Abstract
Mammalian target of rapamycin (mTOR) is a member of the phosphatidylinositol 3-kinase-related kinase (PIKK) family and is a major regulator of translation, cell growth, and autophagy. mTOR exists in two distinct complexes, mTORC1 and mTORC2, that differ in their subunit composition. In this study, we identified KIAA0406 as a novel mTOR-interacting protein. Because it has sequence homology with Schizosaccharomyces pombe Tti1, we named it mammalian Tti1. Tti1 constitutively interacts with mTOR in both mTORC1 and mTORC2. Knockdown of Tti1 suppresses phosphorylation of both mTORC1 substrates (S6K1 and 4E-BP1) and an mTORC2 substrate (Akt) and also induces autophagy. S. pombe Tti1 binds to Tel2, a protein whose mammalian homolog was recently reported to regulate the stability of PIKKs. We confirmed that Tti1 binds to Tel2 also in mammalian cells, and Tti1 interacts with and stabilizes all six members of the PIKK family of proteins (mTOR, ATM, ATR, DNA-PKcs, SMG-1, and TRRAP). Furthermore, using immunoprecipitation and size-exclusion chromatography analyses, we found that knockdown of either Tti1 or Tel2 causes disassembly of mTORC1 and mTORC2. These results indicate that Tti1 and Tel2 are important not only for mTOR stability but also for assembly of the mTOR complexes to maintain their activities.
Keywords: Autophagy, mTOR, mTOR Complex (mTORC), Protein Stability, Protein-Protein Interactions
Introduction
Mammalian target of rapamycin (mTOR)4 is a member of the phosphatidylinositol 3-kinase-related protein kinase (PIKK) family, which also includes ataxia telangiectasia mutated (ATM), ATM- and Rad3-related (ATR), DNA-PKcs, suppressor with morphological effect on genitalia 1 (SMG-1), and the catalytically inactive transformation/transcription domain-associated protein (TRRAP) (1–3). mTOR forms two distinct complexes that differ in their subunit composition, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). mTORC1 includes mTOR, mLST8 (also known as GβL), and Raptor, whereas mTORC2 includes mTOR, mLST8, mSin1, and Rictor (4–7). mTORC1 is activated by several factors, including amino acids. It promotes protein translation through phosphorylation of S6K1 and 4E-BP1 (7), and inhibits autophagy through phosphorylation of ULK1 and Atg13 (8–10). On the other hand, mTORC2 is activated by growth factors and phosphorylates Akt at Ser-473, and plays key roles in cell survival, metabolism, proliferation, and organization of the cytoskeleton (7).
Although growing numbers of mTOR-interacting proteins have been identified, little is known about the assembly of these mTOR complexes. Deletion of the mTORC2 components such as Rictor, mSin1, and mLST8 induces disassembly of mTORC2, impairing Akt phosphorylation (at Ser-473) (11–13), whereas mLST8 is not required for mTOR-Raptor interaction and S6K1 activation (11). Prolonged treatment with rapamycin, which is an mTORC1 inhibitor, inhibits mTORC2 assembly (14). Recently, it was proposed that phosphatidic acid is required for the assembly of mTOR complexes, because depletion of phosphatidic acid caused by treatment with 1-butanol and expression of dominant negative mutant phospholipase D results in the disassembly of both mTORC1 and mTORC2 (15, 16). However, there have been no reports on the existence of any proteins that directly regulate mTOR complex assembly, other than the core subunits of the complexes.
In this study, we identified mammalian Tti1 (KIAA0406) as a novel mTOR-interacting protein. Tti1 constitutively interacts with mTOR and positively regulates the activity of both mTORC1 and mTORC2. As in Schizosaccharomyces pombe (17, 18), Tti1 interacts with telomere maintenance 2 (Tel2, also described as HCLK2), which was recently reported to regulate stability of all six PIKK family proteins (19). Similar to Tel2, Tti1 interacts with and stabilizes all the PIKK proteins. Furthermore, we found that depletion of either Tti1 or Tel2 causes the disassembly of mTOR complexes. Our results demonstrate, for the first time, that Tti1 and Tel2 have a role in mTOR complex formation, which is crucial for maintaining mTOR activity.
EXPERIMENTAL PROCEDURES
Plasmids
The cDNA encoding human Tti1 (KIAA0406) or human Tel2 (HCLK2) was obtained from the Kazusa DNA Research Institute and was cloned into p3xFLAG CMV10 (Sigma). FLAG-mTOR was described previously (20).
Cell Culture and Transfection
HeLa and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 μg/ml penicillin, and streptomycin (complete medium) in a 5% CO2 incubator. For deprivation of serum and amino acids, cells were washed with phosphate-buffered saline and incubated in amino acid-free DMEM (Invitrogen) without FBS. FuGENE 6 (Roche Applied Science) was used for transfection.
Antibodies and Reagents
Anti-Tti1 rabbit polyclonal antibody was produced against the peptide ASGQQNPYTTNVLQLLKELQ (amino acid 1070–1089 of human Tti1) by Immuno-Biological Laboratories. Anti-Tel2 antibody was generated as described below. cDNA encoding the C-terminal region (amino acid 613–838) of human Tel2 was subcloned into pGEX6p1 (Amersham Biosciences). The Tel2 fragment tagged with N-terminal glutathione S-transferase was expressed in Escherichia coli strain BL21(DE3)pLysS (Novagen) and then used to immunize rabbits. Anti-mTOR mouse monoclonal antibody (N5D11) was previously described (21) and used for immunoprecipitation. Anti-LC3 (microtubule-associated protein 1 light chain 3) rabbit polyclonal antibody was previously described (22). Anti-FLAG (M2) antibody (F3165) and M2 agarose beads were purchased from Sigma. Anti-HA antibody (16B12) was purchased from BabCO. Antibodies against 4E-BP1 (#9452), 4E-BP1 pS65 (#9451), S6K1 (#9202), S6K1 pT389 (#9204), Akt (#9272), Akt pS473 (#4051S), mTOR (#2972), mTOR pS2448 (#2971), Raptor (#4978), Rictor (#2114), mLST8 (#2114), ATM (#2873), and TRRAP (#3967) were purchased from Cell Signaling Technology. Anti-ATR antibody (sc-1887) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-DNA-PKcs antibody (MS-423-P0) was purchased from Lab Vision. Anti-SMG-1 antibody (A300–393A) was purchased from Bethyl Laboratories. Anti-Hsp90 antibody (610418) was purchased from BD Biosciences. Rapamycin was purchased from Calbiochem.
Mass Spectrometry
Liquid chromatography/tandem mass spectrometry was performed as previously described (23).
Immunoprecipitation and Immunoblotting
Cell lysates were prepared in a regular lysis buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 10 mm NaF, 0.4 mm Na3VO4, 10 mm sodium pyrophosphate, and Complete EDTA-free protease inhibitor (Roche Applied Science)). The lysates were clarified by centrifugation at 15,000 rpm for 15 min and then boiled in sample buffer. For immunoprecipitation analysis, cell lysates were prepared in a second lysis buffer (40 mm HEPES (pH 7.5), 150 mm NaCl, 2 mm EDTA, 0.3% CHAPS, 10 mm NaF, 0.4 mm Na3VO4, 10 mm sodium pyrophosphate, and Complete EDTA-free protease inhibitor (Roche Applied Science)). The lysates were clarified by centrifugation at 15,000 rpm for 15 min and then subjected to immunoprecipitation using specific antibodies in combination with protein A- or G-Sepharose (Amersham Biosciences). Precipitated immunocomplexes were washed five times in washing buffer (40 mm HEPES (pH 7.5), 150 mm NaCl, 2 mm EDTA, and 0.3% CHAPS) and boiled in SDS-sample buffer. Samples were subsequently separated by SDS-PAGE and transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore). Immunoblot analysis was performed with the indicated antibodies and visualized with SuperSignal West Pico Chemiluminescent substrate (Pierce) or Immobilon Western (Millipore). The signal intensities were analyzed using an LAS-3000mini imaging analyzer and Multi Gauge software version 3.0 (Fujifilm). Contrast and brightness adjustment was applied to the whole images using Photoshop Elements 5.0.
RNA Interference
Stealth RNAi oligonucleotides (Invitrogen) were transfected into the cells using Lipofectamine RNAi MAX (Invitrogen) according to the manufacturer's protocols. The sequences used were as follows (described as sense sequence): Tti1: 5′-UUAUCUGACAACAAGUGGAUGCAGC-3′; Tel2: 5′-AGAACUGUGAGGUCAGAUAGUCGGC-3′. As a negative control, Medium GC duplex of stealth RNAi Negative Control Duplexes (Invitrogen) was used. After 2 days, the cells were transfected again using the same procedure. Another 2 days later, cells were harvested and analyzed.
Gel Filtration Analysis
Cells were lysed in hypotonic buffer (40 mm Tris-HCl (pH 7.5), and Complete EDTA-free protease inhibitor (Roche Applied Science)) by repeated passage (15 times) through a 1-ml syringe with a 27-gauge needle. The lysates were centrifuged at 2,000 rpm for 5 min, and the supernatants were further centrifuged at 43,000 rpm for 30 min. The supernatant fraction was then applied to a Superose 6 column (Amersham Biosciences) and eluted at a flow rate of 0.5 ml/min with 40 mm Tris-HCl (pH 7.5) and 150 mm NaCl. Fractions (0.5 ml) were then analyzed by immunoblotting. The column was calibrated with thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), albumin (67 kDa), ovalbumin (43 kDa), and cytochrome c (12.5 kDa).
RESULTS
Identification of Mammalian Tti1 as a Novel mTOR-binding Protein
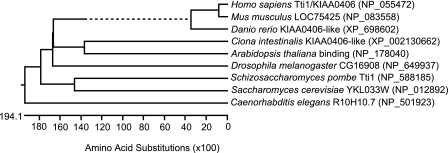
To search for mTOR-interacting proteins, FLAG-tagged mTOR was overexpressed in HEK293T cells, and the co-immunoprecipitates were analyzed by direct nano-flow liquid chromatography/tandem mass spectrometry (23). A protein with unknown function, KIAA0406, was detected. This protein is evolutionally conserved in eukaryotes (Fig. 1). Recently, its homolog in S. pombe was reported with the name Tel two interacting protein 1 (Tti1) (17). Thus, KIAA0406 was named mammalian Tti1.
FIGURE 1.
Evolutional conservation of Tti1/KIAA0406. Homologs of Tti1 were identified by pBLAST search. The protein sequences were aligned by ClustalW method, and a phylogenic tree was described using Lasergene software (version 8.0.2, DNASTAR). The dotted line indicates a negative branch length.
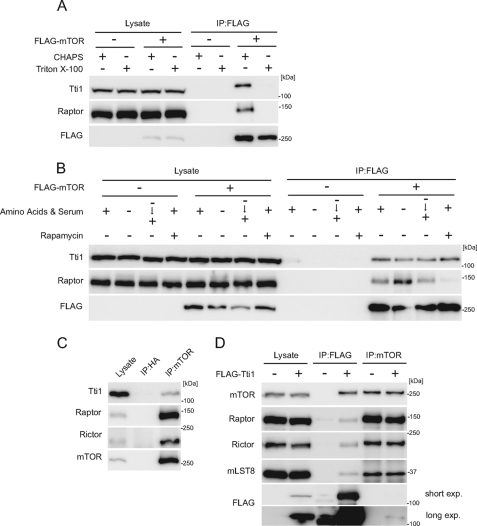
We generated an antibody against Tti1 and confirmed the interaction between FLAG-mTOR and endogenous Tti1 (Fig. 2A). The Tti1-mTOR interaction was observed in CHAPS-containing buffer but was disrupted in Triton X-100-containing buffer (Fig. 2A), as previously reported for mTOR interaction with Raptor and Rictor (24, 25). We thus used the CHAPS-containing buffer in the following immunoprecipitation experiments. This interaction was not affected by nutrient starvation and replenishment, although the starvation treatment enhanced the mTOR-Raptor interaction, as previously reported (Fig. 2B) (24). Likewise, rapamycin did not affect the mTOR-Tti1 interaction, whereas it disrupted the mTOR-Raptor interaction. These data suggest that Tti1 constitutively interacts with mTOR.
FIGURE 2.
Mammalian Tti1 interacts with mTOR. A, HEK293T cells were transfected with an empty vector or FLAG-mTOR. Two days later, cells were harvested and lysed in a buffer containing either 0.3% CHAPS or 0.5% Triton X-100. Immunoprecipitation was then performed with anti-FLAG antibody. The lysate lanes contain one-twelfth of the lysate volume used for immunoprecipitation. B, HEK293T cells were transfected with an empty vector or FLAG-mTOR. Two days later, cells were cultured in an amino acid-free DMEM (without FBS) for 120 min and then cultured in a complete medium for 30 min. Rapamycin treatment was performed by incubation for 30 min with a complete medium containing 20 μg/ml rapamycin. Cells were lysed, and immunoprecipitation was performed with anti-FLAG antibody. The lysate lanes contain one-twelfth of the lysate volume used for immunoprecipitation. C, HeLa cells were lysed, and immunoprecipitation was performed with the indicated antibodies to detect endogenous interactions. The lysate lanes contain one-four-hundredth of the lysate volume used for immunoprecipitation. D, HEK293T cells were transfected with an empty vector or FLAG-Tti1. Two days later, cells were lysed, and immunoprecipitation was performed with anti-FLAG and anti-mTOR antibodies. The lysate lanes contain one-sixth or one-sixtieth of the lysate volume used for immunoprecipitation with anti-FLAG or anti-mTOR antibody, respectively.
The endogenous interaction between mTOR and Tti1 was also detected by immunoprecipitation using an antibody against mTOR, suggesting that their interaction is physiologically relevant (Fig. 2C). However, the ratio of the amount of Tti1 in the precipitate to that in the total cell lysate was very small (Fig. 2C). This was in marked contrast to the result that both Raptor and Rictor were efficiently enriched in the mTOR precipitants, suggesting that only a small population of endogenous Tti1 associates with mTOR. When lysates of cells expressing FLAG-Tti1 were immunoprecipitated with the anti-FLAG antibody, both Raptor and Rictor were detected, suggesting that Tti1 associates with both mTORC1 and mTORC2 (Fig. 2D).
Tti1 Positively Regulates mTOR Activity
We analyzed whether Tti1 regulates the activity of mTORC1 and mTORC2. Small interfering RNA (siRNA)-mediated knockdown of Tti1 was very effective and reduced Tti1 expression to almost undetectable levels (Fig. 3). In these cells, mTOR expression levels were also decreased slightly (discussed in more detail below).
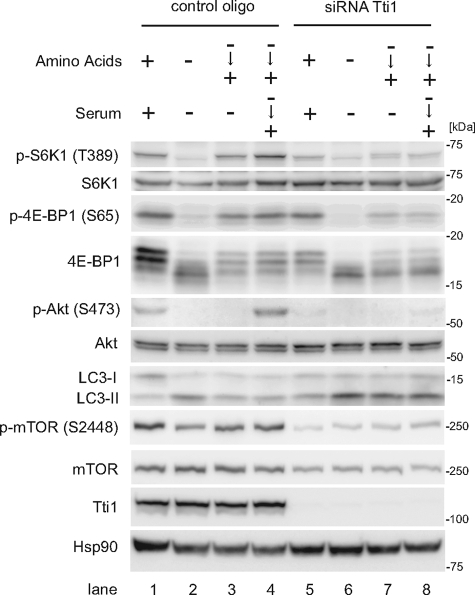
FIGURE 3.
Tti1 positively regulates mTOR activity. HeLa cells transfected with Tti1 siRNA or control siRNA were cultured in amino acid-free DMEM (without FBS) for 120 min, and then cultured in regular DMEM with or without 10% FBS for 30 min.
Amino acid treatment stimulated the phosphorylation of mTORC1 substrates, S6K1 and 4E-BP1 in control cells (Fig. 3, lanes 2–4). However, knockdown of Tti1 suppressed the phosphorylation of S6K1 and 4E-BP1 upon amino acid stimulation (Fig. 3, lanes 6–8). In addition, phosphorylation of mTOR itself was suppressed in these cells. Next we determined whether Tti1 suppression also affected autophagy regulation by the LC3 conversion assay. LC3, which is one of the mammalian homologs of yeast Atg8 (autophagy-related 8), is present in two forms, LC3-I (a cytosolic form) and LC3-II (a membrane-bound phosphatidylethanolamine-conjugated form). The amount of LC3-II correlates with the number of autophagosomes (26). Amino acid replenishment reduced the amount of LC3-II in control cells (Fig. 3, lanes 2–4) but not in Tti1 siRNA-treated cells (Fig. 3, lanes 6–8). Taken together, these data suggest that Tti1 positively regulates mTORC1 activity.
Next, we analyzed the involvement of Tti1 in mTORC2 regulation. Serum replenishment induced the phosphorylation of Akt at Ser-473, which is a well known indicator of mTORC2 activity (Fig. 3, lanes 3 and 4); however, it was abolished in cells treated with Tti1 siRNA (Fig. 3, lanes 7 and 8), suggesting that Tti1 is also important for mTORC2 activity.
Tti1 Interacts with Tel2 and Is Important for Tel2-mTOR Association
S. pombe Tti1 was identified as a protein that interacts with Tor1/2 and Tel2 (17). The Tti1·Tel2 interaction was also reported in Saccharomyces cerevisiae (27). Tel2 was originally identified in S. cerevisiae as a regulator of telomere length (28). In Caenorhabditis elegans, hypomorphic mutants of the Tel2 ortholog (Clk-2/Rad-5) were also reported to show extended life span and hypersensitivity to UV irradiation (29, 30). Subsequent studies showed that Tel2 is essential for growth (31), and its homologs are important for DNA replication checkpoint in S. pombe, C. elegans, and mammalian cells (32–34). Recently, it was reported that mammalian Tel2 interacts with and regulates the stability of all PIKK family proteins (19).
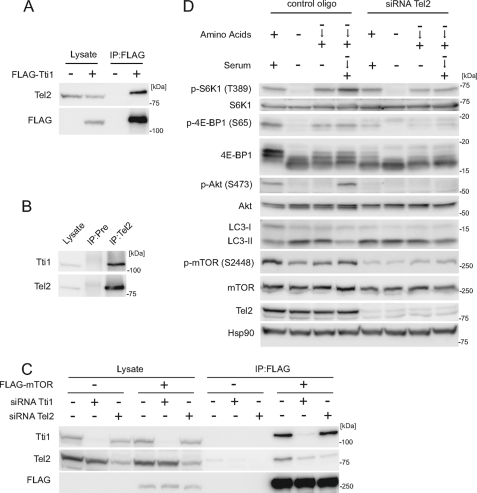
We confirmed that FLAG-Tti1 interacts with Tel2 also in mammalian cells (Fig. 4A) and observed the interaction at endogenous levels as well (Fig. 4B). Next, we investigated how Tti1, Tel2, and mTOR interact with each other. Tti1 was co-precipitated with FLAG-mTOR more efficiently than Tel2 (Fig. 4C). Moreover, knockdown of Tti1 significantly reduced the amount of mTOR-associated Tel2, whereas knockdown of Tel2 did not affect Tti1-mTOR association (Fig. 4C). These results suggest that Tti1 mediates Tel2-mTOR association.
FIGURE 4.
Tti1 interacts with Tel2 and is important for Tel2–mTOR association. A, HEK293T cells were transfected with an empty vector or FLAG-Tti1. Two days later, cells were lysed and immunoprecipitation was performed with anti-FLAG antibody. The lysate lanes contain one-twelfth of the lysate volume used for immunoprecipitation. B, endogenous interaction between Tti1 and Tel2 was determined in HeLa cells. Immunoprecipitation was performed with anti-Tel2 antiserum or preimmune rabbit serum (Pre). The lysate lanes contain one-twentieth of the lysate volume used for immunoprecipitation. C, HEK293T cells were transfected with siRNA against Tti1 or Tel2, and then with an empty vector or FLAG-mTOR. Cells were lysed and immunoprecipitation was performed with anti-FLAG antibody. The lysate lanes contain one-fifteenth of the lysate volume used for immunoprecipitation. D, HeLa cells transfected with Tel2 siRNA or control siRNA were treated as in Fig. 2.
Knockdown of Tel2 suppressed the activity of both mTORC1 (phosphorylation of S6K1, 4E-BP1, and mTOR and autophagy suppression) and mTORC2 (phosphorylation of Akt Ser-473) (Fig. 4D), as previously reported for the suppression of S6K1 phosphorylation (19). These effects of Tel2 knockdown are similar to those of Tti1 knockdown, supporting the idea that Tti1 and Tel2 cooperatively regulate mTOR signaling.
Tti1 and Tel2 Interact with and Stabilize the PIKK Proteins
PIKKs are protein kinases with sequence similarity to phosphatidylinositol 3-kinase. Mammals have six PIKKs: mTOR, ATM, ATR, DNA-PKcs, SMG-1, and TRRAP (1–3). Tel2 was recently reported to interact with all six PIKK proteins, thereby stabilizing them (19). Therefore, we examined whether Tti1 also interacts with and stabilizes other PIKK proteins, in addition to mTOR.
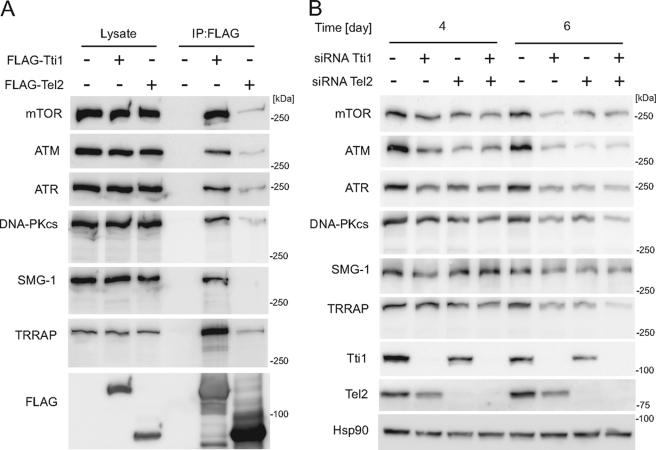
We found that, similar to Tel2, Tti1 interacts with all the PIKK proteins (Fig. 5A). It was noteworthy that FLAG-Tti1 precipitated these PIKK proteins more efficiently than FLAG-Tel2, suggesting that Tti1 directly interacts with these PIKK proteins to mediate Tel2 association (Fig. 5A).
FIGURE 5.
Tti1 and Tel2 interact with and stabilize PIKKs. A, HEK293T cells were transfected with empty vector, FLAG-Tti1, or FLAG-Tel2. Two days later, cells were lysed, and immunoprecipitation was performed with anti-FLAG antibody. The lysate lanes contain one-twenty fourth of the lysate volume used for immunoprecipitation. B, HeLa cells were treated with siRNAs against Tti1 or/and Tel2 for 4 or 6 days. Transfection was performed every other day. Cells were then harvested, and immunoblot analysis was performed with the indicated antibodies.
Prolonged knockdown of Tti1, as well as that of Tel2, clearly decreased the amounts of all the PIKK proteins, indicating that Tti1 is also important for the stability of PIKK proteins (Fig. 5B). In addition, double knockdown of Tti1 and Tel2 did not further reduce the PIKK proteins compared with single knockdown of either of them, suggesting that Tti1 and Tel2 work cooperatively within the same pathway (Fig. 5B).
Tti1 and Tel2 Are Important for mTOR Complex Assembly
Although prolonged depletion of Tti1 or Tel2 caused severe reduction of mTOR protein expression, decreased mTOR activity was clearly observed even when mTOR protein was reduced only slightly following knockdown of Tti1 or Tel2 (Figs. 3 and 4D). These observations prompted us to hypothesize that Tti1 and Tel2 might have functions other than the stabilization of mTOR protein to regulate mTOR activity. One possibility was that Tti1 and Tel2 facilitate mTOR complex formation. This hypothesis may explain both the reduced mTOR expression level and the defect of mTOR activity in Tti1 or Tel2 siRNA-treated cells, because depletion of Raptor or both Raptor and Rictor causes reduction in the mTOR protein level (24, 35, 36).
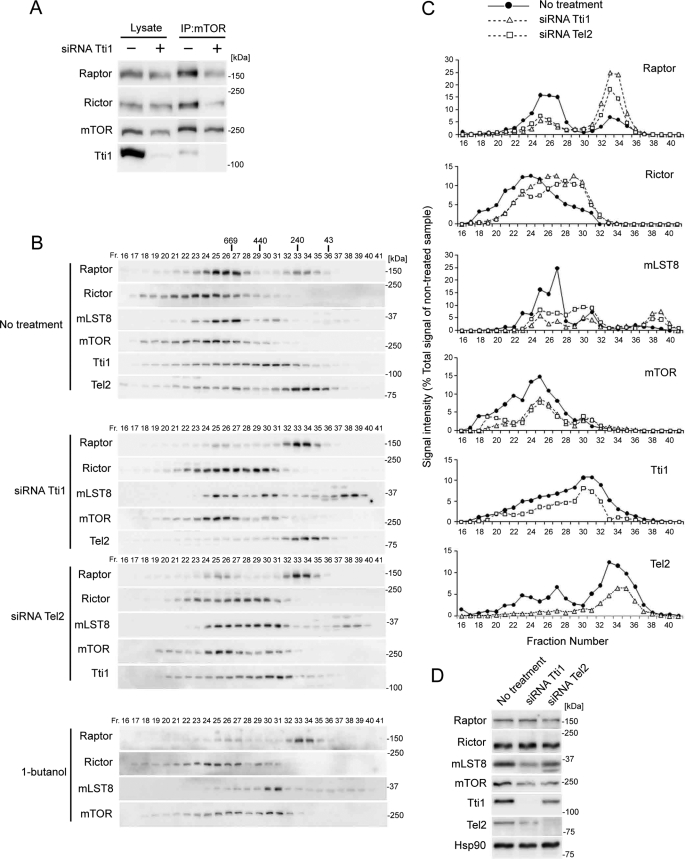
We tested this possibility by immunoprecipitating of the mTOR complex. Although knockdown of Tti1 decreased the amount of mTOR in the mTOR immunoprecipitates only slightly, it markedly reduced the amounts of co-precipitated Raptor and Rictor. This suggests that mTORC1 and mTORC2 are disassembled in Tti1-depleted cells (Fig. 6A).
FIGURE 6.
Tti1 and Tel2 are important for mTOR complex assembly. A, HeLa cells transfected with Tti1 or control siRNAs were lysed, and immunoprecipitation was performed with the indicated antibodies. The lysate lanes contain one-twentieth of the lysate volume used for immunoprecipitation. B–D, HeLa cells transfected with the indicated siRNAs or treated with 0.8% 1-butanol for 3 h were lysed. The cytosolic fractions of the cells were subjected to gel filtration analysis using a Superose 6 column (B) or subjected to Western blot analysis (D). Immunoblot signals in B were quantified with ImageJ and plotted (C). The value of the total signal was adjusted to the amount of total protein of non-treated samples for each protein, as estimated from Western blot analysis of total cell lysates (D).
To further confirm that depletion of Tti1 and Tel2 causes the disassembly of mTOR complexes, we performed size-exclusion chromatography. Consistent with previous reports (13, 15), when cytosolic fractions of HeLa cells were subjected to gel-filtration chromatography, components of mTOR complexes (mTOR, Raptor, Rictor, and mLST8) were mainly detected around fractions corresponding to >600 kDa, suggesting that these fractions contain mTORC1 and mTORC2 (Fig. 6, B and C).
Raptor (150 kDa) distributed in two peaks, which appeared to contain mTORC1-associated Raptor (∼800 kDa) and free Raptor (∼200 kDa) (Fig. 6, B and C). Most Raptor was detected in the higher molecular mass fractions. However, following knockdown of either Tti1 or Tel2, most Raptor was distributed in the lower molecular mass fractions, which were distinct from mTOR-containing fractions, suggesting that mTORC1 was disassembled (Fig. 6, B–D).
Rictor (192 kDa) showed a broad distribution pattern around 300–6000 kDa, with a peak of 1300 kDa (Fig. 6, B and C). Knockdown of Tti1 or Tel2 shifted the distribution to lower molecular mass fractions, suggesting that mTORC2 was also disassembled (Fig. 6, B–D). The distribution shift of Rictor was relatively small compared with that of Raptor. Therefore we tested whether it actually represented the dissociation of Rictor from mTOR using 1-butanol treatment, which is known to disrupt both mTOR complexes (15, 16). Treatment with 1-butanol caused a remarkable shift in Raptor distribution, and a relatively small shift in Rictor distribution, which is similar to the results of knockdown of Tti1 or Tel2 and confirms that Tti1 or Tel2 depletion indeed leads to mTORC2 disassembly (Fig. 6B).
mLST8 (36 kDa) showed three distribution peaks, which appeared to correspond to complete mTOR complexes, mLST8·mTOR complex (without Raptor and Rictor), and free mLST8 (Fig. 6, B and C). Although the majority of mLST8 existed in the largest molecular mass fractions in control cells, the amount of mLST8 in smaller molecular mass fractions was increased by knockdown of Tti1 or Tel2 (Fig. 6, B–D).
mTOR (290 kDa) showed a broad distribution pattern similar to that of Rictor (Fig. 6, B and C). Knockdown of Tti1 or Tel2 caused attenuation of the higher molecular mass peak with a slight enhancement of the lower molecular mass peak (Fig. 6, B–D). Taken together, these data suggest that depletion of Tti1 or Tel2 causes the disassembly of both mTORC1 and mTORC2.
Finally, we also checked the distribution of Tti1 (120 kDa) and Tel2 (80 kDa) and found that both showed a wide distribution. Tti1 and Tel2 in the large molecular mass fractions appeared to represent their population bound to the PIKK family proteins (Fig. 6, B–D). Following knockdown of Tti1, Tel2 disappeared from the higher molecular fractions, whereas knockdown of Tel2 had almost no effect on the distribution of Tti1 (Fig. 6, B–D). This result further confirms that Tti1 is important for the efficient association of Tel2 with all PIKK proteins.
DISCUSSION
In this study, we identified mammalian Tti1 as a novel mTOR-binding protein. We have demonstrated that Tti1 and its binding partner Tel2 are important for the activity of both mTORC1 and mTORC2, likely through maintenance of the complex assembly. We also found that Tti1 is important for the association of Tel2 with mTOR. Finally, we showed that Tti1 could interact with all other PIKK family proteins. During revision of this report, Tti1 was also reported as an SMG-1-interacting protein with the name of SMG-10, which further interacts with RUVBL1 and RUVBL2 (37).
How Tti1 and Tel2 maintain the mTOR complex remains unknown. There are two possible explanations: 1) the Tti1·Tel2 complex interacts with free (unassembled) mTOR and then promotes its assembly with other subunits of mTOR complexes and/or 2) the Tti1·Tel2 complex associates with preformed mTORC1 and mTORC2 and stabilizes them. To test the former possibility that the Tti1·Tel2 complex binds to free mTOR to facilitate its assembly, we have attempted to reconstitute the mTOR complex assembly in vitro. However, addition of Tti1 and Tel2 was not sufficient to restore the complex formation in cell lysates, in which Tti1 and Tel2 were depleted by siRNA treatment.5 Therefore, further unknown factor(s) may be necessary to promote the complex assembly, or it is possible that the primary role of Tti1 and Tel2 is maintenance of preformed mTOR complexes.
As previously reported for Tel2 (19), we also found that Tti1 is important for the stabilization of mTOR. This effect of Tti1 and Tel2 may be a secondary result of the mTOR complex formation. Free mTOR appears to be unstable, because when cells were treated with Tti1- or Tel2-siRNA, we detected a large amount of disassembled Raptor and Rictor, but a much smaller amount of dissembled mTOR in smaller molecular mass fractions (Fig. 6). Furthermore, it was reported that knockdown of Raptor reduces the amount of mTOR protein (24, 35), and double knock-out of Raptor and Rictor causes a large reduction in the amount of mTOR in muscle (36), supporting the idea that free mTOR is unstable. Therefore, the primary role of the Tti1·Tel2 complex would be maintenance of the complex assembly.
This hypothesis could be applied to other PIKK family proteins. Although the precise role of Tti1 and Tel2 on other PIKK proteins remains unclear, one possibility is that Tti1 and Tel2 may be important for complex formation. All members of the PIKK family form protein complexes (3, 38), some of which result in the stabilization of PIKKs. For example, RNAi-mediated suppression of ATR-interacting protein and ATM-interacting protein causes reduction of the ATR and ATM expression levels, respectively (39, 40). Moreover, it was recently reported that knockdown of Tel2 disrupts interaction between ATR and its binding partner, TopBP1 (41). Thus, maintenance of protein complex formation might be a general function of Tti1 and Tel2. Further experiments are needed to address this.
Acknowledgments
We thank Dr. Akiko Kuma for helpful discussion and critical reading of the manuscript.
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, the Toray Science Foundation, and the Takeda Science Foundation (to N. M.).
T. Kaizuka, T. Hara, N. Oshiro, U. Kikkawa, K. Yonezawa, K. Takehana, S.-i. Iemura, T. Natsume, and N. Mizushima, unpublished data.
- mTOR
- mammalian target of rapamycin
- ATM
- ataxia telangiectasia mutated
- ATR
- ATM- and Rad3-related
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- DNA-PKcs
- DNA-dependent protein kinase catalytic subunit
- LC3
- microtubule-associated protein 1 light chain 3
- mTORC1 and -2
- mTOR complexes 1 and 2, respectively
- PIKK
- phosphatidylinositol 3-kinase-related protein kinase
- siRNA
- small interfering RNA
- SMG-1
- suppressor with morphological effect on genitalia 1
- Tel2
- telomere maintenance 2
- TRRAP
- transformation/transcription domain-associated protein
- Tti1
- Tel two interacting protein 1
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- RNAi
- RNA interference.
REFERENCES
- 1.Bakkenist C. J., Kastan M. B. (2004) Cell 118, 9–17 [DOI] [PubMed] [Google Scholar]
- 2.Lempiäinen H., Halazonetis T. D. (2009) EMBO J. 28, 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lovejoy C. A., Cortez D. (2009) DNA Repair 8, 1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wullschleger S., Loewith R., Hall M. N. (2006) Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 5.Guertin D. A., Sabatini D. M. (2007) Cancer Cell 12, 9–22 [DOI] [PubMed] [Google Scholar]
- 6.Yang Q., Guan K. L. (2007) Cell Res. 17, 666–681 [DOI] [PubMed] [Google Scholar]
- 7.Laplante M., Sabatini D. M. (2009) J. Cell Sci. 122, 3589–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., Guan J. L., Oshiro N., Mizushima N. (2009) Mol. Biol. Cell 20, 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung C. H., Jun C. B., Ro S. H., Kim Y. M., Otto N. M., Cao J., Kundu M., Kim D. H. (2009) Mol. Biol. Cell 20, 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganley I. G., Lam D. H., Wang J., Ding X., Chen S., Jiang X. (2009) J. Biol. Chem. 284, 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guertin D. A., Stevens D. M., Thoreen C. C., Burds A. A., Kalaany N. Y., Moffat J., Brown M., Fitzgerald K. J., Sabatini D. M. (2006) Dev. Cell 11, 859–871 [DOI] [PubMed] [Google Scholar]
- 12.Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S. Y., Huang Q., Qin J., Su B. (2006) Cell 127, 125–137 [DOI] [PubMed] [Google Scholar]
- 13.Yang Q., Inoki K., Ikenoue T., Guan K. L. (2006) Genes Dev. 20, 2820–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. (2006) Mol. Cell 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 15.Takahara T., Hara K., Yonezawa K., Sorimachi H., Maeda T. (2006) J. Biol. Chem. 281, 28605–28614 [DOI] [PubMed] [Google Scholar]
- 16.Toschi A., Lee E., Xu L., Garcia A., Gadir N., Foster D. A. (2009) Mol. Cell. Biol. 29, 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi T., Hatanaka M., Nagao K., Nakaseko Y., Kanoh J., Kokubu A., Ebe M., Yanagida M. (2007) Genes Cells 12, 1357–1370 [DOI] [PubMed] [Google Scholar]
- 18.Kanoh J., Yanagida M. (2007) Genes Cells 12, 1301–1304 [DOI] [PubMed] [Google Scholar]
- 19.Takai H., Wang R. C., Takai K. K., Yang H., de Lange T. (2007) Cell 131, 1248–1259 [DOI] [PubMed] [Google Scholar]
- 20.Hara K., Yonezawa K., Kozlowski M. T., Sugimoto T., Andrabi K., Weng Q. P., Kasuga M., Nishimoto I., Avruch J. (1997) J. Biol. Chem. 272, 26457–26463 [DOI] [PubMed] [Google Scholar]
- 21.Nishiuma T., Hara K., Tsujishita Y., Kaneko K., Shii K., Yonezawa K. (1998) Biochem. Biophys. Res. Commun. 252, 440–444 [DOI] [PubMed] [Google Scholar]
- 22.Hosokawa N., Hara Y., Mizushima N. (2006) FEBS Lett. 580, 2623–2629 [DOI] [PubMed] [Google Scholar]
- 23.Natsume T., Yamauchi Y., Nakayama H., Shinkawa T., Yanagida M., Takahashi N., Isobe T. (2002) Anal. Chem. 74, 4725–4733 [DOI] [PubMed] [Google Scholar]
- 24.Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 25.Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 26.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J. F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A. S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabási A. L., Tavernier J., Hill D. E., Vidal M. (2008) Science 322, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lustig A. J., Petes T. D. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 1398–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartman P. S., Herman R. K. (1982) Genetics 102, 159–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakowski B., Hekimi S. (1996) Science 272, 1010–1013 [DOI] [PubMed] [Google Scholar]
- 31.Runge K. W., Zakian V. A. (1996) Mol. Cell. Biol. 16, 3094–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed S., Alpi A., Hengartner M. O., Gartner A. (2001) Curr. Biol. 11, 1934–1944 [DOI] [PubMed] [Google Scholar]
- 33.Shikata M., Ishikawa F., Kanoh J. (2007) J. Biol. Chem. 282, 5346–5355 [DOI] [PubMed] [Google Scholar]
- 34.Collis S. J., Barber L. J., Clark A. J., Martin J. S., Ward J. D., Boulton S. J. (2007) Nat. Cell Biol. 9, 391–401 [DOI] [PubMed] [Google Scholar]
- 35.Kim D. H., Sarbassov D. D., Ali S. M., Latek R. R., Guntur K. V., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2003) Mol. Cell 11, 895–904 [DOI] [PubMed] [Google Scholar]
- 36.Bentzinger C. F., Romanino K., Cloëtta D., Lin S., Mascarenhas J. B., Oliveri F., Xia J., Casanova E., Costa C. F., Brink M., Zorzato F., Hall M. N., Rüegg M. A. (2008) Cell Metab. 8, 411–424 [DOI] [PubMed] [Google Scholar]
- 37.Izumi N., Yamashita A., Iwamatsu A., Kurata R., Nakamura H., Saari B., Hirano H., Anderson P., Ohno S. (2010) Sci. Signal. 3, ra27. [DOI] [PubMed] [Google Scholar]
- 38.Murr R., Vaissière T., Sawan C., Shukla V., Herceg Z. (2007) Oncogene 26, 5358–5372 [DOI] [PubMed] [Google Scholar]
- 39.Cortez D., Guntuku S., Qin J., Elledge S. J. (2001) Science 294, 1713–1716 [DOI] [PubMed] [Google Scholar]
- 40.Kanu N., Behrens A. (2007) EMBO J. 26, 2933–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rendtlew Danielsen J. M., Larsen D. H., Schou K. B., Freire R., Falck J., Bartek J., Lukas J. (2009) J. Biol. Chem. 284, 4140–4147 [DOI] [PubMed] [Google Scholar]