Abstract
The oxazolomycins (OZMs) are a growing family of antibiotics produced by several Streptomyces species that show diverse and important antibacterial, antitumor, and anti-human immunodeficiency virus activity. Oxazolomycin A is a peptide-polyketide hybrid compound containing a unique spiro-linked β-lactone/γ-lactam, a 5-substituted oxazole ring. The oxazolomycin biosynthetic gene cluster (ozm) was identified from Streptomyces albus JA3453 and localized to 79.5-kb DNA, consisting of 20 open reading frames that encode non-ribosomal peptide synthases, polyketide synthases (PKSs), hybrid non-ribosomal peptide synthase-PKS, trans-acyltransferases (trans-ATs), enzymes for methoxymalonyl-acyl carrier protein (ACP) synthesis, putative resistance genes, and hypothetical regulation genes. In contrast to classical type I polyketide or fatty acid biosynthases, all 10 PKS modules in the gene cluster lack cognate ATs. Instead, discrete ATs OzmM (with tandem domains OzmM-AT1 and OzmM-AT2) and OzmC were equipped to carry out all of the loading functions of both malonyl-CoA and methoxymalonyl-ACP extender units. Strikingly, only OzmM-AT2 is required for OzmM activity for OZM biosynthesis, whereas OzmM-AT1 seemed to be a cryptic AT domain. The above findings, together with previous results using isotope-labeled precursor feeding assays, are assembled for the OZM biosynthesis model to be proposed. The incorporation of both malonyl-CoA (by OzmM-AT2) and methoxymalonyl-ACP (by OzmC) extender units seemed to be unprecedented for this class of trans-AT type I PKSs, which might be fruitfully manipulated to create structurally diverse novel compounds.
Keywords: Bacterial Genetics, Fatty Acid Synthase, Gene Knock-out, Metabolic Regulation, Multifunctional Enzymes, Antitumor Antibiotics, Nonribosomal Peptide Synthase (NRPS), Oxazolomycin, Polyketide Synthase (PKS), Trans-acyltransferase (AT)
Introduction
Type I modular polyketide synthases (PKSs)4 or fatty acid synthases usually possess a minimal set of three domains in each module, including a ketosynthase (KS), an acyltransferase (AT), and an acyl carrier protein (ACP), as exemplified by the classical example of the 6-deoxyerythronolide B synthase from Saccharopolyspora erythaea (1). Later it was reported that some modular type I PKSs lack cognate AT domains in each module and instead have one or more discrete AT domains to carry out their functions (2). An example is provided by the leinamycin biosynthetic machinery with two PKSs, LnmI and LnmJ, consisting of six modules but with only one trans-AT unit, LnmG, involved in loading the malonyl-CoA extender units onto all of the modular ACP domains (3). This new class of type I PKSs is known as “AT-less” PKSs, trans-AT PKSs, or discrete AT type I PKSs. Trans-AT PKSs were found to be not as rare as expected because many examples have been identified in recent years (4).
The chemical structural diversity of classical type I polyketides is in part derived from the selective incorporation of various extender units (e.g. malonyl-CoA, methylmalonyl-CoA, ethylmalonyl-CoA, and methoxymalonyl-ACP), which is determined by the specificity of the AT domains in each module, giving rise to different chemical groups extending from the core polyketide backbone of the product (1). Replacement of individual modular ATs with a trans-AT might limit structural diversity. However, polyketides biosynthesized by discrete AT PKSs do not lack α-branched organic groups (methyl-, ethyl-, and methoxy-), which appear to be a common feature of these compounds. It turns out that Me groups are introduced via C-methylation by methyltransferase (MT) domains in trans-AT PKSs, the result of which is equivalent to the use of a methylmalonyl-CoA extender unit. The PKSs from the kirromycin biosynthetic pathway have been proposed to incorporate both malonyl-CoA and ethylmalonyl-CoA (5), whereas the mechanism by which methoxy groups are introduced by trans-AT PKSs is still unknown, because all characterized methoxymalonyl-ACP extender unit-containing polyketide biosynthesis gene clusters encode classical type I modular polyketide synthases, such as those for ansamitocin P-3, geldanamycin, soraphen A, leucomycin, FK506 and FK520, concanamycin, bafilomycin, aflastatin, and tautomycin (6).
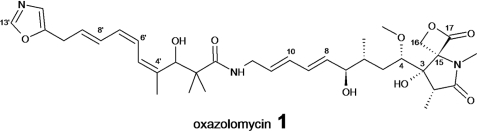
The oxazolomycins (OZMs), represented by oxazolomycin A, B, C, 16-methyl-oxazolomycin, KSM-2690B, KSM-2690C, triedimycin A, B, neooxazolomycin, inthomycins A, B, C and lajollamycin, are a growing family of antibiotics produced by several Streptomyces species that show diverse antibacterial, antitumor, and anti-HIV activity (7, 8). Oxazolomycin A is a peptide-polyketide hybrid compound containing a unique spiro-linked β-lactone/γ-lactam, a 5-substituted oxazole ring, and (E,E)-diene and (Z,Z,E)-triene moieties (Fig. 1). Isotope-labeled precursor feeding experiments have established that the carbon backbone of OZM is derived from three molecules of glycine and nine molecules of acetate, with all five C-methyl groups, one O-methyl group, and one N-methyl group originating from l-methionine (9). Interestingly, propionate, which is commonly incorporated into the polyketide backbone in the form of methylmalonyl-CoA in classical type I PKSs, does not label the OZM molecule or any other products from discrete AT PKSs (9). The origins of C3, C4, C16, and C13′ were unclear from labeling data (9). However, chemical (6), biochemical (10, 11), and genetic investigations (12) on moieties similar to the C3-C4 unit of OZM in other natural products of polyketide origin, like ansamitocin P-3 (13), geldanamycin (14), soraphen (15), tautomycin (16), and FK520 (17), suggested that the two-carbon moiety is probably derived from a metabolic intermediate of the glycolytic pathway, which serves as a precursor for the unusual methoxymalonyl-ACP extender unit. The above information enabled us to isolate the oxazolomycin gene cluster (ozm) from Streptomyces albus JA3453 by cloning the methoxymalonyl-ACP biosynthetic locus and localizing the ozm cluster to a region of about 135 kb. Its involvement in oxazolomycin biosynthesis was confirmed by gene inactivation to generate non-OZM-producing mutants. Sequence analysis of a 12-kb DNA fragment from this region identified six enzymes (OzmB, OzmC, OzmD, OzmE, OzmF, and OzmG) involved in the biosynthesis of the methoxymalonyl-ACP for oxazolomycin production (8). Another related study showed that OzmB serves as a bifunctional glyceryl transferase/phosphatase belonging to the HAD superfamily, which first diverts the primary metabolite 1,3-bisphosphoglycerate into a phosphoglyceryl-S-OzmB intermediate, then removes the phosphate group to afford the 3-glyceryl-S-OzmB (acting as a phosphatase), and finally transfers the glyceryl group to an ACP to set the stage for polyketide biosynthesis (acting as a glyceryl transferase) (10). All of these steps were monitored by nanospray Fourier transform ion cyclotron resonance mass spectrometry. The exact role of OzmC remained unclear, although gene inactivation and complementation experiments have suggested that it is absolutely required for OZM biosynthesis (8).
FIGURE 1.
Chemical structure of oxazolomycin.
In this study, we describe the complete sequencing and in silico annotation of the ozm gene cluster and preliminary delimitation of its boundaries. In addition, the non-ribosomal peptide synthetase (NRPS)-PKS multienzymes (OzmHJKLNOQ) along with discrete ATs (OzmM and OzmC) specifying a novel extender unit were identified. The role of OzmM was also investigated by in vivo site-directed mutagenesis to establish that only the second domain of OzmM (OzmM-AT2) is required for OZM production, whereas OzmM-AT1 may be cryptic. Arguing from bioinformatics and genetic information, we propose a model for the oxazolomycin biosynthetic pathway. Because isotopically labeled precursor feeding experiments have suggested that all of the branch methyl groups of oxazolomycin are derived from methionine rather than methylmalonyl-CoA, we realized that both methoxymalonyl-ACP and malonyl-CoA extender units are evidently incorporated into the oxazolomycin molecular backbone, concluding that ozm polyketide synthase is capable of accommodating two different extender units by discrete ATs (OzmC and OzmM-AT2). These studies thus seem to have provided new insights into the understanding of how trans-AT type I PKS recruits discrete ATs but allows different extender units to be accepted for the biosynthesis of oxazolomycin. This phenomenon could potentially be exploited in combinatorial biosynthesis to generate novel and structurally diverse polyketides.
EXPERIMENTAL PROCEDURES
Sequencing and in Vivo Analysis of the ozm Gene Cluster
DNA sequencing was performed using a set of five cosmids covering the whole oxazolomycin biosynthetic gene cluster: pJTU1059, pJTU1060, pJTU1061, pJTU1062, and pJTU1063 (8). The insert of each cosmid DNA was cut out with DraI and purified using a Plasmid Maxi kit (Qiagen) and sonicated with a 550 Sonic Dismembrator (Fisher). DNA fragments of 1.6–2.0 kb were recovered from 0.7% low melting agarose gel using a Geneclean II reagent kit (Bio 101 Inc., Vista, CA) and subcloned into pUC18. For automated sequencing, plasmid DNA templates were prepared by alkaline lysis using the Prep 96 plasmid kit (Qiagen). Sequencing reactions were carried out with BigDye terminator cycle sequencing kits (PerkinElmer Life Sciences). The sequences of custom-designed sequencing primers were 5′-GTAAAACGACGGCCAGT-3′ (forward) and 5′-GCGGATAACAATTTCACACAGG-3′ (reverse). Sequence reads were obtained from 3730 DNA sequencers (PerkinElmer Life Sciences). Bioinformatic analysis of the sequenced 127,755-bp contiguous DNA and functional assignments of the deduced gene products are summarized in Table 1. The sequence data were analyzed with the Frame-Plot 3.0 online program (18). DNA and deduced protein sequence homology searches were performed using BLAST and FASTA (19). Multiple sequence alignments were done using ClustalW.
TABLE 1.
Deduced ORF functions in the ozm biosynthetic gene cluster
| Gene | Sizea | Protein homologb | Proposed function |
|---|---|---|---|
| aa | |||
| orf(−5) | 335 | Franean1_1604(ABW11043, 52/39) | Transcriptional regulator |
| orf(−4) | 775 | SAV_1033(NP_822208 83/71) | Integral membrane protein |
| orf(−3) | 158 | SAV_1074(NP_822249, 83/79) | Bacterioferritin comigratory protein |
| orf(−2) | 339 | SACE_5658(CAM04844, 59/47)) | Transcriptional regulator |
| orf(−1) | 315 | CMS_2856(YP_001711489, 84/71) | Nucleoside hydrolase |
| Upstream boundary of ozm cluster | |||
| ozmA | 482 | SgcB(AAF13999, 28/43) | Antibiotic efflux protein |
| ozmB | 366 | GdmH(ABI93783, 65/76) | Glyceryltransferase/phosphatase |
| ozmC | 325 | DpsC(AAA65208, 25/39) | Acyltransferase |
| ozmD | 366 | GdmI(ABI93784, 64/75) | Acyl-dehydrogenase |
| ozmE | 93 | GdmJ(ABI93785, 63/78) | ACP |
| ozmF | 221 | TtmC(AAZ08058, 60/77) | O-Methyltransferase |
| ozmG | 287 | TtmB(AAZ08059, 63/72) | 3-Hydroxyacyl-CoA-dehydrogenase |
| ozmH | 7737 | PksP(E69679, 35/51) | Hybrid NRPS/PKS |
| ozmJ | 2926 | ObsC(AAS00421, 52/64) | PKS |
| ozmK | 1202 | BryB(ABK51300, 34/50) | PKS |
| ozmL | 1993 | McyA(AAF00960, 37/55) | NRPS |
| ozmM | 1039 | MmpIII(AAM12912, 47/59) | Acyltransferase/oxidoreductase |
| ozmN | 4971 | LnmJ(AF484556, 38/48) | PKS |
| ozmO | 1196 | PedF(AAS47564, 42/56) | NRPS |
| ozmP | 382 | - | Unknown |
| ozmQ | 842 | NosB(AAF15892, 55/69) | PKS |
| ozmR | 308 | Orf5(BAA32133, 53/64) | Transcriptional regulator |
| ozmS | 214 | SC5F8.18(CAB93746, 71/78) | Transporter |
| ozmT | 439 | SCH63.25(CAC10316, 77/83) | Thr-tRNA synthetase |
| ozmU | 929 | AfsR(BAA14186, 33/45) | Transcriptional activator |
| Downstream boundary of ozm cluster | |||
| orf(+1) | 445 | CypC (ABS73471, 42/59) | Cytochrome P450 |
| orf(+2) | 221 | Unknown | |
| orf(+3) | 282 | SC5F8.24 (CAB93752, 70/81) | RNA polymerase sigma factor |
| orf(+4) | 344 | Unknown | |
| orf(+5) | 700 | Unknown | |
a Sizes are in amino acids (aa).
b Given in parentheses are accession numbers and percentage identity/percentage similarity.
Inactivation of orf(−1), orf(−2), orf(−3)–(−5), orf(+1), orf(+2), and orf(+3)–(+5)
The PCR targeting strategy (20) was adopted to inactivate orf(−1), orf(−2), orf(−3)–(−5), orf(+1), orf(+2), and orf(+3)–(+5), using pIJ778 as a template throughout. The PCR products were transferred into competent cells of BW25113 (pIJ790, pJTU1059) for orf(−1), orf(−2), and orf(−3)–(−5) or BW25113 (pIJ790, pJTU1063) for orf(+1), orf(+2), and orf(+3)–(+5) to isolate apramycin-resistant and spectinomycin-resistant colonies, yielding pJTU1111, pJTU1112, pJTU1113, pJTU1121, pJTU1122, and pJTU1123, respectively. These plasmids were introduced into ET12567 (pUZ8002) and then transferred by conjugation to S. albus JA3453 to screen for apramycin-sensitive and spectinomycin-resistant colonies, resulting in ZH11, ZH12, ZH13, ZH21, ZH22, and ZH23, respectively.
Inactivation of ozmM by In-frame Deletion
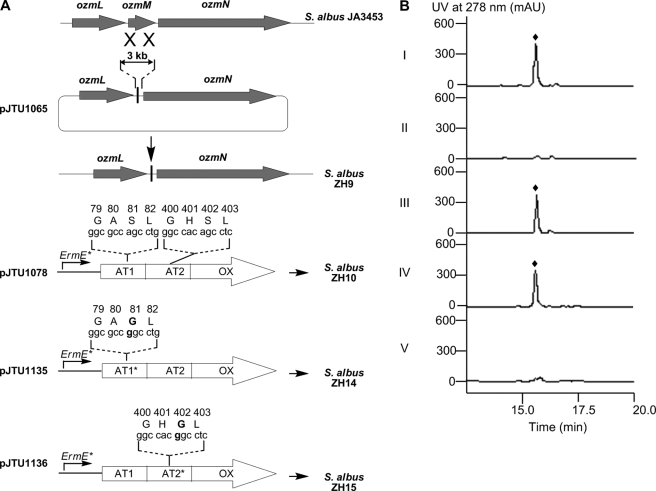
The same PCR targeting strategy (20) described above was used to inactivate ozmM with pIJ778 as the template. The PCR product was introduced by transformation into competent cells of BW25113 (pIJ790, pJTU1061) to screen for apramycin-resistant and spectinomycin-resistant colonies, yielding pJTU1064. To make a construct for in-frame deletion, pJTU1064 was transferred into Escherichia coli DH10B (BT340). The transformant was placed at 42 °C to induce the resistance cassette cut by plasmid BT340. This yielded an 81-bp scar-containing plasmid, pJTU1065, which was transferred by conjugation into S. albus JA3453 and screened for apramycin-resistant colonies in a first round followed by isolating apramycin-sensitive colonies in a second round. An in-frame deletion mutant of ozmM was confirmed by PCR and subsequent sequencing and designated ZH9 (see Fig. 5).
FIGURE 5.
Deletion of ozmM and complementation of the ozmM mutant with either intact ozmM or ozmM-AT1 (S81G) or ozmM-AT2 (S402G) mutant and their effect on OZM biosynthesis. A, schematic representation of constructs for the generation of the ZH9 deletion mutant strain and its genetic complementation strains ZH10, ZH14, and ZH15. Details of site-specific mutagenesis with OzmM-AT1 and OzmM-AT2 are shown. B, HPLC analysis of OZM production in wild-type (I), ZH9 mutant (II), ZH 10 mutant (III), ZH14 mutant (IV), and ZH15 mutant (V) strains. ♦, OZM.
Complementation of the ΔozmM Mutant with Wild Type or Mutated ozmM
The ozmM gene was first amplified by PCR from cosmid pJTU1061 using primers 5′-C CAT ATG ACG ACG GCT GCC GGA GA-3′ (ozmM-FP2, with the NdeI site underlined) and 5′-T GAA TTC CGC CAC AGC GTG TTG TCC AG-3′ (ozmM-RP2, with the EcoRI site underlined). The resultant PCR product was recovered as a 3300-bp NdeI-EcoRI fragment and ligated into the same sites of pBluescript SK to verify PCR fidelity by sequencing and yielding pJTU1088. The ozmM gene was then recovered as a NdeI-EcoRI fragment from pJTU1088 and ligated into the same sites of pJTU1351 (8) to generate pJTU1078 in which the expression of ozmM is under the control of the ErmE* promoter. Introduction of pJTU1078 into S. albus ZH9 by conjugation with apramycin selection afforded the complemented strain named S. albus ZH10 (see Fig. 6).
To mutate the OzmM-AT1 and OzmM-AT2 active sites, pJTU1078 was used as template. The PCR primers for mutating ozmM-AT1 and ozmM-AT2 were 5′-TC GGC GCC GGC CTG GGG GAG TTC AC-3′ (ozmM-AT1-FP)/5′-CCC CAG GCC GGC GCC GAG CAG CAG C-3′ (ozmM-AT1-RP) and 5′-TC GGC CAC GGC CTC GGC GAG TAC GTG-3′ (ozmM-AT2-FP)/5′-GCC GAG GCC GTG GCC GAC GAG GAA G-3′ (ozmM-AT2-RP), respectively (the targeted site for mutation is underlined). A typical PCR (50 μl) consisted of 1.5 mm MgC12, 10 ng of plasmid DNA as template, 5% dimethyl sulfoxide, 200 μm deoxynucleoside triphosphates, 25 pmol of each primer, and 2.5 units of expanded high fidelity polymerase (Roche Applied Science). The PCR reaction was run at 95 °C for 1 min, followed by 16 cycles of amplification (50 s at 95 °C, 50 s at an annealing temperature of 59 °C, 8 min and 40 s for an extension time at 70 °C), and a final postrun extension for 7 min at 72 °C. The PCR products were digested with DpnI and then transformed into competent DH5α cells. This yielded mutated plasmids pJTU1135, where sequence analysis showed that the active site Ser of OzmM-AT1 had been mutated to Gly, and pJTU1136, in which the active site Ser of OzmM-AT2 had been mutated to Gly. Introduction of pJTU1135 and pJTU1136 into ZH9 by conjugation with apramycin selection afforded complementation strains named S. albus ZH14 and ZH15, respectively (see Fig. 5).
Overproduction, Purification, and Amino Acid-dependent ATP-[32P]Pyrophosphate Exchange Assays of the OzmH A Domain
The excised A domain from ozmH was amplified by PCR from pJTU1061 with the following pair of primers: 5′-GGTATTGAGGGTCGCCTCTTCGACGAGGACACCG-3′ (forward) and 5′-AGAGGAGAGTTAGAGCCACCTGCCGCACGCCGGG-3′ (reverse). Purified PCR product was inserted into pET-30Xa using ligation-independent cloning as described by Novagen (Madison, WI), affording pBS7003, and sequenced to confirm PCR fidelity. The plasmid pBS7003 was introduced into E. coli BL21(DE3), and the resultant recombinant was cultured at 18 °C overnight with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside induction. Affinity purification was performed using Ni2+-nitrilotriacetic acid-agarose as described by Qiagen (Valencia, CA). Protein purity (>90%) was assessed by 10% acrylamide SDS-PAGE. Finally, OzmH-A domain was overproduced as an N-His6-tagged protein.
Amino acid-dependent ATP-[32P]pyrophosphate (21) exchange assays were performed following procedures described previously (22) using tetrasodium [32P]pyrophosphate from PerkinElmer Life Sciences. A typical reaction (100 μl) contained 75 mm Tris-HC1, pH 7.5, 5 mm MgC12, 5 mm ATP, 0.9 pm [32P]PPi (54.39 Ci/mmol), a 0.5 mm concentration of the indicated amino acid, and 100 nm OzmH-A protein. Reactions were performed at 30 °C for 10 min and terminated by the addition of 5 volumes of 1% (w/v) activated charcoal and 4.5% (w/v) “cold” tetrasodium pyrophosphate in 3.5% (v/v) perchloric acid. Precipitates were collected with filter paper on sintered glass filters under suction and washed consecutively with 40 mm sodium pyrophosphate in 1.4% (v/v) perchloric acid, water, and ethanol. Filter papers were added to 10 ml of EcoLumeTM scintillation mixture from Fisher, and the radioactivity was quantified using a Packard Tri-Carb liquid scintillation analyzer model 1900TR.
Culture Conditions and OZM Isolation and Analysis
S. albus wild type and mutant strains were grown on ISP4 medium plates at 28 °C for 7 days for spore harvest. Approximately 1 ml of agar containing each strain was used to inoculate 50 ml of fermentation medium consisting of 1.7% malt extract, 1.5% potato starch, 0.75% corn steep liquid, 0.75% Pharmamedia (Southern Cotton Oil Company), and 0.1% CaCO3 in a 250-ml flask. After cultivation on a rotary shaker at 28 °C and 250 rpm for 72 h, the fermentation broth was centrifuged (4000 rpm, 20 min), and the supernatant was extracted twice with ethyl acetate. The extract was evaporated, dissolved in methanol, centrifuged, and subjected to HPLC analysis. HPLC data were obtained using a Prodigy ODS-2 column (150 × 4.6 mm, 5 μm) with a mobile phase gradient of 40–90% CH3OH in H2O at a flow rate of 1 ml/min and UV detection at 278 nm (8, 23). Electrospray ionization-mass spectra were obtained on an Agilent 1100 HPLC-MSD SL quadrupole mass spectrometer. The mobile phase used for liquid chromatography-mass spectrometry comprises buffer A (15% CH3CN, 85% H2O containing 0.1% AcOH) and buffer B (80% CH3CN, 20% H2O containing 0.1% AcOH). Liquid chromatography-mass spectrometry was performed using a Prodigy ODS-2 column (150 × 4.6 mm, 5 μm) eluted with a linear gradient of 30–80% buffer B over 20 min, followed by 5 min at 80% buffer B at a flow rate of 1.0 ml/min with UV detection at 278 nm.
RESULTS
Sequence Analysis of the ozm Gene Cluster
As we described in previous studies (8), the ozm biosynthetic gene cluster was initially identified from S. albus JA3453 by genetic characterization of the methoxymalonyl-ACP biosynthetic locus and confirmed by gene inactivation, affording mutant strains that had lost oxazolomycin production. Also, a preliminary sequence of 12 kb revealed the gene subcluster involved in methoxymalonyl-ACP biosynthesis (8). On the basis of these results (8), shotgun sequencing of the five overlapping cosmids covering the ozm gene cluster yielded a 127,755-bp contiguous DNA sequence (totally 11.7 times coverage through the gene cluster), with an overall GC content of 73.8%, characteristic of Streptomyces DNA (24). Bioinformatic analysis of the sequenced region revealed 50 open reading frames (ORFs), whose functions were predicted by comparing the deduced gene products with proteins of known function in the databases, as noted in Table 1.
Delimitation of Boundaries for the ozm Gene Cluster
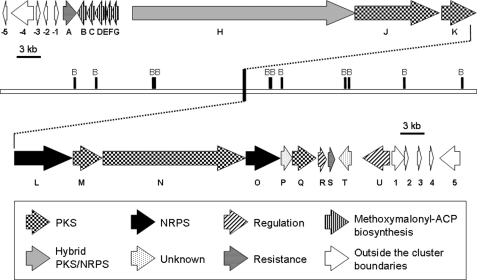
The boundaries of the ozm gene cluster were preliminarily delimitated by gene inactivation of orf(−1), orf(−2), and orf(−3)-orf(−5) for the upstream boundary (8) and orf(+1), orf(+2), and orf(+3)-orf(+5) for the downstream boundary. None of the above gene inactivations exerted any effect on oxazolomycin production, leading to the assignment of these orf genes as outside the cluster. These experiments and bioinformatics analysis established that the ozm gene cluster spans at most 79.5 kb of DNA consisting of 20 ORFs designated ozmA to ozmU (Fig. 2 and Table 1). They encode four trans-AT type I PKSs (OzmJ, OzmK, OzmN, and OzmQ) and discrete AT enzymes (OzmM and OzmC), one hybrid PKS-NRPS (OzmH), two NRPSs (OzmL and OzmO), enzymes for methoxymalonyl-ACP biosynthesis (OzmB, OzmD, OzmE, OzmF, and OzmG) (8), and hypothetical proteins as candidates for resistance (OzmA and OzmS) and regulation or postmodification (OzmR, OzmU, and OzmT), as well as proteins whose functions cannot be predicted from bioinformatics alone (OzmP).
FIGURE 2.
Genetic organization of the ozm biosynthetic gene cluster. B, BamHI. Proposed functions for individual ORFs are summarized in Table 1. Numbers refer to genes outside ozm gene cluster. Letters refer to genes inside ozm gene cluster.
Genes Encoding NRPSs
The ozmO and ozmL genes both encode modular multidomain NRPSs with unusual architectures (Figs. 2 and 3A). The ozmO gene encodes a protein (OzmO) of 1196 amino acids that may serve as the loading module, consisting of a hypothetical formylation (F) domain, an adenylation (A) domain, and a peptide carrier protein (PCP) domain. Isotope-labeling precursor feeding experiments (9) and studies on the specificity-conferring codes of the A domains (22) (Table 2) suggest that glycine is activated by the OzmO A domain and then loaded onto the PCP. Formylation domains as novel tailoring enzymes have been identified in several other NRPSs, such as LgrA from gramicidin biosynthesis (25) and ApdA from anabaenopeptilide biosynthesis (26). The F domain of LgrAl was demonstrated by in vitro assays to be responsible for the formylation of valinyl-S-PCP (25), which transfers the formyl group of formyl-tetrahydrofolate onto the initiation amino acid valine using cofactors N10- and N5-formyl-tetrahydrofolate. Thus, a similar function could be envisioned for the F domain of OzmO for the formylation of glycyl-S-PCP to generate formylglycyl-S-PCP.
FIGURE 3.
A, proposed model for OZM biosynthesis. DH, dehydratase; ER, enoyl reductase; SAM, S-adenosylmethionine; ?, unknown. The upper section shows that OzmM consists of the cryptic AT1 and active AT2 domains that transfer two extender units (malonyl-CoA and methoxymalonyl-ACP) onto corresponding ACPs (OzmC is also involved). The bottom section indicates that the PKS-NRPS assembly line condenses various building blocks by step to biosynthesize oxazolomycin. B, proposed pathway for methoxymalonyl-ACP extender unit biosynthesis in ozm gene cluster.
TABLE 2.
Predictions of substrate specificity of ozm NRPSs based on the specificity-conferring codes of A domains (shown in boldface type)
| Domain | 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | 517 | Similarity |
|---|---|---|---|---|---|---|---|---|---|---|---|
| % | |||||||||||
| Ser | D | V | W | H | L | S | L | I | D | K | |
| OzmL | D | V | W | H | V | S | L | V | D | K | 95 |
| B1mVI | D | V | W | H | V | S | L | V | D | K | 95 |
| Gly | D | I | L | Q | L | G | L | I | W | K | |
| OzmH | D | I | L | Q | L | G | M | I | W | K | 100 |
| OzmO | D | I | L | Q | L | G | M | I | W | K | 100 |
| SafA-1 | D | I | L | Q | L | G | L | V | W | K | 100 |
| Tal | D | I | L | Q | L | G | M | I | W | K | 100 |
The ozmL gene encodes a modular NRPS of 1993 amino acids, probably representing the last module of the OZM hybrid NRPS-PKS multienzyme, for which five domains have been identified as condensation (C), A, MT, PCP, and another C domain (Figs. 2 and 3A). The predicted amino acid specificity for the A domain of OzmL is serine (Table 2) (22), consistent with the results of isotope-labeling experiments (9). The ozmL N-terminal C domain has an intact, highly conserved signature motif, HHXXXDG, which is commonly present in other characterized C domains from gene databases. Mutational analysis (27) and crystallographic studies (28) have indicated that the second histidine (underlined) acts as a crucial catalytic active site for condensation of two aminoacyl substrates or an aminoacyl and peptidyl substrate. Although the C domain residing at the C terminus of OzmL also shows homology with the other C domain, protein sequence alignment revealed an amino acid motif NYFCLDG in register with the conserved motif HHXXXDG, whose function remained elusive.
Genes Encoding a Hybrid NRPS-PKS
The ozmH gene encodes a giant hybrid NRPS-PKS protein of 7737 amino acids with one NRPS module with a predicted specificity for glycine (Table 2) and four PKS modules, together containing five KSs, four ketoreductases (KRs), two dehydratases, two MTs, and five ACPs (Figs. 2 and 3A). This pentamodular complex possesses some unusual features as a polyketide-nonribosomal peptide synthase. For example, tandem KS domains were identified in module 10. The first KS domain of module 10 contains a mutated catalytic triad of Cys-Asn-His, whereas the second KS possesses a full catalytic triad of Cys-His-His (29, 30). The His-His residues are essential for malonyl-ACP decarboxylation to generate a carbon anion, and the Cys residue catalyzes condensation between acyl-S-KS and the resultant carbon anion to form a C–C bond. A single amino acid substitution in the active site (a histidine replaced by Asn) would render the KS inactive. To confirm this argument, in a related paper, we demonstrated experimentally that when catalytic Cys residues of the tandem KSs of OzmH module 10 were site-specifically mutated to Gly, one mutation in the first KS (generating strain SDF7) allowed production of OZM to a level comparable with that of the wild type strain JA3453 (implying a redundancy), but a mutation in the second KS (generating strain SDF8) abolished OZM biosynthesis (23). Also, it was established that both of the C2′-Me groups of oxazolomycin come from methionine (9), but the presence of only one MT in module 6 led to the assumption that this MT domain catalyzes the two C2′ methyl-transfer processes iteratively (31). Furthermore, the OZM structure would suggest that an MT domain should be present in module 10 to account for the introduction of the methyl group at the C6 position; however, the conserved S-adenosyl-l-methionine binding motif EXGXGXG could not be identified in this module. Instead, this motif was unexpectedly found further upstream in module 9, leading to the prediction of a MT domain (31), which is presumably responsible for the introduction of the C6 methyl group.
The substrate specificity of the backbone-assembling NRPS OzmH was predicted to be glycine, based on its A domain specificity-conferring code and in harmony with isotope labeling studies (9). The A domain of OzmH was overproduced in E. coli BL21 (DE3) and purified to homogeneity. Thus, it was possible to further support this substrate specificity prediction by an in vitro amino acid-dependent radiolabel exchange assay between pyrophosphate ([32P]PPi) and ATP. Among the amino acids examined, glycine yielded the most efficient exchange with OzmH (Fig. 4), consistent with biomformatics-based predictions and previous results of the labeling assay.
FIGURE 4.
Determination of OzmH A domain substrate specificities. The ATP-PPi exchange reactions were performed using amino acids Gly, Ala, and Ser as substrates and H2O as a negative control (100% relative activity corresponds to 995,320 cpm).
Genes Encoding PKSs
Four PKS genes were identified in the ozm gene cluster, ozmQ, ozmN, ozmJ, and ozmK, that together encode six PKS modules (Figs. 2 and 3A). The ozmQ gene encodes a KS-ACP bidomain protein of 842 amino acids, constituting module 2. The ozmN gene encodes a protein of 4971 residues that can be subdivided into three modules (modules 3–5), with a total of three KS, three dehydratase, three KR, three ACP, and one MT domain. The ozmJ gene encodes a protein of 2926 residues, forming a complex PKS architecture of KS-ACP-KS-dehydratase-enoyl reductase-KR-ACP, which constitutes module 11, and this module is likely to be responsible for the incorporation of an unusual methoxymalonyl-ACP extender unit to afford the C3-C4 section of oxazolomycin. The first KS in OzmJ has a conserved catalytic triad of Cys-His-His, but the second KS possesses a Cys-Thr-His triad that might be inactive due to the change of a critical histidine residue for malonyl-ACP (or methyl/ethyl-malonyl-ACP) decarboxylation (29, 30). Additionally, two ACPs were identified within this module, both with the conserved signature motif (GX(H/D)S) required for post-translational attachment of the 4′-phosphopantetheine group to the serine residue (underlined). Otherwise, OzmK is characterized with the domain organization ACP-KS-MT-ACP to constitute module 12, featuring two ACPs. The N-terminal ACP contains a leucine substitution for serine at the site for 4′-phosphopantetheine attachment, rendering it inactive, whereas the C-terminal ACP with the conserved motif should be fully functional.
Genes Encoding Enzymes for Methoxymalonyl-ACP Biosynthesis
In previous studies (8), we have demonstrated in detail that the ozmBCDEFG gene subcluster constitutes a co-transcribed operon required for biosynthesis and incorporation of the methoxymalonyl-ACP extender unit into oxazolomycin (Fig. 2) because the numbers of nucleotides between the stop and start codon of the adjacent genes are all too small to code any regulatory elements for transcriptional initiation (8). Five of the six genes (with the exception of ozmC) are absolutely conserved among methoxy malonyl-ACP biosynthetic loci known to date, such as the ansamitocin P-3 (13), geldanamycin (14), FK520 (17), and tautomycin (32) gene clusters. For example, OzmG (287 amino acids) closely resembles members of the HADH family of enzymes, such as Asm13 (61% identity) (13), GdmK (61% identity) (14), FkbK (62% identity) (17), SorD (40% identity) (15), and TtmE (64% identity) (32). Similarly, OzmF (221 amino acids) is a probable O- methyltransferase, OzmE is a probable ACP, and OzmD is an ADH. With regard to OzmB, a related study (10) confirmed by in vitro assays that it acts as a bifunctional glyceryl transferase/phosphatase belonging to the HAD superfamily that diverts the primary metabolite 1,3-bisphosphoglycerate into a phosphoglyceryl-S-OzmB intermediate and then removes the phosphate group to afford 3-glyceryl-S-OzmB (acting as a phosphatase) and finally transfers the glyceryl group to an ACP (acting as a glyceryl transferase) to set the stage for polyketide biosynthesis (Fig. 3B).
As was described before (8), OzmC is a unique enzyme that has not been found in other methoxymalonyl-ACP biosynthesis loci of type I PKSs. OzmC has 25% protein sequence identity to DpsC from Streptomcyes peucetius for doxorubicin biosynthesis, whereas DpsC has been shown in vitro to function both as an acyltransferase transferring the propionyl group from propionyl-CoA to an ACP and as a KS III catalyzing the condensation of propionyl-CoA to malonyl-ACP (33). Previous gene disruption and complementation experiments have confirmed that OzmC plays a crucial role in oxazolomycin biosynthesis (8). OzmC therefore serves as a candidate for the discrete AT loading the methoxymalonyl-ACP extender to module 11 of the OZM AT-less type I PKS.
A hypothesis for the biosynthesis of methoxymalonyl-ACP was proposed as follows. The bifunctional glyceryl transferase/phosphatase OzmB (10), sequesters and dephosphorylates 1,3-bisphospho-d-glycerate, and the resultant glyceryl moiety is transferred to OzmE (ACP), forming glyceryl-ACP, which is then oxidized in steps into hydroxymalonyl-ACP by OzmG, a 3-hydroxyacyl-CoA dehydrogenase, and OzmD, an acyl-CoA dehydrogenase. Finally, a methyl group is transferred onto hydroxymalonyl-ACP by an O-methyltransferase, OzmF, generating the methoxymalonyl-ACP intermediate as an unusual polyketide extender unit (10, 11) (Fig. 3B).
Genes Encoding Resistance and Regulatory Proteins and Proteins of Unknown Functions
As we described before (8), the ozmA gene encodes a putative multidrug transporter that resembles the following proteins: SgcB (accession number AAF13999; 28% identity) in enediyne C-1027 biosynthesis from Streptomyces globisporus (34); RemN (accession number CAE51185; 26% identity) in resistomycin biosynthesis from Streptomyces resistomycificus (35); and EncT (accession number AAF81738; 27% identity) in enterocin biosynthesis from Streptomyces maritimus (36). The deduced gene product of ozmS belongs to the LysE family of transporters (37). OzmA and OzmS together therefore may confer oxazolomycin resistance in S. albus JA3453 via drug transport.
The ozmR and ozmU genes encode hypothetical regulatory proteins. OzmR shows significant similarity to the LysR family of regulators, such as Orf5 (accession number BAA32133; 53% identity) from Streptomyces griseus (38), whereas OzmU belongs to the SARP family of regulators, including Orf31* (accession number CAG15043; 37% identity) involved in glycopeptide teicoplanin biosynthesis in Actinoplanes teichomyceticus (39), AfsR (accession number BAA14186; 33% identity) in Streptomyces coelicolor, a pleiotropic antibiotic regulator (40), and RubS (accession number AAM97369; 30% identity) involved in rubromycin biosynthesis in Streptomyces collinus. Although OzmT resembles other Thr-tRNA synthetases, such as SCH63.25 (CAC 10316, 77/83) in S. coelicolor, its exact roles still need to be determined.
The deduced gene product of ozmP shows little homology to known proteins when searched against gene databases, but its N terminus shows some resemblance to a subfamily of ATP pyrophosphatase and members of the ATP sulfurylase superfamily with a highly conserved SGGKD motif (41). The gene ozmP along with ozmO and ozmQ constitute an operon that may be co-transcribed because the gaps between these three genes are too small to encode a promoter, but the exact role of OzmP for OZM biosynthesis is still elusive.
A Gene Encoding a Discrete AT Enzyme
As described above, the oxazolomycin polyketide synthases OzmHJKNQ lack modular acyltransferases in all 10 PKS modules (Fig. 3A). Instead, a discrete AT enzyme encoded by ozmM was characterized in the ozm gene cluster. OzmM contains tandem AT domains along with an oxidoreductase domain (Figs. 2 and 3A), which was postulated to act as a trans- acyltransferase to load extender units onto the corresponding ACPs. Both the N-terminal domain (OzmM-AT1) and the central domain (OzmM-AT2) of OzmM closely resemble other modular or discrete ATs, and the conserved active site residues (Ser81, Ser402, His182, and His506) were characterized (1, 42). However, two other highly crucial conserved residues vary for OzmM-AT1: Ala80 in place of His and Arg106 in place of Gln. Mutation of these two residues was also observed for the AT1 domains of other OzmM homlogs featuring tandem ATs, such as KirC1 AT1 (accession number CAN89639) and MmpIII AT1 (accession number AAM12912), which are involved in kirromycin and mupirocin biosynthesis, respectively.
The C terminus of OzmM encodes a putative oxidoreductase domain that has high sequence homology to the C terminus of LnmG, a domain of a trans-AT enzyme (accession number AAN85520; 49% identity) (43). Oxidoreductase domains are found associated with AT domains in many examples of trans-AT type I PKSs, exemplified by MmpIII (accession number AAM12912; 47% identity) for mupirocin biosynthesis in Pseudomonas fluorescens NCIMB 10586 (44), PedB (accession number AAS47558; 40% identity) for pederin biosynthesis in the symbiont bacterium of Paederus fuscipes beetles (45), and ChiA (accession number AAY89048; 43% identity) for chivosazol biosynthesis in Sorangium cellulosum So ce56 (46).
To investigate the function of OzmM, the ozmM gene was inactivated by an in-frame deletion via the REDIRECT system, giving a non-OZM-producing mutant strain, ZH9 (Fig. 5, A and B). Furthermore, a construct, pJTU1078, harboring an intact functional copy of ozmM under the control of the ErmE* promoter was made in pJTU1351 (8) and introduced into ZH9 by E. coli-Streptomyces biparental conjugation, yielding S. albus ZH10 with restored OZM production comparable with wild type S. albus JA3453 (Fig. 5, A and B).
Two additional complementation constructs of the ozmM gene in pJTU1351 (8) were made, in which the conserved active site serine residues (employed to form the acyl-O-intermediate before transferring acyl groups from their CoA substrate to the nucleophile recipient ACP) in OzmM-AT1 or OzmM-AT2 were separately mutated to glycine. These plasmids, pJTU1135 (for OzmM-AT1 (S81G)) and pJTU1136 (for OzmM-AT2 (S402G)), were introduced into the non-OZM-producing mutant strain S. albus ZH9 by conjugation, giving two new strains, S. albus ZH14 (with pJTU1135) and S. albus ZH15 (with pJTU1136) (Fig. 5, A and B). Both strains were fermented and analyzed for OZM production by HPLC. All assays were performed under the same conditions as those for wild type JA3453 and mutant ZH10. S. albus ZH15 failed to produce OZM, whereas S. albus ZH14 regained production of OZM, comparable with wild type S. albus JA3453, which was confirmed by liquid chromatography-mass spectrometry analysis (Fig. 5, A and B).
DISCUSSION
A preliminary delimitation of the ozm gene cluster boundaries was carried out by inactivating the genes upstream of ozmA and downstream of ozmU, and the results agreed with bioinformatics analysis that predicted the ozm gene cluster to consist of 20 ORFs (Fig. 2). We also conclude that ozmA and ozmU are in the ozm gene cluster because they highly resemble genes for other natural product biosynthesis, responsible for resistance and regulation, respectively (see “Results”). Detailed analysis of the genes in the ozm cluster led to a model for oxazolomycin biosynthesis, featuring PKSs (OzmJKMNQ), NRPSs (OzmO and OzmL), and a hybrid NRPS-PKS megasynthase (OzmH) (Fig. 2). In this proposed mechanism, oxazolomycin biosynthesis is initiated by OzmO, which functions as an NRPS loading module, and glycine would be activated by the OzmO A domain and formylated to yield formyl-glycyl-S-PCP. This intermediate could then be transferred sequentially to OzmQ (module 2), OzmN (modules 3–5), OzmH (modules 6–10), OzmJ (module 11), OzmK (module 12), and OzmL (module 13), where condensation with five malonyl-CoAs, a glycine, three malonyl-CoAs, a methoxymalonyl-ACP, a malonyl-CoA, and a serine as building blocks takes place, as well as six C- and N-methylation events employing S-adenosyl-l-methionine. In this process, the modules 1, 7, and 13 are NRPSs, and modules 2, 3, 4, 5, 6, 8, 9, 10, 11, and 12 are PKSs. Transitioning of the growing intermediates from an NRPS to a PKS module or vice versa occurs either between two peptides (OzmO/OzmQ and OzmK/OzmL) or between modules on the same peptide (OzmH) (Fig. 2).
Oxazolomycin biosynthesis joins the rapidly growing family of pathways featuring trans-AT type I PKSs (2). However, to the best of our knowledge, our entity is the only one reported to incorporate a methoxymalonyl-ACP extender unit and one of the few examples reported to incorporate two different extender units by stand-alone AT(s). The other PKS that has been proposed to incorporate both malonyl-CoA and ethylmalonyl-CoA is from kirromycin biosynthesis, with a combination of both cis- and trans-acyltransferases (5). Likewise, etnangien biosynthetic gene cluster featuring trans- ATs (EtnB and EtnK) was postulated to recruit both malonyl-CoA and succinyl-CoA extender units. An understanding of how the oxazolomycin trans-PKS is capable of accepting both malonyl-CoA and methoxymalonyl-ACP extender units is highly desirable. Misled by the tandem ATs in OzmM, we initially assumed that the two ATs are specific for the methoxymalonyl-ACP and malonyl-CoA extender unit, respectively. However, only OzmM-AT2 was experimentally proven to be necessary, whereas OzmM-AT1 is dispensable for OZM production (Fig. 5, A and B). When the catalytic active site Ser residue (employed to form the acyl-O-intermediate before transferring acyl groups from their CoA substrate to the nucleophile recipient ACP) in the OzmM-AT2 was specifically mutated (i.e. OzmM-AT2 (S402G)), OZM biosynthesis in the resultant strain ZH15 was completely abolished, as when OzmM was deleted entirely. In contrast, the same mutation in OzmM-AT1 (i.e. OzmM-AT1 (S81G)) caused no change in the level of OZM production in the resultant strain ZH14, suggesting that OzmM-AT1 is cryptic or serves as an inactive domain.
Phylogenetic tree analysis of selected cis- and trans-ATs showed that the tandem AT domains from a single trans-AT enzyme, such as OzmM-AT1 and OzmM-AT2, belong to different subgroups. In fact, the OzmM-AT1 domain forms a clade along with TaV-AT1 and MmpIII-AT1, which may represent inactive AT domains. Thus, TaV, an OzmM homolog from the myxovirescin biosynthetic pathway, was described by Walsh and co-workers (47) as having a PKS featuring tandem AT domains predicted to load variable starter units, but biochemical investigation of TaV could only find activity for the second AT domain. The first AT was characterized to lack a predicted ability to transfer a propionyl group in vitro using succinyl-, methylmalonyl-, or propionyl-CoA as the substrates (and was thus regarded as a cryptic AT), whereas the second AT could transfer malonyl-CoA as predicted (47). Recently, the AT1 domain of KirC1, another homolog of OzmM-AT1 involved in kirromycin biosynthesis, was suggested to be inactive based on bioinformatic analysis (5). Another publication about the migrastatin biosynthetic gene cluster also demonstrated that MgsB may serve as a cryptic acyltransferase, whereas MgsH may act as the sole AT for iso-MGS biosynthesis because the ΔmgsB gene mutant is still capable of producing iso-MGS (48). Here we present the in vivo data to show that these trans-AT domains with the catalytic Ser (amino acid residue 81) and His (amino acid residue 182) (numbering corresponding to OzmM-AT1) but with mutations at another two highly conserved amino acids, His (amino acid residue 80) and Arg (amino acid residue 106), may be inactive.
The fact that two different extender units, malonyl-CoA and methoxymalonyl-ACP, are incorporated into the carbon backbone of the OZM molecular scaffold raises the interesting question of how trans-AT(s) accommodates different extender units for polyketide biosynthesis. Based on its similarity to other discrete AT domains, the OzmM-AT2 domain is most likely responsible for the loading of the malonyl-CoA extender unit to all of the OZM AT-less PKS modules except for module 11 (Fig. 3A). This would also suggest that another AT or AT counterpart either inside or outside the gene cluster is needed for loading the methoxymalonyl-ACP extender unit module 11. A good candidate may be OzmC because it is clustered with other methoxymalonyl-ACP biosynthesis genes (OzmBDEFG). The ozmC gene has been shown to be critical for OZM production by in vivo inactivation experiments, yet the OzmC homolog is absent from all known type I PKSs that incorporate the methoxmalonyl-ACP extender unit with their cognate AT domains (6). In vivo investigations into the role of its homolog DspC revealed that it is required for doxorubicin polyketide chain initiation, and when the gene encoding DspC was knocked out, the mutant became non-selective for the recruitment of malonyl-CoA or methylmalonyl-CoA as the starter unit, whereas the wild type only accepted methylmalonyl-CoA as initiation moiety (33, 49). DpsC has also been shown in vitro to function both as an acyltranferase transferring the propionyl group from propionyl-CoA to an ACP and as a KS III catalyzing the condensation of propionyl-CoA to malonyl-ACP (33). That means that “AT-less” type II PKSs (doxorubicin polyketide) could recruit multiple AT enzymes to load different initiation or extender units, and these AT enzymes may have little or no homology to cognate AT domains that are capable of transferring the same extender units. Thus, we may assume OzmC to be involved in selectively loading methoxymalonyl-ACP (acting as another AT) rather than the usual malonyl-CoA extender unit onto the module 11 ACP of the oxazolomycin trans-AT PKS (Fig. 3A). Pending biochemical confirmation, OzmC represents the first AT enzyme with little sequence homology to known AT enzymes for PKSs to be recruited by AT-less type I PKSs to accommodate a novel extender unit.
The presence of the F domain in OzmO, coupled with the fact that the oxazole ring C11′ can be labeled by [13C]glycine and the predicted Gly specificity for the loading module A domain, suggested that formylglycyl-S-PCP was formed followed by cyclization to generate the oxazole ring moiety for OZM biosynthesis. We propose that the OzmO A domain first activates glycine and loads it onto the OzmO PCP domain, and then the F domain, which is analogous to the loading module LgrAl for gramicidin biosynthesis (25), formylates on glycyl-S-PCP, giving formyl-glycyl-S-PCP. The enzyme responsible for the conversion of formyl-glycyl-S-PCP into the oxazole ring is still unknown, as is the timing of the cyclization process (Fig. 3A). Bioinformatic analysis failed to predict such a cyclase gene or domain within the ozm cluster, although OzmP, adjacent to and co-transcribed with OzmO, whose function could not be assigned hitherto, may be involved in oxazole ring biosynthesis.
Module 13 of OzmL features two C domains, terminating oxazolomycin biosynthesis with the concomitant formation of the spiro-linked β-lactone/γ-lactam moiety (Fig. 3A). First, the amino acid serine is activated by the adenylation domain and covalently bound to the peptidyl carrier protein, and then the N-terminal C domain catalyzes formation of the peptide bond between seryl-S-PCP and the preceding acyl-S-ACP intermediate, followed by N- methylation by the MT domain. The C domain residing at the C terminus was proposed to release the full-length hybrid peptide-polyketide product from the PCP, giving the four-membered β-lactone ring, because no thioesterase domain was identified in the OZM gene cluster. Although the C-terminal C domain lacks a full catalytic triad for condensation (NYFCLDG), it does have the Asp residue that is conserved in both C and heterocyclase (Cy) domains. Cy domains are a subtype of C domains that catalyze not only peptide bond formation but also subsequent cyclization of Cys, Ser, or Thr to afford a thiazoline or oxazoline ring, with the conserved DXXXXD motif rather than the HHXXXDG motif of routine C domains. Although the C-terminal C domain in OzmL has a catalytic motif NYFCLDG and lacks the crucial His amino acid that is believed to be indispensable for condensation of aminoacyl-ACP and peptidyl-ACP, it may still be functional, especially given that this domain is not expected to catalyze peptide bond formation but only cyclization of the Ser side chain to form a β-lactone ring. Some recently characterized C domains exemplified by SgcC5 (50) and Fum14 (51) have been biochemically demonstrated to catalyze ester bond formation. A similar function would be envisioned for the OzmL C-terminal C domain to be involved in β-lactone ring formation. On the other hand, a cyclase may be required for C3–C15 bond formation to afford the heterocyclic γ-lactam ring, but no obvious candidate gene could be identified in the ozm cluster by bioinformatic analysis (Fig. 2).
This oxazolomycin NRPS-PKS megasynthase was also characterized with many features that appear to violate the typical “co-linearity rule” for NRPS or PKS domain organization, including domain redundancy and mispositioning (1, 52), where domain redundancies could be observed by the presence of two ACPs flanking the MT and KR domain in module 9, tandem KSs in module 10, and two ACPs and two KSs in module 11. In addition, an MT domain should be present in module 10 as deduced from the OZM chemical structure, but instead an MT was unexpectedly identified in module 9. This gene organization may be explained as domain mispositioning, reflecting complex domain-domain interactions for polyketide-catalyzed methylation (Fig. 3A). One example of domain redundancy has been confirmed experimentally where the catalytic Cys residues of the tandem KSs of OzmH module 10 were site-specifically mutated to Gly (23).
Acknowledgments
We acknowledge Prof. Sir David A. Hopwood, FRS (John Innes Center, Norwich, UK) for much valuable advice and critical editing of the present manuscript before submission. We thank the Analytical Instrumentation Center of the School of Pharmacy, University of Wisconsin (Madison, WI), for obtaining mass spectrometry data; the John Innes Center for providing the REDIRECT Technology kit; and Werner F. Fleck (Hans Knoell Institute for Natural Product Research, Jena, Germany) for providing the wild type S. albus JA3453 strain.
This work was supported in part by National Institutes of Health Grant CA113297 (to B. S.). This work was also supported by the Ministry of Science and Technology of China 973 (2003CB114205) and 863 programs, the National Science Foundation of China, the Chinese Ministry of Education, and the Shanghai Municipal Council of Science and Technology (to Z. D.).
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) EF552687.
- PKS
- polyketide synthase
- A domain
- adenylation domain
- C domain
- condensation domain
- F domain
- formylation domain
- ACP
- acyl carrier protein
- AT
- acyltransferase
- KR
- ketoreductase
- KS
- ketosynthetase
- MT
- methyltransferase
- PCP
- peptidyl carrier protein
- NRPS
- non-ribosomal peptide synthetase
- HPLC
- high pressure liquid chromatography
- ORF
- open reading frame.
REFERENCES
- 1.Staunton J., Weissman K. J. (2001) Nat. Prod. Rep. 18, 380–416 [DOI] [PubMed] [Google Scholar]
- 2.Nguyen T., Ishida K., Jenke-Kodama H., Dittmann E., Gurgui C., Hochmuth T., Taudien S., Platzer M., Hertweck C., Piel J. (2008) Nat. Biotechnol. 26, 225–233 [DOI] [PubMed] [Google Scholar]
- 3.Cheng Y. Q., Tang G. L., Shen B. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3149–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng Y. Q., Coughlin J. M., Lim S. K., Shen B. (2009) Methods Enzymol. 459, 165–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber T., Laiple K. J., Pross E. K., Textor A., Grond S., Welzel K., Pelzer S., Vente A., Wohlleben W. (2008) Chem. Biol. 15, 175–188 [DOI] [PubMed] [Google Scholar]
- 6.Wenzel S. C., Williamson R. M., Grünanger C., Xu J., Gerth K., Martinez R. A., Moss S. J., Carroll B. J., Grond S., Unkefer C. J., Müller R., Floss H. G. (2006) J. Am. Chem. Soc. 128, 14325–14336 [DOI] [PubMed] [Google Scholar]
- 7.Bagwell C. L., Moloney M. G., Thompson A. L. (2008) Bioorg. Med. Chem. Lett. 18, 4081–4086 [DOI] [PubMed] [Google Scholar]
- 8.Zhao C., Ju J., Christenson S. D., Smith W. C., Song D., Zhou X., Shen B., Deng Z. (2006) J. Bacteriol. 188, 4142–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grafe U., Kluge H., Thiericke R. (1992) Liebigs Ann. Chem. 1992, 429–432 [Google Scholar]
- 10.Dorrestein P. C., Van Lanen S. G., Li W., Zhao C., Deng Z., Shen B., Kelleher N. L. (2006) J. Am. Chem. Soc. 128, 10386–10387 [DOI] [PubMed] [Google Scholar]
- 11.Chan Y. A., Boyne M. T., 2nd, Podevels A. M., Klimowicz A. K., Handelsman J., Kelleher N. L., Thomas M. G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14349–14354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll B. J., Moss S. J., Bai L., Kato Y., Toelzer S., Yu T. W., Floss H. G. (2002) J. Am. Chem. Soc. 124, 4176–4177 [DOI] [PubMed] [Google Scholar]
- 13.Yu T. W., Bai L., Clade D., Hoffmann D., Toelzer S., Trinh K. Q., Xu J., Moss S. J., Leistner E., Floss H. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7968–7973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rascher A., Hu Z., Viswanathan N., Schirmer A., Reid R., Nierman W. C., Lewis M., Hutchinson C. R. (2003) FEMS Microbiol. Lett. 218, 223–230 [DOI] [PubMed] [Google Scholar]
- 15.Ligon J., Hill S., Beck J., Zirkle R., Molnár I., Zawodny J., Money S., Schupp T. (2002) Gene 285, 257–267 [DOI] [PubMed] [Google Scholar]
- 16.Li W., Ju J., Osada H., Shen B. (2006) J. Bacteriol. 188, 4148–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu K., Chung L., Revill W. P., Katz L., Reeves C. D. (2000) Gene 251, 81–90 [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa J., Hotta K. (1999) FEMS Microbiol. Lett. 174, 251–253 [DOI] [PubMed] [Google Scholar]
- 19.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gust B., Challis G. L., Fowler K., Kieser T., Chater K. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar P., Koppisch A. T., Cane D. E., Khosla C. (2003) J. Am. Chem. Soc. 125, 14307–14312 [DOI] [PubMed] [Google Scholar]
- 22.Stachelhaus T., Mootz H. D., Marahiel M. A. (1999) Chem. Biol. 6, 493–505 [DOI] [PubMed] [Google Scholar]
- 23.Song D., Coughlin J., Ju J., Zhou X., Shen B., Zhao C., Deng Z. (2008) Acta Biochim. Biophys. Sin. 40, 319–326 [DOI] [PubMed] [Google Scholar]
- 24.Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000) Practical Streptomyces Genetics, John Innes Foundation Norwich, UK [Google Scholar]
- 25.Schoenafinger G., Schracke N., Linne U., Marahiel M. A. (2006) J. Am. Chem. Soc. 128, 7406–7407 [DOI] [PubMed] [Google Scholar]
- 26.Rouhiainen L., Paulin L., Suomalainen S., Hyytiäinen H., Buikema W., Haselkorn R., Sivonen K. (2000) Mol. Microbiol. 37, 156–167 [DOI] [PubMed] [Google Scholar]
- 27.Stachelhaus T., Mootz H. D., Bergendahl V., Marahiel M. A. (1998) J. Biol. Chem. 273, 22773–22781 [DOI] [PubMed] [Google Scholar]
- 28.Tanovic A., Samel S. A., Essen L. O., Marahiel M. A. (2008) Science 321, 659–663 [DOI] [PubMed] [Google Scholar]
- 29.He M., Varoglu M., Sherman D. H. (2000) J. Bacteriol. 182, 2619–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olsen J. G., Kadziola A., von Wettstein-Knowles P., Siggaard-Andersen M., Lindquist Y., Larsen S. (1999) FEBS Lett. 460, 46–52 [DOI] [PubMed] [Google Scholar]
- 31.Kagan R. M., Clarke S. (1994) Arch. Biochem. Biophys. 310, 417–427 [DOI] [PubMed] [Google Scholar]
- 32.Li W., Ju J., Rajski S. R., Osada H., Shen B. (2008) J. Biol. Chem. 283, 28607–28617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bao W., Sheldon P. J., Hutchinson C. R. (1999) Biochemistry 38, 9752–9757 [DOI] [PubMed] [Google Scholar]
- 34.Liu W., Christenson S. D., Standage S., Shen B. (2002) Science 297, 1170–1173 [DOI] [PubMed] [Google Scholar]
- 35.Jakobi K., Hertweck C. (2004) J. Am. Chem. Soc. 126, 2298–2299 [DOI] [PubMed] [Google Scholar]
- 36.O'Keeffe T., Hill C., Ross R. P. (1999) Appl. Environ. Microbiol. 65, 1506–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vrljic M., Sahm H., Eggeling L. (1996) Mol. Microbiol. 22, 815–826 [DOI] [PubMed] [Google Scholar]
- 38.Klauck E., Böhringer J., Hengge-Aronis R. (1997) Mol. Microbiol. 25, 559–569 [DOI] [PubMed] [Google Scholar]
- 39.Li T. L., Huang F., Haydock S. F., Mironenko T., Leadlay P. F., Spencer J. B. (2004) Chem. Biol. 11, 107–119 [DOI] [PubMed] [Google Scholar]
- 40.Floriano B., Bibb M. (1996) Mol. Microbiol. 21, 385–396 [DOI] [PubMed] [Google Scholar]
- 41.Savage H., Montoya G., Svensson C., Schwenn J. D., Sinning I. (1997) Structure 5, 895–906 [DOI] [PubMed] [Google Scholar]
- 42.Reeves C. D., Murli S., Ashley G. W., Piagentini M., Hutchinson C. R., McDaniel R. (2001) Biochemistry 40, 15464–15470 [DOI] [PubMed] [Google Scholar]
- 43.Tang G. L., Cheng Y. Q., Shen B. (2004) Chem. Biol. 11, 33–45 [DOI] [PubMed] [Google Scholar]
- 44.El-Sayed A. K., Hothersall J., Cooper S. M., Stephens E., Simpson T. J., Thomas C. M. (2003) Chem. Biol. 10, 419–430 [DOI] [PubMed] [Google Scholar]
- 45.Piel J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14002–14007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perlova O., Gerth K., Kaiser O., Hans A., Müller R. (2006) J. Biotechnol. 121, 174–191 [DOI] [PubMed] [Google Scholar]
- 47.Calderone C. T., Iwig D. F., Dorrestein P. C., Kelleher N. L., Walsh C. T. (2007) Chem. Biol. 14, 835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim S. K., Ju J., Zazopoulos E., Jiang H., Seo J. W., Chen Y., Feng Z., Rajski S. R., Farnet C. M., Shen B. (2009) J. Biol. Chem. 284, 29746–29756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rajgarhia V. B., Priestley N. D., Strohl W. R. (2001) Metab. Eng. 3, 49–63 [DOI] [PubMed] [Google Scholar]
- 50.Lin S., Van Lanen S. G., Shen B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4183–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaleta-Rivera K., Xu C., Yu F., Butchko R. A., Proctor R. H., Hidalgo-Lara M. E., Raza A., Dussault P. H., Du L. (2006) Biochemistry 45, 2561–2569 [DOI] [PubMed] [Google Scholar]
- 52.Finking R., Marahiel M. A. (2004) Annu. Rev. Microbiol. 58, 453–488 [DOI] [PubMed] [Google Scholar]