcriptionally activate PUMA after cytokine exposure.
Bcl-XL Inactivation Releases PUMA and Induces β-Cell Demise
PUMA is functionally inhibited by Bcl-XL (30), and we thus evaluated its role in cytokine-induced β-cell apoptosis. Confocal studies demonstrated that Bcl-XL mainly colocalizes with cytochrome c at the mitochondria in both INS-1E and primary β-cells (Fig. 4A and supplemental Fig. S7). Inactivation of Bcl-XL by two different siRNAs (Fig. 4, B and C) triggered the mitochondrial apoptotic pathway as demonstrated by cytochrome c release (supplemental Fig. S8A), caspase-3 cleavage (Fig. 4C), and cell death induction (Fig. 4D). Cytokine exposure augmented apoptosis induced by Bcl-XL inactivation (Fig. 4D). Knockdown of PUMA, but not inactivation of the BH3-only sensitizer DP5 with a previously validated siRNA (16), reversed the proapoptotic effect of Bcl-XL silencing (Fig. 4, E and F and supplemental Fig. S8B). Bcl-XL knockdown did not modify PUMA expression, but inactivation of PUMA decreased Bcl-XL mRNA expression (supplemental Fig. S8C). We detected Bcl-XL and PUMA interaction by immunoprecipitation analysis in INS-1E cells, which was decreased after cytokine exposure (Fig. 4G). Interestingly, double knockdown of DP5 and PUMA did not enhance β-cell protection against cytokines, ruling out an additive effect (supplemental Fig. S9). Taken together, these results suggest that inactivation of Bcl-XL leads to PUMA release, a critical step for cytokine-induced β-cell death.
FIGURE 4.
Apoptosis induced by Bcl-XL inactivation is PUMA-dependent. A, confocal microscopy of Bcl-XL distribution in INS-1E is shown. Single confocal sections show Bcl-XL (green) and mitochondrial localized cytochrome c (red) fluorescence. The profile of fluorescence intensity and colocalization was analyzed using quantification software (Leica). Similar results were obtained in two independent experiments. B, Bcl-XL or control siRNAs were introduced into INS-1E cells, and Bcl-XL mRNA expression was measured by real time RT-PCR after 8-h exposure to cytokines, n = 3. *, p < 0.05; **, p < 0.01. C, INS-1E cells were transfected with control or with two Bcl-XL siRNAs and Western blotting for Bcl-XL, and cleaved caspase-3 was performed at the indicated time points of cytokine exposure, n = 3. D, Bcl-XL knockdown triggered apoptosis and increased IL-1β + IFN-γ-induced INS-1E cell death (24-h exposure), n = 3. *, p < 0.05; **, p < 0.01. E, INS-1E cells were transfected with inactive siRNA (Control), siRNA DP5, siRNA PUMA, and/or siRNA Bcl-XL 1, and cell death was measured by HO/PI after 24 h, n = 5, ***p < 0.001. F, Western blot of cleaved caspase-3 after transfection with siRNAs against PUMA, Bcl-XL, or combination is shown. Quantification of cleaved caspases-3 bands is shown in supplemental Fig. S8B, n = 3. G, following 24-h IL-1β + IFN-γ treatment, INS-1E cells were harvested, and cell lysates were subjected to anti-Bcl-XL antibody immunoprecipitation (IP). The immunoprecipitates were used for Western blotting with anti-PUMA and anti-Bcl-XL antibodies. Band intensity of PUMA was quantified versus Bcl-XL; data are means ± S.E., n = 2.
ABT-737 Triggers PUMA-mediated β-Cell Death
ABT-737 is a novel drug proposed for the treatment of cancer and autoimmune diseases (31, 32). This molecule mimics the BH3-only protein Bad and inhibits Bcl-XL and Bcl-2. Our above described data on Bcl-XL knockdown in β-cells raised the possibility that pharmacological inhibition of Bcl-XL may also lead to β-cell apoptosis. ABT-737 induced cell death in dispersed islet preparation from four human donors, and this effect was augmented by cytokine exposure (Fig. 5A). Similar results were observed in primary β-cells and INS-1E cells (Fig. 5B and supplemental Fig. S10). Knockdown of PUMA partially prevented the proapoptotic action of ABT-737 (Fig. 5C), suggesting that pharmacological inhibition of Bcl-XL/Bcl2 induces PUMA-mediated β-cell apoptosis.
FIGURE 5.
Bcl-XL/Bcl-2 inhibitor ABT-737 aggravates the proapoptotic effects of IL-1β + IFN-γ. A, dispersed human islets were treated for 48 h with ABT-737, IL-1β + IFN-γ, or combination, as indicated. At this time point, cell death was measured by HO/PI, n = 4. *, p < 0.05; ***, p < 0.001. B, primary rat β-cells were treated for 48 h with the inhibitor ABT-737, IL-1β + IFN-γ, or combination, as indicated, and then cell death was measured by HO/PI, n = 3. *, p < 0.05; **, p < 0.01. C, INS-1E cells were transfected with single or smart Pool PUMA siRNAs and exposed to ABT-737 for 24 h. At this time point, cell death was measured by HO/PI, n = 3. *, p < 0.05; **, p < 0.01.
DISCUSSION
The balance and interaction between pro- and antiapoptotic Bcl-2 proteins determines the cell death outcome (11). We show here that pancreatic β-cells induce the BH3-only activator PUMA in response to cytokines and chemical ER stressors, resulting in cell death. These observations contribute to clarify the mechanism by which proinflammatory cytokines induce β-cell demise and the nature of the downstream signals responsible for apoptosis triggering after severe ER stress.
The BH3-only activator subgroup includes Bid, Bim, and PUMA. Although PUMA was first identified as a sensitizer (33), recent evidence based on the use of short peptides and full-length proteins demonstrates that PUMA can bind to Bax and induce its mitochondrial translocation, thus acting as a BH3-only activator similar to Bim and Bid (12, 14, 30). It has been shown that Bid plays a minor if any role in cytokine-induced β-cell death (34), and we thus focused on the putative role of Bim and PUMA. Bim is neither modulated nor activated by IL-1β + IFN-γ and is therefore an unlikely contributor to cytokine-induced β-cell apoptosis. Interestingly, Bim inactivation by two siRNAs increased basal cell death, suggesting that this protein may play a role in the normal physiology of the β-cells. PUMA, on the other hand, is not only released following Bcl-XL inactivation in β-cells but is also transcriptionally induced after cytokine exposure. PUMA inactivation prevents cytokine-induced β-cell apoptosis as shown here by nuclear morphology, Bax translocation, cytochrome c release, and caspase-3 cleavage. In line with these observations, it was recently demonstrated that β-cells lacking PUMA expression are protected against high glucose (35), reinforcing the role of PUMA as an important mediator of different apoptotic stimulus in β-cells.
The transcription factor NF-κB activates both pro- and antiapoptotic signals, with the proapoptotic ones eventually prevailing in cytokine-induced β-cell death (2, 36). Analysis of the PUMA promoter indicates that NF-κB, but not p53, is required for cytokine-mediated PUMA expression in β-cells. In keeping with the promoter study, adenovirus-mediated inactivation of NF-κB decreases PUMA mRNA induction. These results are in agreement with a recent study in colon cancer cells, demonstrating that TNF-α induces p53-independent PUMA expression through NF-κB (25). Inhibition of NF-κB, however, only partially prevented cytokine-induced PUMA activation, suggesting the involvement of additional regulatory mechanisms. An interesting candidate in this respect is the radical NO. This molecule is generated by iNOS and contributes to cytokine-induced β-cell death by modulating the expression of around half of the cytokine-modified genes (3, 21). Inhibition of NO formation by l-NMA decreased the up-regulation of PUMA by cytokine treatment, indicating that this pathway synergizes with NF-κB for PUMA transcriptional activation. An important effect of cytokine-induced NO in β-cells is inhibition of the SERCA2 pump, depleting ER Ca2+ stores and triggering ER stress (8). ER stress activates PUMA in neurons (37, 38), and these cells share the expression of a large number of genes with β-cells (39), raising the possibility that NO mediates PUMA induction via ER stress. To test this possibility, we studied the effect of two chemical ER stressors on PUMA expression. Both CPA, which inactivates the SERCA2 pump leading to ER Ca2+ depletion (40), and tunicamycin, which inhibits N-linked protein glycosylation in the ER (41), activate PUMA up-regulation in pancreatic β-cells. Importantly, PUMA knockdown attenuates chemical ER stress-induced β-cell death. The transcription factor(s) responsible for ER stress-mediated PUMA expression in β-cells and other cell types remains to be clarified. Under our experimental conditions, PUMA up-regulation is independent of the proapoptotic ER stress-activated transcription factor Chop (data not shown).
Bcl-XL plays a major role in β-cell survival as shown recently by the use of a specific β-cell knock-out model (42). Consistently, overexpression of Bcl-XL protects β-cells against cytokines (9). We observed here that Bcl-XL is localized mainly at the mitochondria of β-cells, and its siRNA- or pharmacological-mediated inactivation induces the intrinsic pathway of apoptosis. This effect is prevented by PUMA but not by DP5 knockdown (Fig. 4), suggesting that PUMA activity is downstream of Bcl-XL, as observed previously in other cell types (30). Interestingly, PUMA silencing decreases Bcl-XL mRNA expression, probably as a cell compensatory mechanism to maintain the balance between pro- and antiapoptotic Bcl-2 proteins; this finding deserves further investigation. PUMA release from Bcl-XL is necessary for cytokine-mediated mitochondrial apoptosis. Taking this into account, it is conceivable that the main role of DP5 (and other BH3-only sensitizers, e.g. Bad) in cytokine-induced β-cell death is to repress Bcl-XL, allowing PUMA to induce IL-1β + IFN-γ-mediated mitochondrial Bax translocation (10, 16). The fact that the parallel knockdown of PUMA and DP5 did not confer additive protection against cytokine-induced β-cell death, compared with knockdown of each of these proteins alone (present data), supports this hypothesis. We cannot exclude, however, a direct inhibitory role of Bcl-XL on Bax activation, as was shown in other settings (43). In line with this, we observed that a pharmacological mimic of the BH3-only sensitizer Bad (31), which inhibits Bcl-XL and Bcl-2, induces PUMA-dependent apoptosis and aggravates the effect of proinflammatory cytokines in β-cells. This should be taken into consideration when systemically administering high doses of Bcl-XL-inactivating molecules for the treatment of cancer or autoimmune diseases.
The main conclusions of the present work, integrated with previous data from our group (16), are summarized in Fig. 6. IL-1β combined with IFN-γ induces an early and parallel activation of JNK/c-Jun and NF-κB. This leads to signaling cross-talk responsible for apoptosis induction. JNK/c-Jun activation mediates up-regulation of DP5 after cytokine exposure, sensitizing the β-cells to apoptosis (16). After transcriptional activation, DP5 selectively binds and represses the antiapoptotic Bcl-XL protein (12). Once Bcl-XL is inhibited, the BH3-only activator PUMA is released to bind Bax. This interaction induces conformational changes in Bax and triggers its mitochondrial translocation and oligomerization, mitochondrial membrane pore formation, and cytochrome c release, reaching the point of no return for β-cell apoptosis.
FIGURE 6.
Proposed model for the role of PUMA in cytokine-induced β-cell apoptosis. IL-1β + IFN-γ induce an early JNK activation, leading to c-Jun phosphorylation and DP5 expression. DP5, and other BH3-only sensitizers, inactivate Bcl-XL and release PUMA to induce Bax translocation, cytochrome c release, caspase activation, and apoptosis. In parallel, cytokine-induced NF-κB activation contributes to iNOS expression and PUMA up-regulation. iNOS-mediated NO formation causes ER stress, also contributing to PUMA mRNA induction. These signals converge to activate PUMA and cause β-cell demise.
Dying β-cells may act as a “danger signal” in the context of islet inflammation and, together with the local release of proinflammatory cytokines and chemokines, induce the maturation of dendritic cells, activation of T cells, and amplification of the autoimmune reaction in early T1D (4). We demonstrate here that PUMA plays a key role for pancreatic β-cell death in the context of ER stress and IL-1β + IFN-γ exposure. These findings point to novel avenues to prevent progressive β-cell demise in early T1D.
Supplementary Material
Acknowledgments
We thank Drs. M. Cnop and L. Bakiri for discussions and suggestions; Drs. L. Ladrière and M. Igoillo-Esteve for help with human islet preparations; and M. A. Neef, G. Vandenbroeck, M. Urbain, J. Schoonheydt, R. Leeman, R. Makhnas, A. E. Musuaya, and S. Mertens for technical support.
This work was supported by grants from the Communauté Française de Belgique, Actions de Recherche Concertées and the Fonds National de la Recherche Scientifique Belgium; Belgium Program on Interuniversity Poles of Attraction initiated by the Belgium State, Grant IUAP P6/40, Juvenile Diabetes Research Foundation International Grant 17-2009-106, and the European Union projects Eurodia, in the Framework Programme 6 of the European Community, and Naimit, in the Framework Programme 7 of the European Community.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S10.
- T1D
- type 1 diabetes
- IL
- interleukin
- IFN
- interferon
- ER
- endoplasmic reticulum
- BH3
- Bcl-2 homology 3
- PUMA
- p53 up-regulated modulator of apoptosis
- RT
- reverse transcription
- FACS
- autofluorescence-activated cell sorting
- NO
- nitric oxide
- l-NMA
- NG-methyl-l-arginine
- CPA
- cyclopiazonic acid
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- siRNA
- small interfering RNA
- NF-κB
- nuclear factor-κB
- HO/PI
- Hoechst 33342/propidium iodide
- JNK
- Jun N-terminal kinase
- iNOS
- inducible form of NO synthase.
REFERENCES
- 1.Roglic G., Unwin N., Bennett P. H., Mathers C., Tuomilehto J., Nag S., Connolly V., King H. (2005) Diabetes Care 28, 2130–2135 [DOI] [PubMed] [Google Scholar]
- 2.Cnop M., Welsh N., Jonas J. C., Jörns A., Lenzen S., Eizirik D. L. (2005) Diabetes 54, Suppl. 2, S97–S107 [DOI] [PubMed] [Google Scholar]
- 3.Eizirik D. L., Mandrup-Poulsen T. (2001) Diabetologia 44, 2115–2133 [DOI] [PubMed] [Google Scholar]
- 4.Eizirik D. L., Colli M. L., Ortis F. (2009) Nat. Rev. Endocrinol. 5, 219–226 [DOI] [PubMed] [Google Scholar]
- 5.Hostens K., Pavlovic D., Zambre Y., Ling Z., Van Schravendijk C., Eizirik D. L., Pipeleers D. G. (1999) J. Clin. Invest. 104, 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohara-Imaizumi M., Cardozo A. K., Kikuta T., Eizirik D. L., Nagamatsu S. (2004) J. Biol. Chem. 279, 41271–41274 [DOI] [PubMed] [Google Scholar]
- 7.Eizirik D. L., Sandler S., Welsh N., Cetkovic-Cvrlje M., Nieman A., Geller D. A., Pipeleers D. G., Bendtzen K., Hellerström C. (1994) J. Clin. Invest. 93, 1968–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardozo A. K., Ortis F., Storling J., Feng Y. M., Rasschaert J., Tonnesen M., Van Eylen F., Mandrup-Poulsen T., Herchuelz A., Eizirik D. L. (2005) Diabetes 54, 452–461 [DOI] [PubMed] [Google Scholar]
- 9.Holohan C., Szegezdi E., Ritter T., O'Brien T., Samali A. (2008) J. Cell. Mol. Med. 12, 591–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grunnet L. G., Aikin R., Tonnesen M. F., Paraskevas S., Blaabjerg L., Størling J., Rosenberg L., Billestrup N., Maysinger D., Mandrup-Poulsen T. (2009) Diabetes 58, 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 12.Kim H., Rafiuddin-Shah M., Tu H. C., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2006) Nat. Cell Biol. 8, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 13.Gross A., Jockel J., Wei M. C., Korsmeyer S. J. (1998) EMBO J. 17, 3878–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H., Tu H. C., Ren D., Takeuchi O., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2009) Mol. Cell 36, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Letai A. (2009) J. Cell Biol. 185, 189–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurzov E. N., Ortis F., Cunha D. A., Gosset G., Li M., Cardozo A. K., Eizirik D. L. (2009) Cell Death Differ. 16, 1539–1550 [DOI] [PubMed] [Google Scholar]
- 17.Lupi R., Dotta F., Marselli L., Del Guerra S., Masini M., Santangelo C., Patané G., Boggi U., Piro S., Anello M., Bergamini E., Mosca F., Di Mario U., Del Prato S., Marchetti P. (2002) Diabetes 51, 1437–1442 [DOI] [PubMed] [Google Scholar]
- 18.Moore F., Colli M. L., Cnop M., Esteve M. I., Cardozo A. K., Cunha D. A., Bugliani M., Marchetti P., Eizirik D. L. (2009) Diabetes 58, 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasschaert J., Ladrière L., Urbain M., Dogusan Z., Katabua B., Sato S., Akira S., Gysemans C., Mathieu C., Eizirik D. L. (2005) J. Biol. Chem. 280, 33984–33991 [DOI] [PubMed] [Google Scholar]
- 20.Cunha D. A., Hekerman P., Ladrière L., Bazarra-Castro A., Ortis F., Wakeham M. C., Moore F., Rasschaert J., Cardozo A. K., Bellomo E., Overbergh L., Mathieu C., Lupi R., Hai T., Herchuelz A., Marchetti P., Rutter G. A., Eizirik D. L., Cnop M. (2008) J. Cell Sci. 121, 2308–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutlu B., Cardozo A. K., Darville M. I., Kruhøffer M., Magnusson N., Ørntoft T., Eizirik D. L. (2003) Diabetes 52, 2701–2719 [DOI] [PubMed] [Google Scholar]
- 22.Pirot P., Ortis F., Cnop M., Ma Y., Hendershot L. M., Eizirik D. L., Cardozo A. K. (2007) Diabetes 56, 1069–1077 [DOI] [PubMed] [Google Scholar]
- 23.Gurzov E. N., Ortis F., Bakiri L., Wagner E. F., Eizirik D. L. (2008) PLoS ONE 3, e3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galluzzi L., Aaronson S. A., Abrams J., Alnemri E. S., Andrews D. W., Baehrecke E. H., Bazan N. G., Blagosklonny M. V., Blomgren K., Borner C., Bredesen D. E., Brenner C., Castedo M., Cidlowski J. A., Ciechanover A., Cohen G. M., De Laurenzi V., De Maria R., Deshmukh M., Dynlacht B. D., El-Deiry W. S., Flavell R. A., Fulda S., Garrido C., Golstein P., Gougeon M. L., Green D. R., Gronemeyer H., Hajnóczky G., Hardwick J. M., Hengartner M. O., Ichijo H., Jäättelä M., Kepp O., Kimchi A., Klionsky D. J., Knight R. A., Kornbluth S., Kumar S., Levine B., Lipton S. A., Lugli E., Madeo F., Malomi W., Marine J. C., Martin S. J., Medema J. P., Mehlen P., Melino G., Moll U. M., Morselli E., Nagata S., Nicholson D. W., Nicotera P., Nuñez G., Oren M., Penninger J., Pervaiz S., Peter M. E., Piacentini M., Prehn J. H., Puthalakath H., Rabinovich G. A., Rizzuto R., Rodrigues C. M., Rubinsztein D. C., Rudel T., Scorrano L., Simon H. U., Steller H., Tschopp J., Tsujimoto Y., Vandenabeele P., Vitale I., Vousden K. H., Youle R. J., Yuan J., Zhivotovsky B., Kroemer G. (2009) Cell Death Differ. 16, 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P., Qiu W., Dudgeon C., Liu H., Huang C., Zambetti G. P., Yu J., Zhang L. (2009) Cell Death Differ. 16, 1192–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heimberg H., Heremans Y., Jobin C., Leemans R., Cardozo A. K., Darville M., Eizirik D. L. (2001) Diabetes 50, 2219–2224 [DOI] [PubMed] [Google Scholar]
- 27.Nakano K., Vousden K. H. (2001) Mol. Cell 7, 683–694 [DOI] [PubMed] [Google Scholar]
- 28.Yu J., Zhang L., Hwang P. M., Kinzler K. W., Vogelstein B. (2001) Mol. Cell 7, 673–682 [DOI] [PubMed] [Google Scholar]
- 29.Cazanave S. C., Mott J. L., Elmi N. A., Bronk S. F., Werneburg N. W., Akazawa Y., Kahraman A., Garrison S. P., Zambetti G. P., Charlton M. R., Gores G. J. (2009) J. Biol. Chem. 284, 26591–26602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallenne T., Gautier F., Oliver L., Hervouet E., Noël B., Hickman J. A., Geneste O., Cartron P. F., Vallette F. M., Manon S., Juin P. (2009) J. Cell Biol. 185, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oltersdorf T., Elmore S. W., Shoemaker A. R., Armstrong R. C., Augeri D. J., Belli B. A., Bruncko M., Deckwerth T. L., Dinges J., Hajduk P. J., Joseph M. K., Kitada S., Korsmeyer S. J., Kunzer A. R., Letai A., Li C., Mitten M. J., Nettesheim D. G., Ng S., Nimmer P. M., O'Connor J. M., Oleksijew A., Petros A. M., Reed J. C., Shen W., Tahir S. K., Thompson C. B., Tomaselli K. J., Wang B., Wendt M. D., Zhang H., Fesik S. W., Rosenberg S. H. (2005) Nature 435, 677–681 [DOI] [PubMed] [Google Scholar]
- 32.Bardwell P. D., Gu J., McCarthy D., Wallace C., Bryant S., Goess C., Mathieu S., Grinnell C., Erickson J., Rosenberg S. H., Schwartz A. J., Hugunin M., Tarcsa E., Elmore S. W., McRae B., Murtaza A., Wang L. C., Ghayur T. (2009) J. Immunol. 182, 7482–7489 [DOI] [PubMed] [Google Scholar]
- 33.Kuwana T., Bouchier-Hayes L., Chipuk J. E., Bonzon C., Sullivan B. A., Green D. R., Newmeyer D. D. (2005) Mol. Cell 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 34.McKenzie M. D., Carrington E. M., Kaufmann T., Strasser A., Huang D. C., Kay T. W., Allison J., Thomas H. E. (2008) Diabetes 57, 1284–1292 [DOI] [PubMed] [Google Scholar]
- 35.McKenzie M. D., Jamieson E., Jansen E. S., Scott C. L., Huang D. C., Bouillet P., Allison J., Kay T. W., Strasser A., Thomas H. E. (2010) Diabetes 59, 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melloul D. (2008) Biochem. Soc. Trans. 36, 334–339 [DOI] [PubMed] [Google Scholar]
- 37.Kieran D., Woods I., Villunger A., Strasser A., Prehn J. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20606–20611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith M. I., Deshmukh M. (2007) Cell Death Differ. 14, 1011–1019 [DOI] [PubMed] [Google Scholar]
- 39.Atouf F., Czernichow P., Scharfmann R. (1997) J. Biol. Chem. 272, 1929–1934 [DOI] [PubMed] [Google Scholar]
- 40.Goeger D. E., Riley R. T., Dorner J. W., Cole R. J. (1988) Biochem. Pharmacol. 37, 978–981 [DOI] [PubMed] [Google Scholar]
- 41.Olden K., Pratt R. M., Yamada K. M. (1978) Cell 13, 461–473 [DOI] [PubMed] [Google Scholar]
- 42.Carrington E. M., McKenzie M. D., Jansen E., Myers M., Fynch S., Kos C., Strasser A., Kay T. W., Scott C. L., Allison J. (2009) Diabetes 58, 2316–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Billen L. P., Kokoski C. L., Lovell J. F., Leber B., Andrews D. W. (2008) PLoS Biol. 6, e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.