Abstract
Photosystem I (PSI) is a multiprotein complex consisting of the PSI core and peripheral light-harvesting complex I (LHCI) that together form the PSI-LHCI supercomplex in algae and higher plants. The supercomplex is synthesized in steps during which 12–15 core and 4–9 LHCI subunits are assembled. Here we report the isolation of a PSI subcomplex that separated on a sucrose density gradient from the thylakoid membranes isolated from logarithmic growth phase cells of the green alga Chlamydomonas reinhardtii. Pulse-chase labeling of total cellular proteins revealed that the subcomplex was synthesized de novo within 1 min and was converted to the mature PSI-LHCI during the 2-h chase period, indicating that the subcomplex was an assembly intermediate. The subcomplex was functional; it photo-oxidized P700 and demonstrated electron transfer activity. The subcomplex lacked PsaK and PsaG, however, and it bound PsaF and PsaJ weakly and was not associated with LHCI. It seemed likely that LHCI had been integrated into the subcomplex unstably and was dissociated during solubilization and/or fractionation. We, thus, infer that PsaK and PsaG stabilize the association between PSI core and LHCI complexes and that PsaK and PsaG bind to the PSI core complex after the integration of LHCI in one of the last steps of PSI complex assembly.
Keywords: Chloroplast, Electron Transfer, Photosynthesis, Protein Assembly, Protein Purification, Photosystem, Reaction Center Complex
Introduction
Photosynthetic electron transfer from water to NADP+ in oxygenic photosynthetic organisms is driven by two photosystems, photosystem I (PSI)2 and photosystem II (PSII), operating in series. PSII oxidizes water to O2 and protons, and the electrons extracted from water are transferred to plastoquinone present in the lipid bilayer of the thylakoid membranes, producing plastoquinol. The electrons on the plastoquinol are then transferred to plastocyanin or cytochrome c6 through cytochrome b6f. PSI catalyzes the electron transfer from plastocyanin or cytochrome c6 to NADP+ through ferredoxin and ferredoxin:NADP+ oxidoreductase.
PSI consists of a number of subunits and more than 100 cofactors, including chlorophyll a, β-carotene, naphthoquinone, lipid, and Fe-S clusters (1). PSI harvests light, photochemically converts excitons to redox components, and stabilizes the resulting charge separation. Ten PSI subunits (PsaA-PsaF and PsaI-PsaL) are conserved among plants, algae, and Cyanobacteria (1). In addition to those core subunits, higher plants and algae possess PsaG, PsaH, PsaN, and PsaO, whereas Cyanobacteria contain PsaM, and thermophilic Cyanobacteria additionally contain PsaX (1, 2). The structure of the PSI complex has been determined in the cyanobacterial PSI core complex at 2.5 Å resolution (2) and in the plant PSI-LHCI supercomplex at 3.4 or 3.3 Å resolution (3, 4). Although the basic structure is highly conserved in the cyanobacterial and plant PSI core complexes, higher plant and algal PSI core complexes intimately associate with peripheral light-harvesting complexes (LHCI) to form a PSI-LHCI supercomplex, and the cyanobacterial PSI core is trimeric (2–4).
The PSI reaction center (RC) consisting of a heterodimer of homologous polypeptides PsaA and PsaB is surrounded by several peripheral subunits; PsaC, PsaD, and PsaE are present on the stromal side. PsaC contains two Fe-S terminal electron acceptors, FA and FB, and associates with PsaD and PsaE to form a docking site for ferredoxin. PsaF has a single transmembrane helix and a hydrophilic domain extended on the lumenal side that is required for the efficient electron transfer from plastocyanin to P700 in plants and algae. A small hydrophobic PsaJ with a single transmembrane helix is present in proximity of PsaF, and their two transmembrane helices are present between the PSI RC and LHCI. On the lumenal side, PsaN is also present. PsaH, PsaI, PsaL, and probably PsaO are bound to the PSI RC on the opposite peripheral side of the LHCI oligomer (5). PsaK and PsaG, which contain two transmembrane helices, are pseudo-symmetrically present at either edge of the LHCI oligomer (3, 4).
Because the green alga Chlamydomonas reinhardtii is amenable to biochemical characterization and pulse-chase protein labeling, it is an excellent model organism for the study of structure, function, and biogenesis of photosynthetic components. It contains 4 chloroplast-encoded PSI genes (psaA, psaB, psaC, and psaJ) and 10 nuclear-encoded PSI genes (psaD, psaE, psaF, psaG, psaH, psaI, psaK, psaL, psaN, and psaO) as well as 9 Lhca genes (Lhca1-Lhca9) (6–9). Single particle analysis shows that PSI complexes from C. reinhardtii are larger than those from higher plants but are similarly shaped (10, 11).
Assembly of the PSI complex proceeds in many steps during which a number of subunits and cofactors become integrated into a functional structure (12, 13). The detailed molecular mechanism of PSI complex assembly, however, remains elusive. It is proposed that PsaB is synthesized first and acts as an acceptor for the subsequent PSI subunits integration (14, 15). PsaA integrates with PsaB to form a PSI RC complex, and PsaC is subsequently assembled on the stromal side of PSI RC (16). The three peripheral subunits, PsaC, PsaD, and PsaE, successively assemble on the stromal side (17, 18), but how the other subunits integrate is not clear.
Several factors are involved in PSI complex assembly. Chloroplast-encoded Ycf3, which is conserved in Cyanobacteria, interacts with PsaA and PsaD and is essential for PSI complex assembly (19, 20). Another chloroplast-encoded protein, Yc4, that is also conserved in Cyanobacteria, is essential for PSI complex in Chlamydomonas (19). Ycf4 is also important for cyanobacterial PSI complex assembly (21). Chlamydomonas Ycf4 forms a large complex that associates with newly synthesized PsaA, PsaB, PsaC, PsaD, PsaE, and PsaF (22), and this complex may be involved in an initial step of PSI complex assembly (23). Pyg7-1, a nuclear-encoded factor in higher plants that is conserved in Cyanobacteria as Ycf37, is essential for PSI complex assembly and is important but not essential for cyanobacterial PSI complex assembly (24, 25).
Here, by applying sucrose density gradient ultracentrifugation to C. reinhardtii chloroplasts, we separated the chlorophyll protein complexes from solubilized thylakoid membranes (26). In addition to the PSI complex, which separated as a stable PSI-LHCI supercomplex, we detected a small amount of a PSI subcomplex in the present study. Pulse-chase protein labeling experiments revealed that the PSI subcomplex was a newly synthesized assembly intermediate that was slowly converted to the mature PSI-LHCI complex by integrating PsaG and PsaK. We infer that these two subunits are involved in the efficient integration and stable association of LHCI to the PSI core complex. We also hypothesize a sequential PSI complex assembly mechanism.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
We grew C. reinhardtii wild type 137C cells to logarithmic (2–5 × 106 cells ml−1) or stationary (1.3–1.5 × 107 cells ml−1) growth phase in Tris acetate-phosphate medium (27) at 25 °C in light (50–100 microeinsteins m−2 s−1).
Separation of Chlorophyll-Protein Complexes
We isolated thylakoid membranes as described previously (28). We remove extrinsic proteins with 2 m NaBr from the thylakoid membranes, solubilized them (0.8 mg of chlorophyll ml−1) with 0.8% (w/v) n-dodecyl-β-d-maltoside (DM) and 50 mm Tricine-NaOH, pH 8.0, and separated the thylakoid extracts by sucrose density gradient ultracentrifugation on a swinging bucket rotor (SW41Ti at 200,000 × g for 14 h or SW28 at 100,000 × g for 24 h) (Beckman) as described previously (29) with some modifications. The gradient consisted of linear sucrose concentrations from 0.4 to 1.3 m and 0.05% DM. We then fractionated the entire gradient from the bottom to the top. We performed ion-exchange column chromatography (2.5 × 5 cm) with DEAE Toyopearl 650S (Tosoh, Tokyo) as described previously (7, 26) and eluted the fractions with a linear NaCl gradient of 0–150 mm in buffer containing 50 mm Tris-HCl, pH 8.0, and 0.05% DM. We performed gel filtration column chromatography with an fast protein liquid chromatography system using Superose 6 HR 10/30 (Amersham Biosciences) as described previously (30). The elution buffer contained 50 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 0.05% DM, and the flow rate was 0.2 ml min−1. All procedures were carried out at 4 °C.
SDS-Polyacrylamide Gel Electrophoresis and Immunoblotting
We performed SDS-PAGE as described previously (7) or with 6 m urea in a resolving gel (31). For immunoblotting, polypeptides were electrophoretically blotted onto nitrocellulose filters, probed with antibodies, and visualized by enhanced chemiluminescence (ECL). We detected signals with a LAS-4000 mini luminescent image analyzer (Fujifilm, Tokyo) and quantified them with MultiGauge Version 3.0 software (Fujifilm).
We generated an antibody against PsaG in rabbits using the oligopeptide CKTTGATYFEELQK, conjugated at the cysteine residue with the carrier protein, keyhole limpet hemocyanin (Sigma Genosys). We also generated an antibody against PsaJ obtained from overexpression of the chloroplast psaJ gene product. We amplified the entire coding region of the psaJ by polymerase chain reaction using chloroplast DNA as the template and a pair of oligonucleotides, 5′-CATATGAAAGATTTTACTACTTATTTATC-3′ and 5′-CATATGTTAAAATGAAAATACAAGTGGATC-3′ (underlines denote NdeI restriction sites at both ends of the amplified DNA fragment). We cloned the psaJ into a pET3X-c vector at the NdeI site in the appropriate orientation. We purified the overexpressed protein by preparative SDS-PAGE, homogenized the gel slices containing the protein, and injected the mixture into rabbits as described previously (26).
Pulse-Chase Labeling of Cellular Proteins
We performed pulse-chase labeling experiments of total cellular proteins as described previously (32) with some modification. Cells grown to mid-log phase (4 × 106 cells ml−1) in sulfur-reduced TAP medium were collected by centrifugation, resuspended at 25 μg of chlorophyll ml−1 in sulfur-free TAP medium, and incubated for 2 h. We labeled total cellular proteins with [35S]Na2SO4 (GE Healthcare) at 50 μCi ml−1 under low light for 1 min and stopped the labeling by adding cycloheximide (10 μg ml−1) and chloramphenicol (100 μg ml−1) and immediately freezing the cells. We performed chase experiments in the presence of 10 mm Na2SO4 and chloramphenicol (100 μg ml−1) for 2 h. We lysed the cells by vigorously vortexing them with glass beads and purified the thylakoid membranes by discontinuous sucrose gradient ultracentrifugation. We washed the thylakoid membranes with 2 m NaBr, solubilized them with 0.8% DM, and fractionated the resulting extracts by sucrose density gradient ultracentrifugation (0.1–1.3 m sucrose) with a Beckman TLS-55 (259,000 × g for 3 h) (Beckman).
PSI Activity Measurements
We monitored the continuous light-induced P700 photo-oxidation of PSI preparations (10 μg of chlorophyll ml−1) with a Shimadzu UV-2450 spectrophotometer (Kyoto, Japan) in the presence of 5 nm plastocyanin and 100 μm sodium ascorbate as described previously (33). Actinic light was passed through a 4-96 filter (Corning) at 100 microeinsteins m−2 s−1 with a cut-off VR-68 filter (Toshiba, Tokyo, Japan) and an interference filter (λo = 700 nm, Tmax = 82.4%, Δλ1/2 = 18.4 nm) (Nihonshinku, Tokyo) in front of the photomultiplier. We measured the NADP+ photoreduction activity of the PSI preparations (10 μg of chlorophyll ml−1) in the presence of 10 mm sodium ascorbate, 100 mm dichlorophenolindophenol, 2 mm NADP+, 10 mm MgCl2, ferredoxin, and ferredoxin:NADP+ oxidoreductase with a Shimadzu UV-2450 spectrophotometer. Actinic light was passed through a cut-off VY-50 filter (Toshiba) at 1300 microeinsteins m−2 s−1 with a band pass filter U340 (Hoya, Tokyo, Japan) in front of the photomultiplier. Plastocyanin, ferredoxin, and ferredoxin:NADP+ oxidoreductase were purified from extracts obtained after freeze-thaw lysing of Chlamydomonas cells. We purified plastocyanin and ferredoxin from the extracts by DEAE Toyopearl 650 S column chromatography followed by Macro-Prep ceramic hydroxylapatite Type I (Bio-Rad) column chromatography. We purified ferredoxin:NADP+ oxidoreductase from the extracts by DEAE Toyopearl 650 S chromatography followed by Toyopearl AF-Red-650ML (Tosoh) affinity chromatography.
RESULTS
Detection of a PSI Subcomplex on Sucrose Density Gradient
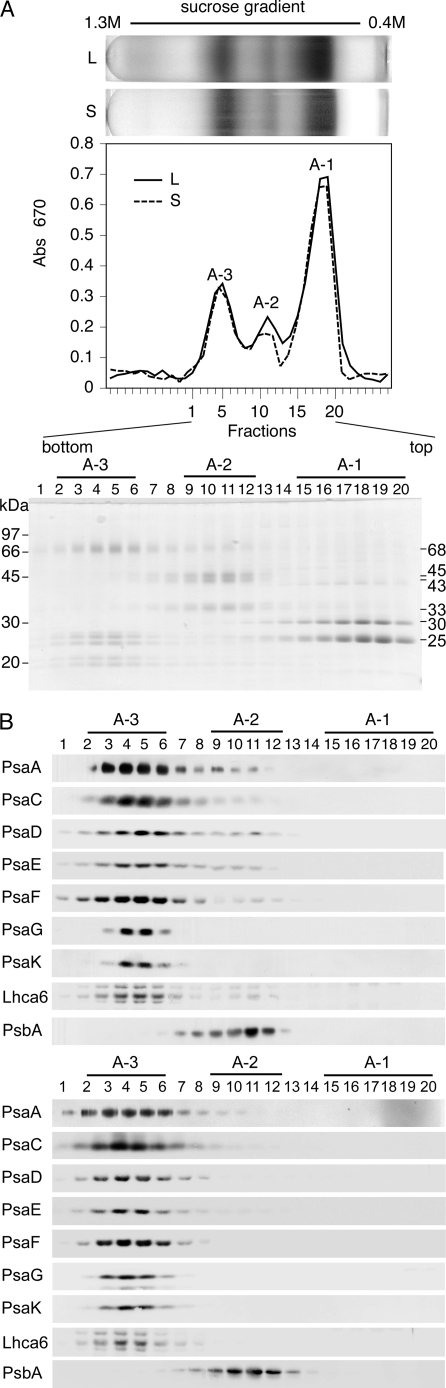
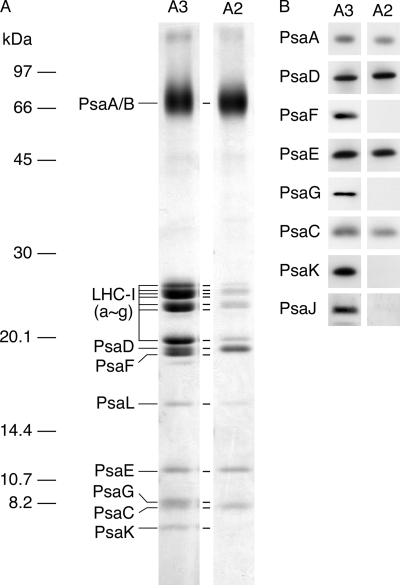
As we observed previously (26), sucrose density gradient ultracentrifugation of the DM extracts of thylakoid membranes separated three major chlorophyll protein complexes, A-1, A-2, and A-3, from the top to the bottom (Fig. 1A, upper and middle panels). The polypeptide profiles of the fractions from cells at logarithmic growth phase revealed that A-1, A-2, and A-3 contained mainly 25- and 30-kDa polypeptides of LHCII, 33-, 43-, and 45-kDa polypeptides of the PSII core complex, and 68-kDa polypeptides of the PSI reaction center (PsaA and PsaB) and several 20–27-kDa LHCI polypeptides (Fig. 1A, lower panel). We also detected a small quantity of PSI polypeptides in A-2, and immunoblotting analysis confirmed the presence of PSI polypeptides, PsaA, PsaC, PsaD, PsaE, and PsaF in A-2 (Fig. 1B, upper panel). We estimated that the amounts of PsaE and PsaF in fractions 8–12 in A-2 were 4 and 5% of PsaE and PsaF in the entire fractions, respectively. Because the PSI complex was smaller in A-2 than PSI-LHCI was in A-3 (see Fig. 3), the PSI complex in A-2 could be a subcomplex derived from partial dissociation of the PSI-LHCI separated in A-3. It is noteworthy that neither PsaG nor PsaK was detected in either A-2 or the upper gradient fractions where monomeric polypeptides were separated.
FIGURE 1.
Separation of chlorophyll-protein complexes. A, shown are separation and fractionation profiles of thylakoid extracts of wild type C. reinhardtii cells at logarithmic (L) and stationary (S) growth phase (upper panel). Polypeptide composition of chlorophyll-protein complexes of logarithmic growth phase cells (lower panel) is shown. The thylakoid membranes were solubilized with DM and separated by sucrose density gradient ultracentrifugation. Polypeptides of fractions 1–20, which included A-3, A-2, and A-1, were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. B, immunoblot analysis of fractions 1–20 from logarithmic (upper panel) and stationary (lower panel) growth phase cells using antibodies against PSI polypeptides (PsaA, PsaC, PsaD, PsaE, PsaF, PsaG, and PsaK), LHCI (Lhca6), and PSII polypeptide (PsbA) is shown. Abs, absorbance.
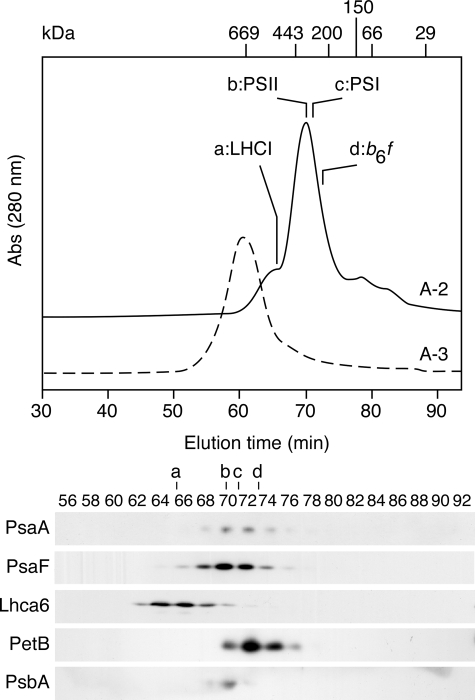
FIGURE 3.
Gel filtration chromatography of A-2 and A-3. A-2 (solid line) and A-3 (dashed line) separated on sucrose density gradient were subjected to gel filtration chromatography, and the resulting fractions were analyzed by immunoblotting using antibodies against PsaA, PsaF, PsbA (D1), Lhca6, and PetB (cytochrome b6). A single peak separated from A-3 (PSI-LHCI supercomplex) and was estimated to be 700 kDa. One peak and several shoulders were separated from A-2. LHCI (a) eluted as an early shoulder and was estimated to be 540 kDa. PSII (b) eluted in the peak and was estimated to be 430 kDa. PSI (330 kDa) (c) and Cyt b6f (300 kDa) (d) eluted late in the peak.
When the thylakoid membranes from cells at stationary phase were solubilized and fractionated, the peak height of A-2 was slightly reduced (Fig. 1A, middle panel) and the PSI polypeptides were barely detected or significantly reduced in A-2 (Fig. 1B, lower panel), indicating that the subcomplex accumulated only in logarithmic growth phase cells. Thus, we inferred that the subcomplex accumulated only in cells actively synthesizing the photosynthetic apparatus.
PSI Subcomplex in A-2 Is Newly Synthesized
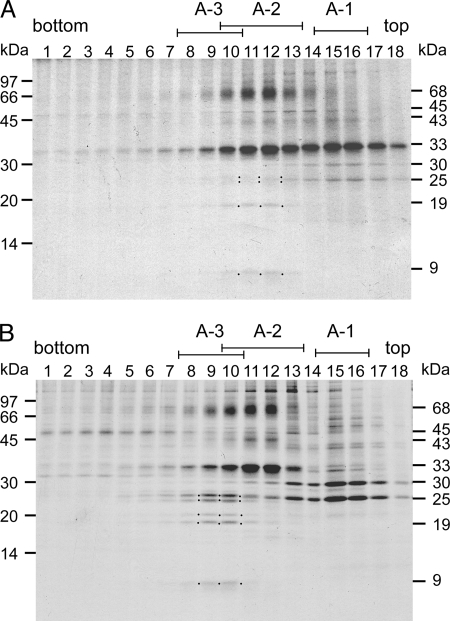
To address whether the PSI subcomplex in A-2 was a dissociated or newly synthesized subcomplex, we pulse-chase labeled total proteins of cells grown at logarithmic growth phase, isolated the thylakoid membranes, solubilized them with DM, and subjected them to sucrose density gradient ultracentrifugation. We fractionated the entire gradient, separated the labeled polypeptides by SDS-PAGE, and visualized them by autoradiography (Fig. 2A, upper panel). We detected a 33-kDa band, which corresponded to the PSII reaction center polypeptides, D1 and D2, mainly in A-1 and A-2. A 43-kDa band, which corresponded to the CP43 apoprotein, and a 45-kDa band, which corresponded to the CP47 apoprotein, were much less intensely labeled and were separated in a similar manner as D1 and D2. These observations suggest that a portion of the newly synthesized PSII polypeptides, which was separated in A-1, might not have been fully assembled in the mature PSII core complex. We detected several PSI polypeptides exclusively in A-2; a 68-kDa band (PSI reaction center polypeptides, PsaA and PsaB), a 19-kDa band (PsaD and PsaF), and a 9-kDa band (PsaC). We also detected weak bands of around 25 kDa, which corresponded to LHCI polypeptides, in A-2. These observations indicate that the newly synthesized PSI and LHCI polypeptides separated exclusively in A-2.
FIGURE 2.
Pulse-chase labeling of cellular proteins. C. reinhardtii wild type cells were pulse-labeled for 1 min with [35S]Na2SO4 (A) and subsequently chased for 2 h. B, thylakoid membranes were isolated, and chlorophyll protein complexes were separated as shown in Fig. 1. The polypeptides of each fraction were separated by SDS-PAGE, and the labeled polypeptides were visualized by autoradiography. Left, positions of molecular size markers. Right, sizes of the major labeled polypeptides: 68 kDa, PsaA and PsaB; 45 kDa, PsbB; 43 kDa, PsbC; 33 kDa, PsbA (D1) and PsbD (D2), 30 and 25 kDa, LHCII polypeptides; 19 kDa, PsaD and PsaF; 9 kDa, PsaC; dots, LHCI polypeptides, PsaD/F, and PsaC.
Fig. 2B shows the pulse-chase-labeled polypeptides. Labeled PSII polypeptides were detected exclusively in A-2, indicating that the pulse-labeled PSII polypeptides were fully integrated into the PSII core complex during the chase period. The 19-kDa band of PsaD and PsaF and the 9-kDa band of PsaC, in contrast, were detected exclusively in A-3. In addition, we observed the labeled LHCI polypeptides in A-3. We detected the 68-kDa band of PsaA and PsaB in A-3, although a similar band was still present in A-2. The 68-kDa band in A-2, however, was slightly up-shifted, corresponding to a D1-D2 aggregation. It is, therefore, possible that the 68-kDa band in A-2 comprised mostly aggregated forms of heavily labeled D1 and D2. These observations lead us to conclude that the newly synthesized PSI polypeptides were rapidly integrated into the PSI subcomplex in A-2 and were converted to the PSI-LHCI supercomplex in A-3, suggesting that the PSI subcomplex in A-2 was an assembly intermediate.
Subunit Composition of the PSI Subcomplex
On gel filtration column chromatography (Fig. 3), A-3 yielded a single peak containing the PSI-LHCI of apparently 700 kDa as we reported previously (30). A-2, in contrast, was heterogeneous; its peak contained the PSII core (430 kDa), the PSI subcomplex (330 kDa), and the cytochrome b6f (300 kDa), and a shoulder contained LHCI (540 kDa), indicating that LHCI remained oligomeric. The separation of the PSI subcomplex and the LHCI complex on gel filtration indicates that they were separate entities. We could not determine, however, whether they were also separate entities in the thylakoid membranes or whether they became dissociated during the solubilization or fractionation.
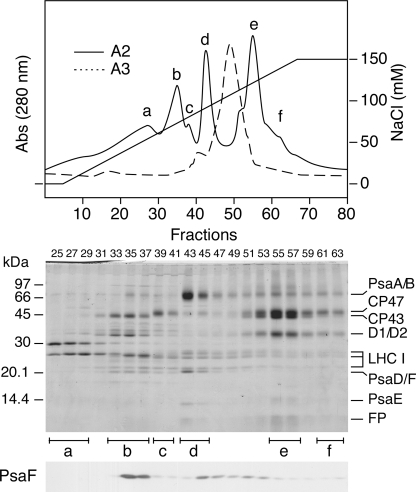
On DEAE chromatography (Fig. 4), A-3 mainly yielded a single peak that contained the PSI-LHCI, as expected. A-2, in contrast, yielded five peaks (a–e) and one shoulder (f), as previously reported (26). Based on their polypeptide composition, we identified the peaks and shoulder as followings; a, LHCII; b, CP43 and cytochrome b6f; c, CP47; d, PSI subcomplex; e, PSII core complex; f, PSII complex lacking CP43 (Fig. 4). As previously reported in PSI-deficient mutants (7), LHCI complex accumulated and remained oligomeric but eluted from the DEAE column at broad NaCl concentrations. Those observations suggest that the oligomeric LHCI complex that disconnected from the PSI complex had heterogeneous structure but was made up of similarly sized entities (Fig. 3). PsaF remained associated with the PSI subcomplex during sucrose density gradient ultracentrifugation (Fig. 1) but dissociated from it during DEAE chromatography and separated into fractions that eluted at NaCl concentrations that were lower (around fraction b) and higher (between fractions d and e) than the concentrations at which the PSI subcomplex was separated (Fig. 3).
FIGURE 4.
Separation of chlorophyll-protein complexes by DEAE chromatography. A-2 and A-3 were subjected to DEAE column chromatography with a buffer containing a linear gradient of NaCl (0–150 mm). One peak and one shoulder were resolved from A-3, whereas five peaks and one shoulder (a–f) were separated from A-2. a, LHCII; b, CP43 (PsbC) and cytochrome b6f; c, CP47; d, PSI subcomplex; e, PSII core complex; f, PSII complex lacking CP43. LHCI eluted broadly. The polypeptides of each fraction were separated by SDS-PAGE and stained with Coomassie Brilliant Blue or detected by immunoblotting using an antibody against PsaF (lower panels). Abs, absorbance. FP, free pigments.
Integration of PsaG and PsaK
Fig. 5 compares the polypeptides of the PSI subcomplex from A-2 with those of the PSI-LHCI supercomplex from A-3. The PSI-LHCI contained most of the PSI and LHCI polypeptides as shown previously (7). In contrast, the PSI subcomplex was deficient in PsaK, PsaG, PsaJ, and PsaF. A-2 lacked PsaK and PsaG and lost PsaF during DEAE chromatography (Fig. 4). The absence of LHCI might have destabilized the binding of PsaF and most probably PsaJ to the PSI subcomplex because the two polypeptides are localized between the PSI core and the oligomeric LHCI complex. Because the LHCI complex eluted broadly, the small amount of LHCI polypeptides in the PSI subcomplex probably represented contamination. Taking those findings together, we infer that the assembly of PSI components into the PSI subcomplex proceeds rapidly, whereas the subsequent integration of PsaG and PsaK proceeds slowly and is among the last assembly steps.
FIGURE 5.
Comparison of polypeptides of PSI complexes from A-2 and A-3. Polypeptides of PSI-LHCI supercomplex and PSI subcomplex purified by DEAE chromatography from A-3 and A-2, respectively, were separated by SDS-PAGE and stained with Coomassie Brilliant Blue (A) and visualized by immunoblotting (B). Each sample was normalized to the quantity of PSI RC proteins, PsaA and PsaB.
Electron Transfer Activity of the PSI Subcomplex
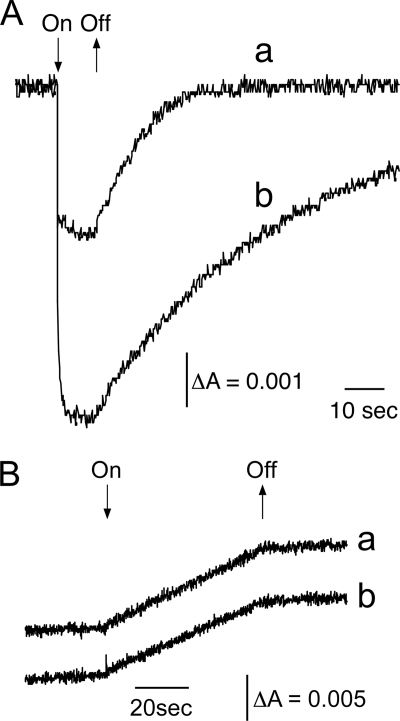
Fig. 6A shows light-induced reversible absorption decreases at 699 nm. The absorption decrease corresponded to the photo-oxidation of P700, whereas the recovery of the absorption decrease corresponded to the re-reduction of P700+ by plastocyanin. We ascribed the smaller signal of the PSI-LHCI from A-3 to the larger antenna contributed by the presence of the LHCI. The rate of P700+ reduction by plastocyanin in the subcomplex was about 10% that of the rate in the PSI-LHCI (Fig. 6A). We ascribed that to the absence of PsaF, which is involved in the efficient electron transfer from plastocyanin to P700+ (34). We also measured NADP+ photo-reduction activity by monitoring light-induced absorption increases at 340 nm, which corresponded to photoreduction of NADP+ (Fig. 6B). Both PSI preparations showed similar activities, indicating that the PSI subcomplex transferred electrons from plastocyanin to ferredoxin in light and was, therefore, fully active.
FIGURE 6.
PSI activities of PSI complexes. A, P700 photo-oxidation activity was monitored via light-induced absorption changes at 699 nm of the PSI-LHCI from A-3 (a) and the PSI subcomplex from A-2 (b) upon illumination in the presence of plastocyanin and ascorbate as electron donors and methyl viologen as an electron acceptor. B, NADP+ photoreduction activity was monitored via light-induced absorption changes at 340 nm of the PSI-LHCI (a) and the PSI subcomplex (b) in the presence of dichloroindophenol and ascorbate as electron donors.
DISCUSSION
PSI Assembly Intermediate Subcomplex
PSI complex assembly proceeds step by step as a number of subunits and cofactors are integrated into the maturing PSI complex (12, 13). Assembly is rapid, however, so the opportunity to detect intermediate subcomplexes is fleeting. Rapid assembly is essential because intermediates are generally unstable and may show only partial photochemical and electron transfer activities and, thus, generate harmful chemical species and strong reductants or oxidants. In the present study we identified a PSI subcomplex that accumulates in logarithmic growth phase cells at 4–5% of total PSI complexes. The separated PSI subcomplex, which was disconnected from the LHCI complex, was appreciably smaller than the mature PSI-LHCI supercomplex (330 versus 700 kDa). The subcomplex was detected in logarithmic growth phase cells but was significantly reduced in stationary phase cells and was specifically pulse-labeled, indicating that it was newly synthesized. In addition, the subcomplex was converted to the PSI-LHCI supercomplex at a relatively slow rate (which is why we were able to detect it). These observations strongly suggest that the subcomplex was an assembly intermediate rather than a partially dissociated subcomplex of the PSI-LHCI supercomplex. The subcomplex was functionally competent and not likely to generate any harmful chemical species despite its accumulation at a detectable level.
Does the PSI Assembly Intermediate Bind LHCI?
Although the purified PSI subcomplex did not bind LHCI, a similar amount of the oligomeric form of LHCI (540 kDa) was present in A-2 and was newly synthesized (Fig. 2A). It seems likely that the newly synthesized PSI subcomplex and LHCI had formed an immature PSI-LHCI supercomplex that was unstable and dissociated during solubilization or fractionation. The structural instability was likely related to the subcomplex lacking two homologous subunits, PsaK and PsaG, as the stable PSI-LHCI supercomplex that separated in A-3 retained the two subunits. In the higher plant PSI-LHCI supercomplex, PsaG and PsaK are located on either edge of the LHCI tetramer that consists of Lhca1, Lhca2, Lhca3, and Lhca4 (3, 4). The LHCIs interact with the PSI core (PsaA and PsaB heterodimer) mainly through small binding surfaces exposed to stroma. Structural details around PsaG reveal that this subunit provides significant contact surfaces for association with Lhca1. This structure must be important for the stabilization of the interaction between PSI and LHCI, although the accumulation of LHCI was barely affected in PsaG-deficient Arabidopsis mutants (35, 36). Although the structure of PsaK is less defined in the crystal structure, it is peripherally located on PsaA near Lhca3. The involvement of PsaK in stabilizing the association between PSI and LHCI has been noted in PsaK-deficient Arabidopsis mutants, where the accumulation of LHCI is partially decreased (35, 37). In addition PsaK might be involved in regulation of LHCI disconnection during iron deficiency (38). Interestingly the additive effect of the absence of PsaG on the accumulation of LHCI was observed in the double mutant in which both PsaG and PsaK were deleted (35). Thus, we infer that PsaG and PsaK stabilize the interaction between PSI core and LHCI by acting together like a buckle and that the instability of the LHCI in the newly synthesized PSI-LHCI supercomplex could follow from the absence of PsaG and PsaK. However, it is possible that the integration of the newly synthesized PSI subcomplex and the oligomeric LHCI complex proceeds slowly, and they were not yet integrated.
Integration of PsaG and PsaK Is the Latest Assembly Step
Fig. 2 revealed that the assembly of the PSI subcomplex and the oligomeric LHCI complex proceeded rapidly, whereas the unusually slow integration of PsaG and PsaK, which may stabilize the association of LHCI to the PSI core, was rate-limiting. Because the presence of PsaG and PsaK in the PSI subcomplex could interfere with the integration of the LHCI oligomer according to the three-dimensional structure of the PSI-LHCI supercomplex, it seems likely that the oligomeric LHCI complex associates with the PSI subcomplex before the integration of PsaG and PsaK, which would make the latter the last assembly step. In vitro experiments indicated that PsaG insertion occurs spontaneously without the assistance of any known chloroplast protein-targeting machinery (39) and that may explain the slow integration of PsaG. Alternatively, PsaG and PsaK synthesis may be slow. Our observation that the purified PSI subcomplex photo-oxidized P700 and participated in electron transfer indicates that the PSI subcomplex that lacked PsaG and PsaK was fully active. Thus, it is likely that accumulation of the PSI subcomplex without PsaG and PsaK generated no light-induced harmful species. This finding is consistent with the result that the PsaG/PsaK-deficient Arabidopsis double mutants show PSI activity and grow photoautotrophically (35). In Cyanobacteria, the PSI complex lacks LHCI and forms a trimeric structure. Interestingly, PsaK is involved in regulation of the trimerization, and the integration of PsaK into the complex is the latest assembly step (40).
Sequential PSI Complex Assembly Process
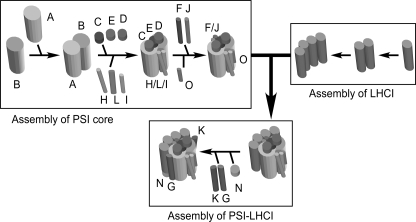
In Fig. 7 we present a working model for the PSI complex assembly based on this and previous studies. The RC complex, consisting of PsaA and PsaB, is the first unit assembled. PsaB is synthesized first and acts as an acceptor for the subsequent sequential integration of PSI polypeptides to form the PSI RC (15). Subsequently, PsaC together with PsaD and PsaE is integrated into the stromal side of the RC complex, forming a docking site for ferredoxin (16). In Chlamydomonas, Ycf4 is part of a large complex and may provide a molecular scaffold for the PSI complex assembly (22). Integration of several newly synthesized PSI subunits such as PsaA, PsaB, PsaC, PsaD, PsaE, and PsaF into a PSI assembly intermediate subcomplex is facilitated on the Ycf4-containing complex. The Ycf4 mutation that substitutes Gln for Glu at positions 179 and 181 completely blocks the integration of all peripheral subunits (23). Thus the Ycf4 complex is involved in initial steps of PSI complex assembly. It is also possible that PsaJ is integrated into the PSI subcomplex together with PsaF because of its proximity and is destabilized when PsaF is deleted (41, 42). Because PsaF and PsaJ have one transmembrane helix and are located between the PSI core and LHCI, it is likely that PsaF and PsaJ are integrated before the LHCI attaches to the PSI subcomplex. The absence of PsaF and PsaJ, however, does not significantly affect the LHCI attachment (42–44).
FIGURE 7.
A working model for PSI assembly. PsaB is synthesized, then dimerizes with PsaA, and the heterodimer then integrates peripheral subunits (PsaC, PsaD, PsaE, PsaF, PsaH, PsaI, PsaJ, and PsaL) to form the PSI core complex. Synthesis and oligomerization of LHCI proceed independently, and then the complex associates with the PSI core complex to form a immature PSI-LHCI supercomplex. In the latest assembly steps, PsaG and PsaK and PsaN are integrated to form the stable mature supercomplex.
Four subunits, PsaH, PsaI, PsaL, and PsaO, are located on the PSI RC on the opposite side of LHCI, and how they are integrated into the PSI intermediate subcomplex remains elusive. According to the crystal structure of PSI complex, PsaH, PsaI, and PsaL form a cluster and are present on the opposite side of the LHCI tetramer in the PSI-LHCI supercomplex. It is likely that PsaI and PsaL are integrated first and then PsaH is assembled. Because PsaO is easily lost from the PSI-LHCI supercomplex without affecting the stability of PsaH, PsaI, and PsaL (Fig. 5), PsaO is integrated as the last step among those four polypeptides.
According to the analyses using PSI-deficient mutants, the PSI core and LHCI complexes are assembled independently (7). As already discussed above, PsaG and PsaK may be integrated after the LHCI is assembled into the PSI core complex. In higher plants, PsaN is located on the lumenal side of LHCI (3, 45). Thus, PsaN integration should also occur in the last assembly step.
Acknowledgments
We thank Drs. Kevin Redding, Michael Hippler, and Masahiko Ikeuchi for kindly providing the antibodies against PsaA, PsaK, Lhca6, and PsbA and Akira Okamuro for technical assistance.
This work was supported by a Japan Society for the Promotion of Science Grant-in-aid for General Research for Creative Scientific Research 18GSO318 (to Y. T.) and Japan Society for the Promotion of Science Research Fellowships for Young Scientists Grant 17003605 (to S. O.).
- PSI
- photosystem I
- PSII
- photosystem II
- DM
- n-dodecyl-β-d-maltoside
- LHC
- light-harvesting complex
- RC
- reaction center
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Nelson N., Yocum C. F. (2006) Annu. Rev. Plant Biol. 57, 521–565 [DOI] [PubMed] [Google Scholar]
- 2.Jordan P., Fromme P., Witt H. T., Klukas O., Saenger W., Krauss N. (2001) Nature 411, 909–917 [DOI] [PubMed] [Google Scholar]
- 3.Amunts A., Drory O., Nelson N. (2007) Nature 447, 58–63 [DOI] [PubMed] [Google Scholar]
- 4.Amunts A., Toporik H., Borovikova A., Nelson N. (2010) J. Biol. Chem. 285, 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen P. E., Haldrup A., Zhang S., Scheller H. V. (2004) J. Biol. Chem. 279, 24212–24217 [DOI] [PubMed] [Google Scholar]
- 6.Stauber E. J., Fink A., Markert C., Kruse O., Johanningmeier U., Hippler M. (2003) Eukaryot. Cell 2, 978–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi Y., Yasui T. A., Stauber E. J., Hippler M. (2004) Biochemistry 43, 7816–7823 [DOI] [PubMed] [Google Scholar]
- 8.Tokutsu R., Teramoto H., Takahashi Y., Ono T. A., Minagawa J. (2004) Plant Cell Physiol. 45, 138–145 [DOI] [PubMed] [Google Scholar]
- 9.Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Maréchal- Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C. L., Cognat V., Croft M. T., Dent R., Dutcher S., Fernández E., Fukuzawa H., González-Ballester D., González-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J. P., Riaño-Pachón D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C. J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martínez D., Ngau W. C., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. (2007) Science 318, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Germano M., Yakushevska A. E., Keegstra W., van Gorkom H. J., Dekker J. P., Boekema E. J. (2002) FEBS Lett. 525, 121–125 [DOI] [PubMed] [Google Scholar]
- 11.Kargul J., Nield J., Barber J. (2003) J. Biol. Chem. 278, 16135–16141 [DOI] [PubMed] [Google Scholar]
- 12.Hippler M., Rimbault B., Takahashi Y. (2002) Protist 153, 197–220 [DOI] [PubMed] [Google Scholar]
- 13.Schöttler M. A., Bock R. (2008) in Progress in Botany (Lüttge U., Beyschlag W., Murata J. eds) Vol. 69, pp. 89–115, Springer Verlag, Berlin [Google Scholar]
- 14.Wollman F. A., Minai L., Nechushtai R. (1999) Biochim. Biophys. Acta 1411, 21–85 [DOI] [PubMed] [Google Scholar]
- 15.Girard-Bascou J., Choquet Y., Schneider M., Delosme M., Dron M. (1987) Curr. Genet. 12, 489–495 [DOI] [PubMed] [Google Scholar]
- 16.Wostrikoff K., Girard-Bascou J., Wollman F. A., Choquet Y. (2004) EMBO J. 23, 2696–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mannan R. M., Pakrasi H. B., Sonoike K. (1994) Arch. Biochem. Biophys. 315, 68–73 [DOI] [PubMed] [Google Scholar]
- 18.Yu J., Smart L. B., Jung Y. S., Golbeck J., McIntosh L. (1995) Plant Mol. Biol. 29, 331–342 [DOI] [PubMed] [Google Scholar]
- 19.Boudreau E., Takahashi Y., Lemieux C., Turmel M., Rochaix J. D. (1997) EMBO J. 16, 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naver H., Boudreau E., Rochaix J. D. (2001) Plant Cell 13, 2731–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilde A., Härtel H., Hübschmann T., Hoffmann P., Shestakov S. V., Börner T. (1995) Plant Cell 7, 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozawa S., Nield J., Terao A., Stauber E. J., Hippler M., Koike H., Rochaix J. D., Takahashi Y. (2009) Plant Cell 21, 2424–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onishi T., Takahashi Y. (2009) Plant Cell Physiol. 50, 1750–1760 [DOI] [PubMed] [Google Scholar]
- 24.Stöckel J., Bennewitz S., Hein P., Oelmüller R. (2006) Plant Physiol 141, 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilde A., Lünser K., Ossenbühl F., Nickelsen J., Börner T. (2001) Biochem. J. 357, 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugimoto I., Takahashi Y. (2003) J. Biol. Chem. 278, 45004–45010 [DOI] [PubMed] [Google Scholar]
- 27.Gorman D. S., Levine R. P. (1965) Proc. Natl. Acad. Sci. U.S.A. 54, 1665–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chua N. H., Bennoun P. (1975) Proc. Natl. Acad. Sci. U.S.A. 72, 2175–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi Y., Goldschmidt-Clermont M., Soen S. Y., Franzén L. G., Rochaix J. D. (1991) EMBO J. 10, 2033–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi H., Iwai M., Takahashi Y., Minagawa J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi Y., Utsumi K., Yamamoto Y., Hatano A., Satoh K. (1996) Plant Cell Physiol. 37, 161–168 [DOI] [PubMed] [Google Scholar]
- 32.Ohnishi N., Takahashi Y. (2001) J. Biol. Chem. 276, 33798–33804 [DOI] [PubMed] [Google Scholar]
- 33.Takahashi Y., Koike H., Katoh S. (1982) Arch. Biochem. Biophys. 219, 209–218 [DOI] [PubMed] [Google Scholar]
- 34.Hippler M., Reichert J., Sutter M., Zak E., Altschmied L., Schröer U., Herrmann R. G., Haehnel W. (1996) EMBO J. 15, 6374–6384 [PMC free article] [PubMed] [Google Scholar]
- 35.Varotto C., Pesaresi P., Jahns P., Lessnick A., Tizzano M., Schiavon F., Salamini F., Leister D. (2002) Plant Physiol. 129, 616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jensen P. E., Rosgaard L., Knoetzel J., Scheller H. V. (2002) J. Biol. Chem. 277, 2798–2803 [DOI] [PubMed] [Google Scholar]
- 37.Jensen P. E., Gilpin M., Knoetzel J., Scheller H. V. (2000) J. Biol. Chem. 275, 24701–24708 [DOI] [PubMed] [Google Scholar]
- 38.Moseley J. L., Allinger T., Herzog S., Hoerth P., Wehinger E., Merchant S., Hippler M. (2002) EMBO J. 21, 6709–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosgaard L., Zygadlo A., Scheller H. V., Mant A., Jensen P. E. (2005) FEBS J. 272, 4002–4010 [DOI] [PubMed] [Google Scholar]
- 40.Dühring U., Ossenbühl F., Wilde A. (2007) J. Biol. Chem. 282, 10915–10921 [DOI] [PubMed] [Google Scholar]
- 41.Farah J., Rappaport F., Choquet Y., Joliot P., Rochaix J. D. (1995) EMBO J. 14, 4976–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischer N., Boudreau E., Hippler M., Drepper F., Haehnel W., Rochaix J. D. (1999) Biochemistry 38, 5546–5552 [DOI] [PubMed] [Google Scholar]
- 43.Haldrup A., Simpson D. J., Scheller H. V. (2000) J. Biol. Chem. 275, 31211–31218 [DOI] [PubMed] [Google Scholar]
- 44.Hansson A., Amann K., Zygadlo A., Meurer J., Scheller H. V., Jensen P. E. (2007) FEBS J 274, 1734–1746 [DOI] [PubMed] [Google Scholar]
- 45.Nielsen V. S., Mant A., Knoetzel J., Møller B. L., Robinson C. (1994) J. Biol. Chem. 269, 3762–3766 [PubMed] [Google Scholar]