Abstract
Importin-α is the nuclear import receptor that recognizes the classic monopartite and bipartite nuclear localization sequences (cNLSs), which contain one or two clusters of basic amino acids, respectively. Different importin-α paralogs in a single organism are specific for distinct repertoires of cargos. Structural studies revealed that monopartite cNLSs and the C-terminal basic clusters of the bipartite cNLSs bind to the same site on importin-α, termed the major cNLS-binding site. We used an oriented peptide library approach with five degenerate positions to probe the specificity of the major cNLS-binding site in importin-α. We identified the sequences KKKRR, KKKRK, and KKRKK as the optimal sequences for binding to this site for mouse importin-α2, human importin-α1, and human importin-α5, respectively. The crystal structure of mouse importin-α2 with its optimal peptide confirmed the expected binding mode resembling the binding of simian virus 40 large tumor-antigen cNLS. Binding assays confirmed that the peptides containing these sequences bound to the corresponding proteins with low nanomolar affinities. Nuclear import assays showed that the sequences acted as functional cNLSs, with specificity for particular importin-αs. This is the first time that structural information has been linked to an oriented peptide library screening approach for importin-α; the results will contribute to understanding of the sequence determinants of cNLSs, and may help identify as yet unidentified cNLSs in novel proteins.
Keywords: Methods/X-ray Crystallography, Peptides/Interactions, Protein/Nuclear Translocation, Protein/Protein-Protein Interactions, Protein/Structure, Protein/Targeting
Introduction
Cellular compartments are the defining feature of a eukaryotic cell. The nucleus allows the separation of the genetic material and transcriptional machinery from the translational and metabolic processes in the cytoplasm. The nucleus is defined by a double membrane called the nuclear envelope, and the transport across this barrier occurs through large macromolecular assemblies called the nuclear pore complexes (1, 2). Transport of proteins through this pore is facilitated by nucleocytoplasmic transport factors termed karyopherins. These proteins recognize special sequences within the cargo proteins and selectively transport them in (importins) or out (exportins) of the nucleus. Most of the transport factors are built from repetitive sequences such as HEAT (huntingtin EF3, A subunit of PP2A, TOR1) repeats or armadillo (ARM)5 repeats, and belong structurally to the class of proteins termed solenoid proteins (3).
The best characterized nuclear import pathway requires a positively charged sequence within the cargo protein, termed the classic nuclear localization sequence (cNLS) (4). This sequence binds to the ARM repeat protein importin-α (Impα). Impα, through its N-terminal sequence termed the importin-β-binding (IBB) domain, in turn binds importin-β (Impβ1) (5), a HEAT-repeat protein. Impβ1 transiently binds to the proteins lining the nuclear pore (nucleoporins) and in this way facilitates the translocation of this trimeric complex through the pore. The principal regulator of importin-dependent nuclear transport pathways is the small GTPase Ran, which cycles between GDP- and GTP-bound states (6, 7), and RanGTP binding to Impβ1 dissociates the cargo complex once it reaches the nucleus (4).
Two classes of cNLSs recognized by Impα have been distinguished: monopartite cNLSs, which contain one cluster of basic amino acids, and bipartite cNLSs, which contain two clusters of basic amino acids separated by a variable linker (8). Structural studies have shown, however, that both these types of cNLSs bind to the same site on Impα (9–16). The cNLS-binding site corresponds to a long groove formed by the ARM repeat structure, and the N- and C-terminal basic clusters of the bipartite cNLS interact with repeats 4–8 and 1–4, respectively, referred to as the minor and major cNLS binding sites (9, 11, 12). By contrast, the monopartite cNLS interacts mainly with the major binding site (10, 11, 13). A recent study proposed subdividing cNLSs further into six different classes, based on the results of screening random peptide libraries using mRNA display (17). The interpretation of the screening results in this work relied on a sequence alignment of the derived peptides. Furthermore, no structural data have been presented to support different binding modes for these different classes, and therefore it is most likely that all six classes use the same binding site, exploiting different features of the site. Class-1 and Class-2-like sequences have already been shown to use an identical binding mode (12, 18).
In animals, duplications and expansions have resulted in several paralogs in the Impα gene family, which are usually classified into three conserved clades (19–23). Humans contain 6 Impαs, with importin-α5, -α6, and -α7 (karyopherin-α1, -α5, and -α6, respectively) representing the α1 clade, importin-α1 (karyopherin-α2) representing the α2 clade, and importin-α3 and -α4 (karyopherin-α4 and -α3, respectively) representing the α3 clade (20). Although members of all clades mediate the import of cNLS-containing cargos, each paralog specializes in distinct repertoires of cargos (21).
Integration of the available structural data with thermodynamic studies of different cNLSs and their mutants (17, 24, 25) defined a consensus for a bipartite cNLS as KRX10–12KRRK, where the Lys in bold and the underlined represents the most important specificity determinant, and the basic residues at the other underlined positions are also critical (12). The monopartite cNLS simply corresponds to the C-terminal basic cluster of a bipartite cNLS. The critical residues in the bipartite cNLS have been termed P1′–P2′ and P2–P5 for the N- and C-terminal clusters, respectively (12). It is clear, however, that the flanking sequences and the composition of the linker in the bipartite cNLS can further modulate the binding affinity (13, 17, 25). An additional complicating factor is that both Impα proteins from different organisms, and different Impα paralogs in a single organism, exhibit differences in specificity (20, 21, 26–34); for example, transcription factors Brn2 (30) and STAT1/STAT2 (29) show specificity for Impα5, whereas RCC1 shows specificity for the Impα3/4 family (28).
Our understanding of the details of the specificity of cNLS·Impα binding and, therefore, the ability to identify cNLSs in protein sequences remains limited. To address these points, we used an oriented peptide library approach to select the optimal sequences for binding to the major cNLS-binding site in representatives of two different clades of Impα. The oriented peptide library approach has been previously used successfully to determine the optimal sequences recognized by, for example, SH2 domains (35), protein kinases (36), and 14-3-3 (37) proteins. We confirmed, using binding assays, that the peptides corresponding to the selected sequences bound to Impα proteins with high affinities, and established the structural basis of their binding to Impα by determining the crystal structure of an Impα·peptide complex. We also demonstrate that these peptides can function as cNLSs, using in vitro nuclear import assays. Thus, linking structural information to an oriented peptide library screening approach for the first time, the results contribute to understanding of the sequence determinants of cNLSs and may help to identify as yet unidentified cNLSs in novel proteins.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Hexa-His-tagged recombinant Impα proteins were used for crystallography and nuclear import assays, whereas GST-tagged proteins were used for peptide library screening and ELISA-based binding assays. The cDNAs encoding sequences of human importin-α1/karyopherin-α2 comprising amino acids 67–503 (hImpα167–503; RefSeq accession number NP_002257) and human importin-α5/karyopherin-α1 comprising amino acids 60–511 (hImpα560–511; NP_002255) were amplified by PCR, and the PCR products were digested with the endonucleases EcoRI and BamHI, purified by agarose gel electrophoresis, and ligated into similarly digested vector pGex-KG (insert:vector molar ratios of 6:1 or 3:1). The ligated DNA was transformed into DH5α Escherichia coli cells. Colonies were screened by endonuclease digestion and sequenced to ensure the fidelity of the constructs. The pGex-KG-hα1 and pGex-KG-hα5 plasmids were transformed into BL21(DE3) host cells, and the GST-tagged proteins were expressed by isopropyl-β-d-thiogalactopyranoside (all chemicals were from Sigma unless stated otherwise) induction at room temperature (23 °C) for 4 h. Protein degradation was observed for the hImpα5 GST fusion protein after inducing with 1 mm isopropyl-β-d-thiogalactopyranoside, but this was minimized by growing the bacteria at 37 °C to A600 of 0.8 without induction. Bacteria carrying mouse importin-α2 (mImpα2; NP_034785) and mouse importin-β (mImpβ1; NP_032405) GST fusion protein-encoding constructs (38) were grown to A600 of 1.0 and induced with 1 mm isopropyl-β-d-thiogalactopyranoside for 4 h at 28 °C. The proteins were purified by affinity chromatography using a glutathione-Sepharose 4B resin. They were eluted using 50 mm glutathione, and the glutathione was removed by overnight dialysis. The purity of the proteins was estimated to be >90% by SDS-PAGE.
Bacteria carrying the hexa-His-tagged hImpα1, hImpα3 (NP_02259), hImpα5, hImpα7 (NP_036448) (28), and hImpαβ1 (NP_002256) (38) protein-encoding constructs were grown to A600 of 1.0 and induced with 1 mm isopropyl-β-d-thiogalactopyranoside for 4 h. The highest level of expression was observed when induction was performed at 28 °C for 4 h. The proteins were purified by affinity chromatography using nickel-nitrilotriacetic acid-agarose (Qiagen). They were eluted with 500 mm imidazole, and the imidazole was removed by dialysis. The purity of the proteins was estimated to be 95% by SDS-PAGE.
Hexa-His-tagged mImpα270–529 (comprising amino acids 70–529) was expressed and purified by nickel affinity chromatography as described previously (12). For crystallization experiments, cation-exchange chromatography was also performed to increase purity. The protein was eluted with 200 mm sodium chloride, and salt was removed by dialysis. The purity was estimated to be 98% by SDS-PAGE.
Peptide Synthesis
Peptides were synthesized according to standard benzatriazole-1-yl-oxy-tris-(dimethylamino)phosphoniumhexafluorophosphate/1-hydroxybenzotriazole coupling protocol. The synthesized peptides were characterized by reversed phase-high-performance liquid chromatography (125 × 4.6 mm, C18 Zorbax column, linear gradients with 0.1% trifluoroacetic acid (Merck) over 25 min, flow 1 ml/min) and by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. The solutions were lyophilized, and the peptides were stored at −80 °C. The sequences of peptides pepTH1, pepTH5, and pepTM are given in Table 1. The peptides were synthesized with a Cys residue at the N terminus to facilitate nuclear import assays.
TABLE 1.
Oriented peptide library screening-derived peptides
The peptide library comprised peptides with the sequence GSEFESPXKXXXXEA (Table 1), where X represents any amino acids except for C and W. Cys and Trp were omitted to avoid oxidation and sequencing problems (39). The total theoretical degeneracy of this library is 185. An aliquot of each library was sequenced by Edman degradation, to confirm that all 18 amino acids were presented in similar amounts at all degenerate positions.
Peptide Library Screening
Impα-GST fusion proteins (1 mg) were immobilized on 100 μl of glutathione-agarose beads. 300 μl of solution containing 1 mg of the peptide mixture was added to the immobilized Impα-GST and incubated for 4 h at 4 °C to allow for peptide binding to Impα. Unbound peptides were subsequently removed by rapid washing twice with 1 ml of ice-cold HBS (Hepes-buffered saline) buffer, followed by 1 ml HBS buffer without Tween 20. To elute the peptides, the beads were boiled for 5 min causing the proteins to denature and release the peptides. The denatured proteins were then separated from the peptides using a 10-kDa cut-off Microcon (Millipore). The flow-through was collected and sequenced on the protein-sequencing system 785A (Applied Biosystems) using Edman degradation. As a control, the same procedure was performed with GST lacking Impα.
The abundance of each amino acid at a given cycle in the sequence of the peptide mixture was divided by the abundance of the same amino acid in the same cycle of the starting mixture. This accounts for variations in the abundance of amino acids at particular residues in the starting library or variations in yield of amino acids during sequencing. Also, the molar percentages of amino acids in each cycle for the sample relative to those in the GST control were normalized. To scale the relative preferences among all amino acids present in the degenerate library, these raw preference values were then summed and normalized to the total number of amino acids in the degenerate position. Selectivity values >2.0 indicate strong selection (37). It should be noted that the level of background noise in both GST-only and GST-importin fusion proteins necessitated a cycle-based approach to amino acid quantification. Individual amino acid intensities were calculated by comparison with the preceding cycle and tracked in the subsequent cycle.
Cross-linking of Peptides with Alexa Fluor 488-Streptavidin
N-terminal Cys-containing peptides (pepTH1, pepTH5, and pepTM) were cross-linked to Alexa Fluor 488-streptavidin (fst, Molecular Probes) using the heterobifunctional cross-linker N-succinimidyl 3-(2-pyridyldithio)propionate (Sigma) in a two-step reaction. 4.2 nmol of Alexa Fluor 488-streptavidin in buffer S2 (50 mm Na3PO4, 0.15 m NaCl, pH 7.2) was mixed with 33.6 nmol (8 molar excess of Alexa Fluor 488-streptavidin) ethanolic solution of N-succinimidyl 3-(2-pyridyldithio)propionate. The mixture was incubated at room temperature overnight. The excess N-succinimidyl 3-(2-pyridyldithio)propionate was removed by running the mixture through a PD10 column. To determine the number of N-succinimidyl 3-(2-pyridyldithio)propionate molecules cross-linked to Alexa Fluor 488-streptavidin, a 5-μl aliquot of the solution was reduced with 5 μl of 0.5 m dithiothreitol and incubated for 1 h, made up to 1 ml with deionized water, and analyzed for pyridine-2-thione production by measuring absorbance at 343 nm. 1.2 mg of peptide (20 molar excess with respect to cross-linked Alexa Fluor 488-streptavidin) was added to 33.6 nmol of cross-linked Alexa Fluor 488-streptavidin and incubated at 20 °C for 24 h in the dark. To determine the number of peptides conjugated to each molecule of cross-linked Alexa Fluor 488-streptavidin, absorbance was again measured at 343 nm to assess the production of pyridine-2-thione. The final product was buffer-exchanged to HBS buffer using a PD10 column. The conjugates were aliquoted and stored at −20 °C in the dark.
ELISA-based Binding Assays
The binding of peptides to bacterially expressed GST-tagged importins was analyzed using an ELISA-based binding assay (40). Peptides were coated into the wells of polystyrene microtiter plates (Nunclon) in triplicate at 5 pmol/well using 50 mm NaHCO3 (pH 9.6) for 16 h at 4 °C. The wells were blocked by adding 400 μl of 1 × IB buffer (110 mm KCl, 5 mm NaHCO3, 5 mm MgCl2, 1 mm EGTA, 0.1 mm CaCl2, 20 mm Hepes, pH 7.4) containing 1% bovine serum albumin for 1 h at room temperature with shaking (Plateform rocker, Bio-Line), and then washed two times with IB buffer, with the second wash including an incubation for 1 h at room temperature with shaking. Serial dilutions of Imp proteins were then added in IB buffer containing 1 mm dithiothreitol and 1% bovine serum albumin to the microtiter plates and incubated for 16 h at 4 °C with shaking. After extensive washing with IB buffer containing 1 mm dithiothreitol and 1% bovine serum albumin, anti-GST antibody (500 ng/well, Amersham Biosciences) was added, and the plates were incubated for 3 h at room temperature with shaking. Non-specifically bound antibodies were removed by washing 10 times with phosphate-buffered saline buffer containing 0.3% Tween 20 (Bio-Rad). Alkaline-phosphatase-conjugated rabbit anti-goat IgG (0.025 unit of enzyme/well, Sigma) was then added, and the plates were incubated for 1 h at room temperature with shaking, and washing was performed as described for the primary antibody. Binding activity was then determined by adding the chromogenic substrate para-nitropheyl phosphate (1 mg/ml, Sigma) dissolved in 10% diethanolamine and 0.5 mm MgCl2 (pH 9.8). The change of absorbance at 405 nm was measured over 90 min using a plate reader (Molecular Devices, Menlo Park, CA).
Processing of the ELISA data was carried out using Microsoft Excel software (40, 41). Binding affinities were evaluated by plotting the corrected absorbance for different concentrations against time using KaleidaGraph 2.13 software. The data in the linear range was used to obtain the absorbance change per min (A/min), which was plotted against the concentration of the protein. Curves obtained were fitted using Equation 1,
where x is the concentration of the protein, B is the level of protein bound, and k is the dissociation constant representing the concentration of the protein yielding half-maximal binding.
Nuclear Import Assays
A method used to mechanically perforate renal epithelial cells (42) has been adapted to cells of the rat hepatoma tissue culture line, a derivative of the Morris hepatoma 7288C cell line (43) (Flow Laboratories, Bonn, Germany). Cells were cultured in Dulbecco's modified Eagle's medium (ICN Biomedical, Inc.) supplemented with 10% heat-inactivated fetal calf serum (from CSL Ltd.) in a humidified incubator (Forma Scientific, Inc.) with 5% CO2 atmosphere at 37 °C (43, 44). The cells were grown in 50-ml flasks (Nunclon, Denmark), and subcultured every 3–4 days at 90–100% confluence in a laminar flow hood (Airpure, Westinghouse Pty. Ltd., Australia). Hepatoma tissue culture cells for in vitro import assays were seeded on coverslips (15 mm x 15 mm) in 12-well culture plates (Nunclon) with Dulbecco's modified Eagle's medium and grown for 36 h to 95% confluence. The Dulbecco's modified Eagle's medium was then replaced with 1 ml of Dulbecco's modified Eagle's medium without phenol red and glutamine (ICN) but containing 1 m Hepes plus 10% fetal calf serum, prior to use for analysis of nuclear import. Coverslips were rinsed with IB buffer and excess liquid drained. To mechanically perforate cells, and a single layer of tissue paper was placed onto the cell monolayer and rapidly removed 5 s later to mechanically perforate cells. Cells were washed once more with IB buffer to remove residual cytosolic material, and the coverslip was then placed with the cell-bearing surface face down onto a 5-μl drop of nuclear import mix containing an ATP-regenerating system (0.125 mg/ml creatine kinase, 30 mm creatine phosphate, 2 mm ATP, and 2 mm GTP), 30 mg/ml bovine serum albumin, and transport substrate (APC-labeled fusion or fst-labeled protein) and Texas Red-labeled dextran (Sigma). Where appropriate, 4 μm RanGDP and 1 μm Impα·Impβ were added. Imaging was performed using a Bio-Rad MRC-600 confocal laser scanning microscope with a 60× magnification oil immersion lens (Nikon) on a confocal laser scanning microscope, with images being stored every 2–4 min up to 30 min. Fluorescein-5-isothiocyanate-labeled dextran or Texas Red labeled-dextran (Sigma, 70-kDa molecular mass) were used to monitor nuclear membrane integrity.
Crystallization and Crystal Structure Determination
For crystallization, mImpα270–529 was concentrated to 19 mg/ml using a Centricon-30 (Millipore) and stored at −20 °C. The crystals were obtained using cocrystallization, by combining 1 μl of protein solution, 0.5 μl of peptide solution (peptide/protein molar ratio of 4), and 1 μl of reservoir solution on a coverslip and suspending over 0.5 ml of reservoir solution. Single crystals were obtained with a reservoir solution containing 0.65–0.70 m sodium citrate (pH 6.0) and 10 mm dithiothreitol after 15–20 days. X-ray diffraction data were collected using a wavelength of 1.46 Å at a synchrotron-radiation source (Laboratório Nacional de Luz Sincrotron, Campinas, Brazil) with a MAR charge-coupled device imaging-plate detector (MAR Research). A crystal was mounted in nylon loops and transiently soaked in reservoir solution supplemented with 25% of glycerol, and flash-cooled at 100 K in a nitrogen stream (Oxford Cryosystems). The data were processed using the HKL2000 package (45) (Table 2). The crystal presented orthorhombic symmetry (space group P212121) and was isomorphous with other mImpα270–529·peptide complexes (a = 78.86 Å, b = 90.90 Å, c = 100.97 Å). The structure of the complex with the bipartite cNLS from the Xenopus laevis phosphoprotein N1N2 (PDB ID 1PJN (12)) with the peptide omitted was used as starting model for crystallographic refinement. After rigid body refinement using the program Refmac (46), electron density maps were inspected confirming the presence of the peptide not only in the major site but also in the minor binding site. The models were improved, as judged by the free R-factor, through rounds of crystallographic refinement (positional and restrained isotropic individual B-factor with an overall anisotropic temperature factor and bulk solvent correction) and manual modeling using the program Coot (47). The final model comprises 427 residues (71–497), two peptide ligands (9 residues could be modeled in the major site, and 6 residues in the minor site), 103 water molecules, and a citrate ion (Table 2). The quality of the model was checked with the programs PROCHECK (48) and MOLPROBITY (49), and the contacts were analyzed with the program LIGPLOT (50). The atomic coordinates and the structure factors have been deposited to the RCSB Protein Data Bank (www.rcsb.org/pdb/) as ID 3L3Q.
TABLE 2.
Structure determination of pepTM·mImpα70–529 complex
| Diffraction data statistics | |
|---|---|
| Resolution (Å) | 40.0-2.3 (2.38-2.30)a |
| Unique reflections | 29,414 |
| Observations | 128,179 |
| Multiplicity | 4.4 (3.9) |
| Completeness (%) | 89.3 (89.6) |
| Rmergeb (%) | 9.4 (55.1) |
| Average I/(I) | 12.89 (2.39) |
| Refinement statistics | |
| Resolution (Å) | 40.0-2.3 (2.38-2.30) |
| Number of reflections | 29,414 |
| Rcryst (%)c | 18.69 |
| Rfree (%)d | 22.66 |
| Number of non-hydrogen atoms: | |
| Protein | 3,253 |
| Peptide | 137 |
| Solvent | 107 |
| Citrate ion | 13 |
| Mean B-factor (Å2) | 52.56 |
| Coordinate errore (Å) | 0.25 |
| Root mean square deviations from ideal values | |
| Bond lengths (Å) | 0.021 |
| Bond angles (°) | 1.92 |
| Ramachandran plot | |
| Residues in most favored (disallowed) regions (%)f | 95 (0.3) |
a Numbers in parenthesis are for the highest resolution shell.
b Rmerge = Σhkl(Σi(|Ihkl,i − 〈Ihkl〉|))/Σhkl,i 〈Ihkl〉, where Ihkl,i is the intensity of an individual measurement of the reflection with Miller indices h, k, and l, and 〈Ihkl〉 is the mean intensity of that reflection. Calculated for I > −3(I).
c Rcryst = Σhkl(‖Fobs hkl| − |Fcalc hkl‖)/|Fobs hkl|, where |Fobs hkl| and |Fcalc hkl| are the observed and calculated structure factor amplitudes.
d Rfree is equivalent to Rcryst but calculated with reflections (5%) omitted from the refinement process.
e Calculated based on a Luzzati plot with the program SFCHECK (61).
f Calculated with the program PROCHECK (48).
RESULTS
Oriented Peptide Library Approach Reveals the Optimal Sequences Binding to the Major cNLS-binding Site of Importin-α
We adapted an oriented degenerate peptide library approach to study the specificity of binding to the major cNLS-binding site of Impα. This approach has been first used to determine the sequence requirements for binding of phosphopeptides to SH2 domains (35) and subsequently used successfully for a number of applications in a variety of biological systems. The key to using this method is that all peptides bind in the same register. To circumvent the problem that cNLS-like sequences contain a number of positively charged residues that could bind in different registers (11), we added specific flanking sequences on each side of the degenerate positions and kept a specific critical Lys residue fixed. The chosen library contained the sequence GSEFESPXKXXXXEA, with five degenerate positions (X) (Table 1). A Lys at the second position (P2) of the cNLS binding cleft on Impα has been shown to be the most important determinant of a monopartite cNLS, its substitution destroying the function of the cNLS from simian virus SV40 large tumor antigen (T-ag) (12, 24, 51). The flanking sequences were taken from the sequences used in an alanine-scanning mutagenesis study (24), where all mutants were established to bind in the same register. Although a monopartite cNLS can bind at both the major and minor sites (11), we did not expect this to complicate matters because of the lower relative affinity of binding of monopartite cNLSs to the minor site, and because the flanking sequences should reduce binding to this site.
A degenerate library of ∼2 million distinct peptides of identical length was incubated with Impα proteins, the bound peptides separated from the bulk of unbound peptides, and the mixture sequenced by Edman degradation. The abundance of amino acids at each position of the bound peptides was compared with the abundance at the same position in the starting mixture. A single screening experiment therefore yields the order of preference for every amino acid at a given degenerate position, and the comparison of enrichment values between different degenerate positions indicates the relative selectivity of each position (39).
We used N-terminally truncated Impα proteins lacking the IBB domain for peptide library screening. The IBB domain has an autoinhibitory function (52) that is reversed by binding to Impβ1, but all evidence suggests that the truncated proteins closely resemble the Impα·Impβ1 complex in terms of cNLS binding (53).
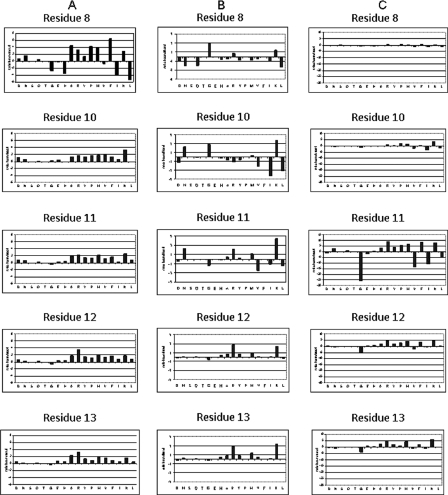
Using mImpα270–529 to screen the peptide library yielded the optimal sequence FKKKRR for the degenerate positions 8–13 in the library (the Lys at position 9 in bold is not degenerate (Fig. 1A and Table 3)). Hydrophobic and positively charged residues were preferred at position 8, with Phe preferred 1.5-fold over Ala and Pro in the second and third place in order of preference. Also strongly enriched were Met, Arg, and Lys. Lys was the only strongly enriched amino acid at position 10, with ∼2-fold preference over other enriched amino acids such as Arg, Pro, Met, Val, and Ala. Lys was also the preferred amino acid at position 11, but followed closely by Arg and Met. Arg was strongly enriched at position 12, preferred almost 2-fold over Met, Ala, Lys, and Phe, with Arg also the optimal amino acid at position 13 (preferred ∼1.5-fold over Ala, Met, Lys, and Tyr).
FIGURE 1.
Oriented peptide library screening. A, specificity of binding to mImpα270–529. A degenerate peptide library (sequence GSEFESPXKXXXXEA, where X indicates any amino acid except Cys and Trp), was used to screen binding to GST-mImpα270–529 bound to beads. Each panel shows the relative abundance of each of the 18 amino acids in a given cycle of sequencing of eluted peptides, compared with control GST beads in the same cycle. B, same as in A, but for hImpα167–503. C, same as in A, but for hImpα560–511.
TABLE 3.
Oriented peptide library screening
| Impα | Position |
||||
|---|---|---|---|---|---|
| 8 | 10 | 11 | 12 | 13 | |
| mImpα270–529 | F (6.4)a | K (3.3) | K (2.6) | R (3.5) | R (3.2) |
| A (4.5) | R (2.2) | M (2.1) | A (2.3) | ||
| P (4.3) | M (2.2) | ||||
| M (3.7) | |||||
| R (3.3) | |||||
| K (2.8) | |||||
| hImpα167–503 | G (2.8) | K (2.7) | K (4.5) | R (2.8) | K (3.4) |
| G (2.8) | N (2.3) | K (2.4) | R (2.9) | ||
| N (2.3) | R (2.1) | ||||
| hImpα560–511 | K (3.7) | R (9.0) | K (4.7) | K (6.1) | |
| F (8.5) | R (4.6) | R (5.1) | |||
| K (8.0) | M (4.0) | M (4.5) | |||
| M (6.9) | F (3.5) | A (2.6) | |||
| P (5.7) | P (2.7) | ||||
| Y (3.9) | Y (2.0) | ||||
| A (3.2) | A (2.0) | ||||
| N (3.1) | |||||
a The most abundant amino acids with enrichment values >2 in each cycle of sequencing are shown (enrichment value is in parentheses).
A similar sequence GKKKRK was obtained for hImpα167–503 (Fig. 1B and Table 3), consistent with the high similarity of the mImpα2 and hImpα1 proteins (95% sequence identity). Gly was the only strongly preferred amino acid at position 8, followed by Lys and Arg. Lys was the optimal residue at position 10, followed by Gly and Asn. Lys was very strongly enriched at position 11, preferred over 2-fold over Asn and Arg. Arg and Lys were significantly enriched at position 12, Arg was preferred ∼1.3-fold over Lys. Lys was slightly (1.2-fold) preferred over Arg at position 13.
hImpα560–511, representing a different Impα subfamily, showed no preference at position 8 but a similar pattern to the other two proteins tested at positions 10–13, yielding the optimal pattern XKKRKK (Fig. 1C and Table 3). Lys was strongly enriched at position 10, followed by Pro and Met; Arg was slightly preferred over Phe and Lys at position 11; Lys and Arg nearly comparable in preference at position 12; and Lys was again preferred followed by Arg and Met at position 13.
Structure of pepTM-mImpα270–529 Complex
To establish the structural basis of binding of the peptide-library derived sequences to Impα, we cocrystallized mImpα270–529 with the pepTM peptide (corresponding to the optimal sequence for binding to mImpα270–529 (Tables 1 and 2)). The structure (Fig. 2) is similar in all respects to the Impα complexes with monopartite cNLS-like peptides reported previously, particularly the T-ag cNLS (11). (i) The structure of mImpα270–529 comprises 10 ARM repeats, each consisting of three α-helices connected by loops (Fig. 2A, after superposition with mImpα270–529 from the T-ag cNLS peptide complex, Protein Data Bank (PDB) ID 1EJL, the root mean square distance for 406 Cα atoms is 0.40 Å). (ii) The electron density maps revealed two peptide ligands binding to one protein molecule in the major (Fig. 2B) and minor cNLS binding sites. Nine and six peptide residues could be modeled in the major and minor binding site, respectively. (iii) The peptides bind in an extended conformation, with their main chain running anti-parallel to the direction of the ARM repeat superhelix, as observed in other cNLS peptide complexes. (iv) The B-factors suggest the peptide binds with higher affinity in the major binding site (57.1 Å2) than the minor binding site (60.0 Å2), as observed for other monopartite cNLS peptides. (v) As observed in other cNLS peptide complexes, most of the hydrogen bonds between the ligand and the protein involve the peptide main chain and the Impα side chains (Fig. 2C). The most favorable contacts involving the peptide side chains occur in positions P2 and P5. Lys in the P2 position has lower B-factors (47.2 Å2) than the rest of the peptide. The side chain of Lys in the P3 position is also well ordered, but the side chain of Lys in the P4 position has poorer electron density, suggesting it forms less favorable interactions. Most importantly, the structure establishes that position 9 in the library corresponds to position P1. This defines the register to interpret the peptide library results.
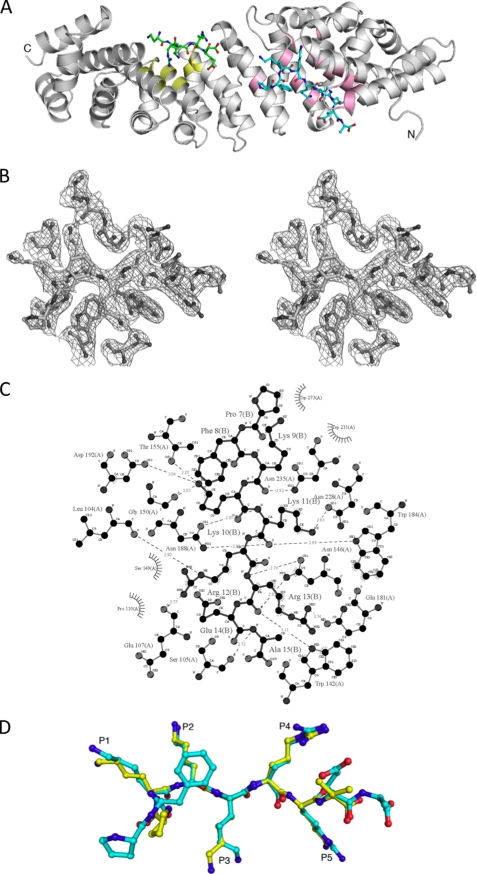
FIGURE 2.
Crystal structure of pepTM-mImpα270–529 complex. A, overall structure of the pepTM·mImpα270–529 complex. mImpα270–529 is shown as a ribbon diagram (pink and yellow colors correspond to the residues defining the major and minor cNLS-binding sites, respectively). The superhelical axis of the repetitive part of the molecule is approximately horizontal. The two cNLS peptides are shown in a stick representation; the peptide bound to the major site is colored cyan, and the peptide bound to the minor site is colored green. B, stereoview of the electron density (drawn with the program PyMOL) in the region of the pepTM peptide bound to the major binding site of mImpα70–529. All peptide residues were omitted from the model, and simulated annealing was run with the starting temperature of 1000 K. The electron density map was calculated with coefficients 3|Fobs| − 2|Fcalc| and data between 40 and 2.3 Å resolution, and contoured at 1.2 standard deviations. Superimposed is the refined model of the peptide. C, schematic diagram of the interactions between the pepTM peptide and the major binding site of mImpα70–529. Polar contacts are shown with dashed lines, and hydrophobic contacts are indicated by arcs with radiating spokes. The cNLS peptide residues are labeled with “R.” Carbon, nitrogen, and oxygen atoms are shown in black, white, and gray, respectively; prepared with the program LIGPLOT. D, superposition of the peptide in the major cNLS binding site in the pepTM- (cyan) and T-ag- (yellow) mImpα70–529 (PDB ID 1EJL) complexes. The Ca atoms of mImpα70–529 in the two complex structures were used in the superposition.
In the minor binding site, the positions P1′ and P2′ are occupied by Lys and Arg, respectively, which appears to be the optimal combination (12). The Arg in particular forms favorable interactions through hydrogen bonds with Ser-360 and Glu-396.
Impα Proteins Bind Their Optimal Peptides with High Affinity
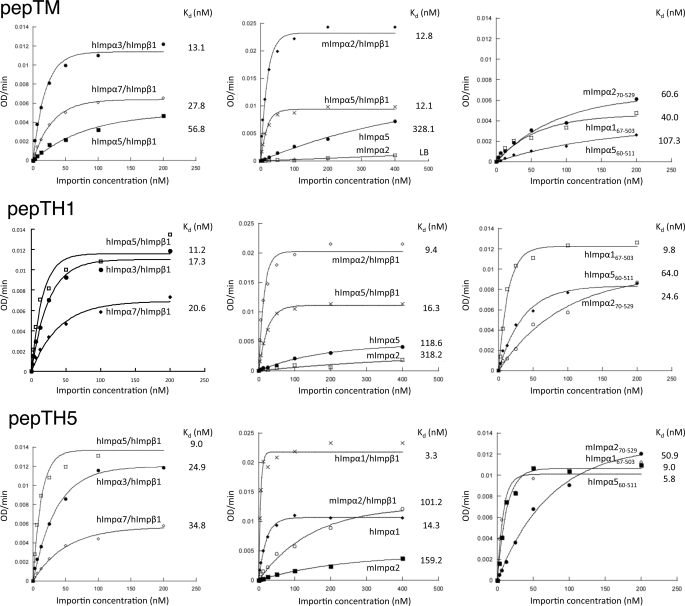
ELISA-based binding assays, as used previously to measure cNLS peptides-Impα affinities (40, 41), were employed to assess the affinities of peptide library-based derived peptides for Impα. mImpα270–529 bound the peptide with its corresponding optimal sequence (pepTM, CGSEFESPFKKKRREA (Table 1); Kd = 53.2 nm) with 1.8-fold higher binding than did hImpα560–511, and with a comparable affinity to hImpα167–503 (Fig. 3 and Table 4). Similarly, hImpα167–503 bound the peptide with its corresponding optimal sequence (pepTH1, CGSEFESPGKKKRKEA (Table 1); Kd = 13.0 nm) with 1.9- and 4.6-fold higher affinity than did hImpα560–511 and mImpα270–529, respectively. Finally, hImpα560–511 bound the peptide with its corresponding optimal sequence (pepTH5, CGSEFESPGKKRKKEA (Table 1); Kd = 9.0 nm) with 5.7-fold higher affinity than mImpα270–529, and with a comparable affinity to hImpα167–503.
FIGURE 3.
Specificity of Impα binding to optimal cNLSs. Binding of Imps to the peptides as determined using an ELISA-based binding assay. Microtiter plates were coated with the peptide and incubated with increasing concentrations of Imps as indicated. Data were fitted to the function B(x) = Bmax(1 − e−kt), where x is the concentration of Imp and B is the level of Imp bound. Results shown are from a single typical experiment, performed in triplicate. See Table 4 for pooled data. Top row, pepTM; middle row, pepTH1; and bottom row, pepTH5.
TABLE 4.
Binding affinities as determined by ELISA-based binding assays
| Peptide | mImpα2 | mImpα2· mImpβ1 | mImpα270–529 | hImpα1 | hImpα1· hImpβ1 | hImpα167–503 | hImpα5· hImpβ1 | hImpα560–511 | hImpα3· hImpβ1 | hImpα7· hImpβ1 |
|---|---|---|---|---|---|---|---|---|---|---|
| pepTM | LBa (2) | 12.8 ± 2.7 (2) | 53.2 ± 14.9 (2) | 328.1 ± 22.8 (2) | 12.1 ± 1.7 (2) | 40.1 ± 0.05 (2) | 56.8 (1) | 97.0 ± 10.4 (2) | 13.1 ± 5.7 (2) | 20.6 ± 4.0 (2) |
| pepTH1 | 203.7 ± 30.9 (2) | 11.4 ± 1.0 (2) | 59.8 ± 0.6 (2) | 118.6 ± 16.7 (2) | 10.7 ± 1.1 (3) | 13.0 ± 4.1 (3) | 16.3 (1) | 24.4 ± 1.0 (3) | 17.3 ± 2.0 (2) | 20.4 ± 14.8 (2) |
| pepTH5 | 158.5 ± 0.75 (2) | 3.7 ± 0.4 (3) | 50.9 (1) | 135.6 ± 68.8 (2) | 13.5 ± 1.6 (3) | 10.7 ± 3.5 (3) | 9.0 (1) | 9.0 + 2.1 (3) | 31.4 + 12.9 (2) | 28.9 + 5.9 (2) |
a Data represent the mean ± S.E. (n in parentheses) for the apparent dissociation constant (Kd). LB, low binding; Kd could not be determined.
The binding affinities of the peptides to the full-length Impα proteins and their Impβ1 complexes were also measured (Fig. 3 and Table 4). As expected, the peptide affinities for Impα·Impβ1 complexes were comparable to the corresponding truncated Impα proteins lacking the IBB domain, and the peptide affinities for full-length Impα proteins were at least 3-fold lower than for the corresponding Impα·Impβ1 complexes or the truncated Impα proteins lacking the IBB domain.
Additionally, the interaction of the peptides with hImpα3·hImpβ1 and hImpα7·hImpβ1 was investigated (Fig. 3 and Table 4). These proteins showed either comparable or up to 3.5-fold lower affinities for the peptides, as compared with the hImpβ1 complexes with the Impα proteins the sequences were originally selected against.
The Peptide Library-derived Sequences Are Functional NLSs
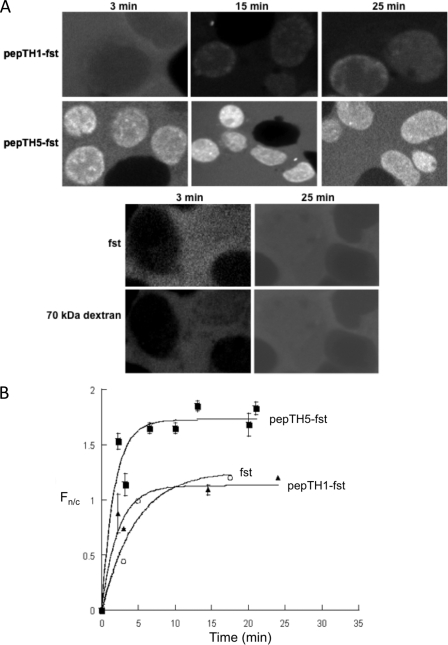
We used an in vitro nuclear import assay to assess the ability of the peptide library-selected Impα-binding sequences to act as NLSs. For this purpose, the peptides were cross-linked to Alexa Fluor 488-streptavidin (fst). hImpα5·hImpβ1 was found to be the most efficient import factor for the corresponding sequence fst-pepTH5 (Fig. 4), with the initial nuclear accumulation rate Fn/c/min of 1.0, and the maximum accumulation Fn/cmax of ∼2.0 (Table 5). Interestingly, hImpα5·hImpβ1 did not import fst-pepTH1 with high efficiency. The importin dependence of nuclear import was indicated by the fact that no nuclear accumulation of the various fst-peptide derivatives was observed in the absence of exogenously added importins. Also, fst itself, lacking the Impα-binding sequence, did not accumulate to any significant extent.
FIGURE 4.
Impα-binding peptides are functional as NLSs in in vitro nuclear import assays. Nuclear import was reconstituted in perforated hepatoma tissue culture cells in the presence of an ATP-regenerating system containing GTP, GDP, and GDP-loaded Ran. Nuclear accumulation of pepTH1- and pepTH5-coupled streptavidin (fst) in the presence of hImpα5·hImpβ1 was followed over time by using a confocal laser scanning microscope. A, confocal laser scanning microscope images are shown for the indicated times at room temperature in the presence of RanGDP. B, results from image analysis are shown, where each data point represents at least five separate measurements for each of Fn, Fc, and background fluorescence. Data are fitted to the function Fn/c = Fn/cmax (1 − e−kt), where Fn/cmax is the maximal level of nuclear accumulation, k is the rate constant, and t is time in minutes. Filled squares, pepTH5-fst; open circles, pepTH1-fst; and filled triangles, fst. Pooled data are presented in Table 5.
TABLE 5.
Nuclear import kinetics of peptide-coupled Alexa Fluor 488-streptavidin (fst) in vitro using purified transport factors
| Componentsa | Nuclear import parameterb |
|
|---|---|---|
| Fn/cmax | Initial rate (Fn/c/min) | |
| pepTH5-fst | ||
| + hImpα3·hImpβ1 | 2.6 | 0.2 |
| + hImpα5·hImpβ1 | 1.7 | 1.0 |
| + hImpα7·hImpβ1 | 1.5 | 0.5 |
| + none | 0.5 | 0.16 |
| pepTH1-fst | ||
| + hImpα3·hImpβ1 | 1.5 | 0.25 |
| + hImpα5·hImpβ1 | 1.1 | UDc |
| + hImpα7·hImpβ1 | 1.5 | 0.6 |
| + none | 0.5 | 0.13 |
| Fst | ||
| + hImpα3·hImpβ1 | 0.8 | 0.1 |
| + hImpα5·hImpβ1 | 1.2 | UD |
| + hImpα7·hImpβ1 | 1.3 | 0.16 |
| + none | 0.6 | 0.12 |
a Transport was performed in the presence or absence of specific exogenously added recombinant nuclear protein import components (hImpα/hImpβ1), and in the presence of GTP and an ATP-regenerating system.
b Raw data were fitted for the function Fn/c (t) = Fn/cmax (1 − e−kt), where Fn/cmax is the maximal level of accumulation at steady state in the nucleus, and t is time in minutes. The Fn/cmax value of 1.0 indicates equal distribution between the nucleus and the cytoplasm, with values below 1 indicating exclusion from the nucleus.
c Unable to be determined due to low level of accumulation.
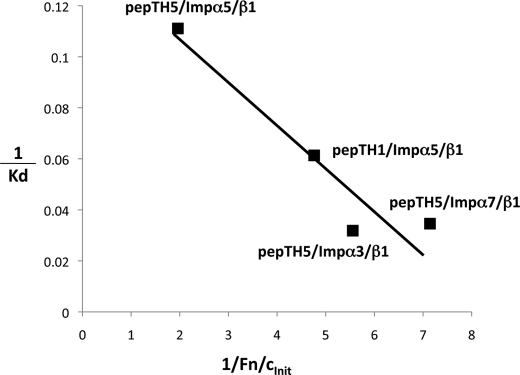
The results for the transport efficiency for the various peptides generally appeared to follow the importin binding affinity (compare the results in Tables 4 and 5). Fig. 5 represents a double-reciprocal plot of selected data for the binding affinity and initial transport rate, highlighting the data for pepTH5 and various Imps, and pepTH1 and Impα5 by way of comparison. As can be seen, there is a direct correlation with the highest binding affinity (that for pepTH5 and Impα5/β1) corresponding with the fastest initial transport rate (for the same combination). This is strong evidence that the Imp-binding affinity is a limiting step in the nuclear import process (41, 54, 55).
FIGURE 5.
Correlation of nuclear import efficiency with importin binding affinity. Reciprocal plot of selected data from Tables 4 and 5 for the binding affinity (nanomolar) (y axis) and initial nuclear import rate (Fn/cinit) in minutes (x axis); the initial import rate value was determined after subtraction of the import rate for fst alone (no NLS). The line represents linear regression (r = 0.8925).
DISCUSSION
The principal sequence determinants of cNLSs and the molecular basis of their action have been known for some time, but understanding of the details of the specificity of binding to the receptor Impα continues to be limited, meaning that the predictive identification of putative cNLSs in novel sequences remains difficult. Here we used an oriented degenerate peptide library approach for the first time to probe the specificity of peptide binding in the major cNLS-binding site in Impα. This approach has been previously used successfully to study peptide binding and modification in a number of biological systems, for example to characterize the binding specificities of SH2 domains (35), PTB domains (56), and 14-3-3 proteins (37), and to determine the specificities of phosphorylation by protein kinases (36) and the specificities of cleavage by proteases (57). An important requirement for this approach is that all peptides bind in the same register, which is established, for example, by the presence of a phosphorylated residue in a fixed position in phosphopeptides when analyzing the binding to phosphopeptide-binding domains. An important limitation of the approach is that the contribution of each residue to overall binding is considered independent, i.e. the presence of a particular amino acid at one position is assumed not to influence the specificity at a neighboring position. Although this assumption is likely incorrect when considering a single isolated peptide, it appears to function well in a positional scanning context and has been validated in the context of peptide binding by major histocompatibility complex molecules, for example (58).
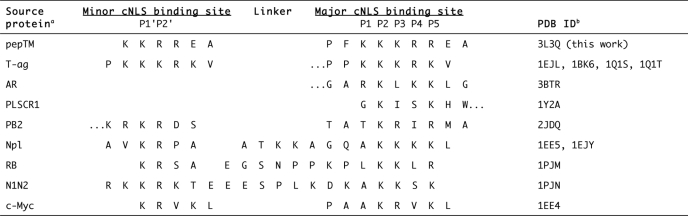
A second potential limitation revolves around the ability of peptides to bind in different registers to Impα. We attempted to achieve binding in the same register by (i) fixing a critical Lys residue in the sequence (12, 24, 51) and (ii) adding specific flanking sequences based on the alanine-scanning mutagenesis study (24), which established that all mutants containing these flanking sequences bound in the same register. The crystal structure of the pepTM·mImpα270–529 complex established that the fixed Lys at position 9 of the library corresponded to position P1, according to the established nomenclature (12). Because all three library experiments identified Lys residues as preferred residues at positions 9 and 10, it was reasonable to assume that the peptides bind in the same register as observed for pepTM in the crystal structure (Table 6).
TABLE 6.
Binding to specific binding pockets of importin-α based on structural data
a AR, androgen receptor; PLSCR1, phospholipid scramblase 1; PB2, influenza virus polymerase PB2; Np1, nucleoplasmin; RB, retinoblastoma protein; N1N2, X. laevis phosphoprotein N1N2; Nup50, nucleoporin Nup50; Nup2, nucleoporin Nup2p.
The sequences selected by the peptide library screening are very similar to already characterized cNLSs, particularly in being rich in positively charged amino acids Lys and Arg (Table 6). The optimal consensus sequences for positions P2–P5 has been previously suggested as K(R/K)X(R/K) (59), K(K/R)X(K/R) (24), KR(R/X)K (12), KRRR (25), and KR(K/R)R or K(K/R)RK (17), all of which are very similar to the sequences selected in our peptide library experiments. There appears to be good agreement that Lys is strongly preferred in the P2 position. The discrimination between Lys and Arg in positions P3–P5 may not be strong, and the preference may indeed depend on the surrounding amino acids in a cNLS and the specific Impα protein used. It is interesting that Arg is selected in our library experiments for hImpα5 in the P3 position, whereas Lys is preferred in hImpα1 and mImpα2. hImpα5 contains an Asp at the position equivalent to Glu-266 in mImpα2 and hImpα1. Crystal structures suggest that this negatively charged residue may be central in determining the preference for a basic side chain in the cNLS at this position. The shorter Asp side chain in hImpα5 could allow more favorable binding of the longer Arg side chain at this position, as suggested by the peptide library experiments. Interestingly, hImpα3 and hImpα7 contain Ser and Asn at the analogous position, respectively, implying that they may display further differences in preference at this position. Nevertheless, Arg appeared to be favored over Lys in the P3 position according to the Ala mutagenesis in the context of c-Myc and T-ag cNLSs, using Saccharomyces cerevisiae Impα (24); this protein contains a Glu at the residue equivalent to mImpα2 Glu-266. Overall, the sequences selected by hImpα1 and hImpα5, representing different subfamilies of Impα, are very similar, indicating that any functional differences between these two paralogs of Impα are unlikely to result from differences in recognition specificity in the major cNLS-binding site.
ELISA-based binding assays confirmed that the optimal sequences selected by the oriented peptide library experiments indeed bind to Impα proteins with high affinity. The measured affinities are similar or higher than measured previously for the functional monopartite cNLS T-ag (53, 60). This is consistent with the fact that, considering positions P1–P5, the sequence of T-ag differs only in 0–2 positions from our library-selected sequences, suggesting that T-ag is already optimized as a monopartite cNLS in terms of these key positions. The implication is that higher affinities can therefore only be achieved by forming favorable interactions using other positions reaching outside the major cNLS binding site (13, 25).
Nuclear import assays show that the sequences selected by the oriented peptide library experiments are functional as NLSs. It has been suggested that there are upper and lower thresholds in terms of binding energy to Impα for functional NLSs (24). The initial rate of protein import has been suggested to be linearly correlated with the cNLS·Impα affinity (41, 54, 55). However, when the affinity becomes too high, the cargo would not be able to be released. Our library experiments did not exceed such as affinity threshold, presumably because we have optimized only a few interactions in the major cNLS binding site.
In summary, we have successfully used an oriented peptide library approach to find optimal sequences for binding to the major cNLS binding site in Impα. Similar sequences consistent with features of known cNLSs were selected for mouse Impα2, human Impα1, and human Impα5. The crystal structure of the peptide·mImpα2 complex showed the expected binding mode, binding assays demonstrating that these peptides bound to Impα proteins with high affinities, and nuclear import assays showed that the sequences acted as functional NLSs. The results contribute to our understanding of the sequence determinants of cNLSs and may help identify cNLSs in novel protein sequences.
Acknowledgments
We thank Matthias Köhler (Damp, Germany) for DNA constructs. We acknowledge the use of the University of Queensland ROCX (Remote Operation Crystallization and X-ray) Diffraction Facility.
This work was supported in part by grants from the Australian Research Council (ARC, to B. K.) and Fundacao de Amparo a Pesquisa do Estado de Sao Paulo and Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq, Brazil, to M. R. M. F.).
The atomic coordinates and structure factors (code 3L3Q) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ARM
- armadillo
- fst
- Alexa Fluor 488-streptavidin
- Impα
- importin-α
- Impβ
- importin-β
- hImp
- human Imp
- mImp
- mouse Imp
- cNLS
- classic nuclear localization sequence
- T-ag
- simian virus 40 large tumor antigen
- IBB
- importin-β-binding
- GST
- glutathione S-transferase
- ELISA
- enzyme-linked immunosorbent assay
- HEAT
- huntingtin, EF3, A subunit of PP2A, TOR1
- HBS
- Hepes-buffered saline.
REFERENCES
- 1.Aitchison J. D., Wozniak R. W. (2007) Nature 450, 621–622 [DOI] [PubMed] [Google Scholar]
- 2.Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Sali A., Rout M. P. (2007) Nature 450, 695–701 [DOI] [PubMed] [Google Scholar]
- 3.Kobe B., Kajava A. V. (2000) Trends Biochem. Sci. 25, 509–515 [DOI] [PubMed] [Google Scholar]
- 4.Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. (2007) J. Biol. Chem. 282, 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cingolani G., Petosa C., Weis K., Müller C. W. (1999) Nature 399, 221–229 [DOI] [PubMed] [Google Scholar]
- 6.Melchior F., Paschal B., Evans J., Gerace L. (1993) J. Cell Biol. 123, 1649–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore M. S., Blobel G. (1993) Nature 365, 661–663 [DOI] [PubMed] [Google Scholar]
- 8.Dingwall C., Laskey R. A. (1991) Trends Biochem. Sci. 16, 478–481 [DOI] [PubMed] [Google Scholar]
- 9.Conti E., Kuriyan J. (2000) Structure 8, 329–338 [DOI] [PubMed] [Google Scholar]
- 10.Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. (1998) Cell 94, 193–204 [DOI] [PubMed] [Google Scholar]
- 11.Fontes M. R., Teh T., Kobe B. (2000) J. Mol. Biol. 297, 1183–1194 [DOI] [PubMed] [Google Scholar]
- 12.Fontes M. R., Teh T., Jans D., Brinkworth R. I., Kobe B. (2003) J. Biol. Chem. 278, 27981–27987 [DOI] [PubMed] [Google Scholar]
- 13.Fontes M. R., Teh T., Toth G., John A., Pavo I., Jans D. A., Kobe B. (2003) Biochem. J. 375, 339–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen M. H., Ben-Efraim I., Mitrousis G., Walker-Kopp N., Sims P. J., Cingolani G. (2005) J. Biol. Chem. 280, 10599–10606 [DOI] [PubMed] [Google Scholar]
- 15.Cutress M. L., Whitaker H. C., Mills I. G., Stewart M., Neal D. E. (2008) J. Cell Sci. 121, 957–968 [DOI] [PubMed] [Google Scholar]
- 16.Tarendeau F., Boudet J., Guilligay D., Mas P. J., Bougault C. M., Boulo S., Baudin F., Ruigrok R. W., Daigle N., Ellenberg J., Cusack S., Simorre J. P., Hart D. J. (2007) Nat. Struct. Mol. Biol. 14, 229–233 [DOI] [PubMed] [Google Scholar]
- 17.Kosugi S., Hasebe M., Matsumura N., Takashima H., Miyamoto-Sato E., Tomita M., Yanagawa H. (2009) J. Biol. Chem. 284, 478–485 [DOI] [PubMed] [Google Scholar]
- 18.Kosugi S., Hasebe M., Tomita M., Yanagawa H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik H. S., Eickbush T. H., Goldfarb D. S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13738–13742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mason D. A., Stage D. E., Goldfarb D. S. (2009) J. Mol. Evol. 68, 351–365 [DOI] [PubMed] [Google Scholar]
- 21.Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. (2004) Trends Cell Biol. 14, 505–514 [DOI] [PubMed] [Google Scholar]
- 22.Tsuji L., Takumi T., Imamoto N., Yoneda Y. (1997) FEBS Lett. 416, 30–34 [DOI] [PubMed] [Google Scholar]
- 23.Köhler M., Ansieau S., Prehn S., Leutz A., Haller H., Hartmann E. (1997) FEBS Lett. 417, 104–108 [DOI] [PubMed] [Google Scholar]
- 24.Hodel M. R., Corbett A. H., Hodel A. E. (2001) J. Biol. Chem. 276, 1317–1325 [DOI] [PubMed] [Google Scholar]
- 25.Kosugi S., Hasebe M., Entani T., Takayama S., Tomita M., Yanagawa H. (2008) Chem. Biol. 15, 940–949 [DOI] [PubMed] [Google Scholar]
- 26.Miyamoto Y., Imamoto N., Sekimoto T., Tachibana T., Seki T., Tada S., Enomoto T., Yoneda Y. (1997) J. Biol. Chem. 272, 26375–26381 [DOI] [PubMed] [Google Scholar]
- 27.Nadler S. G., Tritschler D., Haffar O. K., Blake J., Bruce A. G., Cleaveland J. S. (1997) J. Biol. Chem. 272, 4310–4315 [DOI] [PubMed] [Google Scholar]
- 28.Köhler M., Speck C., Christiansen M., Bischoff F. R., Prehn S., Haller H., Görlich D., Hartmann E. (1999) Mol. Cell. Biol. 19, 7782–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melen K., Fagerlund R., Franke J., Kohler M., Kinnunen L., Julkunen I. (2003) J. Biol. Chem. 278, 28193–28200 [DOI] [PubMed] [Google Scholar]
- 30.Yasuhara N., Shibazaki N., Tanaka S., Nagai M., Kamikawa Y., Oe S., Asally M., Kamachi Y., Kondoh H., Yoneda Y. (2007) Nat. Cell Biol. 9, 72–79 [DOI] [PubMed] [Google Scholar]
- 31.Köhler M., Görlich D., Hartmann E., Franke J. (2001) Virology 289, 186–191 [DOI] [PubMed] [Google Scholar]
- 32.Köhler M., Fiebeler A., Hartwig M., Thiel S., Prehn S., Kettritz R., Luft F. C., Hartmann E. (2002) Cell Physiol. Biochem. 12, 335–344 [DOI] [PubMed] [Google Scholar]
- 33.Jiang C. J., Shoji K., Matsuki R., Baba A., Inagaki N., Ban H., Iwasaki T., Imamoto N., Yoneda Y., Deng X. W., Yamamoto N. (2001) J. Biol. Chem. 276, 9322–9329 [DOI] [PubMed] [Google Scholar]
- 34.Guillemain G., Muñoz-Alonso M. J., Cassany A., Loizeau M., Faussat A. M., Burnol A. F., Leturque A. (2002) Biochem. J. 364, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Songyang Z., Shoelson S. E., Chaudhuri M., Gish G., Pawson T., Haser W. G., King F., Roberts T., Ratnofsky S., Lechleider R. J., Neel B. J., Birge R. B., Fajardo J. E., Chou M. M., Hanafusa H., Schaffhausen B., Cantley L. C. (1993) Cell 72, 767–778 [DOI] [PubMed] [Google Scholar]
- 36.Songyang Z., Blechner S., Hoagland N., Hoekstra M. F., Piwnica-Worms H., Cantley L. C. (1994) Curr. Biol. 4, 973–982 [DOI] [PubMed] [Google Scholar]
- 37.Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. (1997) Cell 91, 961–971 [DOI] [PubMed] [Google Scholar]
- 38.Hu W., Kemp B. E., Jans D. A. (2005) J. Cell. Biochem. 95, 782–793 [DOI] [PubMed] [Google Scholar]
- 39.Songyang Z., Liu D. (2001) Methods Enzymol. 332, 183–195 [DOI] [PubMed] [Google Scholar]
- 40.Efthymiadis A., Shao H., Hübner S., Jans D. A. (1997) J. Biol. Chem. 272, 22134–22139 [DOI] [PubMed] [Google Scholar]
- 41.Hübner S., Xiao C. Y., Jans D. A. (1997) J. Biol. Chem. 272, 17191–17195 [DOI] [PubMed] [Google Scholar]
- 42.Simons K., Virta H. (1987) EMBO J. 6, 2241–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jans D. A., Ackermann M. J., Bischoff J. R., Beach D. H., Peters R. (1991) J. Cell Biol. 115, 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rihs H. P., Jans D. A., Fan H., Peters R. (1991) EMBO J. 10, 633–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 46.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 47.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 48.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 49.Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 51.Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. (1984) Cell 39, 499–509 [DOI] [PubMed] [Google Scholar]
- 52.Kobe B. (1999) Nat. Struct. Biol. 6, 388–397 [DOI] [PubMed] [Google Scholar]
- 53.Catimel B., Teh T., Fontes M. R., Jennings I. G., Jans D. A., Howlett G. J., Nice E. C., Kobe B. (2001) J. Biol. Chem. 276, 34189–34198 [DOI] [PubMed] [Google Scholar]
- 54.Hu W., Jans D. A. (1999) J. Biol. Chem. 274, 15820–15827 [DOI] [PubMed] [Google Scholar]
- 55.Hodel A. E., Harreman M. T., Pulliam K. F., Harben M. E., Holmes J. S., Hodel M. R., Berland K. M., Corbett A. H. (2006) J. Biol. Chem. 281, 23545–23556 [DOI] [PubMed] [Google Scholar]
- 56.O'Bryan J. P., Martin C. B., Songyang Z., Cantley L. C., Der C. J. (1996) J. Biol. Chem. 271, 11787–11791 [DOI] [PubMed] [Google Scholar]
- 57.Turk B. E., Huang L. L., Piro E. T., Cantley L. C. (2001) Nat. Biotechnol. 19, 661–667 [DOI] [PubMed] [Google Scholar]
- 58.Stryhn A., Pedersen L. O., Romme T., Holm C. B., Holm A., Buus S. (1996) Eur. J. Immunol. 26, 1911–1918 [DOI] [PubMed] [Google Scholar]
- 59.Chelsky D., Ralph R., Jonak G. (1989) Mol. Cell. Biol. 9, 2487–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fanara P., Hodel M. R., Corbett A. H., Hodel A. E. (2000) J. Biol. Chem. 275, 21218–21223 [DOI] [PubMed] [Google Scholar]
- 61.Vaguine A. A., Richelle J., Wodak S. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 191–205 [DOI] [PubMed] [Google Scholar]