Abstract
Class 1 cytokines bind two receptors to create an active heterotrimeric complex. It has been argued that ligand binding to their receptors is an ordered process, but a structural mechanism describing this process has not been determined. We have previously described an obligate ordered binding mechanism for the human prolactin/prolactin receptor heterotrimeric complex. In this work we expand this conceptual understanding of ordered binding to include three human lactogenic hormones: prolactin, growth hormone, and placental lactogen. We independently blocked either of the two receptor binding sites of each hormone and used surface plasmon resonance to measure human prolactin receptor binding kinetics and stoichiometries to the remaining binding surface. When site 1 of any of the three hormones was blocked, site 2 could not bind the receptor. But blocking site 2 did not affect receptor binding at site 1, indicating a requirement for receptor binding to site 1 before site 2 binding. In addition we noted variable responses to the presence of zinc in hormone-receptor interaction. Finally, we performed Förster resonance energy transfer (FRET) analyses where receptor binding at subsaturating stoichiometries induced changes in FRET signaling, indicative of binding-induced changes in hormone conformation, whereas at receptor:hormone ratios in excess of 2:1 no additional changes in FRET signaling were observed. These results strongly support a conformationally mediated obligate-ordered receptor binding for each of the three lactogenic hormones.
Keywords: Biophysics, Cell Surface Receptor, Cytokine, Peptide Hormones, Protein-Protein Interactions, Receptor Structure-Function, Growth Hormone, Placental Lactogen, Prolactin
Introduction
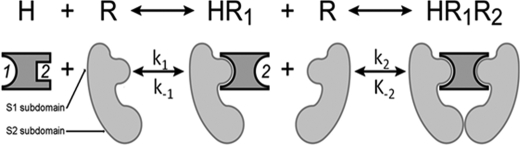
Human prolactin (hPRL),2 growth hormone (hGH), and placental lactogen (hPL) are hormones belonging to the class 1 cytokine family (1). These three proteins share significant sequence homologies; the more closely related hPL and hPRL display 85% sequence identity, and hPRL shares a 23% identity with hGH. Each of these four-helix bundle proteins is recognized as a lactogenic hormone based on their ability to bind and activate primate prolactin receptors (2). The binding reaction that takes place between lactogenic hormones and the prolactin receptor results in the formation of a heterotrimeric complex with a 2:1 ratio of receptor to hormone (3) (Fig. 1). The formation of this ternary complex is believed to proceed in an ordered process in which the N-terminal S1 subdomain of the prolactin receptor first binds at a hormone surface comprised of elements of helices 1 and 4 as well as the sequence connecting helices 1 and 2 (site 1) before subsequently binding a second prolactin receptor by its S1 subdomain at a hormone surface composed of elements of helices 1 and 3 (site 2). During secondary receptor binding, an additional receptor-receptor binding surface is formed between the C-terminal S2 subdomains of the membrane-bound receptors and is distal to the bound hormone. Thus, for site 2 the ΔG is divided between the hormone/receptor interfaces and the two interacting S2 receptor surfaces. The formation of this extracellular trimolecular complex orients the intracellular domains of the prolactin receptors to allow a receptor-associated JAK 2-mediated trans-phosphorylation of intracellular receptor tyrosines that provide docking sites for SH2 proteins and ultimately permit the initiation of intracellular hormone action (4).
FIGURE 1.
Mechanism for lactogen/receptor binding. The reaction and schematic represent the model for obligate ordered binding of lactogens and the hPRLr. Note that site 2 changes its structure as a result of hPRLr binding to site 1 of the hormone. hPRL binds the N-terminal S1 subdomain of the hPRLr. Subsequent formation of the heterotrimeric complex is also associated with binding of the C-terminal S2 subdomains of the two hPRLr.
In recent years solution structures of hPRL have been completed. Two structures of hPRL have been obtained by NMR (5, 6), and an additional two structures of hPRL bound to the extracellular domain of the hPRL receptor have been provided by x-ray crystallography (7, 8). In these x-ray studies the authors compared the receptor-bound and receptor-free hormone structures and concluded that subtle differences exist in the helix bundle structures, but significant restructuring of the N-terminal region of the loop that connects helices 1 and 2 occurs as a result of receptor binding at site 1. Interpretations of the structural information leave open the idea that the two receptor binding sites of the hormone are functionally coupled. Previous surface plasmon resonance and Förster resonance energy transfer (FRET) data from our laboratory indicated that receptor binding induced a conformation change in the hormone associated with a functional coupling of sites1 and 2 (9). The observation that hPRL undergoes a conformational change upon receptor binding is consistent with observations of hGH or hPL binding the extracellular domain of the hPRL receptor (10, 11) or when hGH binds the extracellular domain of the hGH receptor (12, 13). The idea that the receptor binding events are functionally coupled also is consistent with the positive 1.7 Hill coefficient described by Hooper et al. (3) in a heterologous ovine PRL/rat PRL receptor system.
In this work we chose to compare both surface plasmon binding and FRET studies for hPRL, hGH, and hPL binding to the extracellular domain of the hPRL receptor in an effort to compare the binding mechanisms of this family of hormones. We demonstrate that despite sequence homologies of less than 30% between hPRL and either hGH or hPL, all lactogenic cytokines bind the human prolactin receptor in an obligate ordered process. Furthermore we present FRET data supporting a binding mechanism in which receptor binding at site 1 of the hormone conformationally regulates the ability to bind receptor at site 2. Finally, we demonstrate that the presence or absence of ionic zinc uniquely influences the site 1 binding kinetics of each of these three lactogenic hormones.
MATERIALS AND METHODS
Construction of Vectors and Preparation of Proteins
The pT7-7 phagemid expression vectors for methionyl hPRL, hGH, and the extracellular domain of the human hPRL receptor (residues 1–224) were prepared as described by Peterson et al. (14), Duda and Brooks (15), and Sivaprasad et al. (9), respectively. hPL cDNA from the I.M.A.G.E. Consortium (Lawrence Livermore National Laboratory) cDNA Clones (16) was purchased from Open Biosystems (Huntsville, AL) and cloned into pET-28b(+) (Novagen, EMD Chemicals, Darmstadt, Germany). The vector was cut with NcoI and HindIII, removing the vector histidine tag and a portion of the polycloning site. The hPL cDNA was amplified by PCR using a primer that added a methionyl codon to the immediate N terminus of the mature protein and a second primer that added a HindIII site 3′ following the stop codon of hPL. The PCR product was characterized, double-digested with NcoI and HindIII, and ligated into the linearized and modified pET28b(+) plasmid. The hPL expression vector was expanded in XL-10 Gold cells (Stratagene, Cedar Creek, TX), and clones were selected by resistance to kanamycin, purified, and stored in water at −30 °C. The pT7-7 phagemid expression vector for the human prolactin receptor was also modified by the Kunkel procedure (17) to add a stop codon immediately after the codon coding for residue 210, reducing the expressed methionyl protein to the extracellular domain of the human prolactin receptor (hPRLr).
Mutations in the hormones were performed either by the method of Kunkel (17) (hPRL and hGH) or by the QuikChange® method (18) (Stratagene) (hPL). The following three mutants were prepared for hPRL: G129C, M158C, and K181C. Structurally equivalent mutations were prepared in hGH and hPL at positions G120C, N152C, and K172C. The respective G120C and G129C mutations were engineered to exist within site 2 of the hormones, K172C and K181C mutations were engineered to exist within site 1 of the hormones, and the N152C and M158C mutations were designed to exist external to either site 1 or 2 of the hormones. These engineered unpaired cysteine residues were used to thiol-couple hormones to surface plasmon resonance (SPR) chips at the defined positions and selectively corrupt either site 1, site 2, or neither binding site interaction with the receptor. The engineered cysteines at residues 152 or 158 in hGH and hPL or in hPRL also were used for coupling an extrinsic fluorochrome for FRET studies. The complete nucleic acid sequences for the wild-type hormone, the mutant hormones, and hPRLr were confirmed by Sanger di-deoxy sequencing (19). For the purposes of clarity, the N-terminal methionyl residue is regarded as residue 0, so that the sequence numbering for the mature proteins is retained.
Expression of hPRL, hGH, hPL, or hPRLr was performed in BL21 (DE3) Escherichia coli (Novagen). All proteins were expressed as described by Peterson et al. (14). E. coli were induced with isopropyl-β-d-thiogalactopyranoside (0.4 mm for pT7–7 and 1.0 mm for pET-28b), and expression was allowed to continue for more than 4 h. Cells were broken by two passes through a French pressure cell, and inclusion bodies were collected by centrifugation and suspended in 4.5 m urea in 100 mm Tris buffer. Cell particles were removed by centrifugation, the pH of the supernatant was raised to between 11.0 to 11.5 with NaOH, and the protein was stirred at 4 °C open to air for 48 h. The proteins folded during dialysis against 20 mm Tris, pH 7.5. Proteins were purified by DEAE fast-flow Sepharose anion exchange chromatography (2.0 × 5 cm, GE Healthcare) on an Akta 100X chromatograph (GE Healthcare) in 20 mm Tris, pH 7.5, and eluted by a NaCl gradient. Proteins were subsequently desalted and further purified on a 2.6 × 50-cm Superdex 75 (GE Healthcare) gel filtration column run with 10 mm NH4HCO3. Proteins were lyophilized and stored at −30 °C. Protein concentrations were determined by the bicinchoninic assay using bovine albumin as a standard (20) or by using calculated molar extinction coefficients (ϵ280 nm) (21): hPRL = 21,805 cm−1m−1, hGH = 17,670 cm−1m−1, hPL = 17,670 cm−1m−1, and hPRLr = 66,140 cm−1m−1.
Characterization of Proteins
Each protein was characterized by SDS-containing polyacrylamide gel electrophoresis. In addition, all proteins were characterized by absorbance, fluorescence, and circular dichroism spectroscopy for which proteins were in 10 mm Tris, pH 7.4, and 150 mm NaCl buffer. Absorbance, fluorescence, and circular dichroism data were collected using a PerkinElmer Life Sciences Lambda 45 absorbance spectrophotometer, Varian Cary Eclipse fluorescence spectrophotometer (Varian, Inc., Palo Alto, CA), and AVIV circular dichroism spectrometer Model 202 (AVIV Biomedical, Inc., Lakewood, NJ), respectively.
Biological Assays
FDC-P1 cells stably expressing the long form of the human prolactin receptor were provided by Genentech, Inc. (San Francisco, CA) and maintained in RPMI 1640 (Invitrogen) with 10% fetal bovine serum (Hyclone, Logan UT), 10 ng/ml IL-3 (BD Biosciences), and 220 μg/μl G418 sulfate (Hyclone). Hormone dose-response curves were obtained as previously described (22) using a vital dye method (Alamar Blue, Biomed, Cleveland, OH). This method relies on the extent of resazurin reduction (23). Absorbance is measured in each well at 570 and 600 nm, and the percentages of reduced resazurin are calculated.
SPR Studies
SPR is a biophysical technique that measures kinetics of association and dissociation. Proteins used in this work were designed to be thiol-coupled to Biacore CM5 Sensor Chips (GE Healthcare). This coupling method permits the covalent attachment of a protein surface to a chip at a cysteine residue. Proteins were prepared for thiol-coupling by suspension in 10 mm NH4HCO3, pH 7.0, and reduced for 5 min at room temperature by incubation in a 5 μm stoichiometric excess of dithiothreitol (Sigma). This concentration of dithiothreitol was insufficient to reduce the native disulfide bonds but sufficient to reduce the engineered cysteine (24, 25). Dithiothreitol was removed from solution by three centrifugations in YM-10 Centricon Centrifugal Filter Devices (Millipore, Billerica, MA), each time replacing the lost volume with 10 mm Tris, pH 7.4, 150 mm NaCl (FRET buffer). The carboxymethylated dextran surface of the chip was prepared for thiol-coupling by injecting 25 μl of a fresh 1:1 combination of 50 mm N-hydroxylsuccinimide and 20 mm 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride followed by 40 μl of freshly prepared 80 mm 2-(2-pyridinylthio) ethanolamine in 100 mm boric acid, pH 8.5. The remaining free amine binding sites were blocked by injecting 50 μl of 1 m ethanolamine, pH 8.5, across the chip surface. Sequential injections of hormone were then made, coupling protein in a stepwise fashion until the chip surface contained ∼300–800 response units of bound protein. A blank lane was chemically activated but not exposed to experimental protein. Un-reacted thiols on all lanes were blocked by injecting 30 μl of 50 mm cysteine, 20 mm sodium acetate, pH 4.5, and 1 m NaCl. All injections for chip construction were made at 5 μl/min, and all SPR experiments were performed using a Biacore 3000 SPR instrument (GE Healthcare).
Various concentrations of hPRLr (10 nm-100uM) were prepared in each of two separate buffers: 10 mm HEPES, pH 7.4, 150 mm NaCl, 3 mm EDTA, and 0.005% Surfactant P20, (HBS-EP buffer) (GE Healthcare) or 10 mm HEPES, pH 7.4, 150 mm NaCl, 15 μm ZnSO4 (HBS-Zn buffer). The chips were primed with buffer appropriate to the experimental conditions, and changes in response units were recorded as 300-μl injections of various concentrations of hPRLr were made over the thiol-coupled hormones at 50 μl/min before being allowed to dissociate for 1 h. The data were recorded during both these periods. Chips were regenerated between runs with a 25-μl injection of 4.5 m MgCl2, 150 mm NaCl, 3 mm EDTA, 0.005% Surfactant P20, 10 mm HEPES, pH 7.4. Each experiment was performed two to five times, and the results were averaged.
SPR Kinetic Evaluation
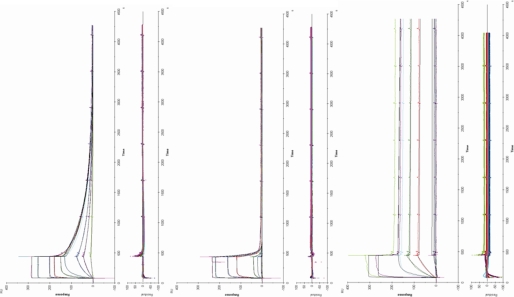
Kinetic binding models for hPRL M158C, hGH N152C, and hPL N152C presume a stoichiometric model where hormone bound to the SPR chip is capable of binding two receptors (Fig. 1). Although previous work has determined that the heterotrimeric complex forms as a result of ordered binding events (hormone binds receptor at site 1 before binding hormone at site 2), an ordered model of dissociation has not been established. To determine meaningful kinetic interpretations for a two-state binding model, it was presumed that receptor dissociation occurs in the opposite order of heterotrimeric formation. Curve-fitting and data analysis were performed using Scrubber 2.0 (David Myszka, Center for Biomolecular Interaction Analysis, University of Utah) and BIAevalutation 3.0 (GE Healthcare). Kinetic studies of hormones coupled to the chip surface through either C129 (hPRL) or C120 (hGH and hPL) record the binding only at site 1; these studies used the 1:1 Langmuir model to calculate site 1 rate constants. In contrast, when the hormones were coupled to the chip through either Cys-158 (hPRL) or Cys-152 (hGH and hPL), hPRLr can bind at both sites 1 and 2. These data were calculated by two methods. First, a 1:1 Langmuir analysis was performed that would treat binding as if all binding was represented by a single site, similar to analyses that were used for decades in studies that use 125I-labeled lactogens. A second analysis used a sequential binding model available in BIAevaluation (Fig. 1). The difficulty with this approach is that two variables, rate constants for site 1 and site 2, need to be simultaneously solved. Unfortunately, unless the rate constants for the two binding events are very different, the iterative calculation for the rate constants for sites 1 and 2 will converge. Kinetic studies were determined from the results from 2 to 5 independent experiments. Residuals from fitting were generally less than 3% of binding, and calculated association and dissociation rate constants were used to determine equilibrium constants (Fig. 2).
FIGURE 2.
Surface plasmon resonance kinetic data and residuals for M158C hPRL (top), N152C hGH (middle), and N152C hPL (bottom) binding to the extracellular domain of the hPRL receptor in the presence of Zn2+. Each of the three lactogens was covalently attached to a CM-5 SPR chip; the fourth lane was activated but not populated with protein. Increasing concentrations of hPRLr (10 nm, 50 nm, 100 nm, 500 nm, 1 μm, 5 μm, 10 μm, 50 μm, and 100 μm) were flowed over the chip surface for 300 s to follow hormone/hPRLr binding. Subsequently, buffer was flowed over the chip surface, and hormone/hPRLr dissociation followed. Residues from the model fit to the data are presented beneath the binding data.
Coumarin Labeling and FRET Assays
Hormones (hPRL M158C, hGH N152C, and hPL N152C) containing a free cysteine distal to receptor binding sites were labeled for FRET by suspension in 10 mm ammonium bicarbonate, pH 7.0, followed by a mild dithiothreitol-mediated reduction as describe previously for SPR studies. 10 mm 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (CPM) (Invitrogen) was prepared in DMSO and added in 10 m excess to the hormone. Labeling reactions were run at 4 °C for 8–12 h in a dark, oxygen-free environment before being filtered through a 1 × 15 cm Sephadex G-50 (Sigma) column to separate protein from unbound CPM. Fractions containing labeled hormone were identified by protein and CPM absorbance at 280 and 340 nm, respectively. Fractions containing protein were pooled and dialyzed in darkness at 4 °C against 150 mm NaCl, 10 mm Tris, pH 7.4 for 8–12 h, and experiments were performed within 12 h of dialysis completion.
FRET studies were performed at room temperature with a total hormone concentration of 1 μm and increasing hPRLr concentrations up to either 5 or 10 μm to obtain saturation curves. 0.25 μm CPM-labeled hormone was combined with 0.75 μm corresponding unlabeled wild-type hormone to obtain a 1 μm concentration yielding an appropriate FRET signal intensity. Reaction mixtures were incubated for 1 h in darkness and assessed using either a Varian Cary Eclipse Fluorescence spectrophotometer (Varian, Inc., Palo Alto, CA) or a FluoroLog-3 fluorimeter (Horiba Jobin Yvon, Inc., Edison, NJ). Samples were illuminated at 295 nm to specifically stimulate tryptophan, and emission spectra were collected between 300 and 550 nm. Tryptophan fluorescence was maximal at ∼340 nm, which overlapped with CPM absorbance. The CMP emission was maximal at ∼470 nm. The 470 nm FRET signals were corrected for overlapping tryptophan emission, the background signal (hormone with no receptor) were subtracted, and the data were plotted to evaluate the titration of receptor binding to hormone.
RESULTS
Protein Characterization
Each of the 13 methionyl proteins prepared for these studies was expressed in E. coli by recombinant DNA techniques. Proteins were >95% pure when evaluated by SDS-containing 15% polyacrylamide gel electrophoresis under reducing conditions (supplemental Fig. 1). Spectroscopy of the hormones showed that the absorbance, fluorescence, or circular dichroism spectra of the three hPRL, hGH, and hPL mutants closely overlaid the spectra of the corresponding wild-type hormones (supplemental Fig. 2), indicating that the engineered cysteine mutations within these hormones produced little or no effect on protein folding and were unlikely to elicit unanticipated effects on receptor binding.
When the ED50 values of the wild-type and mutant hormones were compared in FDC-P1 cells that expressed the complete human prolactin receptor, hormones containing cysteine mutations at Met-158 and Asn-152 were equivalent to the wild-type hormones. Thus, mutations distal to sites 1 and 2 did not affect biological activity and were unlikely to change the basic structure of these hormones. On the other hand, cysteine mutations within site 1 (K172C hGH or K181C hPRL) or site 2 (G120C hGH or G129C hPRL) reduced the biological activities of these hormones between 4- and 100-fold, indicating that our mutations are within sites 1 and 2.
SPR Stoichiometry Measurements
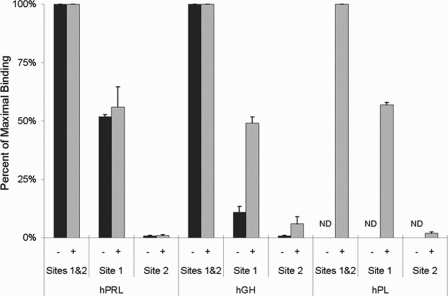
Each of the three lactogenic hormones were attached to a CM5 chip through an engineered free cysteine at a site located within either site 1, site 2, or at a site distal to either binding site. A saturating concentration of hPRLr (100 μm) was flowed over each hormone in buffer containing either 3 mm EDTA (HBS-EP buffer) or 15 μm Zn2+ (HBS-Zn buffer). The total hPRLr bound by each hormone when attached at a site distal to sites 1 and 2 (Cys-158 hPRL, Cys-152 for hGH or hPL) was defined as maximum binding for hPRL, hGH, and hPL (corresponding to a receptor:hormone stoichiometry of 2:1), and the relative binding of hPRLr to the hormones attached through either site 1 or site 2 was expressed as a percentage of this maximum binding value.
When only site 1 of hPRL was available for receptor binding (hPRL coupled through residue 129), slightly greater than 50% (52–56%) of the maximum binding was observed, corresponding to a stoichiometry of ∼1:1. However, when hPRL coupling exposed only site 2 (coupled through residue 181), only background levels (1%) (Fig. 3) of hormone were bound by hPRLr. The presence or absence of Zn2+ failed to meaningfully influence the stoichiometry of receptor binding at site 1of hPRL or remedy the inability of hPRLr to bind site 2 without the availability of site 1 to hPRLr.
FIGURE 3.
Relative binding by hPRL, hGH, or hPL with hPRLr binding only at site 1, site 2, or both site 1 and 2. hPRL, hGH, and hPL were coupled to an SPR chip through residues that expose only site 1 (G129C, G120C, or G120C, respectively), only site 2 (K181C, K172C, or K172C, respectively), or both sites 1 and 2 (M158C, N152C, or N152C, respectively). Relative binding was performed with (+) or without (−) 15 μm ZnSO4. In individual experiments with hormones coupled through a site spatially removed from either sites 1 or 2 (M158C or N152C), the response units corresponding to saturating injections of hPRLr were defined as 100%, and the response units for experiments with hormones coupled within either sites 1 or 2 were expressed as a percentage relative to this value. Data from individual experiments (as well as data not detected, ND) were averaged, and the S.D. among the experiments was calculated.
The relationship between observed stoichiometry and Zn2+ availability for hGH and hPL was quite different from that of hPRL. In the absence of Zn2+, the binding of hPRLr to site 1 of hGH (coupled through site 2 residue 120) was substantially below (11%) the expected 1:1 stoichiometry at saturation, whereas in the presence of Zn2+ a 1:1 stoichiometry (49%) was observed (Fig. 3). Thus, binding of hPRLr to site 1 of hGH is dependent on the presence of Zn2+. When hGH was coupled to the chip through site 1, leaving only site 2 available for binding, meaningful hPRLr binding was not observed in the absence of Zn2+. In the presence of Zn2+ a small but measurable amount of receptor bound (6%), but a 1:1 stoichiometry was not approached. These data indicate that under optimal binding conditions in the presence of zinc, a 1:1 stoichiometry was observed when only site 1 of hGH was available for receptor binding, and very little binding was observed when only site 2 was available.
None of the three forms of hPL bound significant amounts of hPRLr in the absence of Zn2+, but in the presence of Zn2+ hPRLr binding was readily observed (Fig. 3). When only site 1 of hPL was exposed (coupled through site 2), the stoichiometry was slightly greater than 1:1 (57%), but little hPRLr binding occurred (2%) when only site 2 was exposed (coupled through site 1). Thus, hPRLr binding to site 1 of hPL is dependent on Zn2+. These binding stoichiometries obtained with saturating hormone concentrations echo those observed for hPRL and hGH in the presence of Zn2+.
Coupling each of the lactogenic hormones through a cysteine distal from either sites 1 or 2 resulted in the highest stoichiometric ratios of hPRLr binding for each of the three hormones. When receptor binding was limited to site 1, the ratios of hPRLr bound to each of the hormones approached 1:1 under optimal zinc conditions. Finally, when only site 2 was available for binding, the ratios of hPRLr bound to hormone were either close to zero or significantly less than a 1:1 receptor:hormone ratio, independent of Zn2+ availability. The presence or absence of Zn2+ does not influence the receptor binding ratios for hPRL, but the availability of Zn2+ enhances receptor binding for hGH and is necessary for receptor binding to hPL.
SPR Kinetic Studies
Kinetic SPR titration studies were conducted for each hormone in the presence and absence of Zn2+ to determine the hormone site 1 and global affinities for hPRLr. When kinetic studies were performed using hormones coupled to the surface of SPR chips through site 2, significant hPRLr binding was observed at site 1 of the hormones (Table 1, upper data). The affinity of hPRL for hPRLr at site 1 was decreased 35-fold in the presence of 15 μm Zn2+. This Zn2+-induced change in affinity was mediated by a 5-fold decrease in the association rate constant and a 9-fold increase in the dissociation rate constant. The site 1 Kd of hGH for hPRLr was reduced 2166-fold in the presence of Zn2+, brought about by a combination of a 7-fold increase in the association rate constant and a 145-fold increase in the dissociation rate constant. Finally, in the absence of Zn2+ hPL binding to the hPRLr at site 1 was not detected, but in the presence of 15 μm Zn2+, hPL bound the hPRLr with high affinity (Kd = 2.56 × 10−8 m). The association rate constant for hPL was within the range of those observed for hPRL and hGH in the presence of zinc, and the high affinity of hPL was characterized by slow dissociation.
TABLE 1.
Rate and affinity constants of hPRLr binding to hPRL, hGH, and hPL with or without zinc
| Site 1 binding when hormone is coupled through site 2b,c |
||||||
|---|---|---|---|---|---|---|
| [Zn2+] | k1 | k−1 | Kd | Kd CVa | ||
| μm | 1/ms | 1/s | m | % | ||
| hPRL G129C | 0 | 3.42 × 104 | 1.10 × 10−3 | 3.22 × 10−8 | 10 | |
| hPRL G129C | 15 | 6.38 × 103 | 1.01 × 10−2 | 1.58 × 10−6 | 100 | |
| hGH G120C | 0 | 1.77 × 103 | 4.22 × 10−6 | 2.38 × 10−9 | 130 | |
| hGH G120C | 15 | 2.51 × 102 | 2.91 × 10−4 | 1.16 × 10−6 | 80 | |
| hPL G120C | 0 | |||||
| hPL G120C | 15 | 2.46 × 103 | 6.30 × 10−5 | 2.56 × 10−8 | 30 | |
| Composite site 1 and 2 binding when hormone is coupled through distal from sites 1 and 2b,c |
||||||
|---|---|---|---|---|---|---|
| [Zn2+] | ka | kd | KDglobal | KDsite1 | Kd CVa | |
| μm | 1/ms | 1/s | m | m | % | |
| hPRL M158C | 0 | 1.76 × 104 | 1.21 × 10−3 | 6.88 × 10−8 | 6.20 × 10−8 | 40 |
| hPRL M158C | 15 | 2.35 × 103 | 6.11 × 10−3 | 2.60 × 10−6 | 2.58 × 10−6 | 60 |
| hGH N152C | 0 | 3.17 × 103 | 1.97 × 10−2 | 6.21 × 10−6 | 5.76 × 10−6 | 30 |
| hGH N152C | 15 | 2.56 × 103 | 1.06 × 10−4 | 4.14 × 10−8 | 3.88 × 10−7 | 160 |
| hPL N152C | 0 | NA | NA | NA | NA | |
| hPL N152C | 15 | 5.09 × 103 | 9.73 × 10−6 | 1.91 × 10−9 | 1.49 × 10−9 | 60 |
a S.D. and coefficients of variation (CV) were calculated for data from individual experiments and indicate that a several-fold change in KD values are significantly different.
b Kinetic data from two to five independent experiments were used to calculate rate and dissociation constants. Data shown at the top are for hormones that can only bind one hPRLr. Data were analyzed by a 1:1 Langmuir model. Data shown at the bottom are for hormones that can bind two hPRLr. Rate and dissociation constants were calculated by either a 1:1 Langmuir model (ka, kd, and KDglobal) or a sequential affinity model (see KDSite1) (rate constants are not presented).
c The Kd values have been calculated from the ratios of the averaged rate constants. NA = not available.
C158 hPRL coupled to SPR chips permits binding at both receptor binding sites 1 and 2. Data fit to a 1:1 Langmuir binding mechanism describe a global affinity constant. This value combines binding information from sites 1 and 2 and were within severalfold (Table 1, lower data) of the affinity constants obtained for binding at site 1 alone in either the presence or absence of zinc (Table 1, upper data). This suggests that despite sufficient hPRLr concentrations to bind both sites 1 and 2, the binding at site 2 was not sufficiently different from that of site 1 to dramatically change the global affinity constant from that of site 1. Furthermore, when C158 hPRL binding data were analyzed by a model where hPRLr sequentially binds hPRL (Table 1, lower data, KDSite1), the site 1 rate constants and affinity were also similar to those when only measuring site 1 binding (Table 1, upper data) or the global affinity constant.
A different picture is observed for hGH binding to hPRLr. The binding of hPRLr only to site 1 of hGH (Table 1, upper data) shows a strong affinity that is decreased by 22-fold with the addition of 15 μm Zn2+. Thus, the binding of Zn2+ to the half-site residues within site 1 (His-18, His-21, Glu-174) shapes a weaker binding site 1. When Cys-152 hGH is bound by hPRLr, both sites 1 and 2 are available for binding. When these binding data are analyzed by either a 1:1 Langmuir model to calculate a global affinity constant or by a sequential binding model, the affinities are modest in the absence of Zn2+, but both become stronger in the presence of Zn2+, similar to observations made by others. Thus, when binding data from site 2 are included in the rate calculations, a different picture of binding is observed. This suggests that site 2 binding is quite different from that of site 1 for hGH. In the absence of Zn2+, the dissociation rate constant for site 1 of C120 hGH is very slow when compared with the dissociation rate constant observed with Cys-152 hGH where both hPRLr binding sites are available. The addition of Zn2+ removes the differences of dissociation rate constants observed between Cys-120 and Cys-152 hGH.
Finally, in the presence of Zn2+, hPL shows significant binding to hPRLr. Site 1 affinity of Cys-120 hPL was an order of magnitude weaker than that of Cys-152 hPL when data were calculated as a single/global affinity. This difference was largely due to a reduction in the dissociation rate constant when both hPRLr binding sites were available.
FRET Studies
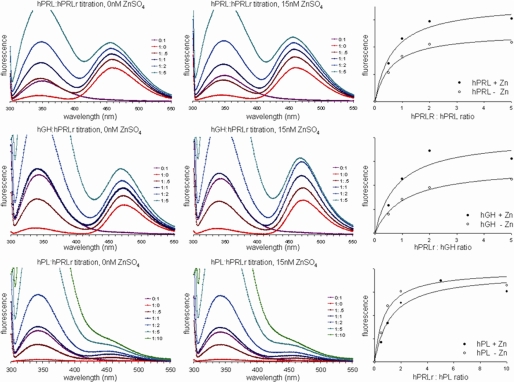
FRET studies were designed to evaluate the effect of hPRLr binding on the conformation of lactogenic hormones as discerned by measuring the change in energy transfer between an internal tryptophan and a fluorochrome coupled at a site structurally removed from either sites 1 or 2 (158C hPRL or 152C hGH and hPL). hPRL contains tryptophan residues at positions 91 and 150, whereas hGH and hPL each contain a single tryptophan at position 86. Based on NMR structures of hPRL (PDB code 1RW5), the distances between the α-carbon of tryptophan 91 and 150 and residue 158 (the site of CPM conjugation) were ∼21 and 16 Å, respectively. In hGH (PDB code 1HGU), the distance between Trp-86 and the site of CPM attachment is ∼23 Å, and the distance between these points in hPL is ∼20 Å. Distances between the hormone-bound CPM and tryptophans in the bound hPRLr are much greater than those found within the hormone and are larger than the 31 Å Förster distance (R°) for CPM. The resonance energy transfer from the hormone tryptophans will be the predominant species contributing to energy transfer and receptor binding, and any subsequent rearrangement of receptor tryptophan will produce little FRET. If hPRLr binding at site 1 is required for the functional organization of site 2 of lactogenic hormones, then a binding-induced conformation change will be observed within the CPM-labeled hormones upon titration with hPRLr.
After a brief incubation to achieve binding equilibrium, measurements were taken from 300 to 550 nm on samples containing molar ratios from 0:1 to either 5:1 or 10:1 (hPRLr:hormone) to measure emission from both tryptophan and CPM (Fig. 4). Increasing CPM emissions at 470 nm, when corrected for the contributions from tryptophan emission, described a dose-dependent signal. When the CPM emissions at 470 nm were plotted against increasing ratios of CPM-labeled hormone to hPRLr, the curves described a saturation process, indicating that these data monitored a saturating binding reaction. In addition, the binding reactions approached saturation at a molar ratio of ∼2:1 (hPRLr:hormone) for each hormone, an observation consistent with the stoichiometry that we and others have predicted for the activated lactogenic hormone-receptor complex. The increases in 470-nm CPM emissions associated with increased hPRLr binding indicated that the mean distances between tryptophan and CPM within each hormone were reduced with increased hPRLr binding up to a molar ratio of ∼2:1 (hPRLr:hormone).
FIGURE 4.
Titration of hPRLr with coumarin-labeled hPRL, hGH, or hPL monitored by FRET measurements in the absence or presence of Zn2+.
DISCUSSION
Binding Stoichiometries
The selection of specific residues for hormone coupling to either the SPR chip or CPM was based upon and supported by previous structural and functional studies. Svensson et al. (7) recently showed that Lys-181 is in the very center of the structural epitopes that comprise site 1 of hPRL. Similar structural work by Somers et al. (11) showed that coupling hGH through residue 172 also links the dextran fiber in the center of site 1. Coupling through these residues would place a dextran fiber adjacent to the critical binding epitopes of Asp-171 and Trp-127 in hGH and hPRL, respectively (26–28). The case for hPL relies on the data of Walsh and Kossiakoff (29) that showed hPL Lys-172 was located in a similar position to that of hGH Lys-172 and when mutated to alanine showed a large ΔΔG for site 1 binding of hPRLr, indicating that the dextran linkage at this residue likely would eliminate site 1 receptor binding within hPL. Taken together, the coupling procedures employed in this work should eliminate receptor binding at site 1. Blocking site 2 by linking the dextran fibers at residues 120 or 129 in hGH and hPL or in hPRL, respectively, was based on structural and functional data. Modification of these residues from glycine to more bulky arginine produced hormone antagonists that blocked binding at site 2 (30, 31). Again, integrating structural and functional information indicates that these residues are located within the functional epitopes that comprise site 2 (26–31). Finally, linkage of the lactogens to the dextran fibers of the chip through Asn-152 or Met-158 in hGH and hPL or in hPRL, respectively, showed that this location is in the C-terminal portion of the loop connecting helices 3 and 4, spatially removed from the functional epitopes that bind the receptor (5, 6, 27, 32). Thus, this is a good location for covalent attachment to the chip or CPM linkage where the modifications will not influence either site 1 or 2 binding by receptor. Finally, spectroscopic data (supplemental Fig. 2) demonstrate that placement of cysteine mutations at these various locations has not altered proper folding of these proteins.
SPR binding experiments demonstrated for all three hormones that when site 1 is blocked (achieved by covalently binding a dextran polymer at either residue 181 of hPRL or 172 of hGH and hPL) and site 2 remains available, no significant hPRLr binding occurs even at concentrations that are orders of magnitude in excess of concentrations found in biological fluids. However, for all three hormones lacking an accessible site 2 (by covalently binding a dextran polymer at either Cys-129 of hPRL or Cys-120 of hGH and hPL) and containing an available site 1, hPRLr binding was readily observed at nanomolar concentrations, and the binding stoichiometry approached 1:1 at saturation. These experiments demonstrate that hPRLr does not independently bind sites 1 and 2 of lactogenic hormones and instead support an obligate ordered binding model (Fig. 1) in which hPRLr binding at site 1 is required for binding to take place at site 2. These observations were consistent for all lactogenic hormones despite significant differences in sequence and structure. Although a previous report from our laboratory described the ordered binding mechanism for hPRL (9), this report expands this model of ordered receptor binding to include all lactogenic cytokines.
The inability of hPRL to bind hPRLr at site 2 when site 1 is unoccupied requires consideration. First, the S2/S2 receptor binding surface is not present when the hormones are blocked at site 1. Data from mutagenic studies for hGH binding to the extracellular domain of the hGH receptor reveals that the free energy for receptor binding at site 2 is approximately equally distributed between the hGH/receptor and the S2/S2 binding interfaces of the receptor (33, 35). Similar data are not available for any of the three hormones when bound to the hPRLr; but assuming a similar free energy distribution between the two site 2 binding interfaces, hPRLr binding should be observed during SPR experiments. This is not observed for any of the three hormones when blocked through site 1 and treated with hPRLr. A second rationalization for the observed data is that hPRLr binding at site 1 of the hormone induces a conformation change that shifts the collection of structural conformers into those favoring hPRLr binding at site 2. Thus, we chose to determine whether hPRLr binding could induce a conformation change within any of the three hormones.
Conformational Changes
We utilized FRET to determine the ability of subsaturating concentrations of hPRLr to induce observable changes in conformation within the structure of lactogenic hormones. FRET data showed an increased signal with increasing subsaturating ratios of hPRLr for each of the three lactogenic hormones. The addition of hPRLr above the predicted saturating 2:1 ratio produced little additional increase in FRET signals (Fig. 4). The increases in FRET intensity indicate that the mean distance between the hormone tryptophan(s) and CPM are reduced as a result of hPRLr binding, revealing a change in conformation of the protein upon receptor binding. Together with the understanding that each of these hormones binds hPRLr at site 1 before binding at site 2, this increasing FRET signal up to stoichiometric saturation reveals a change in conformation upon binding at site 1 and may indicate further conformation change with site 2 hPRLr binding. Both the ordered binding demonstrated in SPR experiments and a binding-induced conformational change as demonstrated by FRET experiments are consistent with a conformationally regulated obligate ordered binding mechanism for each of the three lactogenic hormones. This interpretation extends and strengthens the data we have previously published for hPRL (9). Additionally, Walsh et al. (10) have recently described a similar allosteric coupling of the two human growth hormone receptor binding sites in hGH, suggesting that a similar mechanism may govern hGH receptor binding. We have recently identified contiguous residues in hGH (15)- and hPRL (34)-constituting motifs that, when mutated, uncouple sites 1 and 2. These studies identify the structural features that are necessary for efficient propagation of the site 1 receptor binding-induced conformation change in these two hormones. Taken in aggregate, we believe that these data describe a structurally driven mechanism for the observed ordered binding of lactogenic hormones to the hPRLr that is not contingent upon different affinities for the respective binding sites.
Zn2+ elicited different effects on the hPRLr binding of hPRL, hGH, and hPL. Zn2+ weakened the site 1 affinities of both hPRL and hGH, and this report is the first to demonstrate that zinc directly affects receptor binding in hPRL and is the first to demonstrate that the effects of Zn2+ on site 1 binding of hGH. In contrast, Zn2+ positively affected hGH binding to hPRLr when both sites 1 and 2 were available; this observation is consistent with previous reports (35, 36) where Zn2+ increased the affinity. Finally, Zn2+ was an absolute requirement for hPRLr to bind to hPL. Although our qualitative findings are in line with previous reports, many of the seminal quantitative findings in this area of research were determined by using radiochemical-tagged ligand-based assays to follow binding. Our work determines kinetics and affinities by measuring direct, real-time hormone-receptor interactions through SPR and the hPRL data described here and agrees well with other published findings using a similar experimental design that couples hormone to the surface of an SPR chip (9).
Finally, the absolute requirement for Zn2+ demonstrated for hPL is consistent with previous reports (29, 37). The differences in the effects of Zn2+ on hGH and hPL are striking, particularly when considering that hGH and hPL share ≥85% sequence homology and the residues involved in Zn2+ binding are conserved in both hGH and hPL (His-18, His-21, and Glu-174). Although analogous zinc binding residues are present in hPRL (His-27, His-30, and Asp-183), the effect of zinc on hPRLr binding is opposite that observed for hPL. The amino acids that surround these critical zinc binding residues are not well conserved between hPRL and the more homologous hGH and hPL. Structural comparisons of the hormones also show that the distances between His-27 and Asp-183 in unbound hPRL are large (12 Å) (5, 6) compared with the distance between the analogous His-18 and Glu-174 in unbound hGH and hPL (4.3 and 3 Å, respectively) (29, 32). The exceptional distance between purported zinc binding residues measured in hPRL NMR structural studies was determined in the absence of zinc (5, 6), and any coordination by these residues would necessarily be accompanied by structural rearrangement sufficient to bring these residues adequately near one another to create the zinc binding half-site. In other work we have shown that hPRL,3 hGH (38), and bovine PRL (39) undergo zinc-dependent conformation changes.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01-DK72275.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
J. L. Voorhees and C. L. Brooks, manuscript in preparation.
- hPRL
- human prolactin
- hPRLr
- hPRL receptor
- SPR
- surface plasmon resonance
- hGH
- growth hormone
- hPL
- placental lactogen
- FRET
- Förster resonance energy transfer
- CPM
- 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin.
REFERENCES
- 1.Huising M. O., Kruiswijk C. P., Flik G. (2006) J. Endocrinol. 189, 1–25 [DOI] [PubMed] [Google Scholar]
- 2.Forsyth I. A. (1986) J. Dairy Sci. 69, 886–903 [DOI] [PubMed] [Google Scholar]
- 3.Hooper K. P., Padmanabhan R., Ebner K. E. (1993) J. Biol. Chem. 268, 22347–22352 [PubMed] [Google Scholar]
- 4.Lebrun J. J., Ali S., Sofer L., Ullrich A., Kelly P. A. (1994) J. Biol. Chem. 269, 14021–14026 [PubMed] [Google Scholar]
- 5.Teilum K., Hoch J. C., Goffin V., Kinet S., Martial J. A., Kragelund B. B. (2005) J. Mol. Biol. 351, 810–823 [DOI] [PubMed] [Google Scholar]
- 6.Keeler C., Dannies P. S., Hodsdon M. E. (2003) J. Mol. Biol. 328, 1105–1121 [DOI] [PubMed] [Google Scholar]
- 7.Svensson L. A., Bondensgaard K., Nørskov-Lauritsen L., Christensen L., Becker P., Andersen M. D., Maltesen M. J., Rand K. D., Breinholt J. (2008) J. Biol. Chem. 283, 19085–19094 [DOI] [PubMed] [Google Scholar]
- 8.Jomain J. B., Tallet E., Broutin I., Hoos S., van Agthoven J., Ducruix A., Kelly P. A., Kragelund B. B., England P., Goffin V. (2007) J. Biol. Chem. 282, 33118–33131 [DOI] [PubMed] [Google Scholar]
- 9.Sivaprasad U., Canfield J. M., Brooks C. L. (2004) Biochemistry 43, 13755–13765 [DOI] [PubMed] [Google Scholar]
- 10.Walsh S. T., Sylvester J. E., Kossiakoff A. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17078–17083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Somers W., Ultsch M., De Vos A. M., Kossiakoff A. A. (1994) Nature 372, 478–481 [DOI] [PubMed] [Google Scholar]
- 12.Schiffer C., Ultsch M., Walsh S., Somers W., de Vos A. M., Kossiakoff A. (2002) J. Mol. Biol. 316, 277–289 [DOI] [PubMed] [Google Scholar]
- 13.Kossiakoff A. A. (2004) Adv. Protein Chem. 68, 147–169 [DOI] [PubMed] [Google Scholar]
- 14.Peterson F. C., Anderson P. J., Berliner L. J., Brooks C. L. (1999) Protein Expression Purif. 15, 16–23 [DOI] [PubMed] [Google Scholar]
- 15.Duda K. M., Brooks C. L. (2003) J. Biol. Chem. 278, 22734–22739 [DOI] [PubMed] [Google Scholar]
- 16.Lennon G., Auffray C., Polymeropoulos M., Soares M. B. (1996) Genomics 33, 151–152 [DOI] [PubMed] [Google Scholar]
- 17.Kunkel T. A., Bebenek K., McClary J. (1991) Methods Enzymol. 204, 125–139 [DOI] [PubMed] [Google Scholar]
- 18.Wang W., Malcolm B. A. (1999) Biotechniques 26, 680–682 [DOI] [PubMed] [Google Scholar]
- 19.Sanger F., Nicklen S., Coulson A. R. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 21.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duda K. M., Brooks C. L. (1999) FEBS Lett. 449, 120–124 [DOI] [PubMed] [Google Scholar]
- 23.O'Brien J., Wilson I., Orton T., Pognan F. (2000) Europ. J. Biochem. 267, 5421–5426 [DOI] [PubMed] [Google Scholar]
- 24.Bewley T. A., Li C. H. (1969) Int. J. Protein Res. 1, 117–124 [DOI] [PubMed] [Google Scholar]
- 25.Doneen B. A., Bewley T. A., Li C. H. (1979) Biochemistry 18, 4851–4860 [DOI] [PubMed] [Google Scholar]
- 26.Cunningham B. C., Wells J. A. (1989) Science 244, 1081–1085 [DOI] [PubMed] [Google Scholar]
- 27.Goffin V., Struman I., Mainfroid V., Kinet S., Martial J. A. (1994) J. Biol. Chem. 269, 32598–32606 [PubMed] [Google Scholar]
- 28.Kinet S., Bernichtein S., Kelly P. A., Martial J. A., Goffin V. (1999) J. Biol. Chem. 274, 26033–26043 [DOI] [PubMed] [Google Scholar]
- 29.Walsh S. T., Kossiakoff A. A. (2006) J. Mol. Biol. 358, 773–784 [DOI] [PubMed] [Google Scholar]
- 30.Fuh G., Cunningham B. C., Fukunaga R., Nagata S., Goeddel D. V., Wells J. A. (1992) Science 256, 1677–1680 [DOI] [PubMed] [Google Scholar]
- 31.Chen W. Y., Ramamoorthy P., Chen N., Sticca R., Wagner T. E. (1999) Clin. Cancer Res. 5, 3583–3593 [PubMed] [Google Scholar]
- 32.de Vos A. M., Ultsch M., Kossiakoff A. A. (1992) Science 255, 306–312 [DOI] [PubMed] [Google Scholar]
- 33.Bernat B., Pal G., Sun M., Kossiakoff A. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 952–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uivaprasad U. (2003) The Mechanism of Lactogen Receptor Binding by Human Prolactin Ph.D. dissertation, The Ohio State University [Google Scholar]
- 35.Cunningham B. C., Bass S., Fuh G., Wells J. A. (1990) Science 250, 1709–1712 [DOI] [PubMed] [Google Scholar]
- 36.Dattani M. T., Hindmarsh P. C., Brook C. G., Robinson I. C., Kopchick J. J., Marshall N. J. (1995) J. Biol. Chem. 270, 9222–9226 [DOI] [PubMed] [Google Scholar]
- 37.Lowman H. B., Cunningham B. C., Wells J. A. (1991) J. Biol. Chem. 266, 10982–10988 [PubMed] [Google Scholar]
- 38.Duda K. M., Brooks C. L. (2003) Protein Eng. 16, 531–534 [DOI] [PubMed] [Google Scholar]
- 39.Permyakov E. A., Veprintsev D. B., Deikus G. Y., Permyakov S. E., Kalinichenko L. P., Grishchenko V. M., Brooks C. L. (1997) FEBS Lett. 405, 273–276 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.