Abstract
NOD2 (nucleotide-binding oligomerization domain containing 2) is an important cytosolic pattern recognition receptor that activates NF-κB and other immune effector pathways such as autophagy and antigen presentation. Despite its intracellular localization, NOD2 participates in sensing of extracellular microbes such as Staphylococcus aureus. NOD2 ligands similar to the minimal synthetic ligand muramyl dipeptide (MDP) are generated by internalization and processing of bacteria in hydrolytic phagolysosomes. However, how these derived ligands exit this organelle and access the cytosol to activate NOD2 is poorly understood. Here, we address how phagosome-derived NOD2 ligands access the cytosol in human phagocytes. Drawing on data from Drosophila phagosomes, we identify an evolutionarily conserved role of SLC15A transporters, Drosophila Yin and PEPT2, as MDP transporters in fly and human phagocytes, respectively. We show that PEPT2 is highly expressed by human myeloid cells. Ectopic expression of both Yin and PEPT2 increases the sensitivity of NOD2-dependent NF-κB activation. Additionally, we show that PEPT2 associates with phagosome membranes. Together, these data identify Drosophila Yin and PEPT2 as evolutionarily conserved phagosome-associated transporters that are likely to be of particular importance in delivery of bacteria-derived ligands generated in phagosomes to cytosolic sensors recruited to the vicinity of these organelles.
Keywords: Innate Immunity, Macrophage, Pathogen-associated Molecular Pattern (PAMP), Pattern Recognition Receptor, Phagocytosis, NOD2, Pathogen Processing, Transporter
Introduction
The components of the innate immune system, including cellular defenses and inflammatory pathways, mount a rapid and tightly orchestrated response to pathogen invasion (1). Central to this is the activation of proinflammatory signals that are triggered by the simultaneous ligation of multiple pattern recognition receptors by ligands that are components of microbes and absent from the healthy host (2). The archetypal pattern recognition receptors are the Toll-like receptors (TLRs),2 transmembrane proteins found on the cell surface and in endosomal compartments (3), and the NOD (nucleotide-binding oligomerization domain)-like receptors (NLRs), which reside in the cytosol and recognize bacteria, or their derivatives, that cross the plasma membrane and enter the cell (4, 5).
Despite the TLRs and NLRs surveying two discrete cellular compartments (the extracellular space and the cytosol), these receptors collaborate for optimal host response to many pathogens. However, how this collaboration is coordinated is poorly understood. Previous work has demonstrated that maximal activation of TLR and NOD2-dependent signaling in response to Gram-positive microbes is possible only after delivery into a phagosome and that this event is intimately associated with inflammatory cytokine induction (6, 7). These observations reflect the multifunctional role of the phagosome during innate immune sensing of Gram-positive microbes. An emerging theme is that during maturation, the phagosome enzymatically modifies the cargo to liberate cryptic pattern recognition receptor ligands (8, 9) in a process we refer to as “pathogen processing.” These so-called post mortem PAMPS (10) include ligands for a wide variety of innate immune pattern recognition receptors including NOD2, the focus of this work.
NOD2 was identified less than a decade ago as a NF-κB activating protein (11). Subsequent genetic studies have revealed that mutations in NOD2 are linked to increased risk of development of Crohn's disease (12–14). NOD2 is a multidomain protein containing two CARD domains, a central NOD domain and a C-terminal LRR domain that is thought to be the ligand-binding domain. Peptidoglycan-derived fragments, including the well characterized synthetic ligand muramyl dipeptide (MDP), are known ligands that activate NOD2 signaling pathways. Activation of NOD2 induces the production of multiple NF-kB dependent cytokines, including IL-8 and IL-6, and antimicrobial peptides (15). Because of the association with inflammatory bowel disease, a major focus of work on NOD2 has been in understanding its role and regulation in epithelial cells. However, it is evident that NOD2 is important not only in epithelial immunity but also in other cell types, including phagocytic antigen presenting cells. Supporting this, NOD2 is expressed predominantly in myeloid cells (11), and macrophages derived from both Nod2-deficient mice and from Crohn's disease patients carrying mutations in NOD2 are unable to activate NF-kB and mitogen-activated protein kinase (MAPK) pathways after MDP stimulation while retaining their capacity to respond to TLR ligands (16). Additionally, NOD2 is also involved in microbial sensing in a number of other systems, including a recently described role in sensing of viral pathogens (17), autophagy (18), and induction of adaptive immunity (19).
Intriguingly, despite being a cytosolic sensor, NOD2 participates not only in sensing cytoinvasive microbes but also in the sensing of extracellular pathogens. This fact raises the question of how the NOD2 ligands derived from extracellular microbes access intracellular compartments, and suggest that transport mechanisms must exist to translocate them across membranes to enter the cytosol surveilled by NOD2. As the ligands from extracellular pathogens appear to be generated by digestion and modification in phagolysosomes, we have focused on these organelles and set out to understand MDP transport in phagocytic cells. Specifically, we have addressed how phagosome-derived ligands access the cytosol. Drawing on data from Drosophila phagosomes, we identify an evolutionarily conserved role of SLC15 transporters, Drosophila Yin and PEPT2, as MDP transporters in fly and human phagocytes, respectively. Importantly, we show that these transporters associate with phagosome membranes and hence are likely to be of particular importance in delivery of ligands generated in this compartment to NOD2 recruited to the vicinity of these organelles.
EXPERIMENTAL PROCEDURES
Mice and Cells
C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Nod2−/− mice generated by K. Kobayashi. All mice were kept and handled under a protocol approved by the Subcommittee on Research Animal Care at Massachusetts General Hospital). Bone marrow-derived macrophages were generated by 7 days culture of bone marrow cells in Dulbecco's modified Eagle's medium containing 10% fetal calf serum and 20% L929 cell-conditioned medium and primed with IFNγ for 16 h prior to use. THP-1 and HEK293T cell lines (ATCC, Manassas, VA) were maintained according to ATCC's recommendations. Before use, THP-1 cells were differentiated using phorbol 12-myristate 13-acetate at 5 ng/ml as described previously (20). HEK293T cells were transfected with different constructs by using Lipofectamine 2000 and following Invitrogen's recommendations.
Bacterial Strains and Reagents
Staphylococcus aureus (strain Reynolds capsular serotype 5; provided by Dr. J. C. Lee, Brigham and Women's Hospital, Boston, MA) were grown as described previously (21). The bacteria grown to the midexponential phase were heat-inactivated at 65 °C for 30 min, washed twice (10,000 × g for 10 min), resuspended in PBS, and stored in aliquots at −20 °C before use for cell stimulation in vitro. MDP was from InvivoGen (San Diego, CA).
Constructs
YFP-tagged TLR2 (pcDNA3.1-TLR2-YFP) and CFP-tagged TLR6 (pcDNA3.1-TLR6-CFP) expression vectors were kindly provided by D. Golenbock (University of Massachusetts Medical School, Worcester, MA). GFP-tagged (pEGFPC1-NOD2) NOD2 and nontagged NOD2 mammalian expression vectors were described previously (22). Full-length human SLC15A2 (PEPT2) from THP-1 cells and Drosophila OpT1/Yin from S2 cells were cloned into a Gateway entry vector (Invitrogen) by PCR using primers designed according to the manufacturer's instructions. These entry clones were sequence verified and transferred into a Gateway mammalian expression vector (Invitrogen) by recombination. Human SLC15A1 was subcloned into Gateway vectors from a verified clone purchased from Open Biosystems (Huntsville, AL). Mammalian expression vector containing full-length human SLC15A3 coding sequence was purchased from Open Biosystems (clone ID, 5213632).
Cell Stimulations and Treatments
Macrophages in Dulbecco's modified Eagle's medium with 1% fetal calf serum were stimulated with heat-inactivated bacteria at a multiplicity of infection of 25:1, or MDP at the indicated concentrations, at 37 °C in 5% CO2 for 2 to 4 h for macrophages and 6 h for differentiated THP1 cells, after which culture supernatants were harvested. Cytokine secretion was measured by enzyme-linked immunosorbent assay (DuoSet Enzyme-linked Immunosorbent Assay Development System; R&D System, Minneapolis, MN) in accordance with the manufacturer's protocol.
Phagocytosis and Intracellular TNF-α and Il-6
Macrophages in Dulbecco's modified Eagle's medium with 1% fetal calf serum were incubated with heat-inactivated bacteria, labeled with tetramethylrhodamine (Molecular Probes, Eugene, OR), at a multiplicity of infection of 25:1 for 30 min on ice, allowing the synchronization of bacteria binding onto the cell. In all cases, before incubation with macrophages, bacterial clusters were disrupted by passing the bacteria through a 30-gauge needle. After 30 min on ice, the cells were further incubated for the indicated times at 37 °C in the presence of GolgiStop (BD Bioscience, San Diego, CA) to accumulate intracellular TNF-α. The cells were washed twice with ice-cold PBS containing 5 mm EDTA (PBS/EDTA), detached with scrappers, and fixed in 3% paraformaldehyde. The cells were permeabilized and stained with allophycocyanin-conjugated anti-mouse TNF-α or Il-6 antibody (BD Bioscience) diluted in PBS with 0.2% saponin. After washing, the cells were analyzed by flow cytometry performed on FACSCalibur (Becton Dickinson); analysis was performed with CellQuest Pro software (Becton Dickinson) to determine phagocytosis and intracellular TNF-α production at the single cell level.
NF-κB Luciferase Reporter Assay
Dual luciferase reporter assays for NF-κB activation were performed in HEK293T cells as described previously (23). Briefly, cells were transfected with the indicated expression constructs together with NF-κB luciferase (Firefly) and control luciferase (Renilla) reporter plasmids using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer's protocol. Before assays, the cells were washed with PBS and cultured in fresh Dulbecco's modified Eagle's medium with 1% fetal calf serum. The cells were stimulated with heat-inactivated bacteria or bacterial ligands as described under “Cell Stimulations and Treatments.” Reporter gene activity was measured using the Dual-Glo luciferase assay system (Promega, Madison, WI) in accordance with the manufacturer's protocol. Data were normalized for transfection efficiency with the control reporter activity from the same sample and presented as mean NF-κB fold induction of triplicate samples.
Transporter Analysis
Expression of SLC15 transporters was determined by semiquantitative RT-PCR using a Mastercycler ep realplex thermocyler (Eppendorf). Expression was normalized to glyceraldehyde-3-phosphate dehydrogenase and expressed as a percent of expression in kidney, which was used as a positive control for transporter expression. RT-PCR primers used to estimate relative expression of the different transporters are as follows: SLC15A1-F, 5′-GTTGCTTCTGGTCGTCTGTG-3′; SLC15A1-R, 5′-GCCCCTGACATGAAATATGG-3′; SLC15A2-F, 5′-GCCCTGTCTTGAAGCATTTT-3′; SLC15A2-R, 5′-AGAGTCTCTGGGGCCTTGTT-3′; SLC15A3-F, 5′-GCTTAAGCTCGCTCTCCAAA-3′; SLC15A3-R, 5′-GGCAAGATCTTCACCAGCAC-3′; SLC15A4-F, 5′-AGCGATCCTGTCGTTAGGTG-3′; SLC15A4-R, 5′-AGGAGGCTTGGTGATGAAAA-3′.
Microscopy
HEK293T cells were transiently transfected with pDEST53-PEPT2 to allow expression of a GFP-PEPT2 fusion protein. The cells were than incubated for 2 h with red fluorescent 1 μm diameter latex beads (Estapore, Paris, France) to allow engulfment. Immunofluorescence studies were performed on cells fixed in 4% paraformaldehyde (Sigma-Aldrich) or methanol (for anti-PEPT2 and anti-NOD2 staining) and imaged with a confocal microscope (Bio-Rad/Zeiss Radiance 2000 or Nikon TiE with a Perkin Elmer spinning disc). Nonconfocal images were acquired using a Nikon inverted microscope equipped with a CoolSnap camera. Post-acquisition image analysis was performed with Adobe Photoshop, Openlab, or Velocity software (Improvision).
Statistical Analysis
All experiments were performed in triplicates and repeated at least three times independently. Statistical significance was determined by using a two-tailed unpaired Student's t test.
RESULTS
Response to S. aureus Is Mediated by the Collaboration between TLR2/6 and NOD2 from the Phagosome
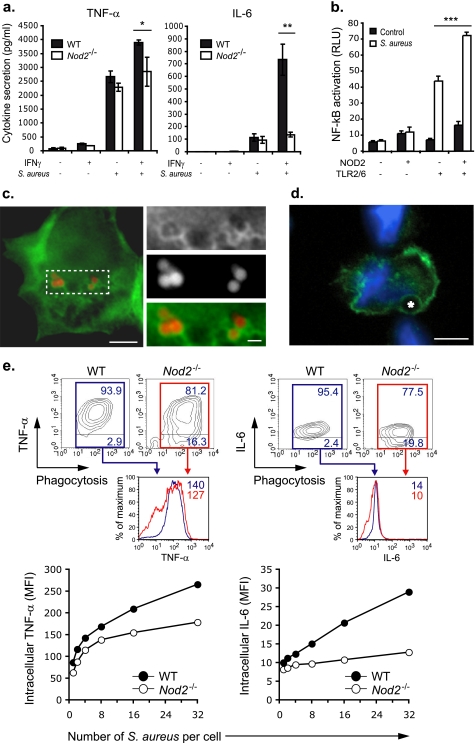
NF-κB driven responses to S. aureus are mediated primarily by membrane-bound TLRs that respond to immunostimulatory components of the Gram-positive bacterial cell wall that include lipoteichoic acid, peptidoglycan, and lipopeptides (24). However, a second family of pattern recognition receptors, the cytosolic NLRs, has also been suggested to participate in response to these primarily extracellular microbes. Specifically, it is believed that NOD2 senses a peptidoglycan derivative, similar to MDP that is released from the bacterial cell wall (25). To further define the role of NOD2 in the response to S. aureus, we isolated macrophages from Nod2−/− mice and measured TNF-α and IL-6. Resting Nod2−/− macrophages showed no defect in response to S. aureus, but when primed with IFNγ, showed impaired response compared with wild-type controls (Fig. 1a). To determine the relative contributions of the TLR and NOD2 pathways to S. aureus sensing, we next used HEK293T transfected with TLR2/6 and/or NOD2 and measured NF-κB activation. Transfection of NOD2 alone was unable to render HEK293T cells responsive to S. aureus (Fig. 1b). However, when coexpressed with TLR2/6, NOD2 enhanced NFkB activation, indicating that although NOD2 is insufficient for full activation, it synergized with TLR2/6 to increase cytokine response. Thus, the collaboration of TLRs with NOD2 is necessary for optimal response to S. aureus in both the mouse and human systems.
FIGURE 1.
NOD2 participates in response to S. aureus from the phagosome. a, cytokine production in macrophages from Nod2−/− mice stimulated with S. aureus before and after IFNγ priming. Graphs are representative of at least three independent experiments; mean ± S.D. are represented. *, p ≤ 0.05 and **, p ≤ 0.01 relative to wild-type. b, NOD2 was expressed in HEK293T cells along with TL2/TLR6 and an NF-κB reporter construct. NF-κB response to S. aureus was measured by luciferase indicate that NOD2 functions primarily by cooperation with TLR2/6. Graph are representative of at least three independent experiments; mean ± S.D. are represented. ***, p ≤ 0.001 relative to TLR2/6 + S. aureus. c, NOD2-GFP was ectopically expressed in HEK293T cells. Phagosomes were generated in those cells by challenged with latex beads. Scale bars correspond to 5 μm in the main picture and to 1 μm in magnified box. d, endogenous NOD2 surrounds phagosomes in THP1 cells. The asterisk denotes a 2 μm latex bead. Scale bar corresponds to 5 μm. e, the relationship of phagosome number to cytokine production in macrophages from Nod2−/− mice stimulated with S. aureus after IFNγ priming demonstrates the contribution of NOD2 to both TNF-α and IL-6 production, representative of at least three independent experiments. RLU, relative light units; WT, wild-type; MFI, mean fluorescent intensity.
NOD2 is normally distributed in the cytosol and weakly associated with the plasma membrane in resting cells (22, 26). However, for signaling to occur, NOD2 must be recruited and tethered to membranes along with its signaling adaptor, RIP2 (26). Our previous work (21, 23) and the work of others (6, 7) have shown that TLR2 and its co-receptors CD36 and MBL are internalized after uptake of particulate ligands (6, 7, 21, 23). We therefore hypothesized that, like other components of the S. aureus sensing machinery, NOD2 may also be recruited to phagosome membranes. Consistent with this possibility, ectopically expressed GFP-NOD2 concentrated in close proximity to phagosomes generated in HEK293T cells (Fig. 1c). Similarly, endogenous NOD2 was also found associated with phagosomes in THP1 human macrophages (Fig. 1d).
As NOD2 is recruited to phagosomes, which also generate the NOD2 ligand (8), we next tested whether these organelles were the origin of NOD2 activation after internalization of S. aureus. Our previous work (21) has shown that TNF-α and IL-6 response to S. aureus is triggered primarily from phagosomes, and, hence, response to these bacteria can be used as a surrogate for phagosome-dependent signaling. Therefore, using a single cell fluorescence-activated cell sorter-based assay in which cytokine production and phagocytosis can be monitored simultaneously (21), we tested whether NOD2-dependent TNF-α and IL-6 production in response to internalized S. aureus was impaired in macrophages from Nod2−/− mice (Fig. 1e). Phagosome-triggered cytokine production, particularly IL-6, was markedly impaired in Nod2−/− macrophages. Together with our data that NOD2 is recruited to phagosomes, these observations support the hypothesis that NOD2 activation occurs primarily from these organelles (27). However, despite recruitment to phagosomes, NOD2 remains on the cytosolic aspect of the membrane. This observation raises the question of how cytosolic NOD2 participates in S. aureus response, as it is an extracellular pathogen that does not normally access the cytosol. We therefore set out to establish how ligands derived from extracellular bacteria such as S. aureus access NOD2, a cytosolic sensor in human cells.
Identification of Drosophila Yin as a Phagosome-associated MDP Transporter
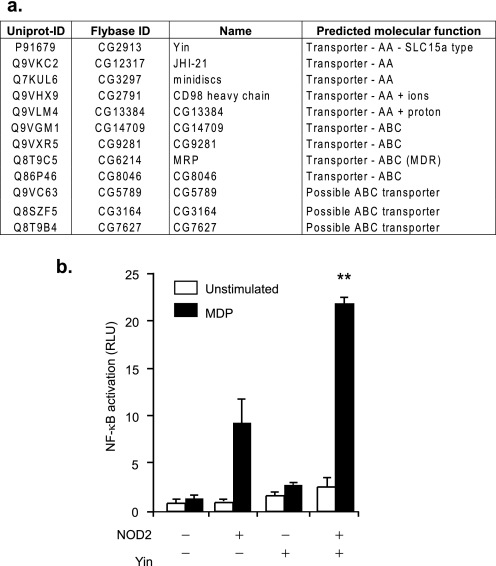
For NOD2 to be activated, the bacteria-derived ligands (such as MDP and other glycopeptides) must first be transported out of the phagosome into the cytosol. To identify potential phagosome-associated transporters, we took advantage of a database of phagosome-associated proteins derived from proteomic analysis of Drosophila phagosomes (23). Interrogation of that database identified a number of candidate transporters (Fig. 2a). One particular protein, Yin (found in the 30 min and 2 h Drosophila phagosome proteome), was identified as a particularly promising candidate. Yin is related to a family of SLC15A transporters that are best known for their role in nutrient acquisition in the gastrointestinal tract and are considered as a promising drug delivery target in colonic and kidney epithelial cells. One member of this family, PEPT1, has been implicated in MDP transport (28, 29) and another (SLC15A4) in Nod1-activating muramyl peptides (Tri-DAP and iE-DAP) sensing in epithelial cells (30). To determine whether Yin was able to transport MDP and hence function as a phagosome-associated MDP exporter, we cloned Drosophila Yin and expressed it along with NOD2 and a NF-κB reporter in mammalian HEK293T cells. Co-transfection of Drosophila Yin and NOD2 increased the sensitivity of HEK293T NF-κB reporter cells to MDP added exogenously to the media (Fig. 2b). These data indicate that Yin is able to transport MDP and suggested that this family of transporters might also be involved in export of MDP from phagosomes in mammals.
FIGURE 2.
Identification of Yin, a phagosome-associated MDP Drosophila transporter. a, the table summarizes all of the proteins identified form our previous proteomic analysis of the Drosophila phagosome (23) that are transporter candidates. Names and predicted molecular function were extracted from the Flybase database. b, NOD2 and an NF-κB -reporter construct were expressed in HEK293T cells with and without Drosophila Yin, a potential MDP transporter. Sensitivity to MDP was determined by luciferase activity. Graph are representative of at least three independent experiments. Mean ± S.D. are represented. **, p ≤ 0.01 relative to NOD2 + MDP. RLU, relative light units.
PEPT2 Is a MDP Transporter Expressed by Human Macrophages
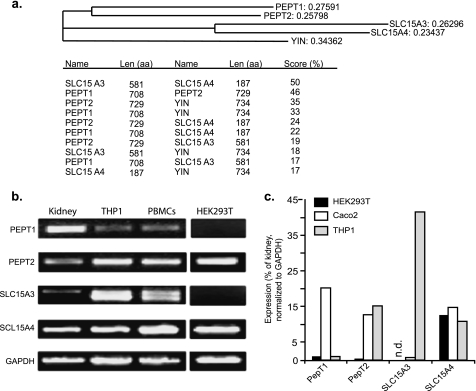
Phylogenetic analysis showed that Yin is related to the four human SLC15A transporters with a common ancestor and that Yin shares the highest amino acid sequence similarity with PEPT2 (35% identity) and PEPT1 (33% identity) but less similarity with SLC15A3 and SLC15A4 (18 and 17% identity, respectively) (Fig. 3a). To determine the predominant transporter in human phagocytic cells, expression of the four mammalian oligopeptide transporters, PEPT1 (SLC15A1), PEPT2 (SLC15A2), SLC15A3, and SLC15A4, was determined by RT-PCR (Fig. 3b). Semiquantitative RT-PCR shows that human monocytes/macrophages expressed high levels of PEPT2 but negligible PEPT1 and also expressed the other two family members SLC15A3 and SLC15A4 (Fig. 3c). The close homology with Yin, along with its high expression in human phagocytes, led us to reason that PEPT2 might be a candidate phagosome-associated MDP transporter in human cells.
FIGURE 3.
PEPT2 is expressed by human phagocytic cells. a, ClustalW analysis of Drosophila Yin and the related human SLC15 oligopepetide transporters, PEPT1, PEPT2, SLC15A3, and SLC15A4. Yin shares 33–35% with identity PEPT1/2 but only 17% with SLC15A3/A4. PEPT1 and PEPT2 are 50% identical. Expression of the SLC15 family members in kidneys, THP1 (human monocyte/macrophage cell line), peripheral blood monocytes, Caco2, (colonic epithelial cell line), and HEK293T (human kidney epithelial cell line) was determined by 40 cycle RT-PCR (b) and quantitative RT-PCR (c). PEPT2, but not PEPT1, is expressed by human phagocytes, which also express SLC15A3 and SLC15A4. RLU, relative light units.
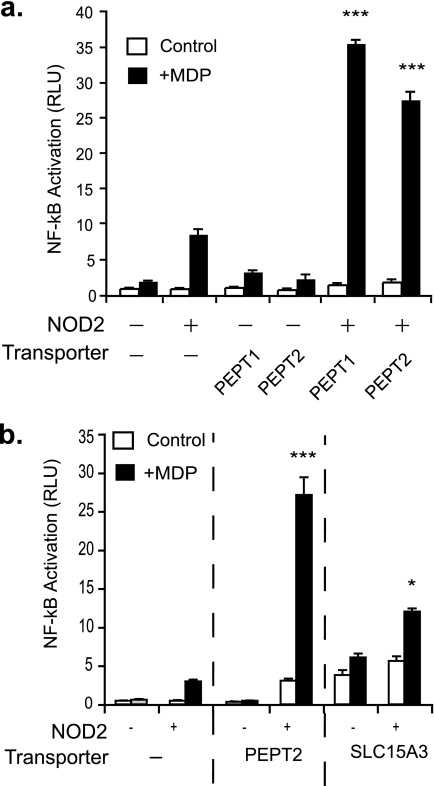
To test whether PEPT2 could transport MDP, we next expressed PEPT2 along with NOD2 and a NF-κB reporter in mammalian HEK293T cells. Similar to Yin, co-transfection of PEPT2 with NOD2 greatly increased the sensitivity of HEK293T NF-kB reporter cells to exogenous MDP (Fig. 4a). Importantly, PEPT2 was as efficient at increasing the sensitivity of HEK cells to MDP as PEPT1, a bona fide MDP transporter expressed by colonic epithelial cells (28). In contrast, the more distantly related transporter, SLC15A3, weakly modified NOD2 activation in the same system (Fig. 4b). Thus, similar to PEPT1 and Yin, PEPT2 is also a MDP transporter.
FIGURE 4.
PEPT2 is an MDP transporter. a, NOD2 and an NF-κB reporter construct were expressed in HEK293T cells with and without PEPT2, a potential phagosome-associated MDP transporter. Sensitivity to MDP was determined by luciferase activity. PEPT1, a known MDP transporter, was expressed as a positive control. b, NOD2 and an NF-κB reporter construct were expressed in HEK293T cells with and without PEPT2 or SLC15A3. Graphs are representative of at least three independent experiments. Mean ± S.D. are represented. ***, p ≤ 0.001 and *, p ≤ 0.05 relative to NOD2 + MDP. RLU, relative light units.
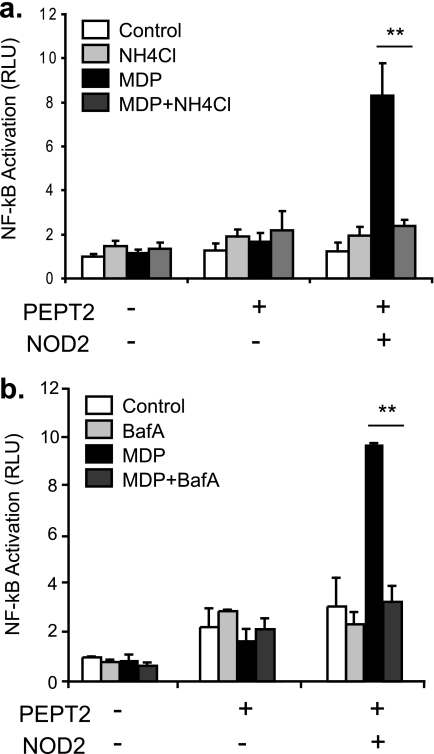
MDP Transport by PEPT2 and NOD2 Activation Is pH-sensitive
SLC15A transporters symport di- and tripeptides with H+ using energy provided by the proton gradient. In vivo, the proton gradients across epithelial cells are maintained by the existence of tight junctions between the cells in monolayers such as the gut. Our observed role for PEPT2 in transporting MDP in nonpolarized, nonconfluent HEK293T cells suggested that an adequate proton gradient for PEPT2 to function could be achieved in the absence of monolayer formation. Recent reports indicate that MDP enters HEK293T cells through the endocytic pathway) (30, 31), and it is possible that in nonconfluent HEK293T cells, acidification of the endolysosomal compartment might be sufficient to provide the energy for MDP transport by PEPT2. To test this hypothesis, we determined whether modifying vesicular acidification in HEK293T cells could perturb PEPT2-dependent activation of NOD2 by MDP. In PEPT2-expressing cells, NOD2-dependent NFkB activation by MDP was blocked with NH4Cl (a weak base) (Fig. 5a) and Baflomycin A, a specific vacuolar-ATPase inhibitor (Fig. 5b), both of which neutralize the endolysosomal pH. These results indicate that the vacuolar proton concentration, established by the vacuolar-ATPase, facilitates PEPT2-mediated transport of MDP out of vesicles such as endosomes or phagosomes to activate NOD2 in the cytosol.
FIGURE 5.
Vacuolar acidification is required for response to MDP. a and b, the role of vacuolar pH in PEPT2-dependent augmentation of MDP sensitivity was determined by blocking acidification of intracellular compartments using NH4Cl (a) or 50 μm Bafilomycin A (BafA) (b). Graphs are representative of at least three independent experiments. Mean ± S.D. are represented, **, p ≤ 0.01 relative to NOD2/PEPT2 + MDP. RLU, relative light units.
PEPT2 Localize to Phagosomes
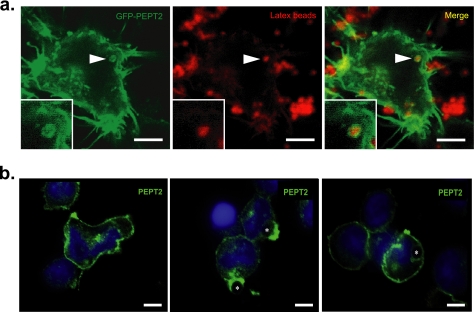
As Drosophila Yin was identified because of its association with phagosomes, we next tested whether PEPT2 also associated with these organelles and thus might locally deliver NOD2 ligands to their receptor. To test this, GFP-PEPT2 was expressed in HEK293T cells and localization determined after phagocytosis. GFP-PEPT2 could be found localized not only to the plasma membrane but also enriched on phagosome membranes (Fig. 6a). Using an anti-PEPT2 antibody, we next determined the localization of endogenous PEPT2 in the human THP1 monocyte-derived cell line during phagocytosis (Fig. 6b). Similar to the localization of GFP-PEPT2 in HEK293T cells, PEPT2 was found associated with the plasma membrane in resting macrophages and also enriched in phagocytic cups and phagosomes after particle internalization. Thus, the high levels of PEPT2 expression in human macrophages, the ability of PEPT2 to increase NOD2 response, and its association with phagosome membranes suggest this as a likely phagosome-associated MDP transporter in human phagocytes.
FIGURE 6.
PEPT2 localize to phagosomes. a, GFP-PEPT2 was ectopically expressed in HEK293T cells that were then allowed to internalize latex beads to generate phagosomes. PEPT2 shows strong membrane association and localization to phagosomes. Latex bead, 0.8 μm; scale bar corresponds to 5 μm. b, localization of endogenous PEPT2 in THP1 cells. PEPT2 localization was determined in resting THP1 cells (left) and during phagocytosis of latex beads (middle and right) by confocal microscopy. Left, PEPT2 localizes to the plasma membrane and intracellular compartments in resting cells. Middle, PEPT2 is enriched in the phagocytic cup. Right, PEPT2 associates with phagosomes in THP1s. An asterisk denotes 2 μm latex beads. Scale bar corresponds to 5 μm.
DISCUSSION
NOD2 is a cytosolic sensor and thus does not readily access extracellular bacteria. For this reason, a number of studies have focused on identifying the mechanism by which NOD2 ligands access the cytosol. Because of the association of NOD2 mutations and gastrointestinal inflammation, these studies have been performed primarily in epithelial cells of the colon or kidney. The consensus is that both NOD1 and NOD2 ligands enter these epithelial cells by endocytosis (30, 32). Additionally, these studies have converged on a family of SLC15A transporters as potential mechanisms to translocate NOD1 and NOD2 ligands across membranes; PEPT1 has been shown to transport MDP across the plasma membrane of colonic epithelial cells to activate cytosolic NOD2, whereas SLC15A4 has been implicated in transport of NOD1 ligands (28–30). However, studies of NLR ligand transport using nonepithelial cells have been limited. One study addressing this in macrophages has shown that NOD2-dependent signaling is unimpaired in PEPT1 deficient macrophages, suggesting that this is not an important transporter in these cells (31). Pannexin-1, a P2X7-interacting hemichannel, has been shown to be an NLR ligand transporter in macrophages and required for MDP to trigger NLRP3-dependent IL-1β secretion (32, 33). However, pannexin-1 does not transport MDP for activation of NFkB by NOD2. Thus, although it has been shown that purified MDP is endocytosed by macrophages or that NOD2 ligands can be derived from bacteria in the phagolysosome (8), the mechanisms of membrane translocation of MDP in professional phagocytes remains to be fully defined. Here, we identify Yin and PEPT2 as evolutionarily conserved transporters of MDP. Importantly, these transporters are highly expressed by professional phagocytes, where they are recruited to phagosomes. These observations lead us to suggest that Yin and PEPT2 may be of particular importance in transporting ligands derived from internalized bacteria to activate the cytosolic sensors recruited to the vicinity of the phagosome.
The study of these transporters in different organisms has been informative. We originally identified PEPT2 as a candidate MDP transporter using proteomic data derived from phagosomes isolated from Drosophila cells. The organelles expressed a number of transporters, including Yin, whose closest mammalian homolog is PEPT2. The functional ability of Yin to transport MDP is notable as flies do not have NOD-like receptors nor do they respond to MDP. However, tracheal cytotoxin, a molecule structurally related to MDP has been shown to be a ligand for peptidoglycan recognition protein-LE, a cytosolic pattern recognition receptor in flies (34). It will be of interest to see whether Yin plays a role in transporting tracheal cytotoxin to activate peptidoglycan recognition protein-LE in a manner analogous to the role we propose for PEPT2 in NOD2 activation in human cells. Additionally, our work studying these transporters in mouse macrophages is also of interest. Despite being sensitive to MDP, we were unable to detect significant levels of either Pept1 or Pept2 in both J774 and elicited peritoneal macrophages (data not shown). These data indicate that mouse phagocytes may use different mechanisms of MDP transport from humans. Of note, these cells did express Slc15a3 and a4, and it is possible that these or other transporters are also able to function as phagosome-associated MDP transporters and partially compensate for the lack of Pept2 in these cells. Intriguingly, when considered with the Drosophila data, these observations suggest that mice have selectively lost expression of this transporter in phagocytes. It is tempting to speculate that the lack of Pept2 might decrease the efficiency of MDP translocation in mouse macrophages and contribute to the relative resistance of these animals to septic shock (35), thus providing them with a survival advantage during certain infectious challenges.
Consistent with redundancy of these transport mechanisms, our gain-of-function experiments clearly show a role of PEPT2 in NOD2 ligand delivery in human cells, whereas our loss of function RNA interference experiments, in which we have attempted to silence PEPT2, have been inconclusive (data not shown). PEPT2 is not the only member of the SLC15 transporter family expressed by phagocytes, and the observed redundancy is most likely due to compensation by other SLC15A transporters. We find that although PEPT1 is expressed only at very low levels it may nonetheless be sufficient to compensate and transport MDP in the absence of PEPT2. Macrophages also express SLC15A3 and A4. However, ectopic expression of SLC15A3 did not increase sensitivity of HEK293T cells to the NOD2 ligand in our system. Although we have not formally tested the role of SLC15A4 in MDP transport, work by Lee et al. (30) has shown that SLC15A4 is a transporter for NOD1 ligands such as M-Tri-DAP but does not have a clear role in transport of MDP. Therefore, it is not clear which of the SLC15A transporters may be compensating for PEPT2 in macrophages. SLC15A transporters have broad substrate specificity for di- and tripeptides and can transport a variety of molecules with varying efficiency. This pleiotropism raises the question of whether PEPT2 functions only to transport MDP or might have additional roles in transport of other innate immune agonists. Work by Swaan et al. (36) compared the relative efficiency of PEPT2 in transport of γ-iE-DAP and MDP and showed that γ-iE-DAP was a better substrate for ectopically expressed PEPT2 in CHO-K1 cells than MDP. Because of our focus on NOD2 we have not tested the role of PEPT2 in transport of γ-iE-DAP but, considering these observations, it is likely that this transporter may also transports NOD1 ligands from the phagosome.
In summary, we identify here Drosophila Yin and human PEPT2 as an evolutionarily conserved mechanism for the transport of MDP into the cytosol of professional phagocytes, where it is recognized by NOD2. PEPT2 transport is pH-dependent and hence is likely to rely on proton gradients generated either in the endolysosomal compartment or during maturation of phagosomes. Importantly, we show that along with NOD2, PEPT2 is enriched around phagosomes. Our results emphasize the importance of considering not only the affinity of the transporters for their substrates but also the patterns of gene expression and the subcellular localization of these proteins when trying to understand how SLC15A transporters function to translocate NOD ligands into cells. The delivery of NOD ligands appears to be mediated by dedicated transporters such as PEPT2, which, therefore, represent potential therapeutic targets to decrease the detrimental effects of MDP and other agonistic fragments derived from bacteria during septic shock. These data indicate that the phagosome compartmentalizes liberation of ligands with the delivery to signaling receptors recruited to the vicinity of the organelle. Moreover, these data show that most of the machinery required for optimal microbial sensing of S. aureus is recruited to the vicinity of this highly hydrolytic organelle, greatly increasing the efficiency of signaling after bacterial internalization.
Footnotes
- TLR
- Toll-like receptor
- NLR
- NOD-like receptor
- MDP
- muramyl dipeptide
- IL
- interleukin
- PBS
- phosphate-buffered saline
- IFNγ
- interferon γ
- YFP
- yellow fluorescent protein
- GFP
- green fluorescent protein
- TNF
- tumor necrosis factor
- CFP
- cyan fluorescent protein
- RT-PCR
- real-time PCR.
REFERENCES
- 1.Hoffmann J. A., Kafatos F. C., Janeway C. A., Ezekowitz R. A. (1999) Science 284, 1313–1318 [DOI] [PubMed] [Google Scholar]
- 2.Janeway C. A., Jr., Medzhitov R. (2002) Annu. Rev. Immunol. 20, 197–216 [DOI] [PubMed] [Google Scholar]
- 3.Takeda K., Kaisho T., Akira S. (2003) Annu. Rev. Immunol. 21, 335–376 [DOI] [PubMed] [Google Scholar]
- 4.Inohara N., Nuñez G. (2003) Nat. Rev. Immunol. 3, 371–382 [DOI] [PubMed] [Google Scholar]
- 5.Viala J., Sansonetti P., Philpott D. J. (2004) C. R. Biol. 327, 551–555 [DOI] [PubMed] [Google Scholar]
- 6.Ozinsky A., Underhill D. M., Fontenot J. D., Hajjar A. M., Smith K. D., Wilson C. B., Schroeder L., Aderem A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13766–13771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Underhill D. M., Ozinsky A., Hajjar A. M., Stevens A., Wilson C. B., Bassetti M., Aderem A. (1999) Nature 401, 811–815 [DOI] [PubMed] [Google Scholar]
- 8.Herskovits A. A., Auerbuch V., Portnoy D. A. (2007) PLoS Pathog. 3, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimada T., Park B. G., Wolf A. J., Brikos C., Goodridge H. S., Becker C. A., Reyes C. N., Miao E. A., Aderem A., Gotz F., Liu G. Y., Underhill D. M.Cell Host Microbe 7, 38–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vance R. E., Isberg R. R., Portnoy D. A. (2009) Cell Host Microbe. 6, 10–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogura Y., Inohara N., Benito A., Chen F. F., Yamaoka S., Nunez G. (2001) J. Biol. Chem. 276, 4812–4818 [DOI] [PubMed] [Google Scholar]
- 12.Ogura Y., Bonen D. K., Inohara N., Nicolae D. L., Chen F. F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R. H., Achkar J. P., Brant S. R., Bayless T. M., Kirschner B. S., Hanauer S. B., Nuñez G., Cho J. H. (2001) Nature 411, 603–606 [DOI] [PubMed] [Google Scholar]
- 13.Hampe J., Cuthbert A., Croucher P. J., Mirza M. M., Mascheretti S., Fisher S., Frenzel H., King K., Hasselmeyer A., MacPherson A. J., Bridger S., van Deventer S., Forbes A., Nikolaus S., Lennard-Jones J. E., Foelsch U. R., Krawczak M., Lewis C., Schreiber S., Mathew C. G. (2001) Lancet 357, 1925–1928 [DOI] [PubMed] [Google Scholar]
- 14.Hugot J. P., Chamaillard M., Zouali H., Lesage S., Cézard J. P., Belaiche J., Almer S., Tysk C., O'Morain C. A., Gassull M., Binder V., Finkel Y., Cortot A., Modigliani R., Laurent-Puig P., Gower-Rousseau C., Macry J., Colombel J. F., Sahbatou M., Thomas G. (2001) Nature 411, 599–603 [DOI] [PubMed] [Google Scholar]
- 15.Franchi L., Warner N., Viani K., Nuñez G. (2009) Immunol. Rev. 227, 106–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi K. S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nuñez G., Flavell R. A. (2005) Science 307, 731–734 [DOI] [PubMed] [Google Scholar]
- 17.Sabbah A., Chang T. H., Harnack R., Frohlich V., Tominaga K., Dube P. H., Xiang Y., Bose S. (2009) Nat. Immunol. 10, 1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Travassos L. H., Carneiro L. A., Ramjeet M., Hussey S., Kim Y. G., Magalhães J. G., Yuan L., Soares F., Chea E., Le Bourhis L., Boneca I. G., Allaoui A., Jones N. L., Nuñez G., Girardin S. E., Philpott D. J. (2010) Nat. Immunol. 11, 55–62 [DOI] [PubMed] [Google Scholar]
- 19.Cooney R., Baker J., Brain O., Danis B., Pichulik T., Allan P., Ferguson D. J., Campbell B. J., Jewell D., Simmons A. (2010) Nat. Med. 16, 90–97 [DOI] [PubMed] [Google Scholar]
- 20.Park E. K., Jung H. S., Yang H. I., Yoo M. C., Kim C., Kim K. S. (2007) Inflamm Res. 56, 45–50 [DOI] [PubMed] [Google Scholar]
- 21.Ip W. K., Takahashi K., Moore K. J., Stuart L. M., Ezekowitz R. A. (2008) J. Exp. Med. 205, 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barnich N., Aguirre J. E., Reinecker H. C., Xavier R., Podolsky D. K. (2005) J. Cell Biol. 170, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuart L. M., Deng J., Silver J. M., Takahashi K., Tseng A. A., Hennessy E. J., Ezekowitz R. A., Moore K. J. (2005) J. Cell Biol. 170, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi O., Hoshino K., Akira S. (2000) J. Immunol. 165, 5392–5396 [DOI] [PubMed] [Google Scholar]
- 25.Girardin S. E., Travassos L. H., Hervé M., Blanot D., Boneca I. G., Philpott D. J., Sansonetti P. J., Mengin-Lecreulx D. (2003) J. Biol. Chem. 278, 41702–41708 [DOI] [PubMed] [Google Scholar]
- 26.Lécine P., Esmiol S., Métais J. Y., Nicoletti C., Nourry C., McDonald C., Nunez G., Hugot J. P., Borg J. P., Ollendorff V. (2007) J. Biol. Chem. 282, 15197–15207 [DOI] [PubMed] [Google Scholar]
- 27.Kapetanovic R., Nahori M. A., Balloy V., Fitting C., Philpott D. J., Cavaillon J. M., Adib-Conquy M. (2007) Infect Immun. 75, 830–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vavricka S. R., Musch M. W., Chang J. E., Nakagawa Y., Phanvijhitsiri K., Waypa T. S., Merlin D., Schneewind O., Chang E. B. (2004) Gastroenterology 127, 1401–1409 [DOI] [PubMed] [Google Scholar]
- 29.Ismair M. G., Vavricka S. R., Kullak-Ublick G. A., Fried M., Mengin-Lecreulx D., Girardin S. E. (2006) Can J. Physiol. Pharmacol. 84, 1313–1319 [DOI] [PubMed] [Google Scholar]
- 30.Lee J., Tattoli I., Wojtal K. A., Vavricka S. R., Philpott D. J., Girardin S. E. (2009) J. Biol. Chem. 284, 23818–23829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marina-García N., Franchi L., Kim Y. G., Hu Y., Smith D. E., Boons G. J., Núñez G. (2009) J. Immunol. 182, 4321–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marina-García N., Franchi L., Kim Y. G., Miller D., McDonald C., Boons G. J., Núñez G. (2008) J. Immunol. 180, 4050–4057 [DOI] [PubMed] [Google Scholar]
- 33.Kanneganti T. D., Lamkanfi M., Kim Y. G., Chen G., Park J. H., Franchi L., Vandenabeele P., Núñez G. (2007) Immunity 26, 433–443 [DOI] [PubMed] [Google Scholar]
- 34.Kaneko T., Yano T., Aggarwal K., Lim J. H., Ueda K., Oshima Y., Peach C., Erturk-Hasdemir D., Goldman W. E., Oh B. H., Kurata S., Silverman N. (2006) Nat. Immunol. 7, 715–723 [DOI] [PubMed] [Google Scholar]
- 35.Warren H. S. (2009) J. Leukoc. Biol. 86, 199–201 [DOI] [PubMed] [Google Scholar]
- 36.Swaan P. W., Bensman T., Bahadduri P. M., Hall M. W., Sarkar A., Bao S., Khantwal C. M., Ekins S., Knoell D. L. (2008) Am. J. Respir. Cell Mol. Biol. 39, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]