Abstract
Rac1 is a member of the Rho family of small GTPases, which regulate cell adhesion and migration through their control of the actin cytoskeleton. Rho-GTPases are structurally very similar, with the exception of a hypervariable domain in the C terminus. Using peptide-based pulldown assays in combination with mass spectrometry, we previously showed that the hypervariable domain in Rac1 mediates specific protein-protein interactions. Most recently, we found that the Rac1 C terminus associates to the ubiquitously expressed adapter protein CMS/CD2AP. CD2AP is critical for the formation and maintenance of a specialized cell-cell contact between kidney podocyte foot processes, the slit diaphragm. Here, CD2AP links the cell adhesion protein nephrin to the actin cytoskeleton. In addition, CMS/CD2AP binds actin-regulating proteins, such as CAPZ and cortactin, and has been implicated in the internalization of growth factor receptors. We found that CD2AP specifically interacts with the C-terminal domain of Rac1 but not with that of other Rho family members. Efficient interaction between Rac1 and CD2AP requires both the proline-rich domain and the poly-basic region in the Rac1 C terminus, and at least two of the three N-terminal SH3 domains of CD2AP. CD2AP co-localizes with Rac1 to membrane ruffles, and small interfering RNA-based experiments showed that CD2AP links Rac1 to CAPZ and cortactin. Finally, expression of constitutive active Rac1 recruits CD2AP to cell-cell contacts in epithelial cells, where we found CD2AP to participate in the control of the epithelial barrier function. These data identify CD2AP as a novel Rac1-associated adapter protein that participates in the regulation of epithelial cell-cell contact.
Keywords: Adaptor Proteins, Cell Junctions, Cell-Cell Interaction, G Proteins, SH3 Domains, Rac1
Introduction
The small GTPases of the Rho family regulate many important cellular processes including cell proliferation, migration, and vesicle trafficking. Rho-GTPases are regulated by guanine nucleotide exchange factors (GEFs)2 that stimulate GTP binding, which renders the GTPase signaling competent (1). GTPase-activating proteins on the other hand, stimulate the intrinsic GTPase activity, i.e. the hydrolysis of GTP to GDP, which results in inactivation of the GTPase (2). The intracellular localization of GTPase-activating and GEF proteins is an important determinant of localized GTPase activation or inactivation. In addition, the hypervariable C-terminal domain of Rho-GTPases has been implicated in the control of localized signaling (3). Previously, we found the Rac1 GEF β-PIX that binds to the Rac1 C terminus to recruit Rac1 to focal adhesions and the peripheral membrane, where Rac1 activity induces the formation of lamellipodia and membrane ruffles (4, 5).
For the formation of lamellipodia and ruffles, remodeling of the actin cytoskeleton is required. Rac1 promotes the formation of a branched actin network via its effectors of the Scar/WAVE family of proteins, which stimulate ARP2/3-mediated actin nucleation (6). Rac1-driven actin remodeling results in lamellipodia formation and ultimately drives cell migration (7). In addition, active Rac1 stimulates cell migration by promoting the translocation of the nuclear oncogene SET toward the plasma membrane (8). In contrast to promoting cell motility, Rac1 activity can also stimulate strengthening of cell-cell contacts via the induction of actin polymerization at sites of cell-cell contact, on the one hand (9, 10), and by promoting the recruitment of β-catenin and E-cadherin to sites of cell-cell adhesion via its effector IQGAP, on the other (11). Finally, Rac1 is activated upon E-cadherin ligation that further stimulates the maturation of cell-cell contacts (12).
To investigate the regulation of these diverse signaling events by Rac1, we searched for novel proteins that selectively associate to the Rac1 hypervariable domain, using a biotinylated peptide encoding the Rac1 C terminus in pulldown assays (4, 13). In the course of these studies, we identified CMS (the human ortholog of mouse CD2AP) as a prominent interaction partner for the Rac1 hypervariable domain. From here on, we refer to this protein in the text as CD2AP. CD2AP is a 639-amino acid adapter protein expressed in all tissues except for brain (14). CD2AP harbors three SH3 domains in its N terminus, a proline-rich domain and a C-terminal leucine-zipper domain via which it can homodimerize (15).
Mutations in the CD2AP gene are associated with kidney problems, e.g. sporadic nephrotic syndrome and focal segmental glomerulosclerosis (16). CD2AP-deficient mice die of renal failure as a result of improper formation of a specialized cell-cell junction called the slit diaphragm (17, 18). The slit diaphragm is localized in the kidney glomerulus between podocyte foot processes, and functions as a protein filtration barrier (19). CD2AP is required for the formation and maintenance of this junction, by linking the cell-cell adhesion protein nephrin to the cytoskeleton (20, 21). In Drosophila, the sole member of the CD2AP/CIN85 family, named Cindr, was also shown to be required for correct localization of adhesion proteins during tissue patterning (22).
Besides the regulation of cell-cell contacts, CD2AP was found to be involved in the formation of the immunological synapse and required for T-cell polarization upon TCR stimulation (14). CD2AP is also involved in down-modulation of the T-cell receptor and together with cortactin and c-Cbl forms a complex that is involved in endocytosis of the epidermal growth factor receptor (23, 24). Finally, CD2AP was also found to interact with actin-associated proteins, such as cortactin, and actin capping proteins CAPZ and capping protein. Moreover, CD2AP is able to directly interact with actin via four actin-binding regions in its C terminus, and promotes actin polymerization by partial inhibition of capping protein and CAPZ (24–26).
We found that the SH3 domains of CD2AP are required for its selective association to the hypervariable C terminus of Rac1. In addition, CD2AP acts as a linker between Rac1 and the actin-binding proteins cortactin and CAPZ. Also, Rac1 and CD2AP co-localize in membrane ruffles but not in focal adhesions. Finally, we show that Rac1 activation induces CD2AP recruitment to epithelial cell-cell contacts where CD2AP participates in the control of the epithelial barrier function. In conclusion, we have identified the adapter protein CD2AP as a novel Rac1-associated protein, which is involved in the control of epithelial cell-cell contact.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The following antibodies were used: anti-CD2AP (B-4, sc-25272), anti-CD2AP (H-290, sc-9137 polyclonal), anti-c-Cbl (C15 sc-170) from Santa Cruz, anti-β-catenin (610154 clone-14), anti-paxillin (610619), and anti-CapZ-α (612460 clone-7) from Transduction Labs, anti-Rac1 (clone 23A8) from Millipore, anti-green fluorescent protein (JL-8) from Clontech, anti-c-Myc from Zymed Laboratories Inc., anti-cortactin (clone 4F11) from Upstate, and F-actin stained with Alexa 633-phalloidin from Invitrogen. 8-(4-Chlorophenylthio)adenosine-3′,5′-cyclic monophosphate (8-CPT-cAMP) was purchased from BioLog and used at 100 μm. GST-CNF1 was isolated as previously described (27) and used at 300 ng/ml. Confocal imaging of immunostained samples was done using a Zeiss LSM510 Meta system in combination with Zeiss Zen software for image analysis and processing.
Cell Culture
HeLa, HEK 293T, H292, and Jurkat T-cells were maintained at 37 °C and 5% CO2 in Iscove's modified Dulbecco's medium containing 10% heat-inactivated fetal calf serum (Bodinco), 2 mm l-glutamine and penicillin/streptomycin, all purchased from PAA Cell Culture Company. Cells were passed by trypsinization. Human umbilical vein endothelial cells were obtained from Lonza and cultured in EGM2 medium (Lonza) prepared according to the manufacturer's instructions.
Freshly obtained peripheral blood mononuclear cells were enriched by density gradient centrifugation over Ficoll-Hypaque (1.077 g/ml; GE Healthcare). Mononuclear cells were resuspended in PBE buffer (phosphate-buffered saline, 0.5% (w/v) bovine serum albumin, and 5 mm EDTA). CD34+ cells were isolated with a hapten-labeled CD34 antibody (QBEND 10) with the VarioMacs System according to the manufacturer's instructions (Miltenyi Biotec GmbH, Gladbach, Germany). At least 95% of the cells isolated from PB-expressed CD34 were determined by FACS analysis (Immunocytometry Systems; BD Biosciences) (28).
Cell Transfection and DNA Constructs
Cells were transiently transfected with TransIT (Mirus) as described by the manufacturer. Briefly, 6 μg of DNA was mixed with 18 μl of TransIT in 600 μl of Opti-MEM (51985026) (Invitrogen) and incubated for 15 min at room temperature, after which the transfection mixture was applied to 50% confluent cells. The following constructs were used: pCMV-Myc-Rac1-Q61L, pCMV-Myc-Rac1-T17N, pcDNA-myr-Myc (8), pEGFP, and GFP-CD2AP. The latter construct (a kind gift from Andrey Shaw, Washington University School of Medicine) was used to generate N-CD2AP, C-CD2AP, and the following SH3 mutants: mSH3-1, mSH3-2, and mSH3-3. Point mutations in the SH3 domains were generated by site-directed mutagenesis (Stratagene) of tryptophan to lysine at positions 37, 144, or 306, respectively. The mSH3-1,2, mSH3-2,3, and mSH3-1,3 mutants were generated by an additional round of site-directed mutagenesis.
siRNA for CD2AP was purchased from Santa Cruz (sc-29984) and control siRNA was from Dharmacon (Perbio, Etten-Leur, The Netherlands). Double strand siRNA oligos were transfected in H292 cells using Interferin transfection reagent (Polyplus) according to the manufacturer's protocol. Briefly, 10.5 pmol of siRNA was diluted in 500 μl of Opti-MEM and 30 μl of Interferin was added to the mixture, which was incubated for 10 min at room temperature. The mixture was then added to ∼70% confluent cells that were incubated for 24 h before seeding on electrical cell impedance sensing (ECIS) electrodes (see below).
Peptide Synthesis
Peptides were synthesized on a peptide synthesizer (Syro II) using Fmoc (N-(9-fluorenyl)methoxycarbonyl) solid phase chemistry. Peptides encoded a biotinylated protein transduction domain (Biotin-YARAAARQARAG) followed by the 10 amino acids proceeding the CAAX domain of the indicated Rho-GTPase peptides (29). The sequences of the Rac1 (PPP) and Rac1 (RKR) mutants are: YARAAARQARAGCAAAVKKRKRK and YARAAARQARAGCPPPVKKAAAK, respectively (13).
Pulldown Assay
Peptide pulldown assays were performed as described previously (4). Cells were lysed in Nonidet P-40 lysis buffer (50 mm Tris-HCl, 100 mm NaCl, 10 mm MgCl2, 10% glycerol, 1% Nonidet P-40). Isolation of proteins and mass spectrometry analysis was performed as described (30) using 5 μg of the indicated biotin-labeled peptide and 25 μl of streptavidin-coated beads (Sigma). GST, GST-Rac, and GST-RacΔC (a kind gift from R. Ahmadian, European Molecular and Cell Biology Laboratory, Heidelberg, Germany (31)) fusion proteins were purified from BL21 bacteria as described earlier (4), after which 100 μg of the purified protein was used per pulldown.
Electrical Resistance Measurements
For ECIS-based measurement of transcellular electrical resistance (4), ECIS electrodes (8W10E; Applied Biophysics) were treated with 10 μm l-cysteine for 15 min and subsequently coated with 10 μg/ml of fibronectin (Sigma) in NaCl (0.9%) for 1 h at 37 °C. 200,000 siRNA-transfected H292 cells were seeded per well in a 350-μl culture medium. Transcellular electrical resistance (TER) was monitored continuously by measuring the impedance at 30 kHz, using either an ECIS model 9600 or ECIS-Zθ. Data were modeled using the ECIS-Zθ software to obtain the barrier function (Rb) (32).
RESULTS
CD2AP Associates to the Small GTPase Rac1
The C-terminal hypervariable domain of Rho-GTPases has previously been shown to control subcellular targeting (3). This targeting is critical for the outcome of downstream signaling, as was recently underscored for the highly homologous Rac1 and Rac3 proteins. The divergent phenotype of these two proteins appeared to be strictly dependent on their C termini (33). To find proteins interacting with the Rac1 hypervariable domain, a biotinylated peptide encoding this region was used in pulldown assays. Bound proteins were resolved by SDS-PAGE, isolated, and subsequently analyzed by mass spectrometry. One of the proteins identified in this screen is the multidomain adapter protein CMS/CD2AP.
To confirm the association between Rac1 and CD2AP and to test the specificity of this interaction, peptides encoding the hypervariable C termini of different Rho family members were used to pull down associating proteins from lysates of various types of cell. As shown in Fig. 1, A–C, CD2AP selectively interacts with the C terminus of Rac1 in lysates from Jurkat T-cells, as well as from primary human umbilical vein endothelial cells and CD34+ hematopoietic stem cells. Only after relatively long exposures of the blots, a weak association of CD2AP with the C termini of other Rho-GTPases; e.g. RhoA, RhoB, and RhoC, could be detected. Thus, in all cell types tested, CD2AP efficiently and selectively associates to the hypervariable C terminus of Rac1.
FIGURE 1.
CD2AP associates specifically to the C terminus of Rac1. A–C, peptides encoding the C-terminal hypervariable domain of different Rho family members were used to pull down proteins from cell lysates of Jurkat T-cells (A), primary human umbilical vein endothelial cell (HUVEC) (B), or CD34+ cells (C). Long and short exposed (Exp.) blots are shown. D and E, to test for association of CD2AP to full-length Rac1, bacterially isolated GST-Rac1, a mutant of Rac1 lacking the C terminus (GST-Rac1-ΔC) and GST as a control, were used to pull down proteins from lysates of HeLa cells transfected with CD2AP-GFP (D), or from Jurkat T-cell lysates (E). Bound proteins were separated by SDS-PAGE and blotted for CD2AP (A–C and E) or GFP (D) and GST (D and E). WB, Western blot.
To test the association of CD2AP to full-length Rac1, HeLa cells were transfected with CD2AP-GFP. Subsequently, full-length Rac1 fused to GST and isolated from Escherichia coli was used to pulldown proteins from cell lysates. These experiments showed that CD2AP-GFP interacts with GST-Rac1 but not with a mutant of Rac1 lacking the C terminus (Fig. 1D). Also endogenous CD2AP from Jurkat T-cells was associated with full-length Rac1 and not with the mutant lacking the C terminus (Fig. 1E). This confirms that endogenous CD2AP interacts with full-length Rac1 and that the C terminus of Rac1 is necessary for this association.
Mapping the Interaction between Rac1 and CD2AP
The Rac1 C terminus encodes at least two protein-binding domains, i.e. a proline-rich stretch and a poly-basic region (4, 13, 34). To investigate which domain was required for the association to CD2AP, we used Rac1-C-terminal peptides in which the proline or basic amino acids in either of these domains were mutated to alanines. The results show that CD2AP neither associates to the Rac1 C-terminal peptide in which the proline-rich domain was mutated, nor to the one in which the poly-basic region was mutated (Fig. 2A). Thus, both protein interaction domains in the hypervariable domain of Rac1 are required for its interaction with CD2AP.
FIGURE 2.
The Rac1-CD2AP interaction requires the Rac1 C-terminal proline-rich and poly-basic regions and the CD2AP SH3 domains. A, the peptide encoding the C-terminal part of Rac1 (Rac1-C), or peptides containing a mutated proline-rich (PPP → AAA) or poly-basic C-terminal region (RKR → AAA) were used to test their association to CD2AP in Jurkat T-cell lysates. Long and short exposed (Exp.) blots are shown. B, full-length CD2AP and its N- and C-terminal fragments, transfected as GFP fusions into HEK293T (human embryonic kidney) cells were tested for their binding in cell lysates to the Rac1 C terminus. C and D, full-length CD2AP with separate point mutations in each of the three SH3 domains (mSH3-1, -2, -3) substituting a tryptophan for a lysine at positions 37, 144, or 306, respectively (C), or mutants with point mutations in two of the three SH3 domains (D) were transfected in HEK293T cells and their binding in lysates to the Rac1 C terminus was tested and analyzed by Western blotting (WB) for GFP.
Earlier we identified the proline-rich stretch in the Rac1 C terminus to bind to the SH3 domain of β-PIX (4). Therefore we tested if the SH3 domains in the N terminus of CD2AP were involved in Rac1 binding. Initially, we generated two deletion mutants of CD2AP: one lacking the N-terminal region that harbors the SH3 domains (C-CD2AP) and a mutant that lacks the C-terminal part of CD2AP (N-CD2AP; Fig. 2B). Whereas the mutant lacking the C-terminal part of CD2AP could still associate to the Rac1 C terminus, the mutant lacking the SH3 domains was no longer able to interact with Rac1 (Fig. 2B). This suggests that one or more of the SH3 domains is required for the binding to Rac1. Therefore, three full-length CD2AP mutants were created, in which point mutations were introduced into each SH3 domain. This resulted in the mSH3-1, mSH3-2, and mSH3-3 mutants in which the tryptophan at amino acid positions 37, 144, or 306, respectively, was mutated to a lysine. Pulldown assays showed that each of these mutants was still capable of associating to the Rac1 C terminus (Fig. 2C). Subsequently, we mutated two of three SH3 domains, in the three possible combinations. When these mutant proteins were tested for binding to the Rac1 C terminus, we found that the association was severely impaired (Fig. 2D). Thus, Rac1 binds to the N-terminal domain of CD2AP and this binding is SH3 domain mediated, albeit that the SH3 domains show redundancy in their binding to the Rac1 C terminus.
CD2AP Links Rac1 to Cytoskeletal Regulators
CD2AP was shown earlier to associate to a series of actin-regulatory proteins including the actin capping protein CAPZ and cortactin (23, 24). These two proteins were also identified in our screen for Rac1-C terminus binding partners. To test whether CD2AP might link Rac1 to these actin regulators, the Rac1-C-terminal peptide was used to perform pulldowns on lysates of cells that were either transfected with control siRNAs or with siRNA to CD2AP (Fig. 3). The results show that in cells with reduced CD2AP levels, cortactin and CAPZ also showed a concomitant reduced association to the Rac1 C terminus (Fig. 3A). This indicates that CD2AP links Rac1 to these actin-regulating proteins. CD2AP also binds c-Cbl (35) and we could identify c-Cbl as a Rac1-C terminus associating protein by specific Western blotting. However, in contrast to CAPZ and cortactin, the association of c-Cbl to Rac1 was not reduced upon siRNA-mediated knockdown of CD2AP. This indicates that CD2AP does not mediate the interaction between Rac1 and c-Cbl.
FIGURE 3.
CD2AP and Rac1 complex with actin-regulating proteins and co-localize in membrane ruffles. A, HeLa cells were either transfected with control siRNA or siRNA for CD2AP, followed by the isolation of Rac1 C terminus binding proteins. Bound proteins were separated by SDS-PAGE and blotted for CD2AP, cortactin, CAPZ, and c-Cbl. B and C, HeLa cells were seeded on fibronectin-coated glass coverslides and either transfected with CD2AP-GFP or co-transfected with either Myc-Rac1-Q61L or Myc-Rac1-T17N (C). D, untransfected HeLa cells were immunostained for endogenous CD2AP and endogenous Rac1. E, HeLa cells were co-transfected with CD2AP-GFP and Rac1-Q61L, and stained for paxillin to visualize peripheral adhesion structures. F, HeLa cells were transfected with CD2AP-GFP, N-CD2AP-GFP, or C-CD2AP-GFP and immunostained for the focal adhesion marker paxillin. Scale bars, 10 μm. WB, Western blot.
Because we found CD2AP and Rac1 in complex with actin-regulating proteins, we tested for co-localization of Rac1 and CD2AP in actin-rich structures. Activated Rac1 is known to induce the formation of, and to localize to, membrane ruffles (36). CD2AP has also been found to localize to membrane ruffles (37). We first confirmed this localization of CD2AP by expression of CD2AP-GFP in HeLa cells (Fig. 3B). In addition to membrane ruffles, CD2AP was also found, to a limited extent, co-localizing with actin stress fibers (Fig. 3, B and F). To test for co-localization with Rac1, CD2AP-GFP was expressed together with constitutively active Rac1 (Rac1-Q61L). The proteins were found to co-localize in membrane ruffles (Fig. 3C). Importantly, endogenous Rac1 as well as endogenous CD2AP were also found to co-localize in membrane ruffles (Fig. 3D). The co-localization of Rac1 and CD2AP was associated with Rac1 activity, because CD2AP did not clearly co-localize with a dominant-negative mutant of Rac1 (Rac1-T17N (Fig. 3C).
Besides membrane ruffles, active Rac1 also localizes to adhesion complexes at the cell periphery (4). In contrast to the co-localization in peripheral membrane ruffles, CD2AP-GFP did not co-localize with Rac1 at these adhesion structures (Fig. 3E). In line with this finding, CD2AP also did not co-localize with the focal adhesion marker paxillin (Fig. 3F). Surprisingly, expression of the N-terminal domain of CD2AP showed that this portion of the protein can associate to paxillin-positive peripheral structures (Fig. 3F). In addition, this mutant of CD2AP showed a partial co-localization with actin stress fibers. In contrast, the C-terminal domain of CD2AP localized prominently to membrane ruffles, but not to paxillin-positive focal adhesions (Fig. 3F).
Rac1 Promotes Localization of CD2AP to Epithelial Cell-Cell Contacts
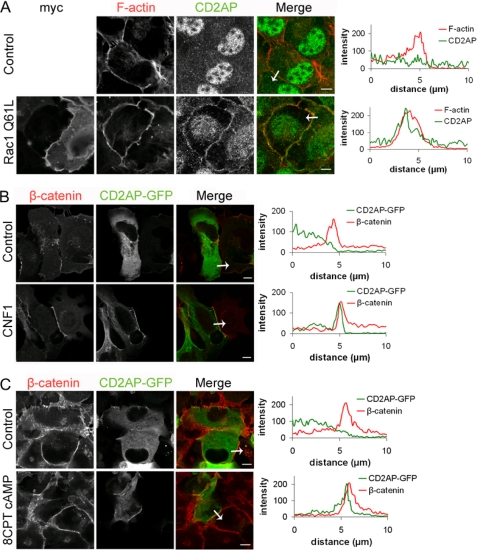
Besides its role in promoting lamellipodia and ruffle formation, Rac1 is activated during cell-cell contact formation and is required for stabilization and maturation of cell-cell contacts (12, 38, 39). CD2AP is also crucial in the formation and maintenance of cell-cell contacts, i.e. between kidney podocyte foot processes (40, 41). Under basal conditions, we found that CD2AP in epithelial cell monolayers localizes to a perinuclear compartment without any apparent localization at cell-cell contacts (Fig. 4). However, following expression of active Rac1, but not upon expression of inactive Rac1, CD2AP-GFP was recruited to cell-cell contacts (Fig. 4A, data not shown). In addition, we observed junctional recruitment of CAPZ and cortactin (Fig. 4, C and D), and an increase in junctional F-actin in the presence of active Rac1 (Fig. 4B).
FIGURE 4.
The presence of active Rac1 promotes recruitment of CD2AP to cell-cell contacts. A–D, HeLa cells were either transfected with Myc-Rac1-Q61L or co-transfected with CD2AP-GFP and stained for F-actin, CAPZ, or cortactin. Cells were co-stained for cell-cell contacts using either β-catenin or F-actin as a marker. For the indicated proteins, intensity profiles across cell-cell contacts and in the direction indicated by the arrows were generated using Zeiss Zen software. Scale bars, 10 (A) or 20 μm (B–D). Asterisks indicate cells that do not express Rac1-Q61L, arrows indicate cell-cell contacts of Rac1-Q61 expressing cells.
Endogenous Rac1 Activity Induces CD2AP Localization to Epithelial Cell-Cell Contacts
Besides CD2AP-GFP, endogenous CD2AP was also recruited to cell-cell contacts following expression of active Rac1 (Fig. 5A).
FIGURE 5.
Endogenous Rac1 activity promotes recruitment of CD2AP to cell-cell contacts. A, HeLa cells transfected with Myc-Rac1-Q61L were stained for endogenous CD2AP. For the indicated proteins, intensity profiles across cell-cell contacts and in the direction indicated by the arrows, were generated using Zeiss Zen software. B and C, HeLa cells transfected with CD2AP-GFP were either treated with 300 ng/ml of CNF1 for 1.5 h or with 100 μm 8-CPT-cAMP for 2 h. Cells were co-stained for cell-cell contacts using either β-catenin or F-actin as a marker. Scale bars, 10 μm.
As an alternative approach, we also tested if the activation of endogenous Rac1 had the same effect on CD2AP localization. To this end we used CNF1 (cytotoxic necrotizing factor 1) from E. coli (27). Upon CNF1 treatment of cells transfected with GFP-CD2AP, we observed translocation of CD2AP toward cell-cell contacts (Fig. 5B). This indicates that endogenous Rac1 activity recruits and/or stabilizes CD2AP at epithelial cell-cell contacts. To test if other GTPases could also recruit CD2AP to cell-cell contacts, active RhoA (RhoV14) was co-transfected together with CD2AP-GFP. However, no translocation of CD2AP to cell-cell contacts was observed (data not shown).
Another small GTPase that positively regulates cell-cell contacts is Rap1 (42–44). To activate Rap1 and test whether this is sufficient to recruit CD2AP to cell-cell junctions, we treated cells with 8-CPT-cAMP, a cAMP analogue that activates Rap1 by directly activating its GEF EPAC1. Similar to the activation of endogenous Rac1, this Rap-1 mediated stimulation of cell-cell contact was accompanied by the recruitment of CD2AP to intercellular junctions (Fig. 5C). In contrast to Rac1, however, the C-terminal domains of Rap1A and Rap1B did not associate to CD2AP (data not shown). This suggests that the Epac1/Rap1-induced recruitment of CD2AP to junctions is induced in an indirect fashion, possibly through the Rap1-mediated activation of Rac1 (45).
CD2AP Is Required for the Maintenance of Epithelial Cell-Cell Contacts
CD2AP is a ubiquitously expressed protein. Therefore the Rac1-dependent translocation of CD2AP to cell-cell contacts was also tested in human lung epithelial cells (H292). In addition to HeLa cells, recruitment of CD2AP-GFP toward cell-cell contacts was also observed in H292 cells, upon co-expression with active Rac1 (Fig. 6A). This shows that the Rac1-induced translocation of CD2AP is not cell-type specific.
FIGURE 6.
Reduction of CD2AP expression in lung epithelial cells reduces TER. A, H292 lung epithelial cells were co-transfected with CD2AP-GFP and Myc-Rac1-Q61L, and subsequently analyzed for localization of CD2AP at sites of cell-cell contact. Intensity plots across cell-cell junctions in the direction of the arrows are shown on the right. Scale bars, 10 μm. B, 24 h post-transfection of CD2AP-GFP, H292 cells were exposed to 2 mm EGTA for 3.5 h. Subsequently, EGTA was washed out and calcium-containing medium was added for 1 h, after which localization of CD2AP at the newly formed cell-cell contacts was analyzed. Scale bars, 10 μm. C, H292 cells were transfected either with control siRNA or siRNA for CD2AP, and seeded in quadruple in ECIS wells to confluence. TER was monitored continuously. TER was normalized at t = 0 and plotted in the bar graph as an average over a period of 2 h, which after 24 h post-seeding corresponds to 48 h post-transfection. Student's t test was performed to calculate significance (n = 4, p < 0.05). After the experiment, cells from each ECIS well were harvested in sample buffer and analyzed by Western blot (WB) for CD2AP expression to confirm the effect of the siRNA.
Because endogenous Rac1 is known to be activated upon cadherin ligation (12), we tested if CD2AP could also be recruited to newly formed cell-cell contacts. EGTA was used in a calcium switch assay to disrupt cell-cell contacts, and a subsequent washout of EGTA with calcium-containing medium was used to allow for reformation of cell-cell contacts. One hour after EGTA washout we observed accumulation of CD2AP-GFP at the newly formed cell-cell contacts (Fig. 6B).
Because CD2AP localization to cell-cell contacts was observed upon Rac1 activation, the contribution of CD2AP to cell-cell adhesion was tested. siRNA to CD2AP was used to reduce CD2AP levels in H292 cells (Fig. 6C), and the effect on TER over confluent monolayers was measured using ECIS (42). Reduction of CD2AP levels resulted in a significant and consistent decrease in TER, indicating that loss of CD2AP is accompanied by a reduction in epithelial integrity (Fig. 6C).
To exclude that the decrease in TER after reducing CD2AP levels was caused a factor other than cell-cell contacts, we measured TER in an ECIS-Zθ system. The ECIS-Zθ is able to define the contribution of cellular membranes (capacitance) to the measured impedance, and is able to extract the resistance that is due to transcellular resistance (supplemental Fig. S1, A and B). The obtained transcellular resistance shows the same decrease in TER, as the effect we observed earlier in the original setup, after reducing CD2AP levels (Fig. 6C and supplemental Fig. S1A). These results match a relatively small contribution of the capacitance, when the impedance was measured at high frequency (30 kHz; supplemental Fig. S1B), as was also done in the experiment in Fig. 6C. The data were modeled using ECIS-Zθ software to determine the effects on the barrier function (Rb) (46). The results show that loss of CD2AP is accompanied by a reduction in the barrier function of H292 cells (supplemental Fig. S1, C and D).
DISCUSSION
The hypervariable domain in Rac1, as in other small GTPases, has been proposed to regulate its subcellular localization and, as a result, Rac1 signaling specificity (33). This is not only based on targeting by this region to specific membrane domains or platforms, but also requires association to specific signaling proteins. Analysis of several of such protein-protein interactions that we identified previously has revealed an increasing complexity as to the role of the hypervariable domain in Rac1 signaling. The GEF β-PIX, which binds the Rac1 C terminus via its SH3 domain, is capable of recruiting Rac1 to peripheral membrane ruffles, and co-localizes with Rac1 in leading edge focal adhesions (4). In contrast, the nuclear oncogene SET, which lacks an SH3 domain, but also associates to the Rac1 C-terminal domain, is recruited by Rac1 activity to the cellular periphery but not to focal adhesions, and serves to promote Rac1-induced cell migration (8). We now describe the interaction between Rac1 and the adapter protein CMS/CD2AP, mediated by the Rac1 C-terminal domain and, as for PIX, the SH3 domains of CD2AP.
The association to CD2AP was specific for the hypervariable C-terminal domain of Rac1, because we detected no significant binding of CD2AP to the corresponding C termini of other Rho family members tested. The interaction required the proline-rich as well as the poly-basic regions in the carboxyl terminus of Rac1, and in addition, the SH3 domains of CD2AP. The individual SH3 domains appeared redundant for Rac1 binding. A similar behavior was described for the SH3 domains of CIN85, an adapter protein that is very homologous to CD2AP. Kowanetz et al. (47) showed that the three SH3 domains in CIN85 are redundant in their binding to the ubiquitin ligase Cbl. Interestingly, the CIN85 and CD2AP SH3 domains were found to bind an a typical polyproline motif that is present not only in c-Cbl and CD2, but also in the Rac1 effector Pak1, and which is in fact similar to the proline-rich region in the Rac1-C terminus (47, 48). Given the fact that CD2AP has actin-binding properties (25), this adapter may play an important role in targeting the Rac1-Pak signaling unit to the actin cytoskeleton. Also, the third SH3 domain of CD2AP is most similar to the SH3 domain of β-PIX, this domain might therefore constitute the primary site of association for full-length Rac1 in vivo.
Besides CD2AP, we found that cortactin and CAPZ also associate to the Rac1-C terminal peptide. Moreover, this interaction appeared to be dependent on the presence of CD2AP. CD2AP thus represents a link between Rac1 and these cytoskeleton remodeling proteins. c-Cbl was also found to complex with Rac1, but this interaction was not CD2AP dependent, although it was shown earlier that CD2AP can associate to c-Cbl (35). This indicates that c-Cbl associates to Rac1 either directly, or through another intermediate protein.
Besides complexing with actin regulators, endogenous CD2AP and endogenous Rac1 were found to co-localize in actin structures such as lamellipodia and membrane ruffles. In addition, full-length CD2AP and the N-CD2AP mutant, but not the C-CD2AP mutant, were found to partially align with actin stress fibers. In turn, C-CD2AP localized extensively to membrane ruffles, but its levels in the cytoplasm appeared lower as compared with full-length CD2AP. This characteristic peripheral localization of C-CD2AP might be explained by the polyproline sequence it contains, which was shown to be involved in association of CD2AP to cortactin (23). A fraction of N-CD2AP was also able to localize to focal adhesions, but we could not detect full-length CD2AP in these structures, under conditions where Rac1 was detected in peripheral adhesions. Earlier CD2AP was found to associate to the focal adhesion protein p130Cas, but in another study it could not be found in paxillin-positive focal adhesions (15, 49). Thus, although CD2AP may localize to focal adhesions under specific cellular conditions, the co-localization of the full-length protein with Rac1 occurs primarily at the peripheral membrane. This is in marked contrast to the Rac1-β-PIX complex which is, next to membrane ruffles, also found in focal adhesions (4).
Under resting conditions, CD2AP did not show prominent localization to cell-cell junctions in cell monolayers, but rather obtains a perinuclear, primarily cytoplasmatic localization. This observation is in line with earlier findings by Mustonen et al. (37). However, when active Rac1 was present, we found CD2AP to become recruited to cell-cell contacts in various types of epithelial cells. To investigate the relevance of Rac1-induced CD2AP translocation, the function of CD2AP was analyzed in the context of epithelial integrity by measurement of TER. Reduction of CD2AP levels by siRNA transfection resulted in a significant reduction in TER and barrier function, indicating reduced cell-cell adhesion. CD2AP thus appears to contribute to epithelial cell-cell contact. This finding is in line with its function in mouse kidney podocytes, where it is required for formation and maintenance of nephrin-based cell-cell contacts (21). Moreover, Rac1 was recently shown to be activated upon nephrin ligation and during differentiation of podocytes (50). We found that active Rac1 not only induced the translocation of CD2AP to cell-cell contacts, but also increased the concentration of F-actin, and of the actin-regulatory proteins CAPZ and cortactin at cell-cell contacts. Moreover, we show that these actin regulators are dependent on CD2AP for their association to Rac1, which suggests that CD2AP could function to link actin-regulating proteins to Rac1 at sites of cell-cell contact.
Our data suggest that CD2AP may have a more general function in epithelial cell-cell contacts, which is also supported by data showing its association to E-cadherin in colon epithelial cells (37). Our observation that CD2AP was concentrated at cell-cell contacts upon the presence of active Rac1 suggests that CD2AP is particularly relevant during transient Rac1-dependent modulation of epithelial cell-cell contacts.
Because we also found CD2AP to localize to newly formed cell-cell contacts, we propose the following model for the role of CD2AP in Rac1 signaling (Fig. 7). Upon formation of cell-cell contact and cadherin engagement, Rac1 becomes recruited to sites of cell adhesion where it is activated (12). Active Rac1 induces actin polymerization, and promotes CD2AP recruitment toward cell-cell contacts. Here, CD2AP might link Rac1 to actin-regulating proteins such as CAPZ and cortactin as well as to its downstream effector PAK. The signaling from this protein complex then serves to further promote actin polymerization and reorganization, required for the stabilization of cell-cell contact.
FIGURE 7.
A model for CD2AP recruitment to cell-cell contacts upon Rac1 activation. Following cadherin engagement, Rac1 becomes recruited to sites of cell adhesion where it is activated. Active Rac1 induces actin polymerization and promotes CD2AP recruitment toward cell-cell contacts. Here, CD2AP contributes to the formation of epithelial cell-cell contacts and barrier function.
The recruitment of CD2AP to cell-cell junctions is so far unique among Rac1-associating proteins. Earlier, we found Rac1 to recruit the nuclear protein SET to the cell membrane. Conversely, β-PIX was found to associate and recruit Rac1 to focal adhesions. But neither of these proteins was found at cell-cell contacts. Therefore, Rac1 molecules that associate to CD2AP probably constitute a different fraction of the total pool of cellular Rac1.
Together, our recent data suggest that protein-protein interaction between Rac1 and a series of cellular proteins is, as has been proposed for Rac1 signaling, compartmentalized. This implies that several interactions may occur simultaneously, but also that a specific interactor, binding the Rac1 hypervariable C terminus, may determine which effector protein is activated following Rac1 GTP-loading at a specific cellular location. To prove this concept warrants further research that may ultimately provide us with a paradigm for the control of localized signaling by the Rho family of small GTPases.
Supplementary Material
Acknowledgments
We thank Prof. Andrey Shaw (Washington University School of Medicine) for the CD2AP-GFP plasmid and Prof. Dr. Gudula Schmidt (Institute of Experimental and Clinical Pharmacology and Toxicology, University of Freiburg) for the CNF1 toxin and CNF1 encoding plasmid.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- GEF
- guanine nucleotide exchange factor
- SH3
- Src homology domain 3
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein
- siRNA
- small interfering RNA
- CNF1
- cytotoxic necrotizing factor 1
- TER
- transepithelial electrical resistance
- ECIS
- electrical cell impedance sensing
- 8-CPT-cAMP
- 8-(4-chlorophenylthio)adenosine-3′,5′-cyclic monophosphate.
REFERENCES
- 1.Rossman K. L., Der C. J., Sondek J. (2005) Nat. Rev. Mol. Cell Biol. 6, 167–180 [DOI] [PubMed] [Google Scholar]
- 2.Del Pozo M. A., Kiosses W. B., Alderson N. B., Meller N., Hahn K. M., Schwartz M. A. (2002) Nat. Cell Biol. 4, 232–239 [DOI] [PubMed] [Google Scholar]
- 3.Michaelson D., Silletti J., Murphy G., D'Eustachio P., Rush M., Philips M. R. (2001) J. Cell Biol. 152, 111–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.ten Klooster J. P., Jaffer Z. M., Chernoff J., Hordijk P. L. (2006) J. Cell Biol. 172, 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ridley A. J. (2001) J. Cell Sci. 114, 2713–2722 [DOI] [PubMed] [Google Scholar]
- 6.Tybulewicz V. L., Henderson R. B. (2009) Nat. Rev. Immunol. 9, 630–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung C. Y., Lee S., Briscoe C., Ellsworth C., Firtel R. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5225–5230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.ten Klooster J. P., Leeuwen I., Scheres N., Anthony E. C., Hordijk P. L. (2007) EMBO J. 26, 336–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamazaki D., Oikawa T., Takenawa T. (2007) J. Cell Sci. 120, 86–100 [DOI] [PubMed] [Google Scholar]
- 10.Popoff M. R., Geny B. (2009) Biochim. Biophys. Acta 1788, 797–812 [DOI] [PubMed] [Google Scholar]
- 11.Noritake J., Fukata M., Sato K., Nakagawa M., Watanabe T., Izumi N., Wang S., Fukata Y., Kaibuchi K. (2004) Mol. Biol. Cell 15, 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovacs E. M., Goodwin M., Ali R. G., Paterson A. D., Yap A. S. (2002) Curr. Biol. 12, 379–382 [DOI] [PubMed] [Google Scholar]
- 13.van Hennik P. B., ten Klooster J. P., Halstead J. R., Voermans C., Anthony E. C., Divecha N., Hordijk P. L. (2003) J. Biol. Chem. 278, 39166–39175 [DOI] [PubMed] [Google Scholar]
- 14.Dustin M. L., Olszowy M. W., Holdorf A. D., Li J., Bromley S., Desai N., Widder P., Rosenberger F., van der Merwe P. A., Allen P. M., Shaw A. S. (1998) Cell 94, 667–677 [DOI] [PubMed] [Google Scholar]
- 15.Kirsch K. H., Georgescu M. M., Ishimaru S., Hanafusa H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 6211–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gigante M., Pontrelli P., Montemurno E., Roca L., Aucella F., Penza R., Caridi G., Ranieri E., Ghiggeri G. M., Gesualdo L. (2009) Nephrol. Dial. Transplant. 24, 1858–1864 [DOI] [PubMed] [Google Scholar]
- 17.Shih N. Y., Li J., Karpitskii V., Nguyen A., Dustin M. L., Kanagawa O., Miner J. H., Shaw A. S. (1999) Science 286, 312–315 [DOI] [PubMed] [Google Scholar]
- 18.Shih N. Y., Li J., Cotran R., Mundel P., Miner J. H., Shaw A. S. (2001) Am. J. Pathol. 159, 2303–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawachi H., Miyauchi N., Suzuki K., Han G. D., Orikasa M., Shimizu F. (2006) Nephrology 11, 274–281 [DOI] [PubMed] [Google Scholar]
- 20.Wolf G., Stahl R. A. (2003) Lancet 362, 1746–1748 [DOI] [PubMed] [Google Scholar]
- 21.Yuan H., Takeuchi E., Salant D. J. (2002) Am. J. Physiol. Renal Physiol. 282, F585–F591 [DOI] [PubMed] [Google Scholar]
- 22.Johnson R. I., Seppa M. J., Cagan R. L. (2008) J. Cell Biol. 180, 1191–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynch D. K., Winata S. C., Lyons R. J., Hughes W. E., Lehrbach G. M., Wasinger V., Corthals G., Cordwell S., Daly R. J. (2003) J. Biol. Chem. 278, 21805–21813 [DOI] [PubMed] [Google Scholar]
- 24.Hutchings N. J., Clarkson N., Chalkley R., Barclay A. N., Brown M. H. (2003) J. Biol. Chem. 278, 22396–22403 [DOI] [PubMed] [Google Scholar]
- 25.Lehtonen S., Zhao F., Lehtonen E. (2002) Am. J. Physiol. Renal Physiol. 283, F734–F743 [DOI] [PubMed] [Google Scholar]
- 26.Bruck S., Huber T. B., Ingham R. J., Kim K., Niederstrasser H., Allen P. M., Pawson T., Cooper J. A., Shaw A. S. (2006) J. Biol. Chem. 281, 19196–19203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pop M., Aktories K., Schmidt G. (2004) J. Biol. Chem. 279, 35840–35848 [DOI] [PubMed] [Google Scholar]
- 28.Voermans C., Kooi M. L., Rodenhuis S., van der Lelie H., van der Schoot C. E., Gerritsen W. R. (2001) Blood 97, 799–804 [DOI] [PubMed] [Google Scholar]
- 29.Ho A., Schwarze S. R., Mermelstein S. J., Waksman G., Dowdy S. F. (2001) Cancer Res. 61, 474–477 [PubMed] [Google Scholar]
- 30.Kanters E., van Rijssel J., Hensbergen P. J., Hondius D., Mul F. P., Deelder A. M., Sonnenberg A., van Buul J. D., Hordijk P. L. (2008) J. Biol. Chem. 283, 31830–31839 [DOI] [PubMed] [Google Scholar]
- 31.Haeusler L. C., Blumenstein L., Stege P., Dvorsky R., Ahmadian M. R. (2003) FEBS Lett. 555, 556–560 [DOI] [PubMed] [Google Scholar]
- 32.Giaever I., Keese C. R. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 7896–7900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hajdo-Milasinović A., Ellenbroek S. I., van Es S., van der Vaart B., Collard J. G. (2007) J. Cell Sci. 120, 555–566 [DOI] [PubMed] [Google Scholar]
- 34.ten Klooster J. P., Hordijk P. L. (2007) Biol. Cell 99, 1–12 [DOI] [PubMed] [Google Scholar]
- 35.Kirsch K. H., Georgescu M. M., Shishido T., Langdon W. Y., Birge R. B., Hanafusa H. (2001) J. Biol. Chem. 276, 4957–4963 [DOI] [PubMed] [Google Scholar]
- 36.Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A. (1992) Cell 70, 401–410 [DOI] [PubMed] [Google Scholar]
- 37.Mustonen H., Lepistö A., Lehtonen S., Lehtonen E., Puolakkainen P., Kivilaakso E. (2005) Biochem. Biophys. Res. Commun. 332, 426–432 [DOI] [PubMed] [Google Scholar]
- 38.Takahashi K., Sasaki T., Mammoto A., Takaishi K., Kameyama T., Tsukita S., Takai Y. (1997) J. Biol. Chem. 272, 23371–23375 [DOI] [PubMed] [Google Scholar]
- 39.Hordijk P. L., ten Klooster J. P., van der Kammen R. A., Michiels F., Oomen L. C., Collard J. G. (1997) Science 278, 1464–1466 [DOI] [PubMed] [Google Scholar]
- 40.Li C., Ruotsalainen V., Tryggvason K., Shaw A. S., Miner J. H. (2000) Am. J. Physiol. Renal Physiol. 279, F785–F792 [DOI] [PubMed] [Google Scholar]
- 41.Khoshnoodi J., Tryggvason K. (2001) Exp. Nephrol. 9, 355–359 [DOI] [PubMed] [Google Scholar]
- 42.Lorenowicz M. J., Fernandez-Borja M., Kooistra M. R., Bos J. L., Hordijk P. L. (2008) Eur. J. Cell Biol. 87, 779–792 [DOI] [PubMed] [Google Scholar]
- 43.Pannekoek W. J., Kooistra M. R., Zwartkruis F. J., Bos J. L. (2009) Biochim. Biophys. Acta 1788, 790–796 [DOI] [PubMed] [Google Scholar]
- 44.Price L. S., Hajdo-Milasinovic A., Zhao J., Zwartkruis F. J., Collard J. G., Bos J. L. (2004) J. Biol. Chem. 279, 35127–35132 [DOI] [PubMed] [Google Scholar]
- 45.Arthur W. T., Quilliam L. A., Cooper J. A. (2004) J. Cell Biol. 167, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becker P. M., Verin A. D., Booth M. A., Liu F., Birukova A., Garcia J. G. (2001) Am. J. Physiol. Lung Cell Mol. Physiol. 281, L1500–L1511 [DOI] [PubMed] [Google Scholar]
- 47.Kowanetz K., Szymkiewicz I., Haglund K., Kowanetz M., Husnjak K., Taylor J. D., Soubeyran P., Engstrom U., Ladbury J. E., Dikic I. (2003) J. Biol. Chem. 278, 39735–39746 [DOI] [PubMed] [Google Scholar]
- 48.Moncalián G., Cárdenes N., Deribe Y. L., Spínola-Amilibia M., Dikic I., Bravo J. (2006) J. Biol. Chem. 281, 38845–38853 [DOI] [PubMed] [Google Scholar]
- 49.Welsch T., Endlich N., Kriz W., Endlich K. (2001) Am. J. Physiol. Renal Physiol. 281, F769–F777 [DOI] [PubMed] [Google Scholar]
- 50.Attias O., Jiang R., Aoudjit L., Kawachi H., Takano T. (2010) Nephron Exp. Nephrol. 114, e93–e106 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.