Abstract
The complex of factor VIIa (FVIIa) with tissue factor (TF) triggers coagulation by recognizing its macromolecular substrate factors IX (FIX) and X (FX) predominantly through extended exosite interactions. In addition, TF mediates unique cell-signaling properties in cancer, angiogenesis, and inflammation that involve proteolytic cleavage of protease-activated receptor 2 (PAR2). PAR2 is cleaved by FVIIa in the binary TF·FVIIa complex and by FXa in the ternary TF·FVIIa·FXa complex, but physiological roles of these signaling complexes are incompletely understood. In a screen of FVIIa protease domain mutants, three variants (Q40A, Q143N, and T151S) activated macromolecular coagulation substrates and supported signaling of the ternary TF·FVIIa-Xa complex normally but were severely impaired in binary TF·FVIIa·PAR2 signaling. The residues identified were located in the model-predicted S2′ pocket of FVIIa, and complementary PAR2 P2′ Leu-38 replacements demonstrated that the P2′ side chain was indeed crucial for PAR2 cleavage by TF·FVIIa. In addition, PAR2 was activated more efficiently by FVIIa T99Y, consistent with further contributions from the S2 subsite. The P2 residue preference of FVIIa and FXa predicted additional PAR2 mutants that were efficiently activated by TF·FVIIa but resistant to cleavage by the alternative PAR2 activator FXa. Thus, contrary to the paradigm of exosite-assisted cleavage of PAR1 by thrombin, the cofactor-associated protease FVIIa recognizes PAR2 predominantly by catalytic cleft interactions. Furthermore, the delineated molecular details of this substrate interaction enabled protein engineering of protease-selective PAR2 receptors that will aid further studies to dissect the roles of TF signaling complexes in vivo.
Keywords: Coagulation Factors, Enzyme Structure, G-protein-coupled Receptors (GPCR), Protein-Protein Interactions, Signal Transduction
Introduction
Coagulation factor VIIa (FVIIa)2 in complex with its cellular receptor tissue factor (TF) mediates activation of two pathways of major physiological importance: (i) initiation of blood coagulation by activation of coagulation factors IX and X (FIX and FX), and (ii) induction of cell signaling through protease-activated receptors (PARs). TF·FVIIa-dependent signaling primarily activates PAR2, which belongs to a family of four G-protein-coupled receptors activated by specific proteolytic cleavage of their N-terminal extracellular domain (1). Cleavage unmasks a new N terminus, which serves as a tethered ligand that induces transmembrane signaling (2). Activation of PAR2 by the TF·FVIIa binary complex involves cellular pools of TF with low affinity for FVIIa, whereas high affinity cell surface TF mediates coagulation activation and the associated cell signaling of the ternary complex of TF·FVIIa·FXa (3). In the latter complex, FXa is the primary activator for PAR2 (4). PAR2 triggers typical G-protein-coupled as well as β-arrestin-dependent signaling and thereby induces cytokine, chemokine, and growth factor expression and regulates protein synthesis, cell motility, proliferation, and apoptosis (5).
Although the TF pathway also induces thrombin-dependent PAR1 signaling and transcriptional responses are often similar following PAR1 and PAR2 activation (6), PAR2 supports biological responses that are distinct from PAR1. For example, TF-dependent angiogenesis in TF cytoplasmic domain-deleted mice requires PAR2, but not PAR1 (7, 8), and breast cancer development was delayed in mice that lack PAR2, but not PAR1 (9). TF·FVIIa binary complex activation of PAR2 can be specifically blocked with an antibody that minimally affects coagulation, and this antibody, but not a coagulation-blocking anti-TF antibody, prevented breast cancer growth and PAR2-dependent pregnancy complication in vivo (9–11). The ternary TF·FVIIa·FXa complex or FXa also activate PAR2, so do proteases other than coagulation proteases (12, 13). The role of these signaling pathways in vivo remains unclear, and improved tools are required to define the specific roles of coagulation protease signaling via PAR2.
Exosite-driven macromolecular substrate recognition is a common mechanism by which proteases of the coagulation system achieve their remarkable specificity (14). This also applies to the activation of macromolecular substrates by TF·FVIIa (15–17) where the initial encounter with FX is governed primarily by interactions involving exosites in FVIIa and TF (18, 19). Subsequent engagement of the FVIIa active site with residues of FX flanking the scissile bond then leads to proteolytic conversion of FX to FXa. Typically, cleavage of macromolecular substrates only requires a proper fit of the primary specificity residues with complementary sites in the catalytic cleft, and these interactions make only minor contributions to macromolecular substrate affinity. Exosite engagement is also a prominent feature in PAR recognition. Cleavage of PAR1 and PAR3 by thrombin is facilitated by charge interactions between exosite I in thrombin and a hirudin-like acidic element located C-terminally in the tethered PAR sequence (20). Disruption of this exosite by mutagenesis severely impairs thrombin binding and the rate of PAR1 activation (21, 22). Although PAR4 does not carry a recognition sequence for thrombin's exosite I, extended interactions between thrombin and the coreceptor PAR3 support efficient thrombin-PAR4 signaling (23, 24). Unlike these PARs, PAR2 lacks distinct exosite recognition sequences, a feature that may be required for its role as a broadly responsive sensor for trypsin-like serine proteases.
Here, we address the role of PAR2 in coagulation protease signaling and define the structural requirements for PAR2 activation by FVIIa. Mutational mapping of FVIIa demonstrates that the S2′ pocket plays a critical role in PAR2 cleavage. Point mutations in this region abolish receptor activation while maintaining essentially normal FIX and FX cleavage, indicating distinctly different modes of recognition of PAR2 and coagulation substrates by TF·FVIIa. Mutations of PAR2 confirm that efficiency of cleavage is primarily determined by complementary interactions of the catalytic cleft and that single point mutations are sufficient to generate PAR2 receptors that loose responsiveness to specific coagulation proteases. These data support novel approaches to further define the role of the promiscuous PAR2 receptor in coagulation signaling pathways.
EXPERIMENTAL PROCEDURES
Proteins
Recombinant FVII and variants and soluble tissue factor (1–219) were produced and purified as previously described (18). FIX and FX were from Enzyme Research Laboratories (Swansea, UK). Plasma-derived FXa was from Hematologic Technologies (Essex Junctions, VT), hirudin was from Calbiochem, and sequencing grade trypsin was from Roche Applied Science (Indianapolis, IN). WEDE15/ATAP2 anti-PAR1 antibodies were kindly provided by Dr. L. F. Brass (25). ERK and phospho-ERK antibodies were from Cell Signaling Technologies (Danvers, MA). Recombinant Nematode Anticoagulant Protein C2 (NAPc2) (4) was kindly provided by Dr. G. Vlasuk (Corvas International, San Diego, CA).
Cell Culture and Transfection
MDA-MB-231 breast carcinoma cells (ATCC, Rockville, MD) and human immortalized keratinocytes (HaCaTs) were maintained in DMEM, 10% fetal bovine serum. Human umbilical vein endothelial cells (HUVECs) were transduced with TF and PAR2 adenoviral constructs as described previously (26). For cell surface detection of PAR2, a FLAG epitope tag (DYKDDDDK) was inserted in front of the mature receptor sequence starting at residue Ile-26 (see Fig. 2C) and cloned into pcDNA3.1/Zeo(+) (Invitrogen). Chinese hamster ovary cells expressing human TF (CHO-TF) (26) were transfected with FLAG-PAR2 expression construct and used in the PAR2 cleavage assay after 24 h. Murine PAR1-deficient M6-11 fibroblasts (27) were transiently transfected with FLAG-PAR2 and human TF for signaling assays 24 h later.
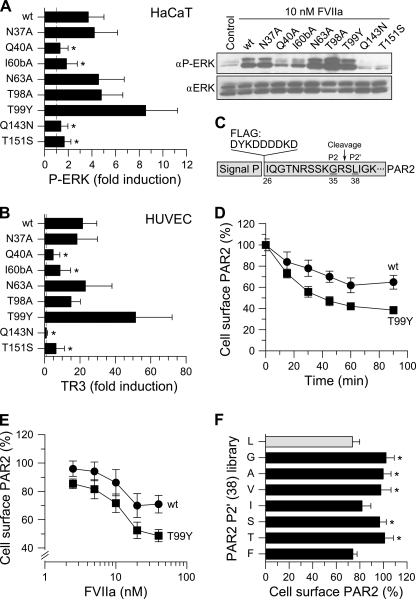
FIGURE 2.
Effect of FVIIa substitutions on PAR2 signaling and receptor cleavage. A, HaCaT cells were stimulated with 10 nm FVIIa variant for 10 min followed by Western blot quantification of phosphor-ERK1/2 (αP-ERK) with total ERK as loading control (αERK, right panel). The -fold induction values (mean ± S.D., n = 5) relative to unstimulated cells (control) are shown (*, p < 0.05 relative to wt FVIIa, unpaired t test). B, TF- and PAR2-transduced HUVECs were stimulated with 10 nm FVIIa variant for 90 min, and PAR2 signaling was evaluated by measuring TR3 mRNA induction (mean ± S.D., n = 5,*, p < 0.01 relative to wt FVIIa, unpaired t test). C, PAR2 cleavage was quantified using FLAG epitope-tagged PAR2. The cleavage site between Arg-36 and Ser-37 and the P2 (Gly) and P2′ (Leu) residues is highlighted. D, CHO-TF cells transiently expressing FLAG-hPAR2 were incubated with 20 nm of wt FVIIa (circles) or T99Y (squares) and cleavage kinetics determined by cell enzyme-linked immunosorbent assay. Untreated cells were set to 100% (mean ± S.D., n = 4). E, PAR2 cleavage by 0–100 nm wt FVIIa (circles) or T99Y FVIIa (squares) measured after 1-h incubation (mean ± S.D., n = 10). F, FLAG-PAR2 variants with the indicated substitutions of P2′ Leu-38 were transiently expressed in CHO-TF cells, and PAR2 cleavage was determined after 1-h incubation with 20 nm wt FVIIa (mean ± S.D., n = 6, *, p < 0.05 relative to P2′ Leu-38, unpaired t test).
FIXa and FXa Generation
FIXa and FXa generation on MDA-MB-231 cells was measured in 10 mm HEPES, 150 mm NaCl, 4 mm KCl, 11 mm glucose, 5 mm CaCl2, 1 mg/ml bovine serum albumin, pH 7.4, with 10 nm FVIIa and 200 nm FIX or 100 nm FX. Following 150 min (FIX) and 15-min incubation (FX) at 37 °C, levels of FIXa and FXa were determined with the chromogenic substrates Spectrozyme FIXa (American Diagnostica) and Chromozyme (Roche Applied Science), respectively. FXa generation on HaCaT cells was measured in serum-free medium with 0.5 nm FVIIa and 100 nm FX after 10-min incubation at 37 °C with the chromogenic substrate Spectrozyme FXa (American Diagnostica).
Signaling Assays
MDA-MB-231 cells were serum-starved in DMEM for 24 h and stimulated with 10 nm FVIIa for 24 h. IL-8 protein levels in the conditioned media were measured using IL-8 Quantikine enzyme-linked immunosorbent assay (R&D Systems, Oxon, UK). For ERK phosphorylation, HaCaT cells or transiently transfected CHO-TF cells were switched for 15 or 120 min, respectively, to serum-free DMEM and stimulated for 10 min. ERK and phospho-ERK were detected by Western blotting and quantified by densitometry. For gene induction, TF- and PAR2-transduced HUVEC cells or transiently transfected M6-11 cells were serum-starved for 5 h in Medium 199 (Lonza) or low glucose DMEM, respectively, and stimulated with agonist for 90 min in the presence of the thrombin inhibitor hirudin (200 nm). mRNA levels of TR3 or the mouse homologue NUR77 were quantified by real-time PCR using TaqMan and normalized to glyceraldehyde-3-phosphate dehydrogenase.
Detection of FLAG-PAR2 Cleavage
CHO-TF cells transiently expressing FLAG-PAR2 were serum-deprived for 2–4 h in DMEM and stimulated with agonist, and surface-exposed FLAG-PAR2 on intact cells was quantified according to Ishii et al. (28). Briefly, after washes with ice-cold PBS, 1 mm CaCl2 and 0.5 mm MgCl2 (PBS-Ca-Mg) cells were incubated with horseradish peroxidase-conjugated anti-FLAG M2 antibody (Sigma-Aldrich) in DMEM with 1 mg/ml bovine serum albumin for 1 h. Plates were then washed four times with PBS-Ca-Mg and developed by incubation with 250 μl of TMB (3,3′,5,5′-tetramethylbenzidine) substrate (Sigma-Aldrich) for 3–10 min at room temperature followed by quenching of the reaction with an equal volume of 1 m sulfuric acid. Reduction in FLAG-PAR2 expression was calculated after background subtraction of untransfected cells.
Profiling the P2 Preference of FXa
A 7-amino-4-carbamoylmethylcoumarin tetrapeptide library was used to determine the P2 amino acid preference of FXa. The library consisted of 18 sub-libraries each containing an Arg at P1, one of the naturally occurring amino acids at P2 (except Met and Cys), and the 18 × 18 possible combinations of amino acids at P3 and P4. The assay was performed as described previously (29) with 50 nm FXa and 3, 6, 9, and 12 μm sub-library in 50 mm HEPES, 100 mm NaCl, 5 mm CaCl2, 0.01% Tween 80, pH 7.4.
Model of PAR2 Bound in the Active Site of FVIIa
Docking of the P5–P5′ (SSKGR↓SLIGK) fragment of PAR2 into the catalytic cleft of FVIIa (based on PDB 1DAN (30)) was guided by the extended β-strand-like backbone interactions characterizing the interaction of proteases with substrates and inhibitors (31). These interactions include the following hydrogen bonds between main-chain atoms (PAR2:FVIIa): R36-N:S214-CO, R36-CO:G193-N, K34-CO:G216N, K34-N:G216-CO, and L38-N:L41-CO, where N and CO refer to nitrogen and carbonyl oxygen atoms, respectively. Side-chain orientations were finally adjusted by energy minimization using the CHARMM force field (32).
RESULTS
Screening for FVIIa Variants with Altered Signaling Properties
Proteases of the trypsin family generally bind substrates and inhibitors in an extended conformation directed by main-chain contacts on both sides of the scissile bond reminiscent of anti-parallel β-strand interactions (31, 33). Guided by these canonical interactions and the crystal structure of FVIIa complexed with a mutant of bovine pancreatic trypsin inhibitor (34), we constructed a FVIIa model in which a 10-amino acid fragment of PAR2, centered around the cleavage site between Arg-36 and Ser-37, was docked into the protease substrate binding cleft (Fig. 1A). FVIIa residues in direct contact with the peptide and/or in the perimeter that could serve as potential exosite contacts were selected for mutagenesis. In a panel of 21 variants, alanine or other amino acid substitutions were introduced based on sequence comparison to related coagulation factors (Fig. 1B). The substitutions D72N, T99Y, and Q217E were chosen by comparison to FIXa and FXa, whereas Q143N and T151S substitutions were based on comparison to thrombin and FIXa, respectively, that both do not cleave PAR2.
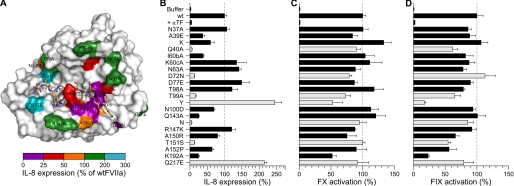
FIGURE 1.
Pro-coagulant and PAR2 signaling activities of FVIIa variants on human MDA-MB-231 breast cancer cells. A, structural model of the protease domain of FVIIa (surface representation) with the P5–P5′ fragment of PAR2 (sticks) bound in the active site. Positions in FVIIa evaluated by mutagenesis are color-coded according to the effect on PAR2-dependent IL-8 expression. B, IL-8 secretion was used as a measure of the PAR2-signaling response after stimulation of MDA-MB-231 cells with 10 nm FVIIa for 24 h. Variants with <25% or >200% of wt FVIIa-signaling activity are shown with light-colored bars. FVIIa Q40A and Q143N were indistinguishable from background levels (p > 0.05, unpaired t test). C and D, the pro-coagulant activity of each variant was measured on MDA-MB-231 cells with 10 nm FVIIa and 100 nm FX (C) or 200 nm FIX (D). Data are shown as mean ± S.D. (n = 3) normalized to wt FVIIa.
Pro-coagulant and signaling properties of FVIIa mutants were first examined using the human breast carcinoma cell line MDA-MB-231, which constitutively expresses TF and PAR2 (6). TF·FVIIa signaling through PAR2 was monitored by cytokine induction after simulation with FVIIa, which induces IL8 expression with an EC50 of ∼10 nm FVIIa (35). Preincubation with an inhibitory anti-TF antibody showed that all activities were strictly TF-dependent. Fig. 1, B–D, shows FVIIa-catalyzed activation of the three substrates, PAR2, FX, and FIX, determined for all 21 variants. PAR2 activation was remarkably sensitive to mutations in the protease domain as 11 of the 21 substitutions reduced PAR2 activation >2-fold (Fig. 1B).
The most significant impact on signaling was observed for the Q40A and Q143N substitutions, which reduced IL-8 expression to background levels, whereas these variants maintained essentially normal pro-coagulant activities (see Fig. 1, C and D). Significantly impaired PAR2 activation was also observed for FVIIa D72N, T99A, and T151A, which were reduced to 13, 17, and 13% of wt FVIIa signaling activity, respectively. Similar results were obtained when Q40 was substituted with Ser or Asn, or Thr-151 with Ala (results not shown), whereas Q143A substitution was less detrimental than Q143D (Fig. 1B). The impact of position 99 replacements was markedly residue-dependent. PAR2 activation was reduced almost to background level by T99A substitution, whereas it was enhanced 2- to 3-fold above the wt FVIIa activity when T99 was replaced by Tyr. Another mutant with enhanced PAR2 activation was Q217E FVIIa, suggesting that interactions in the vicinity of the S3 subsite of FVIIa also contribute to PAR2 recognition (36).
Activation of the macromolecular coagulation substrate FX, in contrast to PAR2, was much less affected by the mutations, and most variants retained ≥80% of wild-type activity (Fig. 1C). Exceptions were T99A and K192A variants with 30–50% reduced activity as previously reported (16, 29, 37). Similarly, FVIIa-catalyzed FIX activation was markedly unaffected by modifications in the FVIIa protease domain (Fig. 1D) as might be expected from the similar binding modes of FIX and FX to the TF·FVIIa complex (38). Taken together, these data support the concept that extended exosite interactions primarily determine macromolecular substrate recognition and indicate that cleavage of PAR2 is governed by recognition at the active site.
PAR2-dependent Signaling in HaCaTs and HUVECs
To confirm signaling defects, eight variants were selected for further characterization of TF·FVIIa signaling with HaCaTs or transduced HUVECs. HaCaTs constitutively express TF and PAR2, and TF signaling in these cells is PAR2-dependent (3). Receptor activation was assessed by measuring phosphorylation of extracellularly regulated kinases (ERK) 1 and 2, which was increased 4-fold upon stimulation with 10 nm wt FVIIa (Fig. 2A). Stimulation with FVIIa variants corroborated the results obtained in MDA-MB-231 cells. Thus, ERK phosphorylation remained at baseline levels (p < 0.05) in the presence of Q40A, I60bA, Q143N, or T151S variants, whereas signaling by FVIIa T99Y was increased 2.3-fold compared with wt FVIIa.
A similar pattern of binary complex signaling was observed with HUVECs transduced with TF and PAR2 (Fig. 2B) as measured by TR3 mRNA induction (10). Again, the response to Q143N FVIIa was not significantly different from the baseline levels, whereas signaling activities of Q40A, I60bA, and T151S FVIIa variants were severely impaired, and T99Y FVIIa was 2.5-fold more active than wt FVIIa. Because PAR1 is abundantly expressed on HUVECs (2) and triggers a transcriptional program partially overlapping with that of PAR2 (6, 27), we verified that the variants maintained PAR2 selectively. To test this each FVIIa variant was evaluated for TR3 gene induction in cells transduced solely with TF and treated with an antibody to block residual endogenous PAR2. PAR1 signaling capability was not acquired by these mutations as indicated by results showing that neither variant induced TR3 signaling, whereas in contrast 50 nm FXa triggered a 147-fold TR3 gene induction that could be 94% blocked by a combination of 50 μg/ml WEDE15 and 25 μg/ml ATAP2 anti-PAR1 antibodies.
Contributions of S2 and S2′ Pockets of FVIIa to PAR2 Cleavage
To directly measure cleavage of PAR2, a full-length PAR2 construct fused to an N-terminal FLAG epitope tag was transiently expressed in CHO cells stably expressing human TF (CHO-TF). Receptor cleavage was determined by measurement of residual surface-exposed PAR2 after defined times of incubation with FVIIa. As shown in Fig. 2D cleavage was not running to completion, but reached a final steady-state level. This may indicate that a fraction of PAR2 was inaccessible to the relatively slow cleavage by TF·FVIIa, or alternatively that cleavage was counteracted by mobilization of intact receptor from intracellular Golgi pools (39). We consider it unlikely that PAR2 mobilization had a major impact on the cell surface cleavage assay under our experimental conditions, because addition of brefeldin A (a known inhibitor of vesicular trafficking to the membrane) during the cleavage assay was without an appreciable effect on the relative exposure of residual FLAG-PAR2 epitope remaining after FVIIa stimulation (results not shown).
We first addressed the improved signaling of T99Y FVIIa. Dose titrations showed similar EC50 values of ∼10 nm for wt FVIIa and T99Y FVIIa (Fig. 2E), excluding that the mutant bound TF with higher affinity. However, compared with wt FVIIa, the T99Y variant mediated more efficient cleavage indicated by a lower final level of surface-exposed PAR2 (Fig. 2F).
According to the model presented in Fig. 1A, FVIIa residues Gln-143, Gln-40, and Thr-151 are part of the S2′ pocket accommodating the P2′ L38 side chain of PAR2. To confirm the crucial role of P2′-S2′ interactions suggested by FVIIa mutagenesis, we constructed a limited P2′ library of PAR2 and examined receptor cleavage in the CHO-TF cell model at 20 nm FVIIa ensuring complete TF saturation (see Fig. 2E). Consistent with a requirement for extended interactions at the S2′ subsite, wt FVIIa was highly discriminatory and cleaved only variants with large P2′ side-chains (Leu, Ile, and Phe), whereas variants with smaller P2′ residues (Gly, Ala, Val, Ser, and Thr) were not appreciably cleaved during the observation period (Fig. 2F). Collectively the data confirmed that interactions at both the PAR2 P2 and P2′ positions were required for efficient cleavage by TF·FVIIa.
Binary Complex Signaling-defective FVIIa Variants Are Normal in Ternary Complex Signaling
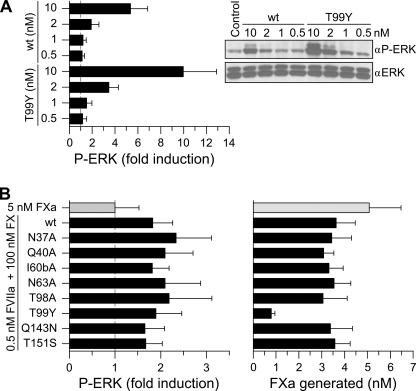
In the ternary TF·FVIIa·FXa initiation complex, FXa efficiently cleaves PAR2 (4). This is illustrated in Fig. 3 where a concentration of 0.5 nm FVIIa was too low to stimulate binary complex signaling in HaCaT cells (Fig. 3A), whereas stimulation with 0.5 nm FVIIa in the presence of 100 nm FX produced a ∼2-fold increase in ERK phosphorylation (Fig. 3B). Under these conditions, the eight variants with impaired TF·FVIIa signaling activated FX efficiently and were also indistinguishable in ternary TF·FVIIa·FXa-mediated ERK1/2 phosphorylation (Fig. 3, A and B). Thus, the binary complex signaling-deficient mutants are not only essentially normal in the coagulant response, but also in coagulation initiation phase signaling of the ternary TF·FVIIa·FXa complex.
FIGURE 3.
Comparison of binary TF·FVIIa and ternary TF·FVIIa·FXa PAR2 signaling in HaCaT keratinocytes. A, TF·FVIIa-mediated ERK1/2 phosphorylation was determined after stimulation of HaCaT cells for 10 min with the indicated concentrations of wt FVIIa or T99Y (mean ± S.D., n = 4). B, ternary complex signaling was evaluated by measuring phospho-ERK1/2 levels (left panel) after 10 min of stimulation with 100 nm FX and 0.5 nm FVIIa. At this concentration FVIIa-TF binary signaling did not contribute to PAR2 activation. The amount of FXa generated at this time (right panel) was measured by chromogenic assay (mean ± S.D.; n = 5). The signaling induced by 5 nm FXa is included for comparison (light-colored bar).
Two points were noteworthy when comparing HaCaT ternary complex signaling (Fig. 3B) with FX activation (Fig. 3C). Firstly, signaling from accumulating free FXa could not account for the response observed as a result of ternary complex formation, because no signaling was observed by 5 nm FXa, corresponding to the highest final concentration achieved. Secondly, despite very low pro-coagulant activity of the T99Y variant, signaling of the ternary complex was very similar to wt FVIIa. Although FVIIa T99Y showed improved binary TF·FVIIa signaling, this required concentrations >2 nm FVIIa T99Y and therefore unlikely caused the signaling observed in the presence of FX. A slow release of FXa from the mutant ternary complex could explain the high ternary complex signaling activity of this mutant. Consistent with this conclusion, kinetic measurement of FX activation with relipidated TF showed a 2-fold lower Km and a 13-fold slower kcat for the mutant relative to wt FVIIa.
Engineering Protease-selective PAR2 Mutants
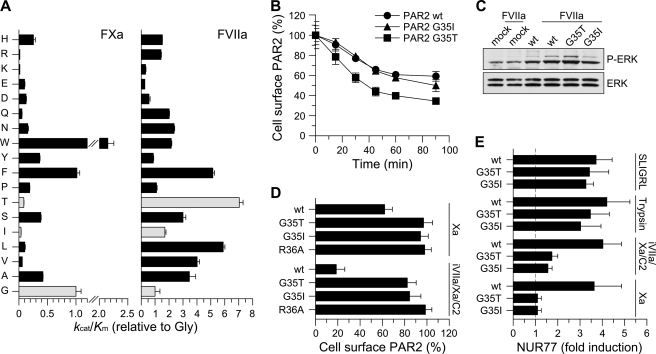
Although previous antibody inhibition experiments showed that signaling of the binary TF·FVIIa complex can be selectively inhibited to reduce pathological TF signaling (9, 11), the biological roles of ternary complex signaling through PAR2 are incompletely understood. To this end, we asked if the delineated molecular recognition of PAR2 by FVIIa could be exploited to generate PAR2 variants capable of discriminating between FVIIa and FXa as PAR2 activating coagulation proteases. We focused on the P2-S2 interaction known from FVIIa T99A and T99Y variants to modulate the efficacy of PAR2 cleavage while still preserving PAR2-tethered ligand interactions following activation. The P2 preference of TF·FVIIa has previously been determined using combinatorial fluorogenic tetrapeptide substrate libraries (29). To allow for direct comparison we determined the P2 preference of FXa under identical conditions (Fig. 4 A). Inspection of the two profiles revealed notable differences. Although FVIIa had a preference for aliphatic and β-branched side chains at P2, these were among the least preferred by FXa.
FIGURE 4.
Substitution of the P2 position in PAR2 generates TF·FVIIa signaling selectivity. A, the P2 preference of FXa (left panel) was determined as described under “Experimental Procedures.” The apparent kcat/Km value for hydrolysis of each sub-library by FXa is shown relative to that for the P2-Gly sub-library (error bars represent ±S.D.). The P2 preference of sTF·FVIIa described in Larsen et al. (29) and determined under identical conditions is included for comparison (right panel). Light-colored bars highlight the preference of sTF·FVIIa for P2-Gly, Thr, and Ile. B, cleavage of wt PAR2 (circles) or P2 variants G35I (triangles) and G35T (squares) was determined by measuring residual receptor exposure in CHO-TF cells transiently transfected with FLAG-tagged receptors following incubation with 20 nm wt FVIIa for the indicated times (mean ± S.D., n = 4). No difference in PAR2 surface exposure was observed between wt and mutant receptors (surface expression of unstimulated cells 75 ± 6 (wt), 75 ± 4 (G35I), and 81 ± 10 (G35T) arbitrary absorbance units). C, FVIIa-induced signaling by wtFLAG-PAR2, G35I, and G35T was determined by stimulating transiently transfected CHO-TF cells with 10 nm wt FVIIa for 10 min followed by Western blot quantification of phosphor-ERK1/2 (αP-ERK), (mean -fold increases, n = 2). D, the effect of G35I and G35T substitutions on the cleavage of FLAG-PAR2 by free FXa (100 nm) or the NAPc2-stabilized ternary complex formed by 5 nm FVIIa S195A FVIIa (iFVIIa), 50 nm FXa, and 150 nm NAPc2. The cleavage-resistant PAR2 variant R36A was included for comparison (light-colored bar). Transiently transfected CHO-TF cells were incubated with FXa (1 h) or NAPc2-stabilized ternary complex (30 min) followed by measurement of residual surface exposed FLAG-PAR2 relative to unstimulated cells (mean ± S.D., n = 9). E, FXa- and ternary complex-induced signaling by FLAG-PAR2 G35I and G35T was determined in serum-starved, transiently transfected mouse PAR1-deficient M6-11 fibroblasts. Cells were stimulated for 90 min with FXa (100 nm), the ternary complex (iFVIIa, FXa, and NAPc2 at 5, 50, and 150 nm, respectively), trypsin (10 nm), or the PAR2 agonist peptide (SLIGRL, 100 μm). Induction of NUR77 mRNA was quantified by real-time PCR (mean ± S.D., n = 11).
Taking advantage of the differences in P2 preferences, we constructed two PAR2 variants carrying G35I and G35T substitutions and tested them in the CHO-TF cell model with the PAR2 P1 R36A mutant as an uncleavable control (Fig. 4B). Consistent with the established P2 preference, 20 nm wt FVIIa cleaved the G35T variant more rapidly than wt PAR2, whereas cleavage of G35I PAR2 was very similar to cleavage of wt PAR2 (Fig. 4B). Measurements of ERK phosphorylation showed that the PAR2 mutants also signaled normally and with an intensity expected from the cleavage measurements. In contrast, incubation of the cells with 100 nm FXa for 1 h failed to show any decrease in cell surface-expressed FLAG-PAR2, although 100 nm FXa was as least as efficient as 20 nm wt FVIIa in cleaving the wild-type receptor (Fig. 4, B and D). Because FXa cleaves PAR1, PAR1-deficient mouse fibroblasts were utilized to verify that these mutants did not mediate FXa signaling. PAR1-deficient fibroblasts transiently transfected with wt PAR2 or the two PAR2 variants showed similar mouse TR3 (NUR77) mRNA induction when stimulated with either the PAR2 agonist peptide (SLIGRL) or trypsin (Fig. 4E). In contrast, only transfection with wt PAR2, but not with either of the mutants induced maximal gene induction when FXa was the activating protease.
We further tested whether FXa-mediated PAR2 cleavage and signaling was prevented when FXa was localized in the ternary TF·FVIIa·FXa complex. The ternary complex is transient in nature under physiological conditions but can be stabilized by the nematode anticoagulant protein C2 (NAPc2). Binding of NAPc2 to the ternary complex generates a highly efficient PAR2 activator by retaining its FXa activity while inhibiting FVIIa (4). Stimulation of CHO-TF cells with the NAPc2-stabilized ternary complex led to an 80% reduction of cell surface wt PAR2. Under these conditions the two variants receptors (PAR2 G35I and PAR2 G35T) were highly resistant to activation, and <20% of the membrane-exposed receptors disappeared after prolonged incubation (Fig. 4D). Inefficient cleavage of the PAR2 variants by the NAPc2-stabilized complex was also reflected in the signaling response in M6-11 cells, which showed a 4-fold up-regulation of NUR77 mRNA in cells expressing the wild-type receptor as compared with a ∼1.5-fold induction with the two mutant receptors (Fig. 4E). These data confirm the crucial role of primary specificity site recognition of PAR2 by coagulation proteases and demonstrate that single point mutations generate PAR2 receptors that are essentially resistant to activation of FXa.
DISCUSSION
Here, we provide a mutational mapping of the protease domain of FVIIa to identify structural determinants for recognition and cleavage of its cognate substrates, FIX, FX, and PAR2. TF·FVIIa-catalyzed FIX and FX activation was largely unaffected by mutations in the catalytic cleft of FVIIa, consistent with the previously established concept that binding of the macromolecular substrate is highly dependent on exosite interactions that facilitate primary substrate recognition at the active site (15, 17). In contrast, cleavage of PAR2 was highly sensitive to substitution of FVIIa residues in the active site cleft and its perimeter. Results with three independent signaling readouts for TF·FVIIa concordantly identified residues Gln-40, Asp-72, Gln-143, and Thr-151 as key determinants for PAR2 recognition, whereas Ala-39 and Ile-60b were found to play minor but significant roles. An unaffected FIX or FX activation is indicative of an intact structural arrangement of the active site, and the mutational effect on PAR2 activation can be interpreted as a specific effect on substrate recognition. Remarkably, the Q143N FVIIa variant did not activate PAR2, but had essentially normal pro-coagulant activity, demonstrating that these two activities can be completely dissected by protein engineering.
PAR2 activation with FVIIa and PAR2 variants agrees well with the predictions made by the model in Fig. 1A, which describes the docking of the P5-P5′ fragment of PAR2 into the active cleft of FVIIa. The model identifies FVIIa residues Gln-40, Gln-143, and Thr-151 as essential elements of the S2′ pocket, which accommodates the side chain of PAR2 Leu-38 (P2′). Consistent with the model, recognition of PAR2 was shown to be sensitive both to modifications of FVIIa at the S2′ subsite and to complementary mutations of PAR2 at the P2′ subsite. PAR2 signaling was diminished by FVIIa mutations that eliminated direct hydrophobic contacts and even by subtle changes, such as the T151S mutation. We also showed a strict requirement for large bulky side chains Leu, Ile, or Phe in P2′ of PAR2 and that cleavage-resistant receptors were produced by mutations of P2′ Leu-38 to Ser, Thr, or Val. In addition, PAR2 activation was abolished by the D72N substitution in the 70–80 Ca2+-binding loop. Recent characterization of this variant and its inhibition by AT showed that it exhibited normal catalytic activity and inhibitor interaction. Furthermore, the crystal structure of the D72N FVIIa-AT complex showed that Asn-72 adopted a conformation very similar to that of Asp-72 in wt FVIIa (40). No significant perturbation of the Ca2+-binding loop or the catalytic cleft region was observed, which could suggest that Asp-72 interacts with P5′ Lys-41 in PAR2 as predicted by the model.
Additional evidence for critical contributions to PAR2 recognition comes from the Q217E and T99Y FVIIa variants that exhibited >2-fold enhanced PAR2 binary complex signaling. Gln-217 is located adjacent to the S3/4 binding pocket, and Q217E replacement may stabilize the interaction with PAR2 P3 Lys-34. Thr-99 is located at the entrance to the S2 pocket, and substrate profiling (29, 41, 42) indicated that the small Thr-99 residue in wt FVIIa allows access of large side chains to the open S2 pocket and thereby creates a preference for β-branched and hydrophobic amino acids (Thr, Leu, and Phe). The bulkier Tyr-99 in FVIIa T99Y likely restricts such access and shifts the P2 specificity toward Gly (29) explaining the increased rate of PAR2 cleavage and enhanced wt PAR2 signaling.
The delineated critical complementary contacts of FVIIa and PAR2 allowed us to engineer the PAR2 cleavage by FXa and still maintain efficient activation by TF·FVIIa. Two such PAR2 mutants (P2 G35T and G35I) were remarkably resistant to activation by free and ternary-complexed FXa. Contrary to the exosite interaction-driven FIX and FX activation, TF·FVIIa binary complex signaling is highly dependent on direct interactions on the cell surface between the extracellular domain of PAR2 and the catalytic cleft of FVIIa. This also contrasts with the activation of PAR1 by thrombin, which is greatly facilitated by thrombin's exosite 1 interaction with a hirudin-like C-terminal element of PAR1 involving charge complementarities (21, 22, 43). Disruption of this exosite interaction by mutagenesis in contrast to mutations in the primed subsite region of the active site (e.g. Arg-35 and Glu-39) severely reduced the rate of PAR1 activation (21, 22, 43). PAR1 or PAR2 recognition by thrombin or TF·FVIIa thus depends on different mechanisms and structural regions of the respective proteases.
In conclusion, the results presented herein provide new insight into the structural requirements for PAR2 activation by TF·FVIIa and offer clues to how specific protein-protein interactions contribute to TF·FVIIa-mediated PAR2 signaling that triggers a diversity of cellular responses in cancer, inflammation, and angiogenesis. By exploiting the predominantly active-site-driven interaction between PAR2 and its activating proteases, we demonstrate the feasibility of rationally optimizing the receptor recognition sequence to selectively reduce PAR2 activation by FXa to a minimum. The physiological relevance of TF·FVIIa·FXa or other FXa-mediated PAR2 signaling is currently incompletely understood. The FVIIa and PAR2 variants generated in this study provide important new tools to dissect the role of FVIIa-mediated PAR2 activation and to further characterize the relative importance of TF coagulation complex signaling pathways in vivo.
Acknowledgments
We thank Anette W. Bruun, Elke Gottfriedsen, Jennifer Royce, and Pablito Tejada for technical support.
This work was supported, in whole or in part, by National Institutes of Health Grant P01-HL31950 (to W. R.).
- FVIIa
- factor VIIa
- FIX
- factor IX
- FX
- factor X
- TF
- tissue factor
- PAR
- protease-activated receptor
- ERK
- extracellular-regulated kinase
- TR3
- TR3 orphan receptor
- NAPc2
- nematode anticoagulant protein C2
- wt
- wild type
- HaCaT
- human immortalized keratinocyte
- HUVEC
- umbilical vein endothelial cell
- DMEM
- Dulbecco's modified Eagle's medium
- PBS
- phosphate-buffered saline
- CHO
- Chinese hamster ovary
- IL-8
- interlurkin-8.
REFERENCES
- 1.Camerer E., Huang W., Coughlin S. R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5255–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coughlin S. R. (2000) Nature 407, 258–264 [DOI] [PubMed] [Google Scholar]
- 3.Ahamed J., Versteeg H. H., Kerver M., Chen V. M., Mueller B. M., Hogg P. J., Ruf W. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13932–13937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riewald M., Ruf W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7742–7747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaffner F., Ruf W. (2008) Semin. Thromb. Hemost. 34, 147–153 [DOI] [PubMed] [Google Scholar]
- 6.Albrektsen T., Sørensen B. B., Hjortø G. M., Fleckner J., Rao L. V. M., Petersen L. C. (2007) J. Thromb. Haemost. 5, 1588–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belting M., Dorrell M. I., Sandgren S., Aguilar E., Ahamed J., Dorfleutner A., Carmeliet P., Mueller B. M., Friedlander M., Ruf W. (2004) Nat. Med. 10, 502–509 [DOI] [PubMed] [Google Scholar]
- 8.Uusitalo-Jarvinen H., Kurokawa T., Mueller B. M., Andrade-Gordon P., Friedlander M., Ruf W. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 1456–1462 [DOI] [PubMed] [Google Scholar]
- 9.Versteeg H. H., Schaffner F., Kerver M., Ellies L. G., Andrade-Gordon P., Mueller B. M., Ruf W. (2008) Cancer Res. 68, 7219–7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Versteeg H. H., Schaffner F., Kerver M., Petersen H. H., Ahamed J., Felding-Habermann B., Takada Y., Mueller B. M., Ruf W. (2008) Blood 111, 190–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Redecha P., Franzke C. W., Ruf W., Mackman N., Girardi G. (2008) J. Clin. Invest. 118, 3453–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ossovskaya V. S., Bunnett N. W. (2004) Physiol. Rev. 84, 579–621 [DOI] [PubMed] [Google Scholar]
- 13.Ruf W., Mueller B. M. (2006) Semin. Thromb. Hemost. 32, Suppl. 1, 61–68 [DOI] [PubMed] [Google Scholar]
- 14.Krishnaswamy S. (2005) J. Thromb. Haemost. 3, 54–67 [DOI] [PubMed] [Google Scholar]
- 15.Baugh R. J., Dickinson C. D., Ruf W., Krishnaswamy S. (2000) J. Biol. Chem. 275, 28826–28833 [DOI] [PubMed] [Google Scholar]
- 16.Dickinson C. D., Kelly C. R., Ruf W. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14379–14384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shobe J., Dickinson C. D., Edgington T. S., Ruf W. (1999) J. Biol. Chem. 274, 24171–24175 [DOI] [PubMed] [Google Scholar]
- 18.Dickinson C. D., Kelly C. R., Ruf W. (1996) Circulation (8 suppl. S) 94, 1272 [Google Scholar]
- 19.Norledge B. V., Petrovan R. J., Ruf W., Olson A. J. (2003) Proteins 53, 640–648 [DOI] [PubMed] [Google Scholar]
- 20.Liu L. W., Vu T. K., Esmon C. T., Coughlin S. R. (1991) J. Biol. Chem. 266, 16977–16980 [PubMed] [Google Scholar]
- 21.Ayala Y. M., Cantwell A. M., Rose T., Bush L. A., Arosio D., Di Cera E. (2001) Proteins 45, 107–116 [DOI] [PubMed] [Google Scholar]
- 22.Vu T. K., Hung D. T., Wheaton V. I., Coughlin S. R. (1991) Cell 64, 1057–1068 [DOI] [PubMed] [Google Scholar]
- 23.Bah A., Chen Z., Bush-Pec L. A., Mathews F. S., Di Cera E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11603–11608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss E. J., Hamilton J. R., Lease K. E., Coughlin S. R. (2002) Blood 100, 3240–3244 [DOI] [PubMed] [Google Scholar]
- 25.O'Brien P. J., Prevost N., Molino M., Hollinger M. K., Woolkalis M. J., Woulfe D. S., Brass L. F. (2000) J. Biol. Chem. 275, 13502–13509 [DOI] [PubMed] [Google Scholar]
- 26.Ahamed J., Ruf W. (2004) J. Biol. Chem. 279, 23038–23044 [DOI] [PubMed] [Google Scholar]
- 27.Riewald M., Petrovan R. J., Donner A., Mueller B. M., Ruf W. (2002) Science 296, 1880–1882 [DOI] [PubMed] [Google Scholar]
- 28.Ishii K., Hein L., Kobilka B., Coughlin S. R. (1993) J. Biol. Chem. 268, 9780–9786 [PubMed] [Google Scholar]
- 29.Larsen K. S., Østergaard H., Bjelke J. R., Olsen O. H., Rasmussen H. B., Christensen L., Kragelund B. B., Stennicke H. R. (2007) Biochem. J. 405, 429–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banner D. W., D'Arcy A., Chène C., Winkler F. K., Guha A., Konigsberg W. H., Nemerson Y., Kirchhofer D. (1996) Nature 380, 41–46 [DOI] [PubMed] [Google Scholar]
- 31.Bode W., Huber R. (1992) Eur. J. Biochem. 204, 433–451 [DOI] [PubMed] [Google Scholar]
- 32.Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) J. Comput. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyndall J. D., Nall T., Fairlie D. P. (2005) Chem. Rev. 105, 973–999 [DOI] [PubMed] [Google Scholar]
- 34.Zhang E., St Charles R., Tulinsky A. (1999) J. Mol. Biol. 285, 2089–2104 [DOI] [PubMed] [Google Scholar]
- 35.Hjortoe G. M., Petersen L. C., Albrektsen T., Sorensen B. B., Norby P. L., Mandal S. K., Pendurthi U. R., Rao L. V. M. (2004) Blood 103, 3029–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang Y. J., Hamaguchi N., Chang S. C., Ruf W., Shen M. C., Lin S. W. (1999) Biochemistry 38, 10940–10948 [DOI] [PubMed] [Google Scholar]
- 37.Neuenschwander P. F., Morrissey J. H. (1995) Biochemistry 34, 8701–8707 [DOI] [PubMed] [Google Scholar]
- 38.Zhong D., Bajaj M. S., Schmidt A. E., Bajaj S. P. (2002) J. Biol. Chem. 277, 3622–3631 [DOI] [PubMed] [Google Scholar]
- 39.Roosterman D., Schmidlin F., Bunnett N. W. (2003) Am. J. Physiol. Cell Physiol 284, C1319–C1329 [DOI] [PubMed] [Google Scholar]
- 40.Bjelke J. R., Olsen O. H., Fodje M., Svensson L. A., Bang S., Bolt G., Kragelund B. B., Persson E. (2008) J. Biol. Chem. 283, 25863–25870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Backes B. J., Harris J. L., Leonetti F., Craik C. S., Ellman J. A. (2000) Nat. Biotechnol. 18, 187–193 [DOI] [PubMed] [Google Scholar]
- 42.Harris J. L., Backes B. J., Leonetti F., Mahrus S., Ellman J. A., Craik C. S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7754–7759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myles T., Le Bonniec B. F., Stone S. R. (2001) Eur. J. Biochem. 268, 70–77 [DOI] [PubMed] [Google Scholar]