Abstract
Caspase activating and recruitment domain 8 (CARD8) has been implicated as a co-regulator of several pro-inflammatory and apoptotic signaling pathways. In the present study, we demonstrate a specific modulation of NOD2-induced signaling by CARD8 in intestinal epithelial cells. We show that CARD8 physically interacts with NOD2 and inhibits nodosome assembly and subsequent signaling upon muramyl-dipeptide stimulation. Furthermore, CARD8 inhibits the direct bactericidal effect of NOD2 against intracellular infection by Listeria monocytogenes. Thus, CARD8 represents a novel molecular switch involved in the endogenous regulation of NOD2-dependent inflammatory processes in epithelial cells.
Keywords: Immunology, Inflammation, NF-κB, Protein-Protein Interactions, Signal Transduction, CARD8, Caspase Recruitment Domain, Crohn Disease, NOD2
Introduction
NOD-like receptor (NLR)3 proteins form a family of intracellular pathogen-recognition receptors with more than 24 members in humans and over 200 members in lower metazoans (1–3). The NLR NOD2 is constitutively expressed in monocytes and intestinal Paneth cells (4, 5) and can be induced in various cell types including intestinal epithelial cells (6, 7). NOD2 serves as an intracellular pathogen sensor that requires muramyl dipeptide (MDP) as a minimal motif to trigger activation of the transcription factor NF-κB (4). Mutations in the gene encoding for NOD2 have been associated with a genetic predisposition to chronic inflammatory disorders (8–11) including Crohn disease, a human chronic inflammatory bowel disease. It is hypothesized that ligand recognition by the LRR domain induces a conformational change of the molecule resulting in the oligomerization of NOD2 via its nucleotide binding and oligomerization domain (NBD) domain, and subsequent recruitment of the serine threonine kinase RIP2 (4) through homotypic CARD-CARD domain interaction and induced proximity signaling.
In addition to this nodosome, multiprotein module, another major type of NLR complex, the inflammasome, is formed by pyrin domain-containing NLR proteins, e.g. NALP1 and NALP3 and several adaptor molecules, e.g. ASC and CARD8 (12–14). The protein complex forms a large scaffold, which is required for caspase-1-dependent IL-1β processing and secretion (15, 16).
CARD8 (also known as TUCAN and CARDINAL) is comprised of a C-terminal CARD domain and a N-terminal FIIND (domain with function to find) domain (17). CARD8 was first described in the context of NALP-independent NF-κB activation and apoptosis (17, 18). It has been reported to form dimers and bind to procaspase-9 suppressing caspase-9 activation via Apaf1-dependent mechanisms (19). However, CARD8 overexpression has been shown to induce apoptosis (20). Interestingly CARD8 expression is elevated in colonic carcinoma cells (19) and correlates with shorter patient survival (19, 21) indicating an involvement in cancer progression.
CARD8 has been postulated to serve as a molecular bridge recruiting an additional caspase-1 molecule to the NALP3 inflammasome through homotypic CARD-CARD-interaction (13). Thus, upon activation CARD8 may bind with its FIIND domain to the central NBD part of NALP3. Based on structural homology of NALP3 and NOD2, we investigated the possible interaction of CARD8 and NOD2. We hypothesized that the interaction of CARD8 with NOD2 may have an impact on cellular regulation of caspase-1 and NF-κB activation in the context of innate immune reactions.
EXPERIMENTAL PROCEDURES
Construction of Plasmids
The generation of FLAG-tagged constructs containing full-length wild-type NOD2, the NBD, the LRR, and the CARD domains of NOD2 have been described previously (22). Expression constructs for Myc- and CFP-tagged NOD2, YFP-tagged CARD8, and GFP-tagged CARD8 (1–320) were generated using standard cloning techniques (see supplemental Table S1). Plasmids pcDNA3.1-CARD8 and pEGFPC3-CARD8 have been described elsewhere (17). The plasmids pFLAG-CMV-caspase1 and pcDNA3-murine-proIL-1β were a kind gift from Dr. Junying Yuan.
RNAi Knockdown of CARD8
Control and specific siRNAs against CARD8 were purchased from Invitrogen (Carlsbad, CA; supplemental Table S3). Transfection of HeLaS3 cells with a pool of three different siRNAs was performed using siPort Amine (Applied Biosystems, Foster City, CA). Knockdown efficiency was evaluated by semi-quantitative RT-PCR.
Immunoprecipitation and Western Blot
Transfected HEK293 cells were subjected to co-immunoprecipitation according to standard protocols (23). Total protein lysates were prepared as described previously (24, 25). Proteins were transferred onto 0.45 μm polyvinylidene difluoride membranes (Millipore, Billerica, MA) and probed with respective antibodies. The antibodies used for immunoprecipitation and Western blotting were monoclonal mouse anti-FLAG antibody (M2, Sigma-Aldrich), anti-Myc antibody (Clontech, Palo Alto, CA), anti-GFP antibody (Clontech, Palo Alto, CA); anti-CARD8 antibody (ProSci, Poway, CA), and β-actin (Sigma Aldrich).
Transfection, Dual Luciferase Reporter Gene Assay, and ELISA
HEK cells were plated in 96-well plates and transfected using Fugene 6TM (Roche, Basel, Switzerland) with 15 ng/well of pNF-κB-Luc plasmid (Stratagene) in combination with 5 ng/well of pRL-TK (Promega) and 30 ng/well of expression plasmids as indicated. NF-κB-dependent luciferase activity was determined using a Dual Luciferase Reporter gene kit (Promega). For determination of human IL-8 and murine IL-1β supernatants of transfected HEK cells were analyzed by ELISA (R&D Systems, Rochester, MN).
Patient Samples
Mucosal colonic biopsies were taken from 10 healthy subjects and 10 CD patients following embedding in cryomatrix as described (26). Sections were incubated with primary antibodies (NOD2, 1:50, 2D9 rabbit polyclonal (Novus) and CARD, 1:50 (ProSci)) and FITC- and Cy3-conjugated secondary antibodies (1:100, both ImmunoResearch Laboratories Inc., West Grove, PA). Controls were included using secondary antibodies only. Samples were counterstained using DAPI and subjected to fluorescence microscopy.
RESULTS AND DISCUSSION
The aim of the present study was the molecular dissection of the putative interaction between the CARD-containing protein CARD8 and the NLR protein NOD2. We further investigated the effects of the interaction on NOD2-induced signal transduction and regulatory events that determine CARD8 expression levels in epithelial cells. For the expression profile of CARD8 mRNA and protein in human cell lines and tissues, see supplemental Fig. S1.
CARD8 Physically Interacts with NOD2
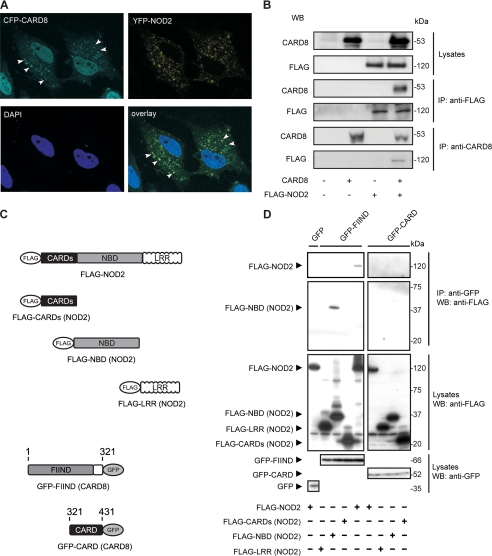
Co-expression of CFP-CARD8 and YFP-NOD2 in HeLa cells revealed a similar mainly granular cytoplasmic distribution pattern of both proteins (Fig. 1A). Furthermore, CARD8 was also found in the nucleus, which is in line with previous findings (17). Although it does not harbor any nuclear localization sequences (NLS) (21) CARD8 may shuttle to the nucleus through binding to NLS-containing proteins such as IKKγ/NEMO (27). The overlay of the CFP and YFP channel (Fig. 1A and supplemental Fig. S2) revealed a significant co-localization of CARD8 and NOD2 in the cytosol. Several studies have reported a partial membrane localization of NOD2 (22, 28–30). However, when co-expressed both NOD2 and CARD8 were nearly exclusively observable within the cytoplasm suggesting that the putative CARD8/NOD2 interaction may hinder the membrane localization of NOD2. Co-immunoprecipitation studies in HEK showed that CARD8 physically interacts with NOD2 (Fig. 1B).
FIGURE 1.
CARD8 co-localizes and physically interacts with NOD2. A, HeLa cells expressing CFP-CARD8 and YFP-NOD2 were subjected to fluorescence microscopy. Regions of co-localization are indicated by white arrows. B, HEK cells were co-transfected with expression constructs encoding for FLAG-NOD2 and CARD8. 24 h after transfection NOD2 complexes were immunoprecipitated with anti-FLAG or anti-CARD8 antibody. Co-precipitating CARD8 (or FLAG-NOD2) was detected by immunoblotting with the respective antibodies. C and D, HEK cells were co-transfected with GFP-CARD8 and the indicated FLAG-NOD2 expression constructs encoding for full-length NOD2, LRR, NBD, or the CARDs. Co-precipitating FLAG-NOD2 was detected with anti-FLAG monoclonal antibody. Expression of GFP-CARD8 and FLAG-NOD2 constructs is shown in the lower insets.
Further experiments on identification of relevant domains involved in CARD8 and NOD2 binding demonstrate a constitutive interaction between the FIIND domain of CARD8 with the NBD domain of NOD2 whereas no interaction was detectable between NOD2 and the CARD domain of CARD8 (Fig. 1D). This is molecularly similar to the interaction of CARD8 with NALP3 where the FIIND (CARD8) domain binds to the NBD domain of NALP3 (13, 31).
CARD8 and NOD2 Co-localize in Intestinal Inflammation
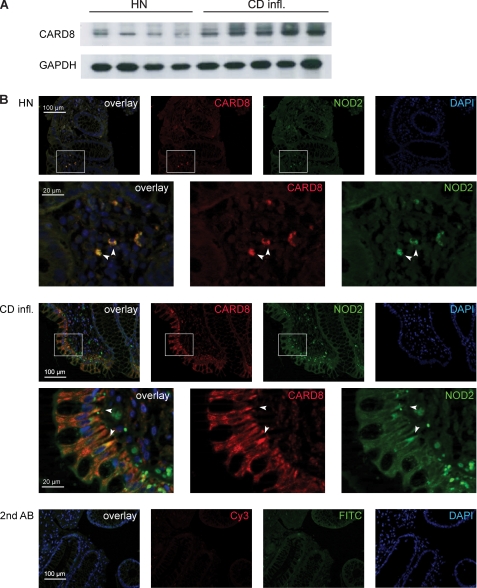
To investigate the in vivo relevance of these findings localization of endogenous CARD8 and NOD2 was analyzed in human colonic tissue biopsies of patients with Crohn disease. As shown in Fig. 2A CARD8 expression is elevated in the latter group and strong co-localization of NOD2 and CARD8 can be detected in epithelial cells in the inflamed colonic mucosa of CD patients (Fig. 2B). This may argue for a potential role of the CARD8/NOD2 interaction on NOD2 signaling in intestinal epithelial cells. Moderate overlapping expression can also be detected in mononuclear cells in the lamina propria in both the uninvolved colon of healthy individuals and CD patients. No signal was detected when omitting the respective primary antibodies.
FIGURE 2.
CARD8 is up-regulated in intestinal inflammation and co-localizes with NOD2 in the mucosa of Crohn disease patients. A, protein extracts were prepared from biopsies from the colonic mucosa of healthy individuals (HN) and patients with Crohn Disease (CD infl.). B, cryosections of mucosal biopsies were stained using appropriate antibodies to detect endogenous NOD2 (green channel, FITC) and CARD8 (red channel, Cy3), respectively. In addition, nuclei were stained using DAPI (blue channel). The merged pictures show overlapping expression patterns of endogenous NOD2 and CARD8 (indicated by arrows).
CARD8 Negatively Influences NOD2-mediated Responses
Because NOD2 is involved in the defense of bacterial invasion and intracellular bacterial killing (28, 32–35) the impact of CARD8 on NOD2-mediated defense against bacterial infection with Listeria monocytogenes was investigated. Expression of CARD8 and co-expression of CARD8 and NOD2 resulted both in an increased cellular bacterial invasion (Fig. 3A), indicating that CARD8 is able to abolish the protective effect of NOD2 against L. monocytogenes. This is likely due to the inhibition of nodosome assembly by CARD8. However, it is also conceivable that CARD8 is masking binding sites of NOD2 critical for bacterial killing, which have been identified previously (32) and thereby disabling bactericidal action.
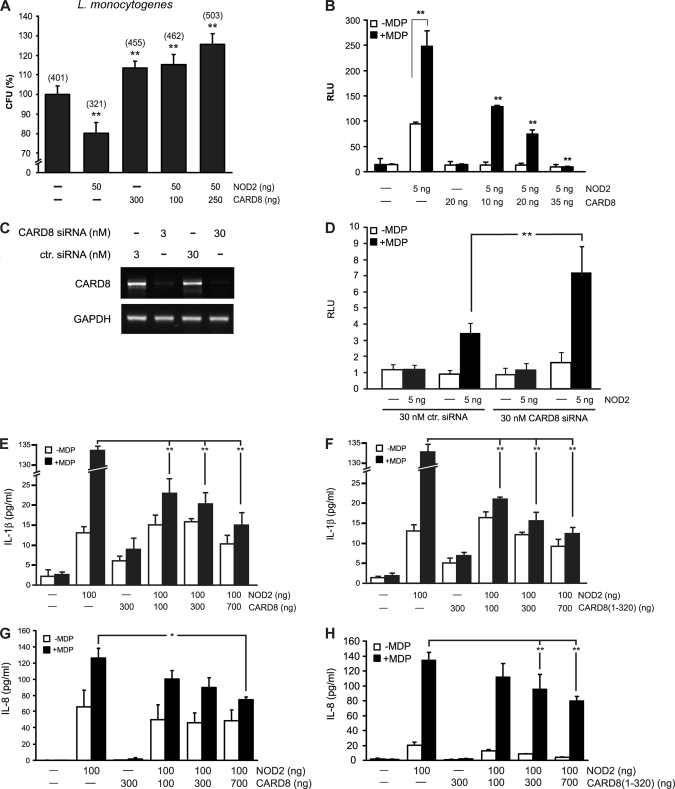
FIGURE 3.
CARD8 increases bacterial cytoinvasion and suppresses NOD2-induced NF-κB activity and secretion of IL-1β and IL-8. A, CARD8 and/or NOD2 were expressed in HEK cells following infection with L. monocytogenes and gentamicin-mediated elimination of extracellular bacteria. Relative cytoinvasion was calculated as percentage of CFUs found in lysates of untransfected cells. Actual numbers of CFU are shown in parentheses. **, p < 0.01; data are representative of three independent experiments (n = 3). B, NOD2 and CARD8 were expressed as indicated in HEK cells and stimulated with MDP (24 h, 1 μg/ml). Data are expressed as relative luciferase activity. C, knockdown of CARD8 gene expression by siRNA. Cells were transfected with a pool of specific siRNAs targeting CARD8 or irrelevant control siRNA with the indicated amounts. Efficiency of knock-down was analyzed using RT-PCR. D, cells were transfected with siRNA in combination with a NOD2 expression construct. After stimulation with MDP (20 μg/ml) for 24 h relative luciferase activity was determined by normalization of luciferase activity to protein content. E–H, FLAG-NOD2, GFP-CARD8, and GFP-CARD8-(1–320) (FIIND-domain) were expressed in HEK cells as indicated and treated with MDP (24 h, 10 μg/ml) before levels of cytokines were determined. All data represent the mean ± S.D. (n = 3). *, p < 0.05; **, p < 0.01.
To further dissect the influence of CARD8 on NOD2-mediated signaling, MDP-induced NF-κB activation and IL-1β and IL-8 release were measured. Co-expression of CARD8 and NOD2 in HEK cells significantly reduces MDP-induced NF-κB-activation in a dose-dependent manner (Fig. 3B). Consistently, siRNA-mediated knockdown of endogenous CARD8 (Fig. 3C) resulted in an increased NF-κB promoter transactivation in HeLa cells compared with controls (Fig. 3D).
To test the hypothesis that CARD8 could block NOD2-induced activation of caspase-1 and subsequent IL-1β, we transiently transfected a murine pro-IL-1β expression plasmid into HEK cells together with plasmids encoding for human CARD8, human pro-caspase-1, and NOD2. The murine pro-IL-1β expression plasmid was used to monitor secretion of this cytokine independent from altered processing of endogenous pro-IL-1β. Expression of full-length CARD8 or FIIND domain of CARD8 diminishes NOD2-induced release of IL-1β into the culture medium of HEK cells in a dose-dependent manner (Fig. 3, E and F).
Because NOD2-induced NF-κB activation has previously been shown to induce the expression and secretion of chemotactic cytokine IL-8 (6), we also tested the influence of transient transfection with NOD2 and/or full-length CARD8 and/or the FIIND domain of CARD8 on IL-8 secretion in MDP-stimulated HEK cells (Fig. 3, G and H). NOD2-dependent IL-8 secretion was significantly reduced in the presence of the full-length CARD8 protein and the FIIND domain of CARD8. The expression of the respective proteins was confirmed by Western blot analysis (supplemental Fig. S3).
The FIIND Domain Inhibits Nodosome Assembly
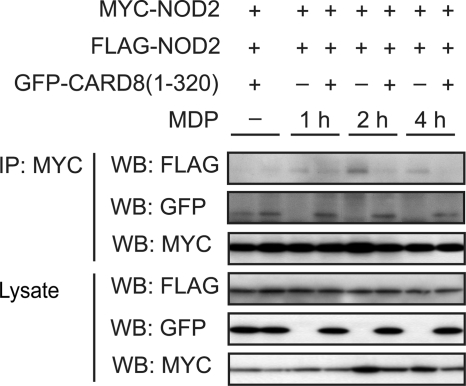
As CARD8 negatively affects NOD2-dependent signaling and NOD2-mediated antibacterial effects, we investigated whether CARD8 may disturb the assembly of the nodosome complex via an inhibition of the oligomerization of NOD2. For this reason we transiently expressed Myc- and FLAG-tagged NOD2 together with increasing amounts of the FIIND domain of CARD8 and subsequently studied the MDP-induced oligomerization of NOD2. Through the use of differently tagged NOD2 proteins self-oligomerization could be detected via co-immunoprecipitation studies. The FIIND domain of CARD8 is able to inhibit the oligomerization of NOD2 upon MDP stimulation (Fig. 4). These data suggest that binding of CARD8 and NOD2 negatively interferes with the oligomerization of NOD2 thereby diminishing MDP-induced NOD2 signaling. Co-expression of GFP alone, which was used as a tag in the FIIND expression plasmid, did not affect the MDP-induced oligomerization (data not shown). Interestingly CARD8 interacts with NALP3, another member of the NOD-like receptor family (13). According to the molecular mechanisms proposed in this study, CARD8 represents an adaptor of the inflammasome complex by binding to the NBD domain of NALP3 and enabling it to recruit a second caspase-1 protein via homotypic CARD-CARD interactions.
FIGURE 4.
CARD8 interferes with MDP-induced NOD2 oligomerization. FLAG- and Myc-tagged full-length NOD2 and the CARD8-(1–320) (FIIND domain of CARD8) were expressed in HEK cells and self-association of NOD2 was determined in absence or presence of MDP (10 μg/ml) by immunoprecipitation. Total lysates were immunoblotted with anti-FLAG, anti-GFP, and anti-Myc antibodies (lower three blots). Each gel is representative of at least three independent experiments.
This study demonstrates novel aspects on the function of CARD8 in epithelial cells. Our data indicate that CARD8 acts as a negative regulator of NOD2 signaling by inhibiting the oligomerization process of NOD2 itself. In the context of the pathophysiology of Crohn disease, where loss-of function variants of NOD2 have been shown to play a causative role in disease etiopathogenesis, a suppressor of NOD2 activity such as CARD8 may be of special interest. In vivo an inhibition of NOD2 function via CARD8 could lead to decreased epithelial responsiveness to MDP and to an increased secondary inflammatory response. This is in contradiction to findings for NALP3, where CARD8 represents an adaptor of the inflammasome complex by binding to the NBD domain of NALP3 and enabling recruiting of a second caspase-1 protein via homotypic CARD-CARD interactions. Thus, CARD8 seems to act as a molecular controller regulating different inflammatory pathways by either assembling or disassembling protein scaffolds.
Our study may highlight another putative consequence of the NOD2/CARD8 interaction. Studies have reported on an increased concentration of intracellular bacteria in the mucosal epithelia of patients with colonic cancer (36). Because CARD8 has been shown to be overexpressed in colonic cancer cells and overexpression correlates with shorter patient survival (18), elevated CARD8 protein levels, and resulting inhibition of NOD2 function could contribute to this phenomenon.
Supplementary Material
Acknowledgment
We greatly appreciate the kind gift of plasmids from Junying Yuan.
This work was supported by the DFG (Deutsche Forschungsgemeinschaft) SCHR512/11-1, the Clusters of Excellence Inflammation at Interfaces and The Future Ocean and by the German Federal Ministry of Education and Research BMBF through the NGFN plus Network on Environment-related diseases.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1–S3.
- NLR
- NOD-like receptor
- DAPI
- 4′,6-diamidino-2-phenylindole
- GFP
- green fluorescent protein
- NLS
- nuclear localization signal
- CARD
- caspase activating and recruitment domain
- ELISA
- enzyme-linked immunosorbent assay
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- IL
- interleukin.
REFERENCES
- 1.Ting J. P. Y., Davis B. K. (2005) Annu. Rev. Immunol. 23, 387–414 [DOI] [PubMed] [Google Scholar]
- 2.Inohara N., Nuñez G. (2003) Nat. Rev. 3, 371–382 [DOI] [PubMed] [Google Scholar]
- 3.Rast J. P., Smith L. C., Loza-Coll M., Hibino T., Litman G. W. (2006) Science 314, 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogura Y., Inohara N., Benito A., Chen F. F., Yamaoka S., Nunez G. (2001) J. Biol. Chem. 276, 4812–4818 [DOI] [PubMed] [Google Scholar]
- 5.Lala S., Ogura Y., Osborne C., Hor S. Y., Bromfield A., Davies S., Ogunbiyi O., Nuñez G., Keshav S. (2003) Gastroenterology 125, 47–57 [DOI] [PubMed] [Google Scholar]
- 6.Rosenstiel P., Fantini M., Bräutigam K., Kühbacher T., Waetzig G. H., Seegert D., Schreiber S. (2003) Gastroenterology 124, 1001–1009 [DOI] [PubMed] [Google Scholar]
- 7.Gutierrez O., Pipaon C., Inohara N., Fontalba A., Ogura Y., Prosper F., Nunez G., Fernandez-Luna J. L. (2002) J. Biol. Chem. 277, 41701–41705 [DOI] [PubMed] [Google Scholar]
- 8.Hugot J. P., Chamaillard M., Zouali H., Lesage S., Cézard J. P., Belaiche J., Almer S., Tysk C., O'Morain C. A., Gassull M., Binder V., Finkel Y., Cortot A., Modigliani R., Laurent-Puig P., Gower-Rousseau C., Macry J., Colombel J. F., Sahbatou M., Thomas G. (2001) Nature 411, 599–603 [DOI] [PubMed] [Google Scholar]
- 9.Ogura Y., Bonen D. K., Inohara N., Nicolae D. L., Chen F. F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R. H., Achkar J. P., Brant S. R., Bayless T. M., Kirschner B. S., Hanauer S. B., Nuñez G., Cho J. H. (2001) Nature 411, 603–606 [DOI] [PubMed] [Google Scholar]
- 10.Hampe J., Cuthbert A., Croucher P. J., Mirza M. M., Mascheretti S., Fisher S., Frenzel H., King K., Hasselmeyer A., MacPherson A. J., Bridger S., van Deventer S., Forbes A., Nikolaus S., Lennard-Jones J. E., Foelsch U. R., Krawczak M., Lewis C., Schreiber S., Mathew C. G. (2001) Lancet 357, 1925–1928 [DOI] [PubMed] [Google Scholar]
- 11.Rosenstiel P., Till A., Schreiber S. (2007) Microbes Inf./Institut. Pasteur 9, 648–657 [DOI] [PubMed] [Google Scholar]
- 12.Martinon F., Tschopp J. (2004) Cell 117, 561–574 [DOI] [PubMed] [Google Scholar]
- 13.Agostini L., Martinon F., Burns K., McDermott M. F., Hawkins P. N., Tschopp J. (2004) Immunity 20, 319–325 [DOI] [PubMed] [Google Scholar]
- 14.Martinon F., Burns K., Tschopp J. (2002) Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 15.Faustin B., Lartigue L., Bruey J. M., Luciano F., Sergienko E., Bailly-Maitre B., Volkmann N., Hanein D., Rouiller I., Reed J. C. (2007) Mol. Cell 25, 713–724 [DOI] [PubMed] [Google Scholar]
- 16.Gattin Z., van Gunsteren W. F. (2008) Chembiochem 9, 923–933 [DOI] [PubMed] [Google Scholar]
- 17.Bouchier-Hayes L., Conroy H., Egan H., Adrain C., Creagh E. M., MacFarlane M., Martin S. J. (2001) J. Biol. Chem. 276, 44069–44077 [DOI] [PubMed] [Google Scholar]
- 18.Stilo R., Leonardi A., Formisano L., Di Jeso B., Vito P., Liguoro D. (2002) FEBS Lett. 521, 165–169 [DOI] [PubMed] [Google Scholar]
- 19.Pathan N., Marusawa H., Krajewska M., Matsuzawa S., Kim H., Okada K., Torii S., Kitada S., Krajewski S., Welsh K., Pio F., Godzik A., Reed J. C. (2001) J. Biol. Chem. 276, 32220–32229 [DOI] [PubMed] [Google Scholar]
- 20.Razmara M., Srinivasula S. M., Wang L., Poyet J. L., Geddes B. J., DiStefano P. S., Bertin J., Alnemri E. S. (2002) J. Biol. Chem. 277, 13952–13958 [DOI] [PubMed] [Google Scholar]
- 21.Checinska A., Oudejans J. J., Span S. W., Rodriguez J. A., Kruyt F. A., Giaccone G. (2006) Anticancer Res. 26, 3819–3824 [PubMed] [Google Scholar]
- 22.Till A., Rosenstiel P., Bräutigam K., Sina C., Jacobs G., Oberg H. H., Seegert D., Chakraborty T., Schreiber S. (2008) J. Cell Sci. 121, 487–495 [DOI] [PubMed] [Google Scholar]
- 23.Lipinski S., Till A., Sina C., Arlt A., Grasberger H., Schreiber S., Rosenstiel P. (2009) J. Cell Sci. 122, 3522–3530 [DOI] [PubMed] [Google Scholar]
- 24.Rosenstiel P., Huse K., Till A., Hampe J., Hellmig S., Sina C., Billmann S., von Kampen O., Waetzig G. H., Platzer M., Seegert D., Schreiber S. (2006) Proc. Natl. Acad. Sci. U.S.A. 0505423103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valentonyte R., Hampe J., Huse K., Rosenstiel P., Albrecht M., Stenzel A., Nagy M., Gaede K. I., Franke A., Haesler R., Koch A., Lengauer T., Seegert D., Reiling N., Ehlers S., Schwinger E., Platzer M., Krawczak M., Müller-Quernheim J., Schürmann M., Schreiber S. (2005) Nat. Genet. 37, 357–364 [DOI] [PubMed] [Google Scholar]
- 26.Schreiber S., Rosenstiel P., Hampe J., Nikolaus S., Groessner B., Schottelius A., Kühbacher T., Hämling J., Fölsch U. R., Seegert D. (2002) Gut 51, 379–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verma U. N., Yamamoto Y., Prajapati S., Gaynor R. B. (2004) J. Biol. Chem. 279, 3509–3515 [DOI] [PubMed] [Google Scholar]
- 28.Barnich N., Aguirre J. E., Reinecker H. C., Xavier R., Podolsky D. K. (2005) J. Cell Biol. 170, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kufer T. A., Kremmer E., Banks D. J., Philpott D. J. (2006) Infect. Immun. 74, 3115–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald C., Chen F. F., Ollendorff V., Ogura Y., Marchetto S., Lécine P., Borg J. P., Nuñez G. (2005) J. Biol. Chem. 280, 40301–40309 [DOI] [PubMed] [Google Scholar]
- 31.Dinarello C. A. (2004) Immunity 20, 243–244 [DOI] [PubMed] [Google Scholar]
- 32.Hisamatsu T., Suzuki M., Reinecker H. C., Nadeau W. J., McCormick B. A., Podolsky D. K. (2003) Gastroenterology 124, 993–1000 [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi K. S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nuñez G., Flavell R. A. (2005) Science 307, 731–734 [DOI] [PubMed] [Google Scholar]
- 34.Swidsinski A., Ladhoff A., Pernthaler A., Swidsinski S., Loening-Baucke V., Ortner M., Weber J., Hoffmann U., Schreiber S., Dietel M., Lochs H. (2002) Gastroenterology 122, 44–54 [DOI] [PubMed] [Google Scholar]
- 35.Barnich N., Hisamatsu T., Aguirre J. E., Xavier R., Reinecker H. C., Podolsky D. K. (2005) J. Biol. Chem. 280, 19021–19026 [DOI] [PubMed] [Google Scholar]
- 36.Swidsinski A., Khilkin M., Kerjaschki D., Schreiber S., Ortner M., Weber J., Lochs H. (1998) Gastroenterology 115, 281–286 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.