Abstract
Galactitol 2-dehydrogenase (GatDH) belongs to the protein superfamily of short-chain dehydrogenases. As an enzyme capable of the stereo- and regioselective modification of carbohydrates, it exhibits a high potential for application in biotechnology as a biocatalyst. We have determined the crystal structure of the binary form of GatDH in complex with its cofactor NAD(H) and of the ternary form in complex with NAD(H) and three different substrates. The active form of GatDH constitutes a homo-tetramer with two magnesium-ion binding sites each formed by two opposing C termini. The catalytic tetrad is formed by Asn116, Ser144, Tyr159, and Lys163. GatDH structurally aligns well with related members of the short-chain dehydrogenase family. The substrate binding pocket can be divided into two parts of different size and polarity. In the smaller part, the side chains of amino acids Ser144, Ser146, and Asn151 are important determinants for the binding specificity and the orientation of (pro-) chiral compounds. The larger part of the pocket is elongated and flanked by polar and non-polar residues, enabling a rather broad substrate spectrum. The presented structures provide valuable information for a rational design of this enzyme to improve its stability against pH, temperature, or solvent concentration and to optimize product yield in bioreactors.
Keywords: Carbohydrate Processing, Crystal Structure, Dehydrogenase, Ligand-binding Protein, X-ray Crystallography, Biotechnology, SDR Enzyme
Introduction
Dehydrogenases represent an important class of enzymes in biotechnological processes increasingly used in the chemical or pharmaceutical industry due to their enantio- and stereoselective oxidative and reductive catalytic properties (1–4). A dehydrogenase with a promising catalytic potential is galactitol dehydrogenase (galactitol:NAD+ 5-oxidoreductase (GatDH)3), an enzyme originally isolated from a galactitol-utilizing mutant of the bacterium Rhodobacter sphaeroides (5). GatDH is a homotetrameric protein that requires Mg2+ for maintenance of its quaternary structure and enzymatic activity. It catalyzes the dehydrogenation of a variety of polyvalent aliphatic alcohols and polyols to the corresponding ketones and ketoses, respectively, and in the reverse reaction it reduces prochiral ketones with high stereoselectivity yielding the corresponding S-configured secondary alcohols (5–7). Based on these catalytic capabilities, GatDH was used with cofactor recycling (8, 9) for several preparative conversions. Oxidation at C5 of galactitol gave the rare sugar l-tagatose in almost quantitative yields (6). l-Tagatose is a precursor for the synthesis of the glycosidase inhibitor 1-deoxygalactonojirimycin (10) and is a substrate of pyranose 2-oxidase yielding the interesting synthon 5-keto-psicose (11). Similarly, xylitol is oxidized at C4 to give l-xylulose (5). We also demonstrated the preparation of several (R)-1,2-diols by racemic resolution with GatDH as well as the synthesis of several S-configured aliphatic alcohols by reducing corresponding prochiral ketones (7). Furthermore, the suitability of GatDH for conversions with electrochemical cofactor regeneration was demonstrated with GatDH-bound covalently to the surface of a gold electrode (12). These examples indicate that GatDH has a considerable application potential, which is only limited because of intrinsic deficiencies of the enzyme such as low thermal and operational stability and restricted pH tolerance (5). A question that has not been addressed so far is its tolerance toward organic solvents in the case of alcohol conversions. There are also open questions concerning the region- and stereoselectivity of GatDH which, apparently, are influenced by the chain length and position of the carbonyl group of the substrate. With regard to polyol conversions, the starting materials for rare sugar syntheses, it seems that GatDH preferentially accepts polyvalent alcohols with d-threo-configuration adjacent to the primary alcohol group (Fig. 1). Thus, GatDH is not only an attractive biocatalyst for the synthesis of certain rare ketoses but also for asymmetric synthesis of enantiomerically pure diols (7). To provide a basis for a thorough understanding of the enzymatic properties of GatDH, structure determination of the substrate-free and substrate-bound enzyme in complex with its cofactor was performed. The presented structures provide insight into the properties of the active site of the enzyme and represent the prerequisite for optimization of this and similar dehydrogenases (13–19) that are available for biocatalytic processes, especially in electrochemical enzyme reactors.
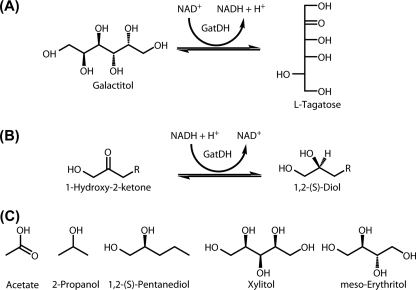
FIGURE 1.
Some substrates and reactions of GatDH. A, GatDH performs the NAD+-dependent regioselective dehydrogenation of the sugar alcohol galactitol at C5 producing l-tagatose and NADH. Similarly, xylitol is oxidized at C4 to l-xylulose (reaction not shown) (5). B, hydrogenation of short, aliphatic 1-hydroxy-ketones preferably yields the S-enantiomer of the 1,2-diol (5, 7). C, structures of substances co-crystallized with GatDH or used for soaking experiments of GatDH crystals.
EXPERIMENTAL PROCEDURES
Expression and Purification of GatDH
For production of recombinant GatDH from R. sphaeorides strain D, the respective gene was cloned into the vector pET24a (Novagen Inc.) with an N-terminal (His)6 affinity tag. For removal of the affinity tag, a cassette coding for the tobacco etch virus-protease cleavage site was ligated into the plasmid. The new plasmid was transformed into Escherichia coli strain BL21(DE3)gold (Novagen). A 6-liter culture was grown in lysogeny broth (LB) medium supplemented with the antibiotic kanamycin (50 μg ml−1) at 310 K until an A600 of 1.5 was reached. The production of (His)6-tagged GatDH was induced by addition of isopropyl-β-d-thiogalactopyranoside (final concentration, 0.5 mm) and expressed for 5 h at 303 K. The cells were harvested by centrifugation (10 min, 6,000 × g, 277 K), washed in buffer (20 mm Bis-Tris, pH 6.5, 1 mm MgCl2, 0.1 mm phenylmethylsulfonyl fluoride), frozen in liquid nitrogen, and stored at 193 K. For purification, cells were thawed, resuspended in 120 ml of lysis buffer (20 mm phosphate buffer, pH 7.4, 500 mm NaCl, 0.1 mm phenylmethylsulfonyl fluoride), and disrupted at 16,000 p.s.i. by a cell homogenizer (Avestin Inc.). To digest the DNA, small amounts of DNase were added. After centrifugation (120 min, 75,000 × g, 277 K) the supernatant was applied onto a nickel-nitrilotriacetic acid-superflow matrix (Qiagen), and the column was washed to baseline with 20 mm phosphate buffer, pH 7.4, 500 mm NaCl, 0.1 mm phenylmethylsulfonyl fluoride and 20 mm imidazole. His-tagged proteins were eluted in the same buffer in multiple gradient steps from 20 mm to 500 mm imidazole. Fractions containing (His)6-GatDH (identified by SDS-PAGE analysis, stained with Coomassie-Blue) were pooled and dialyzed against 50 mm Tris-HCl, pH 8.0, supplemented with 1 mm dithiothreitol and 10 mm EDTA. The affinity tag was removed with tobacco etch virus-protease S219V in a molar ratio of 1:10 (protease to protein). For the removal of uncleaved (His)6-GatDH and (His)6-tagged tobacco etch virus-protease, the cleavage reaction mixture was treated with nickel-nitrilotriacetic acid-matrix (Qiagen Inc.), equilibrated with 20 mm Bis-Tris, pH 6.5, and 1 mm imidazole. The supernatant, containing pure GatDH without the affinity tag (SDS-PAGE analysis, staining with Coomassie Blue) was dialyzed against 20 mm Bis-Tris, pH 6.5, and concentrated to 12 mg ml−1 (0.45 mm). The homogeneity of the purified protein was further verified by matrix-assisted laser desorption ionization time-of-flight. The activity of GatDH toward the used polyols was determined with an assay described by Schneider et al. (5).
Multiple Angle Light-scattering Measurements
For protein separation, the asymmetric flow field-flow fractionation technique (AFFF, Eclipse, Wyatt Technology) with a spacer of 490 nm and a cellulose membrane with 5-kDa cut-off was used. The system was connected to a UV-detector (Agilent), a multi-angle light-scattering detector (miniDAWN, λ = 690 nm, Wyatt Technology) and a refractive index detector (Agilent). The mobile phase was 20 mm Bis-Tris buffer, pH 6.5, with and without 1 mm MgCl2, respectively. The analysis was carried out at a cross flow rate of 1.5–3 ml/min at room temperature (∼298 K). The protein concentration was 1 mg/ml in 20 mm Bis-Tris, pH 6.5, with or without 1 mm MgCl, and with or without 1 mm NAD+ or NADH, respectively. Finally, the molecular weight was calculated using the ASTRA® software (Wyatt Technology).
Crystallization
All crystallization setups were performed at 291 K using the vapor-diffusion method with hanging drops. For crystallization of the holoenzyme, 1 mm NAD+ or NADH was added to the protein solution (0.45 mm) prior to crystallization. 1 μl of protein solution (in 20 mm Bis-Tris, pH 6.5) was mixed with 1 μl of reservoir solution and equilibrated against 1 ml of reservoir solution. Crystal screening was carried out using Crystal ScreenTM, Crystal Screen 2TM, and PEG/Ion ScreenTM (Hampton Research). The best crystallization condition identified was used for further optimization using additive screens 1–3TM (Hampton Research). The first crystallization condition contained 200 mm sodium acetate, 100 mm sodium cacodylate, pH 6.5, and 30% (w/v) PEG 8000. Under this condition crystals in space group P41212 (with unit cell dimensions a = b = 109 Å, c = 125 Å) grew within 1 week. Adding 4% (v/v) 2-propanol to the reservoir solution improved the crystal quality in respect to x-ray diffraction and resulted in crystals grown in the orthorhombic space group P212121 (with unit cell dimensions a = 98 Å, b = 107 Å, and c = 109 Å). These crystals were used for an initial crystal structure determination. For soaking and co-crystallization experiments, a second crystallization condition was optimized containing 100 mm MES, pH 5.5–5.9, 200 mm MgCl2, and 10–20% (w/v) methoxy poly(ethylene glycol) 5000. Crystals produced under these conditions grew in the orthorhombic space group P212121 (with unit cell dimensions a = 65 Å, b = 115 Å, and c = 124 Å). Structures of GatDH with bound substrate were obtained utilizing two different strategies: co-crystallization with excess of substrate as well as soaking of protein crystals. For soaking of crystals, the reservoir solution from the second crystallization condition was supplemented with 1 mm NAD+ as well as with the substrate or putative inhibitor in concentrations in the range of 10–100 mm. Crystals were soaked in increasing concentrations of MPEG 5000 (until 30–35% (w/v)) within ∼60 min. For co-crystallization the substrate was added to the protein solution in a final concentration of 5–50 mm prior to crystallization. To identify potential binding sites for bivalent metal ions, a protein solution containing 0.75 mm GatDH was dialyzed against a solution containing 10 mm EDTA to remove Mg2+. In the corresponding crystallization setups, MgCl2 was replaced by 200 mm CoCl2 in the protein and reservoir solution.
Data Collection, Processing, and Structure Refinement
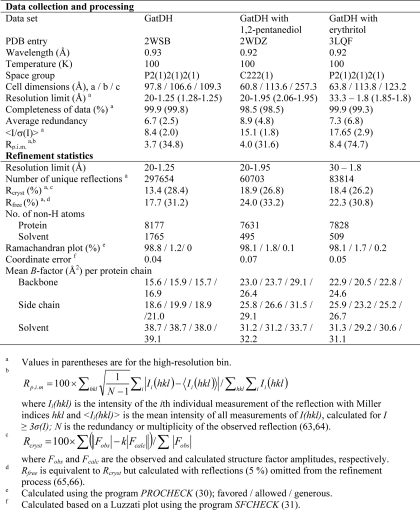
Data collection was performed at cryogenic temperatures (100 K) after flash cooling of the crystals in liquid nitrogen. Depending on the crystallization condition either PEG 8000 or MPEG 5000 up to a final concentration of 30% (w/v) were used as cryoprotectants. X-ray diffraction data sets were collected on a sealed tube X-ray generator (IμS, Incoatec Inc.) attached to a MARdtb goniostat and a MAR345 image plate detector (MAR Research, Norderstedt, Germany) or at various synchrotron beamlines (see Table 1). All data sets were indexed, integrated, and scaled using the program package XDS/XSCALE (20, 21) or Mosflm/Scala (22). The parameters for data collection and data processing are summarized in Table 1. For the phase determination of the first data set the molecular replacement method was used. Multiple sequence alignments with related proteins from the short-chain dehydrogenases/reductases family were performed with ClustalW (23). A homology model was generated as a starting search structure using the Swiss Model internet server (24). The modeling was based on the structure of gluconate 5-dehydrogenase from Thermotoga maritima (TM0441) (PDB entry 1VL8). The rotational and translational searches were performed with the program MOLREP (25). This first refined structure of GatDH was used as a search model to solve all subsequent structures of GatDH by molecular replacement. All structures were inspected and manually adjusted with the programs O (26) and COOT (27). Refinement was performed with REFMAC5 (28). Omit maps were generated by using the randomized omit map procedure (29). The coordinates of the questioned peptide regions were removed from the model, and each of the remaining coordinates was randomly translated up to 0.2 Å. This altered model was subjected to 10 rounds of restrained refinement with REFMAC5, and omit electron density maps with coefficients 2Fobs − 1Fcalc were calculated. The quality of the refined structure was verified with the programs PROCHECK (30) and SFCHECK (31). The final refinement statistics for all structures are given in Table 1. Assignment of secondary structure elements was performed with DSSP (32). Identification of structurally related proteins in the Protein Data Bank (PDB) (33), was performed with the DALI server (34, 35). Graphical representations were designed in PyMOL (36).
TABLE 1.
Diffraction data collection and refinement statistics
RESULTS
Overall Structure
The gene of GatDH extended by an N-terminal (His)6 affinity tag followed by a tobacco etch virus protease cleavage site was heterologously expressed in E. coli, and the produced protein was purified to homogeneity via affinity chromatography and size-exclusion chromatography. The final amount of pure (His)6-GatDH was 80 mg/liter cell culture. The structure of GatDH with its cofactor NAD(H), but without substrate, was solved to 1.25-Å resolution (Table 1). All 254 amino acid residues of the polypeptide chain and the cofactor NAD+ in the cofactor binding site were well defined in the electron-density maps.
GatDH consists of a seven-stranded β-sheet, surrounded by three α-helices on either side (Fig. 2). In its core it displays a Rossmann fold (37), built of two βαβαβ motifs (βA-αB-βB-αC-βC and βD-αE-βE-αF-βF). The cofactor is bound in a deep cleft above the β-sheets. This central motif is C-terminally extended through a seventh β-sheet (βG) and one α-helix (αG), separated from the Rossmann motif through a left-handed helix-turn-helix motif built out of two small helices (αFG1 und αFG2). A search for structurally related proteins in the PDB (33), identified 2-(2-(R)-hydroxypropylthio)ethanesulfonate dehydrogenase (PDB entry 2CFC (38)) from Xanthobacter autotrophicus (with root mean square deviation of 1.2 Å (243 of 254 Cα atoms), 39% sequence identity, and a Z-score of 37.8), gluconate-5-dehydrogenase (PDB entry 1VL8) from Thermotoga maritima (with root mean square deviation of 1.4 Å (248 of 254 Cα atoms), 36% sequence identity, and a Z-score of 37.4) and 3-oxoacyl-reductase (PDB entry 2UVD (39)) from Bacillus anthracis (with root mean square deviation of 1.2 Å (242 of 254 Cα atoms), 33% sequence identity, and a Z-score of 37.2) as the closest structural homologues of GatDH (supplemental Fig. 1).
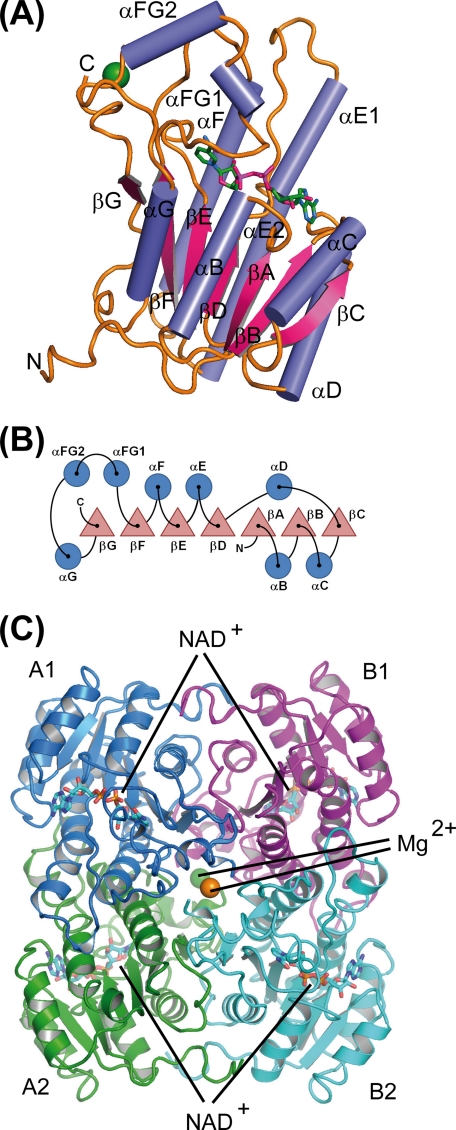
FIGURE 2.
Schematic representation of the secondary structure elements and their arrangement within GatDH. A, ribbon representation of one molecule of GatDH in complex with the cofactor NAD(H) (stick representation, carbon atoms colored in green) and one magnesium ion (green sphere). B, sketch of the arrangement of the secondary structure elements. Helices are represented by circles and β-strands by triangles. The nomenclature of secondary structure elements is according to 3α/20β-hydroxysteroid dehydrogenase (60). C, quaternary structure of GatDH. GatDH forms a homo-tetramer of point group symmetry D2. The individual protein chains are differently colored. Two magnesium ions (green and orange spheres) are coordinated each by two opposing C termini (A1–B2 and A2–B1, respectively). The active sites are positioned toward the surface of the protein as indicated by the NAD(H) molecules in ball-and-stick representation. The tetramer is oriented according to the PQR coordinate system (61) and displayed along the R-axis.
Independent from the bound cofactor or the concentration of MgCl2, GatDH was found to be tetrameric in solution, as analyzed by size-exclusion chromatography as well as by static (dynamic light scattering) and dynamic (multiple-angle laser light scattering) light scattering (data not shown). In line with this finding, GatDH crystals displayed a homo-tetramer with point-group symmetry D2 formed by a dimer of two homo-dimers (Fig. 2C). The interaction interfaces between monomer A1/A2 and B1/B2, coordinated through the helices αE and αF, are 1581 Å2, between monomer A1/B1 and A2/B2, coordinated through αF, αG, βF, the N terminus, and αB, are 1786 Å2. The contact area between A1/B2 and A2/B1 (452 Å2) is formed through the last four amino acids of the C terminus and the loop preceding helix αF. One element of this interface is one magnesium ion binding site per dimer. Each magnesium ion is complexed by the two carboxyl groups of the C termini of two opposing peptide chains. Therefore, the homo-tetramer contains two magnesium binding sites not associated with the bound cofactor (Fig. 3).
FIGURE 3.
Stereo representation of the Mg2+ binding site. The blue dashed lines mark the coordinating bonds between the magnesium ion and the two symmetry-related C-terminal carboxylate groups. The six shortest distances are in the range of 2.0–2.1 Å.
NAD(H) Binding Site
The cofactor binding site in SDR enzymes is characterized by the conserved TGXXXGXG cofactor binding motif. In GatDH the corresponding TGAGSGIG segment (residues 18–24) is located between the first β-strand (βA) and the first α-helix (αB) (Figs. 2 and 4A). This sequence is identical to the one in human estradiol 17-β-dehydrogenase type 8 (PDB entry 2PD6) (40). Further fingerprints are the motifs NNAG (at position 86–89) and PG (at position 189–190), both involved in cofactor binding at the active site (reviewed in Refs. 41, 42). In 70% of the cases, SDR enzymes with NAD(H) dependence comprise an aspartate residue at the C terminus of the second β-strand, and SDRs with NADP(H) dependence comprise an arginine residue (43). GatDH contains at this end of βB the sequence Asp42-Arg43. SDR enzymes containing this rare sequence are NAD-dependent (43), which is in accordance with the observed NAD(H) dependence of GatDH (5, 7). Asp42 of GatDH discriminates against a phosphate group attached to the 2′-OH of the nicotinamide cofactor, whereas the side chain of Arg43 interacts via π-interactions with the adenine base NAD(H). The cofactor is bound in a deep cleft and is protected against the environment through the helix αFG1 (Figs. 2 and 4A). The cofactor binds via 22 H-bonds directly and via six well coordinated water molecules with the protein. One of these water molecules is hydrogen bonded to the first and to the last conserved glycine residues within the TGXXXGXG signature motif and to the side-chain hydroxyl group of Ser92 at the end of β-strand βD. An equivalent water molecule is described as highly conserved in dinucleotide-binding proteins (44).
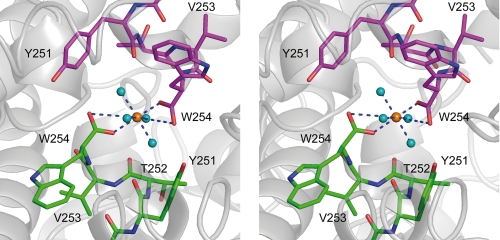
FIGURE 4.
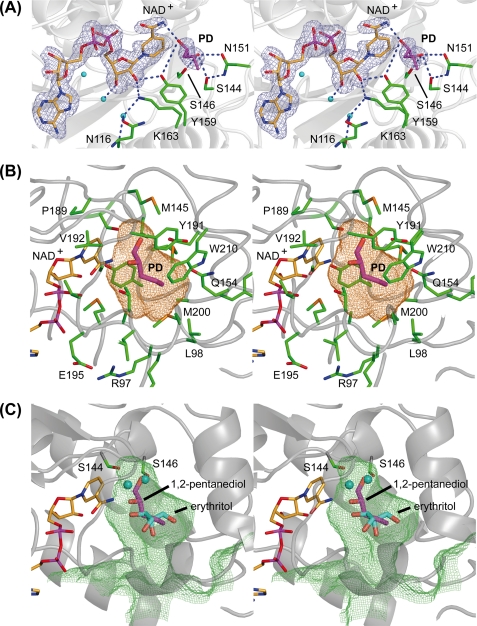
Stereo representation of the substrate binding pocket of GatDH. The main-chain trace of the protein is displayed as a ribbon in gray. The carbon atoms of the cofactor NAD+ are colored in orange, and those of the amino acid residues are in green. One-letter codes are used to identify amino acids. A, electron density map around the cofactor and the substrate 1,2-(S)-pentanediol (PD, carbon atoms in magenta). The final σA-weighted (2Fobs − Fcalc) electron density omit map drawn in blue and contoured at 2σ was calculated after removal of the substrate from the model. Side chains in the surrounding neighborhood are displayed as sticks. The blue dashed lines mark the coordinating hydrogen bonds related to substrate binding and the catalyzed redox reaction. B, stick representation of the cofactor NAD+ and substrate 1,2-(S)-pentanediol (PD) superimposed with the mesh representation of the substrate binding pocket. The substrate binding pocket was analyzed with the program VOIDOO (62) and displayed with PyMOL (36). The cavity is displayed in a mesh representation and colored in orange for a probe radius of 1.4 Å (inner part of the binding pocket). The side chain of those amino acids that define the substrate binding pocket are displayed as sticks and are superimposed on the cofactor, the substrate, and the binding cavity. C, superposition of meso-erythritol (carbon atoms in cyan) together with its two coordinating water molecules (cyan spheres) onto the structure of GatDH with bound 1,2-(S)-pentanediol (carbon atoms in magenta) and the mesh representation of the substrate binding pocket in lime green for a probe radius of 1.1 Å (indicating the access path of the substrate). The two serine residues Ser144 and Ser146 are represented as sticks indicating their importance for the positioning of the substrate (in the case of 1,2-(S)-pentanediol) or the well coordinated water molecules within the substrate binding pocket. For clarity, the other amino acid residues of the binding pocket are not shown.
Mapping of the Substrate Binding Site
The active site in SDR enzymes is characterized by the sequence motif Tyr-X-X-X-Lys, with tyrosine as the most frequently conserved and the lysine second highly conserved residue in all members of the SDR family (45). In GatDH, Tyr159 and Lys-163 form the catalytic tetrad together with Ser144 and Asn116, which superimpose very well with the catalytic tetrad of human 3β/17β-hydroxysteroid dehydrogenase, suggesting a similar kind of reaction mechanism (41, 46). The putative active site in GatDH is formed by a pocket at the end of the deep cleft in which the cofactor is bound. This cleft is predominantly shielded against the environment via helix αFG1 (Figs. 2 and 4A).
In GatDH without bound substrate, one large cavity next to the nicotinamide ring of the cofactor could be identified (Fig. 4 (B and C) and supplemental Fig. 3). This cavity has a volume of ∼250 Å3, a length of ∼14 Å, and a diameter of ∼8–10 Å with a small oval opening to the solvent environment of a diameter of ∼2–5 Å. It is formed by residues 96–98, 144–146, 151, 154–156, 159–160, 189–191, 196–197, 200, 210, and the nicotinamide moiety of the cofactor. It displays mostly an apolar surface. Aliphatic molecules with up to eight carbon atoms and even cyclic compounds might fit within this cavity.
In crystals grown under crystallization condition I, a nearly tetragon-shaped electron density was found within the putative substrate binding pocket next to the nicotinamide ring. The interpretation of this electron density was ambiguous. One possible interpretation of this electron density is the two different binding modes of one acetate molecule averaged over the sampled crystal volume. Based on this interpretation, the carbonyl group of the acetate molecule is strongly bound via five H-bonds to side chains in the active site pocket (Ser144, Ser146, Asn151, and Tyr159). To gain further insight into the mode of substrate binding we tried to map the substrate binding pocket with different substances. Because bound acetate might prevent potential substrates and/or inhibitors from binding, a second crystallization condition was optimized, which does not contain acetate or similar negatively charged substances. Based on the kinetic studies and identified substrates (5), we have performed soaking and co-crystallization experiments with 20 different compounds.
In co-crystallization experiments with a racemic (R,S)-mixture of 1,2-pentanediol (Km = 1.2 mm) the electron density was of sufficient quality to verify that only the S-enantiomer bound to the active site (Fig. 4). Forming a hydrogen-bonding network, Ser144 of the catalytic tetrad together with the side chains of Ser146 and Asn151 stabilize the primary, non-transformed hydroxyl-group of 1,2-(S)-pentanediol (Fig. 5).
FIGURE 5.
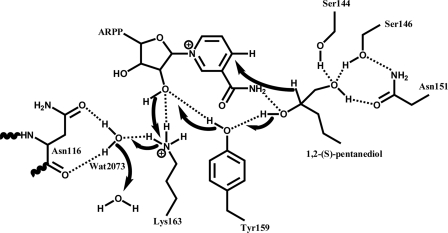
Postulated reaction mechanism of GatDH for the oxidation of 1,2-(S)-pentanediol according to Filling et al. (41). A charge relay system is formed by a hydrogen bond network involving Tyr151-OH, the 2′-OH group of the nicotinamide connected ribose moiety, Lys163-NH3+, Asn116-Oδ1, Asn116-O, and a well conserved water molecule, Wat2073. Catalysis is initiated by proton transfer from the secondary hydroxyl-group of the substrate onto Tyr151-OH followed by a hydride transfer to the nicotinamide ring C4 atom. The proton from the secondary hydroxyl group is transmitted to the solvent via the hydrogen network.
The secondary, to be oxidized hydroxyl-group, forms two hydrogen bonds with Tyr159-OH and the amide group of the nicotinamide moiety of the cofactor. The aliphatic tail is positioned within the larger part of the binding pocket. A second binding site for 1,2-pentanediol could be identified within the crystal-packing interface between two GatDH tetramers. This binding induced a small relative rotation of the protein molecules causing a small change in the overall crystal packing and as a consequence a different unit cell compared with the other crystal structures of GatDH (Table 1).
Further complex structures were derived from crystals soaked with 10 mm meso-erythritol (displays 4% relative activity for oxidation compared with galactitol (5); Km could not be determined) or 10 mm xylitol (displays 410% relative activity for oxidation compared with galactitol (5); Km = 22 mm). The electron density for these two polyols showed heterogeneity in the occupation of the cofactor binding pocket and the associated protein environment. This reflects the possibility of incomplete soaking. Therefore, the interpretation of the electron density within the substrate binding pocket was performed after the final refinement of the remaining protein, cofactor, and water structure. The final omit maps were of sufficient quality to unambiguously interpret the remaining electron density as one substrate (or product) molecule per binding pocket with additional water molecules in close proximity (Fig. 4).
Metal Dependence
The activity of GatDH was reported to be strictly dependent on the presence of divalent metal ions (5). Although the alcohol dehydrogenase members of the medium-chain dehydrogenases need Zn2+ for their enzymatic activity (47), only a few members of the SDR enzymes are found to be dependent of divalent metals, e.g. the R-specific alcohol dehydrogenase of Lactobacillus brevis (48) and within the active site of gluconate 5-dehydrogenase of Streptococcus suis a Ca2+ was identified (49). For identification of further binding sites for bivalent metals in GatDH, Mg2+ was replaced by Co2+. Bound Co2+ could be identified in electron density maps via their anomalous contribution to the x-ray scattering. A single-wavelength anomalous dispersion data set was collected, and an anomalous difference Patterson map was calculated. Although two strong electron density peaks at the C termini verified the cobalt ions bound to the magnesium binding sites in the interface of the tetramer, further binding sites could not be identified.
DISCUSSION
Based on typical sequence motifs (the NAD-binding TGXXXGXG motif, the active site YXXXK motif, and a conserved serine residue located in the active site) GatDH can be grouped into the subfamily of “classic” short-chain dehydrogenases (45, 50–56).
Mapping the Substrate Binding Pocket
For the structural elucidation of the substrate binding site, we could solve the structures of GatDH in complex with its cofactor NAD+ and its substrates xylitol, meso-erythritol, and 1,2-S-pentanediol (Fig. 1C). Interestingly, these three compounds presented two different binding modes within the substrate binding pocket. Whereas in the case of 1,2-S-pentanediol, the substrate is bound in close proximity to the nicotinamide ring, in the case of meso-erythritol and xylitol the substrate molecules are positioned further away and well ordered water molecules are located next to the nicotinamide ring forming hydrogen bonds with Ser144 and Ser146 (Fig. 4). In the case of 1,2-S-pentanediol no water molecules could be observed at these positions. Even though the substrates 1,2-S-pentanediol, meso-erythritol, and xylitol are differently bound within the substrate binding pocket, the surrounding protein environment is not significantly rearranged. The observed binding mode for meso-erythritol and xylitol are presumably not productive, because no hydroxyl group of the substrate is in close proximity to the nicotinamide ring. For proton and hydride transfer, structural arrangements would have to occur, which are more profound compared with those for 1,2-pentanediol. Because co-crystallization was used for 1,2-pentanediol, and soaking was used for erythritol and xylitol, and based on the electron density, in the last two cases substrate and derived product cannot be distinguished from each other. Therefore, an alternative interpretation could be that the observed mode of binding is that of the bound product prior to release from the substrate binding pocket.
Postulated Mode of Action Is Similar to Other SDR Dehydrogenases
The active site of SDR redox enzymes is constituted by a tetrad formed of Asn-Ser-Tyr-Lys residues (positions Asn116, Ser144, Tyr159, and Lys163 in GatDH, Fig. 4). The general reaction mechanism postulated for SDR enzymes is based on an observed hydrogen bond network involving the side chains of Lys, Tyr, and Ser, the nicotinamide ribose 2′-OH group, and a well coordinated water molecule bound to the main-chain carbonyl of Asn. This extended hydrogen bond network functions as a proton relay system and facilitates the positioning and polarization of the Tyr-OH next to the processed carbonyl or hydroxyl group of the substrate. Through this hydrogen bond network it is assumed that the pKa of the Tyr-OH is lowered to promote proton transfer from or to the substrate. Along the reaction pathway the Tyr-OH group functions as a general base and transfers the proton from the substrate OH-group via the 2′-OH group of the nicotinamide ribose moiety onto the side-chain Lys-NH3 group from which it is passed by the well positioned water molecule to the bulk solvent environment (41, 57, 58). In GatDH, the reduction of 1-hydroxy-2-ketones to 1,2-diols requires substrate binding to the residues Ser144, Ser146, and Asn151 (Figs. 4 and 5). The side chains of these residues form a hydrogen bond network with the OH-group of the substrate, which will not be oxidized by the enzyme. The secondary OH-group to be oxidized is in hydrogen-bonding distance with the amide group of the nicotinamide moiety of the cofactor and the OH-group of Tyr159 (Fig. 4). Based on this observed binding mode of 1,2-(S)-pentanediol and in analogy to 3β/17β-hydroxysteroid dehydrogenase (41, 46) a similar proton relay system can be postulated for the oxidation reaction (Fig. 5). In a first binding event the substrate is oriented by forming the hydrogen bond network with Ser144, Ser146, and Asn151 with the OH-group or carbonyl-group adjacent to the OH-group to be oxidized. In GatDH the conserved water molecule is coordinated by Asn116-Nδ1 and Asn116-O. Lys163 forms hydrogen bonds with the nicotinamide ribose moiety. In a concerted manner the oxidation reaction is completed by the transfer of the hydride ion from the carbon atom to the nicotinamide moiety to form NADH out of NAD+ (Fig. 5). This proposed mode of action is in agreement with the observed pH optimum at ∼9 for the oxidation reaction and pH 6.5 for the reduction reaction (59). Based on the 1,2-pentanediol complex structure it is obvious that only the S-configured OH-group vicinal to the hydrogen-bound OH-group of the substrate can be converted. The R-configuration points the OH-group away from the general base Tyr159. GatDH performs the oxidation reaction as well as the reduction. For the reduction of a carbonyl group in α-position to a hydroxyl (α-hydroxy ketone) group or a carbonyl-group (α-diketone) the transfer path of the proton from the bulk solvent environment and the hydride ion from the nicotinamide moiety are reversed.
Substrate Spectrum Can Be Rationalized
The substrate binding pocket can be subdivided into two regions: one smaller binding pocket and one larger binding pocket. The smaller binding pocket is defined by residues Ser144, Met145, Ser146, Asn151, Pro189, Gly190, Tyr191, Met196, and Trp210 and the nicotinamide ring moiety of the cofactor NAD(H). Within this part the amino acid residues Ser144 and Ser146 form hydrogen bonds with the not converted polar group in α- or β-position to the processed hydroxyl- or carbonyl-group. The larger binding pocket is formed by residues Ala96, Arg97, Leu98, Gln154, Pro155, Ala156, Tyr159, Met160, Met197, and Met200. Within the smaller binding pocket only a few methylene groups (around 3–4) can fit, whereas the larger pocket can accommodate much larger and bulkier alkyl groups, even cyclic or aromatic groups might fit. Thus, asymmetric substrates with a small and a large substituent can bind to the substrate binding pocket in two alternative modes. The one with the smaller substituent being oriented within the smaller binding pocket and the larger substituent within the larger binding pocket should be the more processable in respect of turnover. This interpretation will help to predict the regio- and stereochemistry of formed products from asymmetric substrates with two processable functional groups such as 1,2-dihydroxy alkanes, 1-hydroxy-2-keto alkanes, or 1-hydroxy-2,3-diketones.
Based on this proposed mode of action and the functional topology of the substrate binding pocket the following statements are deduced: 1) aldehydes or ketones with no α- or β-positioned hydrogen bond-forming group will be not or barely converted; 2) the binding pocket is asymmetric such that the α-positioned hydrogen bond-forming group is positioned in the smaller part, and the moiety of the substrate on the other side from the transformed carbonyl- or hydroxyl-group is bound in the larger part of the binding pocket; 3) in the small part of the binding pocket only up to three carbon atoms can be accommodated, whereas in the larger half even an aromatic ring system might fit; 4) as a consequence, preferred substrates are asymmetric with the hydrogen bond-forming group at the short part; and 5) the shape and size of the binding pocket make it unlikely that cyclic polyols or cyclic α-diketones are favored substrates.
Based on the observed alternative binding modes for the different substrates (or products), it can be assumed that an unproductive mode of binding of substrate molecules might influence the overall kinetic characteristics of the enzyme. It might even be that the enzyme displays for some substrates substrate- or product-induced inhibition such that a second bound molecule has to diffuse out of the binding pocket to clear the way for the converted molecule. However, such a mode of inhibition could not be detected in the case of meso-erythritol and xylitol, respectively. Further kinetic characterizations with a broader spectrum of substrates will clarify this point. Based on the elongated shape of the binding pocket, it seems to be more likely that acyclic sugar molecules are easier to be transformed than their bulky cyclic counterparts.
Conclusion
Kinetic characterization of the enzyme GatDH revealed a high degree of stereoselectivity within a widespread substrate spectrum covering sugars, sugar alcohols, secondary alcohols, or corresponding ketones. These characteristics make GatDH a very interesting enzyme in industrial biotechnology for the production of optically pure building blocks and the bioconversion of bioactive compounds. Here, we have presented different structures of GatDH in complex with its cofactor NAD(H). One high resolution structure represents the holoenzyme with cofactor but without bound substrate. Three structures could additionally be solved with different bound substrates. Together, these structures provide insight into the substrate binding pocket to rationalize the observed substrate spectrum and reaction selectivity. This information will help to further optimize the substrate specificity, the catalytic activity, and the stability toward pH, temperature, and solvents by directed evolution or rational design of enzyme variants. In addition it will help to rationalize the results of kinetic characterization of the enzyme and in silico docking experiments.
Supplementary Material
Acknowledgments
We gratefully acknowledge access to the core facilities of the ZBM/LMB of the Christian-Albrechts-University Kiel, access to the EMBL beamlines at the DORIS storage ring (DESY, Hamburg), beamline BL14.2 at BESSY (Berlin) and ID 14-2 at the European Synchrotron Radiation Facility (Grenoble, France). We thank Joachim Bräutigam for performing the mass spectrometry experiments and Dr. Markus Greiner and Mario Schu for help with multiple-angle laser light scattering.
This work was supported by the Seventh Framework Programme of the European Union for the Small Scale Collaborative Project Electrochemical Reactors Using Dehydrogenases for Enantiopure Synthon Preparations (ERUDESP) under Contract NMP3-SL-2008-213487.
The atomic coordinates and structure factors (codes 2WSB, 2WDZ, and 3LQF) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3, Table 1, and references.
- GatDH
- galactitol 2-dehydrogenase
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PEG
- polyethylene glycol
- MES
- 4-morpholineethanesulfonic acid
- SDR
- short-chain dehydrogenase.
REFERENCES
- 1.Panke S., Held M., Wubbolts M. (2004) Curr. Opin. Biotechnol. 15, 272–279 [DOI] [PubMed] [Google Scholar]
- 2.Hummel W. (1997) Adv. Biochem. Eng. Biotechnol. 58, 145–184 [DOI] [PubMed] [Google Scholar]
- 3.Goldberg K., Schroer K., Lütz S., Liese A. (2007) Appl. Microbiol. Biotechnol. 76, 237–248 [DOI] [PubMed] [Google Scholar]
- 4.Goldberg K., Schroer K., Lütz S., Liese A. (2007) Appl. Microbiol. Biotechnol. 76, 249–255 [DOI] [PubMed] [Google Scholar]
- 5.Schneider K. H., Jäkel G., Hoffmann R., Giffhorn F. (1995) Microbiology 141, 1865–1873 [DOI] [PubMed] [Google Scholar]
- 6.Huwig A., Emmel S., Jäkel G., Giffhorn F. (1998) Carbohydr. Res. 305, 337–339 [Google Scholar]
- 7.Kohring G. W., Wiehr P., Jeworski M., Giffhorn F. (2003) Commun. Agric. Appl. Biol. Sci. 68, 309–312 [PubMed] [Google Scholar]
- 8.Lee L. G., Whitesides G. M. (1985) J. Am. Chem. Soc. 107, 6999–7008 [Google Scholar]
- 9.Schüte H., Flossdorf J., Sahm H., Kula M. R. (1976) Eur. J. Biochem. 62, 151–160 [DOI] [PubMed] [Google Scholar]
- 10.Furneaux R. H., Tyler P. C., Whitehouse L. H. (1993) Tetrahedron Lett. 34, 3609–3612 [Google Scholar]
- 11.Freimund S., Huwig A. (2000) Carbohydr. Lett. 3, 1585–1587 [DOI] [PubMed] [Google Scholar]
- 12.Kornberger P., Gajdzik J., Natter H., Wenz G., Giffhorn F., Kohring G. W., Hempelmann R. (2009) Langmuir 25, 12380–12386 [DOI] [PubMed] [Google Scholar]
- 13.Stein M. A., Schäfer A., Giffhorn F. (1997) J. Bacteriol. 179, 6335–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philippsen A., Schirmer T., Stein M. A., Giffhorn F., Stetefeld J. (2005) Acta Crystallogr. D. Biol. Crystallogr. 61, 374–379 [DOI] [PubMed] [Google Scholar]
- 15.Bastian S., Rekowski M. J., Witte K., Heckmann-Pohl D. M., Giffhorn F. (2005) Appl. Microbiol. Biotechnol. 67, 654–663 [DOI] [PubMed] [Google Scholar]
- 16.Bannwarth M., Heckmann-Pohl D., Bastian S., Giffhorn F., Schulz G. E. (2006) Biochemistry 45, 6587–6595 [DOI] [PubMed] [Google Scholar]
- 17.Kühn A., Yu S., Giffhorn F. (2006) Appl. Environ. Microbiol. 72, 1248–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dambe T. R., Kühn A. M., Brossette T., Giffhorn F., Scheidig A. J. (2006) Biochemistry 45, 10030–10042 [DOI] [PubMed] [Google Scholar]
- 19.Gajdzik J., Szamocki R., Natter H., Kohring G. W., Giffhorn F., Hempelmann R. (2007) J. Solid State Electrochem. 11, 144–149 [Google Scholar]
- 20.Kabsch W. (1988) J. Appl. Crystallogr. 21, 916–924 [Google Scholar]
- 21.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 22.Leslie A. G. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography 26 [Google Scholar]
- 23.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 25.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 26.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard. (1991) Acta Crystallogr. 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 27.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28.Murshudov G. N., Vagin A., Dodson E. J. (1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29.McRee D. E. (1993) Practical Protein Crystallography, pp. 223–226, Academic Press, Inc., San Diego, CA [Google Scholar]
- 30.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 31.Vaguine A. A., Richelle J., Wodak S. J. (1999) Acta Crystallogr. D. Biol. Crystallogr. 55, 191–205 [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W., Sander C. (1983) Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 33.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm L., Sander C. (1996) Science 273, 595–603 [DOI] [PubMed] [Google Scholar]
- 35.Holm L., Park J. (2000) Bioinformatics 16, 566–567 [DOI] [PubMed] [Google Scholar]
- 36.DeLano W. L. (2004) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 37.Rossmann M. G., Moras D., Olsen K. W. (1974) Nature 250, 194–199 [DOI] [PubMed] [Google Scholar]
- 38.Krishnakumar A. M., Nocek B. P., Clark D. D., Ensign S. A., Peters J. W. (2006) Biochemistry 45, 8831–8840 [DOI] [PubMed] [Google Scholar]
- 39.Zaccai N. R., Carter L. G., Berrow N. S., Sainsbury S., Nettleship J. E., Walter T. S., Harlos K., Owens R. J., Wilson K. S., Stuart D. I., Esnouf R. M. (2008) Proteins 70, 562–567 [DOI] [PubMed] [Google Scholar]
- 40.Rotinen M., Celay J., Alonso M. M., Arrazola A., Encio I., Villar J. (2009) J. Endocrinol. 200, 85–92 [DOI] [PubMed] [Google Scholar]
- 41.Filling C., Berndt K. D., Benach J., Knapp S., Prozorovski T., Nordling E., Ladenstein R., Jörnvall H., Oppermann U. (2002) J. Biol. Chem. 277, 25677–25684 [DOI] [PubMed] [Google Scholar]
- 42.Jörnvall H., Persson B., Krook M., Atrian S., Gonzàlez-Duarte R., Jeffery J., Ghosh D. (1995) Biochemistry 34, 6003–6013 [DOI] [PubMed] [Google Scholar]
- 43.Duax W. L., Pletnev V., Addlagatta A., Bruenn J., Weeks C. M. (2003) Proteins 53, 931–943 [DOI] [PubMed] [Google Scholar]
- 44.Bottoms C. A., Smith P. E., Tanner J. J. (2002) Protein Sci. 11, 2125–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kavanagh K. L., Jörnvall H., Persson B., Oppermann U. (2008) Cell Mol. Life Sci. 65, 3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benach J., Filling C., Oppermann U. C., Roversi P., Bricogne G., Berndt K. D., Jörnvall H., Ladenstein R. (2002) Biochemistry 41, 14659–14668 [DOI] [PubMed] [Google Scholar]
- 47.Persson B., Hedlund J., Jörnvall H. (2008) Cell Mol. Life Sci. 65, 3879–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niefind K., Müller J., Riebel B., Hummel W., Schomburg D. (2003) J. Mol. Biol. 327, 317–328 [DOI] [PubMed] [Google Scholar]
- 49.Zhang Q., Peng H., Gao F., Liu Y., Cheng H., Thompson J., Gao G. F. (2009) Protein Sci. 18, 294–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jörnvall H. (2008) Cell Mol. Life Sci. 65, 3873–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kallberg Y., Oppermann U., Jörnvall H., Persson B. (2002) Protein Sci. 11, 636–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kallberg Y., Oppermann U., Jörnvall H., Persson B. (2002) Eur. J. Biochem. 269, 4409–4417 [DOI] [PubMed] [Google Scholar]
- 53.Klasen R., Bringer-Meyer S., Sahm H. (1995) J. Bacteriol. 177, 2637–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oppermann U., Filling C., Hult M., Shafqat N., Wu X., Lindh M., Shafqat J., Nordling E., Kallberg Y., Persson B., Jörnvall H. (2003) Chem. Biol. Interact 143–144, 247–253 [DOI] [PubMed] [Google Scholar]
- 55.Persson B., Krook M., Jörnvall H. (1991) Eur. J. Biochem. 200, 537–543 [DOI] [PubMed] [Google Scholar]
- 56.Persson B., Kallberg Y., Bray J. E., Bruford E., Dellaporta S. L., Favia A. D., Duarte R. G., Jörnvall H., Kavanagh K. L., Kedishvili N., Kisiela M., Maser E., Mindnich R., Orchard S., Penning T. M., Thornton J. M., Adamski J., Oppermann U. (2009) Chem. Biol. Interact 178, 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao D. I., Thompson J. E., Fahnestock S., Valent B., Jordan D. B. (2001) Biochemistry 40, 8696–8704 [DOI] [PubMed] [Google Scholar]
- 58.Tanaka N., Nonaka T., Tanabe T., Yoshimoto T., Tsuru D., Mitsui Y. (1996) Biochemistry 35, 7715–7730 [DOI] [PubMed] [Google Scholar]
- 59.Schneider K. H., Giffhorn F. (1989) Eur. J. Biochem. 184, 15–19 [DOI] [PubMed] [Google Scholar]
- 60.Ghosh D., Weeks C. M., Grochulski P., Duax W. L., Erman M., Rimsay R. L., Orr J. C. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10064–10068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rossmann M. G., Adams M. J., Buehner M., Ford G. C., Hackert M. L., Liljas A., Rao S. T., Banaszak L. J., Hill E., Tsernoglou D., Webb L. (1973) J. Mol. Biol. 76, 533–537 [DOI] [PubMed] [Google Scholar]
- 62.Kleywegt G. J., Jones T. A. (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 178–185 [DOI] [PubMed] [Google Scholar]
- 63.Weiss M. (2001) J. Appl. Crystallogr. 34, 130–135 [Google Scholar]
- 64.Diederichs K., Karplus P. A. (1997) Nat. Struct. Biol. 4, 269–275 [DOI] [PubMed] [Google Scholar]
- 65.Brünger A. T. (1992) Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 66.Tickle I. J., Laskowski R. A., Moss D. S. (2000) Acta Crystallogr. D. Biol. Crystallogr. 56, 442–450 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.