Abstract
Plague, one of the most devastating diseases in human history, is caused by the bacterium Yersinia pestis. The bacteria use a syringe-like macromolecular assembly to secrete various toxins directly into the host cells they infect. One such Yersinia outer protein, YopJ, performs the task of dampening innate immune responses in the host by simultaneously inhibiting the MAPK and NFκB signaling pathways. YopJ catalyzes the transfer of acetyl groups to serine, threonine, and lysine residues on target proteins. Acetylation of serine and threonine residues prevents them from being phosphorylated thereby preventing the activation of signaling molecules on which they are located. In this study, we describe the requirement of a host-cell factor for full activation of the acetyltransferase activity of YopJ and identify this activating factor to be inositol hexakisphosphate (IP6). We extend the applicability of our results to show that IP6 also stimulates the acetyltransferase activity of AvrA, the YopJ homologue from Salmonella typhimurium. Furthermore, an IP6-induced conformational change in AvrA suggests that IP6 acts as an allosteric activator of enzyme activity. Our results suggest that YopJ-family enzymes are quiescent in the bacterium where they are synthesized, because bacteria lack IP6; once injected into mammalian cells by the pathogen these toxins bind host cell IP6, are activated, and deregulate the MAPK and NFκB signaling pathways thereby subverting innate immunity.
Keywords: Acetyl Coenzyme A, Bacterial Toxins, Covalent Regulation, Inositol Phosphates, NF-κB, AvrA, MEK, Salmonella, Activator
Introduction
The genus Yersinia of Gram-negative bacteria comprises eleven species of which three are pathogenic to humans. While Yersinia pestis is the causative agent of bubonic plague, the common enteric pathogens Y. enterocolitica and Y. pseudotuberculosis are responsible for gastroenteritis and lymphadenitis. All pathogenic Yersinia species harbor a 70-kb plasmid (pCD1 in Y. pestis and pYV in Y. enterocolitica and Y. pseudotuberculosis) that is essential for virulence. This plasmid encodes a Type III secretion system comprising a secretion apparatus, chaperones, and secreted effectors. The Type III secretion system mediates host-cell contact-dependent delivery of bacterial virulence proteins known as Yops (Yersinia outer proteins) (1). Whereas some Yops are required to form the translocation apparatus and to control the process of translocation, a bouquet of six effector Yops is directly injected into the cytosol of target mammalian cells. These secreted toxins YopE, YopT, YopO, YopH, YopM, and YopJ act synergistically to rapidly overwhelm the host immune response (2).
Innate immunity provides the first line of defense against infectious diseases; however, many pathogens are able to avoid host recognition or to diminish the subsequent immune activation through interactions with host response molecules. All three pathogenic species of Yersinia share a marked tropism for lymphoid tissue where they use the arsenal of effector Yop proteins to resist uptake by phagocytic cells thus enabling extracellular replication. The effector Yops interfere with critical signaling processes of the host immune response.
YopJ (from Y. pestis and Y. pseudotuberculosis; homologous to YopP from Y. enterocolitica) abrogates the innate immune response by inhibiting the MAPK2 and NFκB signaling pathways, preventing the production of protective cytokines such as tumor necrosis factor-α and interleukin 8 (3, 4). YopJ has been shown to possess acetyltransferase activity (5, 6). The acetyltransferase activity of AvrA and VopA, the YopJ homologues from Salmonella typhimurium and Vibrio parahemeolyticus, respectively, has also been recently demonstrated (7, 8).
We have earlier shown that the enzymatic activity of YopJ results in the acetylation of serine and threonine residues on the activation loops of the kinases MEK1/2 in the MAPK pathway and of the IKKα/β kinases in the NFκB pathway (6). These kinases are part of signaling cascades and are activated by upstream kinases that phosphorylate them, and they, in turn, propagate signaling by phosphorylating downstream molecules. YopJ transfers the acetyl group from the ubiquitous acetyl donor, acetylcoenzyme A (AcCoA) to serine, threonine, and lysine residues on target molecules (5, 6). Acetylation of an activation loop threonine residue in IKKα/β prevents their phosphorylation and thus inhibits their ability to phosphorylate IκBα: the cytosolic regulator of the transcription factor NFκB (6). Phosphorylation of IκBα would normally lead to its ubiquitination and subsequent proteasomal degradation, freeing up NFκB for translocation to the nucleus where it would regulate the transcription of pro-inflammatory genes (9). Similarly, acetylation of MEK2 activation loop residues Ser-222 and Ser-226 by YopJ prevents its phosphorylation by upstream kinases and thus inhibits the signal-transducing ability of MEK (6). In this report we establish the requirement of a host-cell factor for the activation of the acetyltransferase activity of YopJ and identify this activating cofactor to be inositol hexakisphosphate (IP6).
EXPERIMENTAL PROCEDURES
Reagents
IP3, IP5, IP6, IS6, AcCoA, ubiquitin, and phytase were from Sigma. Alkaline phosphatase was from Roche Applied Science. The anti-MEK1/2 antisera CST9122 and 47E6 were from Cell Signaling Technology. [14C]AcCoA was from Amersham Biosciences.
Plasmid Constructs
The mammalian cell expression plasmids encoding WT YopJ (pSFFV-YopJ) and the inactive C172A mutant (pSFFV-YopJ-C172A) were the kind gifts of K. Orth (University of Texas Southwestern Medical Center, Dallas, TX); pCMV-MEK2 was provided by K.-L. Guan (University of Michigan, Ann Arbor, MI).
Expression of Proteins in E. coli
GST-YopJ was expressed in Escherichia coli using the plasmid pGEX6P; as was GST-MEK2. GST fusion proteins were purified on glutathione-Sepharose, and bead-bound fusion proteins were digested with PreScission protease to liberate YopJ and MEK2, which were then purified by gel filtration. Using the primers 5′-atatggatccatgatattttcggtgcaggagctatcatgtgg-3′ and 5′-atatctcgagttacggtttaagtaaagacttatattcagc-3′ AvrA was amplified from genomic DNA isolated from the wild-type strain 12023 of S. typhimurium (a generous gift of Dr. E. Boucrot, MRC, Laboratory of Molecular Biology, Cambridge, UK). DNA sequencing showed a frameshift after amino acid 265 of the AvrA cDNA. This was corrected by site-directed mutagenesis. The coding sequence of wild-type AvrA was ligated into the plasmid pGEX6P and purified as described above. During the course of these studies Du and Galan (10) published the correct start methionine for AvrA. Both AvrA constructs (one with the corrected start and the one with 15 extra N-terminal residues) displayed autoacetylation.
Acetyltransferase Assays
In a 25-μl reaction volume, MEK2 (5 μg) was incubated with varying amounts (0.3–1.2 μg) of YopJ in the presence of 60 μm [1-14C]AcCoA (54 mCi/mmol, Amersham Biosciences) (1 Ci = 37 GBq) at 37 °C for 1 h. IP6, when included, was typically present at a final concentration of 100 nm. Reaction products were resolved on 4–12% SDS-PAGE gels. Gels were stained with Coomassie Blue, de-stained, dried, and subjected to autoradiography.
Preparation of HeLa Cytosol
20 confluent 90-mm dishes of HeLa cells (∼6 × 107 cells) were harvested in 6 ml of buffer containing 20 mm Tris, pH 8.0, 2 mm EDTA, 150 mm NaCl and 1 mm dithiothreitol (TEND Buffer) and lysed by sonication. The lysate was centrifuged at 250,000 × g for 20 min using a TLA 100.2 rotor in a Beckman benchtop ultracentrifuge. The clarified supernatant (3 mg/ml protein) was harvested; 1 ml of which was dialyzed overnight against 2 liters of TEND buffer (to remove endogenous AcCoA) and used in acetyltransferase assays.
Size-exclusion Chromatography of HeLa Cytosol
The remaining 5 ml of the HeLa cytosol described above was concentrated to 2 ml using a 10-kDa molecular weight cut-off filter and loaded onto a HiLoad 16/60 Superdex 75 gel-filtration column (Amersham Biosciences) equilibrated with TEND buffer, and 2-ml fractions were collected. 15 μl of each fraction were then included in acetyltransferase reactions using 5 μg of MEK2, 1 μg of YopJ, and [1-14C]AcCoA to identify the fractions that contained cofactor activity.
Preparation of Acid Extracts from Cells
Initially, acid extracts were made from HeLa cells by precipitating proteins from the cleared lysate using either 10% trichloroacetic acid or 5% perchloric acid. Subsequently, a modification of the method described by Azevedo and Saiardi (11) was used. Briefly, frozen cell pellets were thawed and resuspended directly in 1 m perchloric acid/3 mm EDTA and kept on ice for 10 min with intermittent vortexing. The extract was then centrifuged at 20,000 × g for 20 min at 4 °C in a tabletop microcentrifuge. The clear supernatant was then neutralized using 1 m potassium carbonate/3 mm EDTA. Precipitated salt was removed by brief centrifugation. The clear supernatant recovered at the end of the procedure represented the acid extract of cells and was verified to be at near neutral pH before use in assays. This protocol was used to make extracts from the different cell types described in this study. Yeast cells were vortexed in the presence of glass beads.
Purification of the Cofactor by Ion-exchange Chromatography
A HeLa cell pellet corresponding to ∼2 × 108 cells was thawed and resuspended in 10 ml of 1 m perchloric acid/3 mm EDTA solution and vortexed. This extract was centrifuged at 20,000 × g for 20 min at room temperature in a tabletop microcentrifuge, and the clear supernatant was harvested and heated at 95 °C for 5 min. The extract was then neutralized using 1 m potassium carbonate/3 mm EDTA and centrifuged. The clarified, neutralized extract containing the cofactor was filtered through a 0.22-μm filter and mixed with a 1-ml slurry of AG1-X8 strong anion-exchange resin (Bio-Rad) that had been pre-equilibrated with acetate. After overnight incubation at room temperature the mixture was poured into an empty chromatography column; the flow-through was discarded and the resin washed with 30 ml of water. Bound material was eluted from the resin using 10 ml of 2 n hydrochloric acid, lyophilized to remove the acid, and taken up in 200 μl water. This preparation was assayed for the presence of cofactor activity (Fig. 3A) and then subjected to analysis by mass spectrometry to help identify the cofactor.
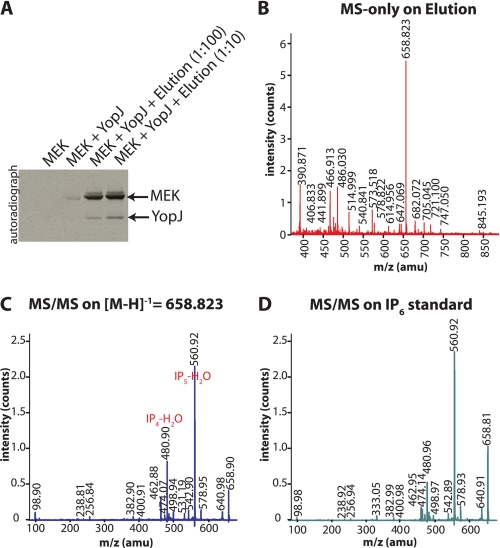
FIGURE 3.
Cofactor activity could be purified by anion exchange chromatography and was identified by MS to be IP6. A, cofactor activity present in the HeLa cell acid extract was eluted from AG1-X8 resin, lyophilized, and resuspended in water for inclusion in acetyltransferase assays. The stimulation of YopJ activity by two different dilutions (1:100 and 1:10) of the elution is shown here. The elution was then subjected to mass spectrometric analysis. B, MALDI-MS spectrum acquired in the reflectron negative ion mode revealed a prominent m/z peak of 658.823 Da. This peak was manually selected for further MS/MS fragmentation (C), and the resulting ion series corresponds to the breakdown products expected for IP6. D, MS/MS fragmentation ion series obtained for an IP6 standard.
Mass Spectrometry
The elution described above was subjected to MALDI-MS, in the reflectron negative ion mode, to measure the m/z of the substance(s) present. 0.7 μl of the sample was applied onto a stainless steel target, and 0.7 μl of 2′,4′,6′-trihydroxyacetophenone matrix was added. The matrix had been prepared by dissolving 10 mg of 2′,4′,6′-trihydroxyacetophenone matrix in 1 ml of acetonitrile/5 mg/ml diammonium citrate (1:1, v/v). All spectra were acquired with an Ultraflex III TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). A single major species with m/z = 658.823 was observed and was selected for MALDI-MS/MS fragmentation in negative LIFT mode. The fragmentation ion series of [M-H]−1 = 658.823 showed the loss of H3PO4 and HPO3 ions thereby indicating that the elution contained IP6. This identification was confirmed by comparing with the MS/MS fragment spectrum of commercially available IP6 acquired under the same conditions.
Deletion Yeast Strains
The following strains of Saccharomyces cerevisiae were generously provided by A. Saiardi (MRC Laboratory for Molecular and Cell Biology, University College London, London, UK). Wild-type strains: BY4741 (MATa; his3D1; leu2D0; met15D0; ura3D0) and DDY1810 (MATa, leu2, ura3–52, trp1, prb1–1122, pep4–3, pre1–451). Mutant strains: ipk1Δ and ipk1Δkcs1Δ (in the BY4741 background) and ipmkΔ and kcs1Δ (in the DDY1810 background). Mutants in the BY4741 background were grown in the presence of 200 μg/ml G418, whereas those in the DDY1810 background were cultured in the absence of leucine. The growth of ipk1Δ yeast is comparable to that of wild-type yeast, whereas the remaining mutant strains grow slower. The optical densities of the cultures were monitored, and an equal number of yeast cells from each culture was used for the preparation of yeast acid extracts.
Tryptophan Fluorescence of AvrA
Protein tryptophan fluorescence of AvrA (5.6 μm in 20 mm Tris, pH 8.0, 150 mm NaCl) was excited at 280 nm, and emission at 330 nm was monitored at 20 °C over time in a 200-μl volume using a FluoroMax-2 spectrofluorometer (Horiba Yvon Spex). The addition of IP6 (10 μm) resulted in a saturable decrease of fluorescence.
CD Spectroscopy
Far UV (from 195 to 250 nm) CD spectra of AvrA (10 μm) in 20 mm Tris, pH 8.0, 150 mm NaCl were recorded at 20 °C using a 1-mm path length quartz cuvette (200-μl sample volume) in a JASCO J-815 CD spectrometer. Successive spectra were recorded after the addition of different amounts of IP6 (0 μm, 0.5 μm, 1 μm, 2 μm, 3 μm, 4 μm, 5 μm, and 10 μm). Spectra recorded after the addition of IP6 showed a drop in signal at 208 nm and 222 nm indicating a gain in helicity by AvrA. There was no further decrease after addition of IP6 at concentrations higher than 10 μm.
Time course of change in ellipticity of AvrA upon addition of IP6 was measured in an Aviv 202SF spectrometer. AvrA (5 μm) in 20 mm Tris, pH 8.0, 150 mm NaCl was taken in a stirred 1- × 1-cm cuvette (2.5-ml sample volume), and, upon establishment of a stable baseline, IP6 was added to a final concentration of 10 μm.
RESULTS
Mammalian Cells Contain an Activator of YopJ
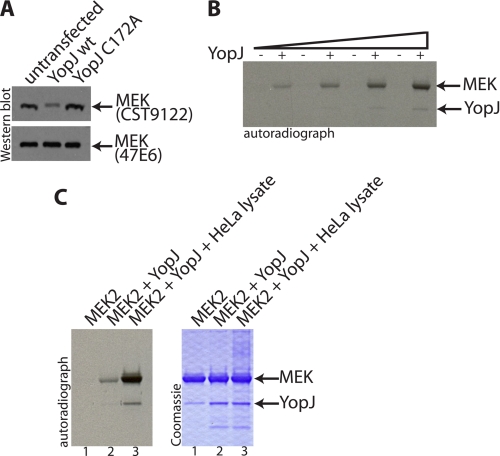
Expression of YopJ in cultured mammalian cells results in the acetylation of cellular MEK1/2. We discovered that acetylation of the activation loop of MEK1/2 resulted in the loss of immunodetection of MEK1/2 by the antiserum CST9122 (Fig. 1A) (6). This antiserum, raised against a peptide comprising the residues of the activation loop of MEK1/2, efficiently recognizes unmodified MEK1/2 but does not recognize MEK1/2 that has been acetylated by YopJ on the serine and threonine residues within the activation loop. Loss of signal on a Western blot thus provided us a stringent readout for the assessment of the extent of YopJ-mediated modification of MEK1/2. The 47E6 antibody, on the other hand, was not sensitive to the modification and allowed visualization of total amounts of MEK1/2. Using the antiserum CST9122 as a readout of MEK modification, we had earlier (6) concluded that even very small amounts of YopJ expressed in mammalian cells were sufficient to cause the acetylation of most, if not all, of the MEK1/2 in cells, suggesting that YopJ is a very efficient acetyltransferase. A similar inference has also been made by others, wherein it has been reported that as little as 1 ng of transfected YopJ or VopA (the YopJ homologue from V. parahemeolyticus) results in inhibition of MAPK signaling (12). Most bacterial toxins are very active in terms of their enzymatic activities; YopH, another type III effector from Yersinia that manifests a protein-tyrosine phosphatase activity, has been recognized as one of the most potent protein-tyrosine phosphatases isolated to date (13).
FIGURE 1.
Assays for the acetylation of MEK; eukaryotic HeLa cells contain an activating cofactor for YopJ. A, HeLa cells were transfected with wild-type YopJ (pSFFV-YopJ wt) or with the enzymatically inactive C172A mutant (pSFFV-YopJ C172A). Cell lysates were immunoblotted for endogenous MEK using an antiserum that is sensitive to acetylation of MEK (CST9122) and another that is not sensitive to the modification (47E6). It is seen that expression of wild-type YopJ leads to modification of the majority of endogenous MEK. B, an in vitro acetylation assay carried out in the presence of [14C]AcCoA shows that progressively higher amounts of YopJ (0.3–1.2 μg) led to an increase in the acetylation of MEK (5 μg). C, inclusion of HeLa cell cytosol (1 μl of a 3 mg/ml preparation) results in the stimulation of YopJ acetyltransferase activity as evidenced by the increased acetylation of MEK and also the increased autoacetylation of YopJ (in lane 3 compared with lane 2).
The acetyltransferase activity of YopJ can also be observed in vitro using purified recombinant proteins expressed in bacteria (5, 6). Acetylation of purified MEK2 by YopJ could be monitored by the inclusion of radioactive [14C]-AcCoA in the reaction mixture (Fig. 1B).
However, analysis of the same reactions by Western blotting showed that only a small percentage of the total MEK molecules had been modified when enzymatic concentrations of YopJ were used. Increasing the concentration of AcCoA, raising the temperature or duration of incubation of the reaction mixture did not result in any significant improvement in the extent of acetylation. The poor efficiency of acetylation in vitro, which contrasted with the extremely efficient modification of MEK in mammalian HeLa cells, led us to wonder if there might be a mammalian cell cofactor required for the activation of the acetyltransferase activity of YopJ.
Indeed, inclusion of dialyzed HeLa cytosol resulted in a marked stimulation of the acetyltransferase activity of YopJ (Fig. 1C). Not only was there a significantly higher acetylation of MEK but also an increase in YopJ autoacetylation. These observations suggested that mammalian cell cytosol contains a cofactor that stimulates the acetyltransferase activity of YopJ.
This observation is not without precedent. Another type III effector protein from Yersinia, YpkA (YopO), requires eukaryotic cell actin as an activator. Binding of G-actin to YpkA stimulates the serine-threonine kinase activity of YpkA (14). Actin is also reported to be a cofactor required for maximal activity of the adenovirus protease adenain (15).
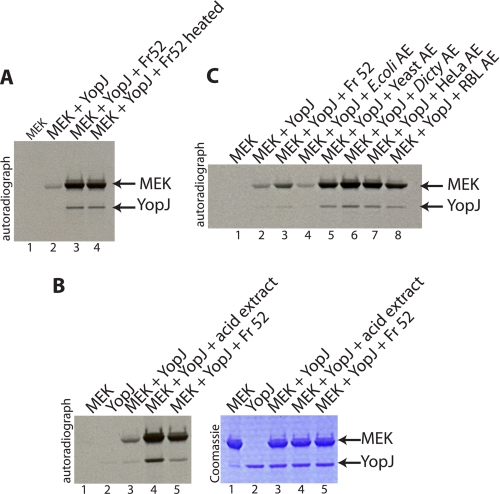
Purification of a Heat-stable Activatory Cofactor Present Only in Eukaryotic Cells
As a first step toward identifying the cofactor required for YopJ we decided to fractionate HeLa cytosol by size-exclusion chromatography. HeLa cytosol corresponding to 30 mg of total protein was resolved over S75 Superdex resin, and 2-ml fractions were collected. 15 μl of each fraction was analyzed for the presence of the cofactor. Cofactor activity was observed (supplemental Fig. S1) to elute from the gel-filtration column as a relatively broad peak starting at Fraction 52, indicating that the cofactor was likely a low molecular mass molecule (smaller than the 12.4-kDa cytochrome c standard).
YopJ has been suggested to act as a deubiquitinase (16, 17), although we have not been able to observe any significant deubiquitinase activity associated with YopJ (expressed as recombinant protein in bacteria or in insect cells). We thus examined whether ubiquitin (molecular mass of 8.5 kDa) might be serving as the low molecular weight activating cofactor for YopJ, but this was not the case. Inclusion of various amounts of ubiquitin in YopJ-catalyzed acetyltransferase assays did not result in any stimulation of YopJ activity.
We then tried various treatments on Fraction 52 (obtained from size-exclusion chromatography described above) to better understand the nature of the cofactor. Significantly, heating Fraction 52 at 95 °C for 30 min did not destroy the cofactor activity (Fig. 2A). Protease, nuclease, and phosphatase treatments of Fraction 52 also did not significantly affect cofactor activity (except at very high doses). The results obtained thus far suggested that YopJ required a mammalian cell, low molecular weight, heat-stable cofactor for activation of its acetyltransferase activity. Furthermore, the cofactor did not seem to be proteinaceous or composed of nucleic acids. Consistent with these observations, an acid extract of HeLa cells was found to contain cofactor activity (Fig. 2B). This treatment removes most protein and nucleic acid components and leaves intact only small molecules and metabolites.
FIGURE 2.
Analysis of the cofactor activity from HeLa cytosol; absence of cofactor activity in bacteria. A, Fraction 52 obtained from size-exclusion chromatography of HeLa cytosol (supplemental Fig. S1) was used in the in vitro acetyltransferase assay and displayed significant stimulation of YopJ activity (compare lane 3 to lane 2). Heating Fraction 52 at 95 °C for 30 min did not destroy the cofactor activity (compare lane 4 to lane 3). B, the activating cofactor for YopJ was also present in an acid extract prepared from HeLa cells (lane 4). C, acid extracts (AE) prepared from the bacteria E. coli, yeast S. cerevisiae, amoeba (D. discoideum), and mammalian sources, HeLa cells and rat brain lysate (RBL), were tested for the presence of cofactor activity by assessing their ability to stimulate the acetylation of MEK by YopJ in vitro. Of the extracts tested, only the bacterial extract (lane 4) was devoid of cofactor activity whereas the extracts prepared from different eukaryotic sources displayed presence of the activity (lanes 5–8). Fraction 52 (lane 3) was used as the positive control.
We next examined whether the cofactor was specific to mammalian cells. Acid extracts were prepared from bacteria, yeast, slime mold, HeLa cells and from rat brain, neutralized, and tested for the presence of cofactor. Interestingly, it was discovered that only bacteria lacked the cofactor (Fig. 2C). The activatory cofactor for YopJ was thus exclusively eukaryotic and not found in bacteria. This observation immediately suggested that the powerful acetyltransferase YopJ is likely kept quiescent in Yersinia by virtue of the lack of a eukaryotic cofactor.
Isolation of the Cofactor and Its Identification as IP6
Having narrowed down the cofactor to an acid extract of eukaryotic cells, we next employed the ion-exchange resin AG1-X8 to further fractionate the extract. Neutralized acid extracts of HeLa cells were passed over acetate-equilibrated AG1-X8 resin, and the resin was subsequently washed with water. Bound material was eluted with hydrochloric acid and tested for cofactor activity in YopJ-catalyzed acetyltransferase assays. Fig. 3A shows that the stimulatory cofactor was, indeed, enriched on AG1-X8 resin.
The eluate from the AG1-X8 resin was then analyzed by mass spectrometry (MS) to identify the molecule(s) contained therein. MS analysis in the negative ion mode revealed the presence of a predominant species with m/z = 658.823 in the elution (Fig. 3B). This moiety was further subjected to fragmentation analysis using MALDI-MS/MS and displayed peaks corresponding to the loss of H3PO4 and HPO3 ions (Fig. 3C). The MS analyses presented in Fig. 3 (B and C) suggested that the cofactor present in the elution was likely IP6. The presence of fragments corresponding to breakdown products of IP6, namely IP5 (m/z = 578.9 Da), IP4 (m/z = 498.9 Da), and other fragments resulting from the loss of H2O from these species, were also seen (Fig. 3C). The identification was confirmed by comparing with the MS/MS fragment spectrum of commercially available IP6 acquired under the same conditions (Fig. 3D). These results establish that the activatory cofactor for YopJ purified from mammalian cells using AG1-X8 resin is IP6, also known as phytic acid or phytate.
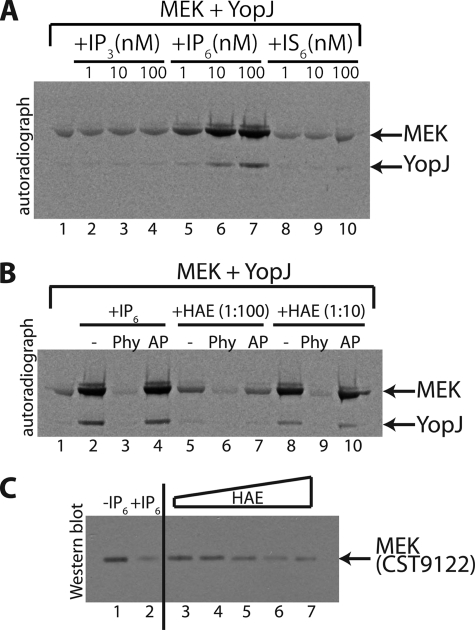
Efficient analysis of inositol phosphates is difficult, because the compounds do not absorb visible or UV light nor can they easily be identified using specific colorimetric reagents. To verify that IP6 is indeed the activatory cofactor we performed in vitro acetyltransferase assays using chemically pure IP3, IP6, and inositol hexakissulfate (IS6). IS6 has similar structure and charge density compared with IP6 and can be used to examine the specificity of the requirement for IP6 (18). It is seen in Fig. 4A that IP3 and IS6 could not substitute for IP6 as cofactors of YopJ. This observation indicates that YopJ is selective in its requirement for phosphate groups and that three phosphates, as in IP3, are insufficient to stimulate the activity of YopJ.
FIGURE 4.
Validation of IP6 as the activating cofactor. A, autoradiograph examining acetylation (using [14C]AcCoA) of MEK by YopJ in the presence of increasing doses (1, 10, and 100 nm) of IP3, IP6, and IS6. In A and B, lane 1 depicts the acetylation of MEK by YopJ in the absence of any added stimulatory factor. It is seen in panel A that only IP6 (lanes 5–7) causes a stimulation of YopJ activity. B, the stimulation caused by IP6 (lane 2) is reversed by the addition of phytase (Phy, lane 3) but not by alkaline phosphatase (AP, lane 4). Phytase sensitivity is also seen for the stimulatory activity present in the HeLa cell acid extract (HAE) shown here for two different dilutions (1:100 and 1:10) of the extract. C, a YopJ-catalyzed acetyltransferase assay was performed (using non-radioactive AcCoA) with MEK as substrate in the absence or presence of IP6 or in the presence of increasing doses of HeLa acid extract (HAE). The reactions were then visualized by Western blot using the modification-sensitive anti-MEK antiserum (CST9122). It is seen that acetylation of MEK by YopJ in the presence of IP6 results in almost complete loss of detection by CST9122 (compare lanes 1 and 2). Also, inclusion of progressively increasing amounts of the HAE in the acetylation reaction result in successively greater modification of MEK as evidenced by the loss of CST9122 immunodetection (lanes 3–7).
The enzyme phytase (myo-inositol-hexakisphosphate 6-phosphohydrolase), naturally present in many plants and microorganisms, breaks down IP6 (phytate) releasing phosphate (19). We observed (Fig. 4B) that phytase destroyed the cofactor activity of both IP6 and of the mammalian cell extract (two dilutions); alkaline phosphatase, on the other hand, did not. This result constitutes further proof of IP6 being the activatory cofactor.
We then examined the extent of MEK acetylation using the discriminatory CST9122 antiserum. As seen in Fig. 4C inclusion of IP6 in acetyltransferase reactions resulted in very high levels of modification of MEK as inferred by the significant loss of immunodetection by the CST9122 antiserum (lanes 1 and 2); also, inclusion of progressively higher amounts of the HeLa cell extract led to progressively higher modification of MEK within the duration of the assay (1 h). This result shows that inclusion of IP6 or HeLa extract can stimulate the acetyltransferase activity of YopJ to achieve near complete acetylation of MEK.
Taken together, these results identify IP6 as the mammalian cell cofactor required by YopJ for activation of its acetyltransferase activity. This is manifested in increased autoacetylation as well as increased substrate acetylation by YopJ.
Our results establish YopJ as the second Yersinia effector, after YpkA, which requires a host cell molecule for stimulation of its enzymatic activity. Binding of eukaryotic cell actin has been shown to result in increased autophosphorylation of YpkA resulting in elevated kinase activity of YpkA toward substrates (20). However, we observed that prior autoacetylation of YopJ was not required for acetylation of MEK by YopJ (supplemental Fig. S2). Thus, unlike the autoacetylation of transcription factor IIB (21) and of p300/cAMP-responsive element binding protein association factor (22) that activates their enzymatic activities, the autoacetylation of YopJ has likely no effect on its substrate acetyltransferase activity.
We demonstrated above (Fig. 4A) that IP3 did not act an activator of YopJ but that the higher phosphorylated inositol polyphosphate IP6 did. The common precursor of all soluble inositol phosphates in most eukaryotic cells is IP3 (which is produced when phospholipase C cleaves phosphatidylinositol 4,5-bisphosphate yielding IP3 and diacylglycerol). IP3 is then processed by a sequence of enzymes to produce a number of more highly phosphorylated inositol species. Inositol pentakisphosphate (IP5) and inositol hexakisphosphate (IP6) are the two most abundant inositol polyphosphates in mammalian cells (23). They are also the precursors of inositol pyrophosphate molecules that contain one or more pyrophosphate bonds (24). We thus examined the absolute requirement for IP6 to be an activator. We have established in Fig. 2C that an acid extract from wild-type yeast S. cerevisiae contained cofactor activity. We thus examined extracts from yeast deletion strains that lack one or more enzymes of the inositol phosphate pathway. The inositol polyphosphate content of yeast has been very well characterized (25).
The major inositol phosphate present in wild-type yeast is IP6 with modest levels of IP7 and IP8 also present (11). In the budding yeast, S. cerevisiae, the enzyme IPK1 converts IP5 to IP6 (26). The ipk1Δ deletion strain thus lacks IP6 but instead accumulates IP5 and diphosphoinositol tetrakisphosphate (PP-IP4), a pyrophosphate resulting from the action of the inositol pyrophosphate-forming enzyme, KCS1. Yeast bearing the double deletion ipk1Δkcs1Δ thus show accumulation of IP4 and IP5 and do not contain any higher polyphosphates. The enzyme inositol phosphate multikinase, IPMK, metabolizes IP3 to IP4 and IP5; ipmkΔ yeast thus accumulate only IP2 and IP3 (27). The yeast strain kcs1Δ lacks the inositol pyrophosphate-forming enzyme, KCS1 (which synthesizes the pyrophosphates IP7 and IP8 from IP6). Therefore, kcs1Δ yeast contain predominantly IP6 but no IP7 or IP8.
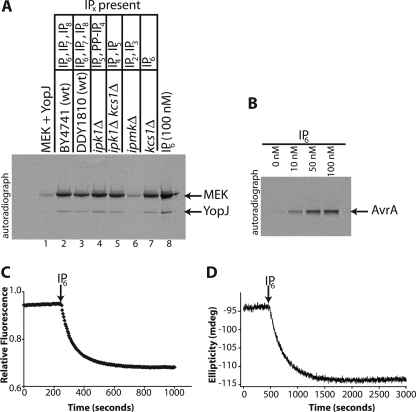
We examined extracts from the wild-type yeast strains BY4741 and DDY1810 and from the deletion strains ipk1Δ (BY4741), ipk1Δkcs1Δ (BY4741), ipmkΔ (DDY1810), and kcs1Δ (DDY1810). The results presented in Fig. 5A show that of the extracts analyzed only those from ipmkΔ yeast (lane 6) were deficient in providing the cofactor for YopJ activity. These yeast contain IP2 and IP3 as the only inositol phosphates. The extract from ipk1Δ yeast (lane 4) that lack IP6 but still contain IP5 was able to activate YopJ. Similarly, the ipk1Δkcs1Δ yeast extract (lane 5) that contains IP4 and IP5 retained activity as well. Inositol pyrophosphates were clearly not required for cofactor activity because kcs1Δ yeast extract (lane 7) supported YopJ activation as well. In control experiments it was verified that purified IP5 was indeed able to support the activation of YopJ (supplemental Fig. S3).
FIGURE 5.
An inositol phosphate-deficient deletion strain of yeast lacks the cofactor; AvrA is stimulated by IP6 undergoing conformational change in response to IP6 addition. A, neutralized acid extracts were prepared from various deletion strains of the yeast S. cerevisiae and included in an acetyltransferase assay using [14C]AcCoA. Lane 1 shows the basal level of MEK acetylation catalyzed by YopJ. Inclusion of yeast extracts results in stimulation of the acetyltransferase activity of YopJ. Extract from the yeast strain ipmkΔ, deleted for the enzyme inositol phosphate multikinase that accumulates IP2 and IP3, does not have the ability to stimulate YopJ (lane 6). The predominant inositol polyphosphate species present in the extracts of the yeast strains used are also indicated in panel A. The stimulation resulting from the inclusion of 100 nm IP6 is shown as a positive control (in lane 8). As in earlier panels, note the concurrent increase in autoacetylation upon stimulation of YopJ. B, inclusion of IP6 (0–100 nm) results in increased autoacetylation of AvrA (1 μg), the YopJ homologue from S. typhimurium. C, change in the emission of protein tryptophan fluorescence of AvrA upon the addition of IP6 (final concentration, 10 μm) to AvrA (5.6 μm). D, change in ellipticity at 222 nm in the far UV CD spectrum of AvrA (5 μm) upon addition of IP6 (10 μm).
We thus conclude that IP5 can also support the activation of YopJ. However, because IP6 is the predominant IPx species present in mammalian cells and was the principal species found in the AG1-X8 purification, it is likely to be the natural activator of the acetyltransferase activity of YopJ. It is interesting to note that, although IP5 is accepted as an efficient activator of YopJ, IS6 was not (Fig. 4A).
IP6 Activates the Acetyltransferase Activity of AvrA and Causes Conformational Change in AvrA
We have noted above that autoacetylation of YopJ provides a convenient readout of its activity. We used this feature as a readout of activation of another member of the YopJ family. We analyzed the activation of AvrA, the YopJ homologue from S. typhimurium. The autoacetylation of AvrA was also found (Fig. 5B) to be activated by IP6. Our results thus establish the generality of the requirement of IP6 as a eukaryotic cell activator for the YopJ family of type III secreted effectors.
We wondered about the nature of the association of IP6 with YopJ and AvrA. The binding of IP6 to TIR1, a receptor for the plant hormone auxin (28) and to the RNA deaminase ADAR2 (18), is shown by their crystal structures to occur deep within the core of the protein molecule in a cavity lined by basic residues. In each of these cases IP6 is found to co-purify with the recombinant protein that has been expressed in a eukaryotic expression system. The binding of the deeply embedded IP6 molecule likely happens during the folding of the protein, which is why the IP6 is carried through the various steps of purification (in IP6-free buffers) to be revealed in the crystal structure. YopJ did not display such an irreversible association with IP6, because in control experiments it was observed by us that YopJ purified in a single column step from a eukaryotic expression system (baculovirus-mediated expression in insect cells) still required activation by externally added IP6. Rather, we believe that IP6 binds to basic residues on or close to the surface of YopJ resulting in allosteric activation of the acetyltransferase activity of YopJ (and other YopJ-like molecules). Crystal structures of the cysteine protease domains of the Vibrio cholerae RTX toxin (29) and of the Clostridium difficile Toxin A (30) show IP6 binding to such basic surface cavities on the proteins distant from the active sites.
Because YopJ was difficult to produce in quantities sufficient for biophysical analyses we chose to examine AvrA by fluorescence and CD spectroscopy. A change in the intrinsic tryptophan fluorescence of a protein upon ligand binding is a very sensitive measure of conformational changes in the environment of the reporter tryptophan residue. AvrA contains a single tryptophan residue (tryptophan 44). We thus examined the effect of IP6 addition on the tryptophan fluorescence of AvrA. As seen in Fig. 5C, addition of IP6 resulted in a decrease in the intensity of emission of intrinsic tryptophan fluorescence of the protein. IP6 binding therefore results in a conformational change in AvrA.
We examined the nature of this IP6-induced conformational change using CD spectroscopy. The phenomenon of circular dichroism is very sensitive to the secondary structure of polypeptides and proteins and is a particularly powerful technique for monitoring conformational changes. Measurement of CD spectra of AvrA in the far-UV spectral region (190–250 nm) in the presence of increasing amounts of IP6 showed a decrease in ellipticity at 208 nm and 222 nm indicative of an increase in α-helical content of AvrA (supplemental Fig. S4A).
We then measured the change in ellipticity of AvrA upon IP6 addition as a function of time (Fig. 5D). It was observed that addition of IP6 resulted in a saturable decrease of the CD signal at 222 nm. Thus, binding of IP6 resulted in an increase of total helicity of AvrA. In control experiments it was verified that addition of AcCoA to AvrA either before or after the addition of IP6 did not result in any significant conformational change. The most likely interpretation of these observations is that IP6 binds in a basic pocket on the surface of AvrA and induces the formation of a helix leading to allosteric activation of the acetyltransferase activity of AvrA. Furthermore, addition of IP6 to the catalytically inactive C172A mutant of AvrA also resulted in a similar conformational change (supplemental Fig. S4B). This suggests that the binding site for IP6 on AvrA is likely distant from the active site for catalysis.
Taken together, our results demonstrate that the YopJ family of type III effector proteins, which includes AvrA from S. typhimurium, requires the eukaryotic host cell molecule IP6 for activation of their acetyltransferase activity. This mechanism suggests that YopJ-like molecules are quiescent in the bacterium where they are synthesized, because bacteria do not contain IP6. Upon injection into mammalian cells by the pathogen type III secretion system these molecules bind host cell IP6 and become activated, thereby dampening the host immune response by covalently modifying host cell-signaling proteins.
DISCUSSION
YopJ has been shown to inhibit the MAPK kinase superfamily (31); the MAPK kinase, MKK6 is acetylated on Ser-207, Lys-210, and Thr-211 by YopJ (5) and, additionally on Lys-172 by VopA, the YopJ homologue from V. parahemeolyticus (8). It is interesting to note that, in these studies the acetylated MKK6 samples (for mass spectrometric identification of the sites of modification) had been prepared from bacteria coexpressing MKK6 and YopJ/VopA. Clearly, under those conditions, the level of overexpressed YopJ/VopA was high enough that even the small amount of basal acetyltransferase activity of YopJ/VopA was sufficient to cause detectable acetylation of MKK6.
Our identification of IP6 as an activating cofactor for YopJ-like acetyltransferases suggests that other members of the YopJ family should also be assayed for activity in the presence of this cofactor. For example, recombinant Y4lO, the YopJ homologue from Rhizobium sp. NGR234, has been reported to lack acetyltransferase activity when assayed in vitro (32). Furthermore, the requirement of a “low molecular mass and heat-stable cofactor” for the complete activity of the eukaryotic sialate-O-acetyltransferases has been reported (33, 34). Based on the properties described for the cofactor we speculate that it might possibly be IP6.
Examination of literature reveals that IP6, whose relative molecular mass is only about 660 Da, has been reported to display properties of a much larger molecule eluting from size-exclusion columns with an apparent size of 9 kDa (35). Fortuitously, IP6 can be retained by 10-kDa molecular mass cutoff dialysis tubing (36) as well, thereby allowing us to observe (in Fig. 1C) the presence of cofactor activity in dialyzed HeLa cytosolic extract.
We have shown that IP6 efficiently stimulates the acetyltransferase activity of YopJ and AvrA. Both molecules have also been proposed to have deubiquitinating activity (16, 17, 37). However, in the absence or presence of IP6, we did not observe any deubiquitinating activity associated with either YopJ or AvrA using Lys-48-linked polyubiquitin chains, Lys-63-linked polyubiquitin chains or the fluorogenic substrate Ubiquitin-AMC.
C. difficile Toxin A and Toxin B possess a glucosyltransferase domain at their N termini. Only the N-terminal fragment having the glucosyltransferase activity reaches the cytosol and acts on its targets the Rho family GTPases (38). This autocatalytic cleavage activity is dependent on host cytosolic inositol phosphate cofactors with highest activity shown by IP6 (39). In Toxin A, IP6 has been shown to be bound in a highly basic pocket on one face of the molecule distant from the active site of the cysteine protease domain (30). Auto-processing of the multifunctional autoprocessing RTX toxin from V. cholerae is also stimulated by IP6 (40), and IP6-induced allosteric activation of protease activity has been proposed (29). YopJ-family proteins have also been proposed to contain a catalytic triad similar to those found in cysteine proteases (41). Members of the cysteine protease superfamily possibly share a common catalytic mechanism in which an activated cysteine residue acts as a catalytic nucleophilic group to achieve chemically distinct outcomes. It is likely that other cysteine protease fold proteins may also be stimulated in their activities by IP6. A slight twist in the tale is provided by the effector cysteine protease AvrRpt2 of the plant pathogen Pseudomonas syringae, which requires a eukaryotic cofactor for autocatalytic cleavage. This eukaryotic factor was identified as the 18-kDa single domain cyclophilin peptidylprolyl-cis/trans-isomerase, rotamase cyclophilin from Arabidopsis (42). This activation required the prolyl isomerization activity of the cyclophilin and required that rotamase cyclophilin be bound to AvrRpt2 for it to be structured and active (43).
IP6 has also been shown to be a cofactor required for inducing conformational change in the Ku70/80 subunits of DNA-dependent protein kinase (44). Additionally, IP6 binding is also important for the activity of the eukaryotic mRNA export factor Gle1 (45) and is involved in maintaining the structural integrity of the human enzyme RNA deaminase (18).
Outbreaks of plague have been reported as recently as 2006 in the Democratic Republic of Congo and somewhat earlier in Algeria (in 2003), Malawi (in 2002), India (in 2002), and Zambia (in 2001). Also, worryingly, a multidrug-resistant strain of Y. pestis has been identified (46). It is thus imperative to gain an understanding of host-pathogen interactions at the molecular level. Our finding that the acetyltransferase activity of the bacterial toxin YopJ of Yersinia is activated by eukaryotic host cell inositol hexakisphosphate is a step toward that goal.
Supplementary Material
Acknowledgments
We acknowledge K. Orth (University of Texas Southwestern Medical Center, Dallas, TX) for plasmids encoding YopJ, K.-L. Guan (University of Michigan, Ann Arbor, MI) for pCMV-MEK2, and A. Saiardi (MRC-Laboratory for Molecular Cell Biology, University College London, UK) for all the yeast strains used in this study. Finally, we thank E. Boucrot (MRC-Laboratory for Molecular Biology, Cambridge, UK) for S. typhimurium genomic DNA and for his careful reading of the manuscript. This work was supported by the Medical Research Council, UK.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- MAPK
- mitogen-activated protein kinase
- MKK
- MAPK kinase
- MEK1,2
- MAPK/extracellular signal-regulated kinase kinase 1 and 2
- AcCoA
- acetylcoenzyme A
- MRC
- Medical Research Council
- CMV
- cytomegalovirus
- MALDI
- matrix-assisted laser desorption ionization
- MS
- mass spectrometry
- MS/MS
- tandem MS
- IPMK
- inositol phosphate multikinase
- IP3
- inositol 1,4,5-trisphosphate
- IP5
- inositol pentakisphosphate
- IP6
- inositol hexakisphosphate
- IP7
- diphosphoinositol pentakisphosphate
- IP8
- bis-diphosphoinositol-tetrakisphosphate
- PP-IP4
- diphosphoinositol-tetrakisphosphate
- IS6
- inositol hexakissulfate.
REFERENCES
- 1.Mueller C. A., Broz P., Cornelis G. R. (2008) Mol. Microbiol. 68, 1085–1095 [DOI] [PubMed] [Google Scholar]
- 2.Trosky J. E., Liverman A. D., Orth K. (2008) Cell Microbiol. 10, 557–565 [DOI] [PubMed] [Google Scholar]
- 3.Monack D. M., Mecsas J., Ghori N., Falkow S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10385–10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmer L. E., Hobbie S., Galán J. E., Bliska J. B. (1998) Mol. Microbiol. 27, 953–965 [DOI] [PubMed] [Google Scholar]
- 5.Mukherjee S., Keitany G., Li Y., Wang Y., Ball H. L., Goldsmith E. J., Orth K. (2006) Science 312, 1211–1214 [DOI] [PubMed] [Google Scholar]
- 6.Mittal R., Peak-Chew S. Y., McMahon H. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18574–18579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones R. M., Wu H., Wentworth C., Luo L., Collier-Hyams L., Neish A. S. (2008) Cell Host Microbe 3, 233–244 [DOI] [PubMed] [Google Scholar]
- 8.Trosky J. E., Li Y., Mukherjee S., Keitany G., Ball H., Orth K. (2007) J. Biol. Chem. 282, 34299–34305 [DOI] [PubMed] [Google Scholar]
- 9.Skaug B., Jiang X., Chen Z. J. (2009) Annu. Rev. Biochem. 78, 769–796 [DOI] [PubMed] [Google Scholar]
- 10.Du F., Galán J. E. (2009) PLoS. Pathog. 5, e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azevedo C., Saiardi A. (2006) Nat. Protoc. 1, 2416–2422 [DOI] [PubMed] [Google Scholar]
- 12.Trosky J. E., Mukherjee S., Burdette D. L., Roberts M., McCarter L., Siegel R. M., Orth K. (2004) J. Biol. Chem. 279, 51953–51957 [DOI] [PubMed] [Google Scholar]
- 13.Guan K. L., Dixon J. E. (1993) Semin. Cell Biol. 4, 389–396 [DOI] [PubMed] [Google Scholar]
- 14.Juris S. J., Rudolph A. E., Huddler D., Orth K., Dixon J. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9431–9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown M. T., McBride K. M., Baniecki M. L., Reich N. C., Marriott G., Mangel W. F. (2002) J. Biol. Chem. 277, 46298–46303 [DOI] [PubMed] [Google Scholar]
- 16.Zhou H., Monack D. M., Kayagaki N., Wertz I., Yin J., Wolf B., Dixit V. M. (2005) J. Exp. Med. 202, 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sweet C. R., Conlon J., Golenbock D. T., Goguen J., Silverman N. (2007) Cell Microbiol. 9, 2700–2715 [DOI] [PubMed] [Google Scholar]
- 18.Macbeth M. R., Schubert H. L., Vandemark A. P., Lingam A. T., Hill C. P., Bass B. L. (2005) Science 309, 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haefner S., Knietsch A., Scholten E., Braun J., Lohscheidt M., Zelder O. (2005) Appl. Microbiol. Biotechnol. 68, 588–597 [DOI] [PubMed] [Google Scholar]
- 20.Trasak C., Zenner G., Vogel A., Yüksekdag G., Rost R., Haase I., Fischer M., Israel L., Imhof A., Linder S., Schleicher M., Aepfelbacher M. (2007) J. Biol. Chem. 282, 2268–2277 [DOI] [PubMed] [Google Scholar]
- 21.Choi C. H., Hiromura M., Usheva A. (2003) Nature 424, 965–969 [DOI] [PubMed] [Google Scholar]
- 22.Santos-Rosa H., Valls E., Kouzarides T., Martínez-Balbás M. (2003) Nucleic Acids Res. 31, 4285–4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Losito O., Szijgyarto Z., Resnick A. C., Saiardi A. (2009) PLoS One 4, e5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett M., Onnebo S. M., Azevedo C., Saiardi A. (2006) Cell Mol. Life Sci. 63, 552–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarmah B., Latimer A. J., Appel B., Wente S. R. (2005) Dev. Cell 9, 133–145 [DOI] [PubMed] [Google Scholar]
- 26.York J. D., Odom A. R., Murphy R., Ives E. B., Wente S. R. (1999) Science 285, 96–100 [DOI] [PubMed] [Google Scholar]
- 27.Saiardi A., Caffrey J. J., Snyder S. H., Shears S. B. (2000) FEBS Lett. 468, 28–32 [DOI] [PubMed] [Google Scholar]
- 28.Tan X., Calderon-Villalobos L. I., Sharon M., Zheng C., Robinson C. V., Estelle M., Zheng N. (2007) Nature 446, 640–645 [DOI] [PubMed] [Google Scholar]
- 29.Lupardus P. J., Shen A., Bogyo M., Garcia K. C. (2008) Science 322, 265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruitt R. N., Chagot B., Cover M., Chazin W. J., Spiller B., Lacy D. B. (2009) J. Biol. Chem. 284, 21934–21940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orth K., Palmer L. E., Bao Z. Q., Stewart S., Rudolph A. E., Bliska J. B., Dixon J. E. (1999) Science 285, 1920–1923 [DOI] [PubMed] [Google Scholar]
- 32.Yang F. J., Cheng L. L., Zhang L., Dai W. J., Liu Z., Yao N., Xie Z. P., Staehelin C. (2009) J. Bacteriol. 191, 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Iwersen M., Dora H., Kohla G., Gasa S., Schauer R. (2003) Biol. Chem. 384, 1035–1047 [DOI] [PubMed] [Google Scholar]
- 34.Lrhorfi L. A., Srinivasan G. V., Schauer R. (2007) Biol. Chem. 388, 297–306 [DOI] [PubMed] [Google Scholar]
- 35.Ali N., Craxton A., Shears S. B. (1993) J. Biol. Chem. 268, 6161–6167 [PubMed] [Google Scholar]
- 36.Van der Kaay J., Van Haastert P. J. (1995) Anal. Biochem. 225, 183–185 [DOI] [PubMed] [Google Scholar]
- 37.Ye Z., Petrof E. O., Boone D., Claud E. C., Sun J. (2007) Am. J. Pathol. 171, 882–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Just I., Selzer J., Wilm M., von Eichel-Streiber C., Mann M., Aktories K. (1995) Nature 375, 500–503 [DOI] [PubMed] [Google Scholar]
- 39.Reineke J., Tenzer S., Rupnik M., Koschinski A., Hasselmayer O., Schrattenholz A., Schild H., von Eichel-Streiber C. (2007) Nature 446, 415–419 [DOI] [PubMed] [Google Scholar]
- 40.Prochazkova K., Satchell K. J. (2008) J. Biol. Chem. 283, 23656–23664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukherjee S., Hao Y. H., Orth K. (2007) Trends Biochem. Sci. 32, 210–216 [DOI] [PubMed] [Google Scholar]
- 42.Coaker G., Falick A., Staskawicz B. (2005) Science 308, 548–550 [DOI] [PubMed] [Google Scholar]
- 43.Coaker G., Zhu G., Ding Z., Van Doren S. R., Staskawicz B. (2006) Mol. Microbiol. 61, 1485–1496 [DOI] [PubMed] [Google Scholar]
- 44.Hanakahi L. A., West S. C. (2002) EMBO J. 21, 2038–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolger T. A., Folkmann A. W., Tran E. J., Wente S. R. (2008) Cell 134, 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galimand M., Guiyoule A., Gerbaud G., Rasoamanana B., Chanteau S., Carniel E., Courvalin P. (1997) N. Engl. J. Med. 337, 677–680 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.