Abstract
The bacterial pathogen Vibrio parahemeolyticus manipulates host signaling pathways during infections by injecting type III effectors into the cytoplasm of the target cell. One of these effectors, VopS, blocks actin assembly by AMPylation of a conserved threonine residue in the switch 1 region of Rho GTPases. The modified GTPases are no longer able to interact with downstream effectors due to steric hindrance by the covalently linked AMP moiety. Herein we analyze the structure of VopS and its evolutionarily conserved catalytic residues. Steady-state analysis of VopS mutants provides kinetic understanding on the functional role of each residue for AMPylation activity by the Fic domain. Further mechanistic analysis of VopS with its two substrates, ATP and Cdc42, demonstrates that VopS utilizes a sequential mechanism to AMPylate Rho GTPases. Discovery of a ternary reaction mechanism along with structural insight provides critical groundwork for future studies for the family of AMPylators that modify hydroxyl-containing residues with AMP.
Keywords: AMP, Enzyme Kinetics, Enzyme Mechanisms, G Proteins, Signal Transduction, AMPylation, Vibrio, VopS
Introduction
Pathogenic bacteria possess a mechanism of virulence called the type III secretion system (T3SS)4 that enables pathogens to evade and disrupt host signaling systems during infection (1). The T3SS apparatus contains a base that spans across bacterial membranes and a needle-like structure that extends through the cell wall and across the host cell membrane. Bacterial proteins, called effectors, are translocated through this structure into the host cytosol (2). These effectors are quiescent in the pathogen but mimic or capture an endogenous eukaryotic activity inside the host cell to promote the progress of infection, disease, and survival of the bacteria (1).
The pathogenic strains of the Gram-negative bacterium Vibrio parahemeolyticus, a leading cause of global seafood-associated food poisoning in humans from the consumption of raw or undercooked shellfish, encode two T3SSs, each translocating a unique repertoire of effectors (3). Infection with V. parahemeolyticus causes gastroenteritis with clinical symptoms resulting in diarrhea, nausea, vomiting, fever, and headache (4, 5). The second secretion system, T3SS2, has been associated with enterotoxicity, whereas the first, T3SS1, is associated with cytotoxicity (6). T3SS1 uses a unique mechanism to kill cells that first involves induction of autophagy followed by cell rounding and, finally, cell lysis (7). The first step, autophagy, is caused by the effector VopQ (VP1680), whereas cell rounding appears to be mediated by the effector VopS (VP1686) (8, 9).
Bioinformatic analysis revealed that the C terminus of VopS contains a protein domain called Fic (filamentation induced by cyclic adenosine monophosphate) (8). Fic domains are evolutionarily conserved from prokaryotes to eukaryotes and fall into a domain superfamily called Fido, which comprises three protein domain families: Fic domains, Doc (death on curing) domains, and AvrB (avirulence protein B) (8, 10). All members of the Fido superfamily have similar tertiary topology. Fic and Doc domains have the conserved central HPFX(D/E)GN(G/K)R motif, whereas AvrB members lack the conserved histidine. Until recently the function of Fic domains was not known (11).
Initial infection studies with wild-type laboratory strain and a VopS mutant strain of V. parahemeolyticus demonstrated inhibition of Rho family GTPase activation. Rho family GTPases are small G proteins that have essential roles in actin cytoskeletal dynamics (12). Yarbrough et al. (8) discovered that the Vibrio Fic domain-containing protein, VopS, uses ATP to directly transfer an AMP moiety to a conserved threonine residue located on the switch 1 region of Rho family GTPases. The newly discovered functional activity of Fic domains was elucidated, and the posttranslational modification was named AMPylation (8). The resulting AMPylation prevented interaction of Rho GTPases with downstream effectors, thereby inhibiting actin assembly in the infected cell. The inhibitory AMPylation activity of VopS was abolished when the conserved histidine from the central Fic motif was mutated to an alanine (8).
These recent discoveries indicated that the function of Fic domain-containing proteins is to mediate the novel posttranslational modification. The newly discovered posttranslational modification of AMPylation is similar to many other posttranslational modifications, including phosphorylation, which uses ATP as a substrate (11). Phosphorylation occurs also on tyrosine residues, and subsequently, a Fic domain-containing bacterial virulence factor was shown to AMPylate a tyrosine residue on host GTPases (13). In this paper we present the structural analysis of a Fic domain for the first functionally proven AMPylator, VopS. We performed kinetic analyses on mutants of the protein to determine the catalytic contribution and molecular role of evolutionarily conserved Fic domain residues. Also, mechanistic kinetic studies provide evidence of a sequential (ternary complex) mechanism used by VopS to mediate AMPylation of its eukaryotic signaling targets.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
VopS-(75–387) was cloned into pGEX-TEV vector to generate a (GST)-tagged VopS-(75–387) construct for purification using GST affinity chromatography (Sigma). The construct was transformed and expressed in Escherichia coli BL21 (DE3) cells with 0.4 mm final isopropyl β-d-thiogalactopyranoside (Roche Applied Science) for 20 h at 20 °C. Cell pellets were stored at −80 °C. Cells were then lysed using a cell disrupter (EmulsiFlex-C5, Avenstin Inc.) and affinity purified via GST chromatography (Sigma). The GST tag was removed with overnight cleavage at 4 °C using recombinant His6-tobacco etch virus protease. The protein was then loaded onto a 1-ml HiTrapQ HP column. VopS fractions were pooled and subsequently loaded onto a HiLoad 16/60 Superdex 75 (GE Healthcare) column in a final buffer containing 10 mm Tris, pH 7.5, 50 mm NaCl, 1 mm DTT. Selenomethionine-labeled VopS-(75–387) protein was expressed in the methionine auxotroph E. coli strain B834 (DE3) (Novagen). Cells were grown using SelenoMetTM medium supplemented with selenomethionine (Molecular Dimensions Ltd), and VopS was expressed and purified similarly to the native protein. Wild-type GST-VopS and mutant GST-VopS-(31–387) constructs were cloned, expressed, and purified with GST affinity chromatography as described above. VopS mutants were made using site-directed mutagenesis that was performed according to the manufacturer's instructions (Stratagene). For kinetic studies the wild-type and mutant GST-VopS-(31–387) constructs were expressed and purified using GST affinity chromatography as previously above. The GST tag was cleaved as described above, and the protein was loaded onto a 1-ml HiTrapQ HP column to collect cleaved wild-type or mutant VopS-(31–387). Protein was buffer exchanged in 10 mm Tris, pH 8.0, 100 mm NaCl, 1 mm DTT, 10% glycerol with Amicon 10K concentrators and stored at −80 °C. The His6-Cdc42-(1–179)-Q61L construct was generated using a pET28a vector (Novagen). The clone was transformed in BL21 (DE3) cells, and protein expression was similar as described above. His6-Cdc42-(1–179)-Q61L pellets were purified using immobilized metal ion affinity chromatography (Sigma). Protein substrate was buffer-exchanged and stored at −80 °C as described previously. Protein concentration was assayed by the Bradford method (Bio-Rad) and purity by SDS-PAGE analysis.
Limited Proteolysis
Stock trypsin dissolved at 1 mg/ml was serially diluted (1:10, 1:100, and 1:1000) in 10 mm HCl. 1 μl of trypsin dilution was added to recombinant VopS-(31–387) in a 50-μl volume. Each reaction was incubated at room temperature for 30 min and 5 μl of 0.1 mg/ml Pefabloc (Roche Applied Science) to stop the proteolytic reaction. Reactions were analyzed by SDS-PAGE and stained with Colloidal Blue (Invitrogen), and samples were N-terminal-sequenced. Samples subjected to matrix-assisted laser desorption ionization time-of-flight to determine the total molecular weight.
Protein Crystallization
Initial screening of purified VopS-(75–387) at 12 mg/ml was performed in sitting-drop vapor diffusion mode in 96–3 well Intelli-Plates (Art Robbins Instruments) at 20 °C. Initial hits were seen in the PEGs Suite (Qiagen). Crystal condition #37 with 0.1 m HEPES, pH 7.5, and 25% (w/v) polyethylene glycol 3000 was selected for optimization. Optimized crystallization conditions were 0.1 m HEPES, pH 7.25–7.5) and 21% polyethylene glycol 3500 at 4 °C with 6 mg/ml protein.
Data Collection, Structure Determination, and Refinement
Diffraction data were collected at the Structural Biology Center at the Advanced Photon Source (Argonne National Laboratory). Data sets were indexed, integrated, and scaled using the HKL-3000 program package (14). The native VopS-(75–387) crystal had the symmetry of space group P21 with unit cell parameters a = 66.67 Å, b = 62.32 Å, c = 75.76 Å, and β = 91.3°, diffracted X-rays to a minimum Bragg-spacing, dmin, of 1.80 Å, and contained two molecules in the asymmetric unit. Phases were obtained from a selenomethionyl-substituted protein crystal by the single-wavelength anomalous dispersion method using X-rays with energy near the selenium K-shell absorption edge. Selenium sites were located using the program SHELXD (15). Phases were refined with MLPHARE (16), resulting in an overall figure of merit of 0.17 for data between 30.0 and 2.28 Å. Phases were improved via density modification and 2-fold symmetry averaging in the program dm (17), resulting in a figure of merit of 0.65. An initial model containing ∼93% of all residues was constructed automatically using the program ARP/wARP (18). Manual model building was performed with the program Coot (19). Refinement was performed with native data to a resolution of 1.8 Å using the program PHENIX (20) with a random 5.07% of all data set aside for Rfree calculation. Molprobity was used during refinement to check on the model quality (21). All protein structure figures were constructed using the MacPyMOL program (22).
Filter Binding Assays
VopS-(31–387) activity was assayed by AMPylation of His6-Cdc42-(1–179)-Q61L with [α-32P]ATP purchased from PerkinElmer Life Sciences. P81 Whatman filters were used for filter binding. Buffer conditions were 20 mm Hepes, pH 7.4, 5 mm MgCl2, 100 mm NaCl, 1 mg/ml bovine serum albumin, and 1 mm DTT. ATP was dissolved in 20 mm Hepes, pH 7.4. Reactions were performed in triplicate at 25 °C and initiated with [α-32P]ATP (30–120 cpm/pmol), and time points were taken at 15, 30, 45, 60, and 75 for mutants and an additional time point at 90 s for the wild-type protein. A 30-μl reaction volume per time point collected was used. Reactions were stopped by pipetting 20 μl of the reaction onto P81 Whatman filters. Filters were then immediately immersed in 75 mm phosphoric acid and stirred in a beaker on a platform shaker. Filters were subsequently washed 3 times with 75 mm phosphoric acid, rinsed with acetone, and allowed to air dry. Each filter was placed in a vial with scintillation fluid, and counts per minute were measured using a Beckman LS6500 scintillation counter. To determine apparent steady-state kinetic values for ATP with VopS-(31–387), His6-Cdc42-(1–179)-Q61L, protein substrate was kept at a constant concentration of 600 μm while varying ATP concentrations (40, 80, 120, 160, 250, 400, 700, 1000, 1500, 2000 μm) were studied. Apparent kinetic values for ATP with mutant VopS constructs were performed at 600 μm protein substrate varying ATP with concentrations of 100, 200, 300, 500, 750, 1000, 1500, and 2000 μm. Assays of the R299A mutant had the same ATP concentrations as other VopS mutants, but an additional measurement at 150 μm ATP was taken. To determine apparent steady-state kinetic values for His6-Cdc42-(1–179)-Q61L, the ATP concentration was fixed at 2000 μm while varying His6-Cdc42-(1–179)-Q61L concentrations (50, 75, 100, 150, 200, 300, 500, 800, 1200 μm). The R299A mutant was assayed at 2000 μm ATP, whereas the following concentrations were used for the protein substrate: 50, 75, 100, 200, 300, 500, 800, and 1200 μm. To determine the reaction mechanism, ATP concentrations were fixed (50, 75, 100, 200, 1000 μm) while varying the concentration of His6-Cdc42-(1–179)-Q61L (50, 75, 100, 200, 500 μm). pH-dependent VopS assays were performed in triplicate with 5 nm VopS (31–387), 100 μm His6-Cdc42-(1–179)-Q61L, and 200 μm [α-32P]ATP. Assay were incubated for 75 s at 25 °C and spotted onto P81 Whatman filters and washed as described previously. 150 mm Bis-Tris buffer was used for pH ranges 5.5, 6.0, and 6.5. 150 mm Hepes buffer was used for pH 7.0 and 7.5, and 150 mm Tris-HCl buffer was used for pH 8.0, 8.5, and 9.0. All reactions contained 5 mm MgCl2, 100 mm NaCl, 0.1 mg/ml bovine serum albumin, and 1 mm DTT.
Kinetic Data Analysis
Single-substrate kinetic measurements were fitted into the Michaelis-Menten equation (Equation 1) using GraphPad Prism 5,
Bisubstrate kinetic studies fitted best to a (sequential) random, rapid-equilibrium model using Sigma Plot 11.0 Enzyme Kinetics 1.3. Equations for random rapid-equilibrium (Equation 2), ordered rapid-equilibrium (Equation 3), and ping pong (Equation 4) are listed below, respectively.
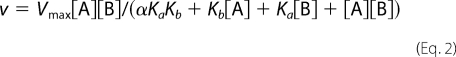 |
where Vmax is maximum velocity, and A and B represent substrates. Ka and Kb are Michaelis-Menten constants of the substrates, and α is the interaction factor between A and B.
In Vitro AMPylation Assays
AMPylation assays used purified recombinant protein. Each reaction contained 200 μm cold ATP with 0.1–0.4 μm [α-32P]ATP. 100 μm His6-Cdc42-(1–179)-Q61L was incubated with 5 nm GST-VopS-(31–387) wild-type or mutant protein. Assays were performed at 25 °C in buffer containing 20 mm Hepes, pH 7.4, 5 mm MgCl2, 100 mm NaCl, 0.1 mg/ml bovine serum albumin, 1 mm DTT. Reactions were stopped with SDS sample buffer. Samples were boiled and separated by SDS-PAGE and visualized by autoradiography.
RESULTS
VopS-(75–387) Is Proteolytically Stable and Catalytically Active
To identify the optimal fragment of VopS protein for crystallization trials, we used limited proteolysis with trypsin on the catalytically active VopS-(31–387) that has the first 30 amino acid residues of the protein removed to increase protein solubility. Limited proteolysis with trypsin uncovered a relatively stable cleavage product. N-terminal sequencing and mass spectrometry revealed that the cleavage after lysine 74 yielded the stable product. VopS-(75–387) and VopS-(75–387)-H348A, a catalytically inactive mutant where the conserved histidine 348 of the Fic motif was mutated to an alanine, were cloned, purified, and assayed for in vitro enzymatic activity. Consistent with previous observations, VopS-(75–387), but not the mutant VopS-(75–387)-H348A, was catalytically active in an in vitro AMPylation assay using [α-32P]ATP and dominant active His6-Cdc42-(1–179)-Q61L (supplemental Fig. 1) (8).
Structure of Vops
The stable, enzymatically active, purified VopS-(75–387) was used for crystallization trials. After optimization of crystallographic conditions, VopS-(75–387) crystals grew overnight at 4 °C in 0.1 m HEPES, pH 7.25–7.5) and 21% polyethylene glycol 3500. The structure was phased by single-wavelength anomalous dispersion with selenomethionine-substituted VopS-(75–387) protein crystals that diffracted to 2.30 Å resolution. Refinement was performed on native data to a resolution of 1.8 Å, and the model contains two VopS-(75–387) monomers (molecule A with residues 88–387 and molecule B with residues 80–387) and 514 waters. The Rwork is 17.2%, and the Rfree is 22.4%. Phasing and model refinement statistics are provided in supplemental Table 1.
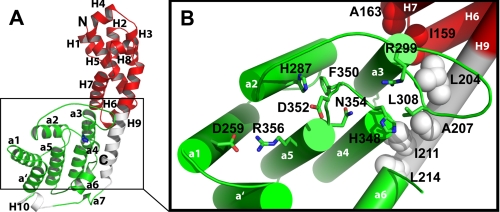
The VopS-(75–387) structure is predominantly α-helical and is divided into two subdomains, an N- and C-terminal (Fig. 1A). The N-terminal subdomain (red) comprises 9 α-helices (H1-H9), is not conserved in other solved structures of Fic domains, and does not appear to be homologous to any existing structures. Hydrophobic side chains (depicted as spheres) on the N-terminal subdomain α-helices H6, H7, and H9 pack against hydrophobic side chains of a3 and a4 to stabilize and maintain Fic domain tertiary structure (Fig. 1B). The N-terminal subdomain is required for full activity; the removal of 150 N-terminal residues results in the loss of catalytic activity in vitro (data not shown). With nine turns, the H9 helix is the longest helix and serves as a backbone that spans both N- and C-terminal subdomains (Fig. 1A).
FIGURE 1.
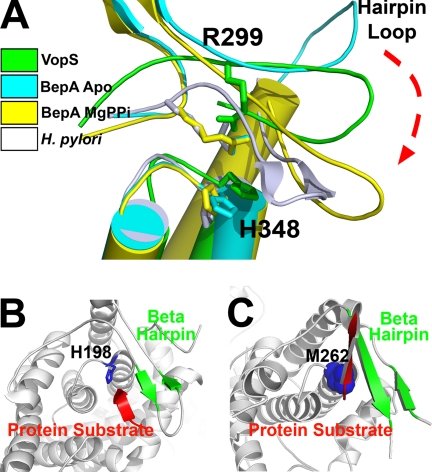
Crystal structure of VopS. A, shown is a ribbon diagram of VopS-(75–387) determined at 1. 8 Å resolution. The N-terminal subdomain of VopS contains helices H1-H9 (red) that are not conserved among Fic-domain-containing proteins. The C-terminal subdomain includes the later half of H9 and the π-helix H10 (white) and the structurally conserved Fic domain (green). Highly conserved histidine 348 (blue) is shown in the core of the Fic domain in a stick representation. B, shown is a detailed view of VopS Fic domain residues. N-terminal subdomain hydrophobic residues from H6, H7, and H9 (spheres) structurally stabilize the C-terminal Fic domain of VopS. Fic motif (HPFX(D/E)GN(G/K)R) residues and residues His-287 and Asp-259 are displayed as sticks. Leu-308 and Arg-299 are positioned in the structurally conserved hairpin loop of the Fic domain. In this figure and all that follow, nitrogen atoms are colored blue, and oxygen atoms are red.
The C-terminal subdomain possesses 8 α-helices (green) that are structurally similar in Fic domains (Fig. 1A, green, see “Discussion”) (10). The evolutionarily conserved core of the VopS Fic domain comprises two internal helices, a4 and a5, that are encircled by helices a′, a1, a2, a3, a6, and a7. The VopS Fic domain is decorated by an additional helix H10 and the later part of the backbone helix H9.
VopS Fic Domain Comparisons to Existing Structures
Crystal structures have been determined of several different Fic domain family proteins by the Midwest Center for Structural Genomics and by the Joint Center for Structural Genomics (8, 23). Tilman Schirmer's group solved additional structures of a Fic family protein, BepA.5 The sequence identity of Fic domains is lower than 20% among the known structures, providing a wide evolutionary sampling of Fic domains for comparison.
The structures of Fic family members have a conserved core topology (described above and in Kinch et al. (10)) that is decorated by various additional secondary structure elements and domains (supplemental Fig. 2). Structure superpositions of Fic proteins with the VopS Fic domain (DaliLite) show a similar tertiary structure (Z-scores ranging from 3.0–9.0). A circular permutation is seen in the a′ helix of Fic domains (supplemental Figs. 2 and 3) (10). The a′ helix in all Fic domains occupies the same location in the tertiary structure but not in the sequence; it may be located at either the N or C terminus of the protein (10). VopS has an N-terminal a′ helix and a second permutation corresponding to the Fic helix a7 (supplemental Fig. 3).
Crystal structures of Fic domains, such as Helicobacter pylori Fic protein include a β-hairpin between helix a2 and a3 that is positioned near the Fic motif loop and has been proposed to mediate peptide substrate binding (10). In the VopS model, the corresponding loop does not form the hydrogen bonding pattern of a β-hairpin and is, therefore, referred to as the hairpin loop (Fig. 1A). The lack of a well defined β-hairpin may possibly result from the absence of an interaction with a substrate such as ATP or a Rho GTPase.
VopS Active Site Fic Motif Residues Contribute to Catalysis
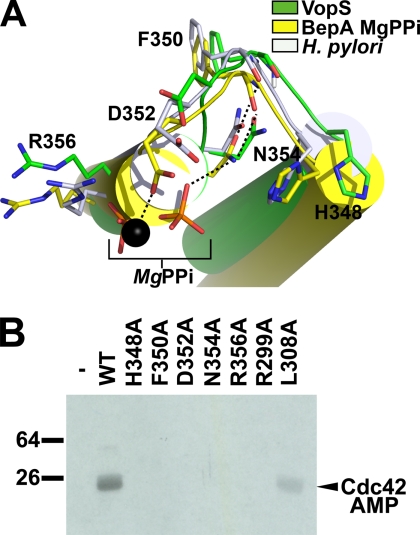
Fic domain family members are characterized by a consensus sequence motif HPFX(D/E)GN(G/K)R. The evolutionarily conserved histidine from the motif is seen in the central core of the VopS Fic domain, located in the loop between helix a4 and a5 (Fig. 2A) (8, 11, 13).
FIGURE 2.
Residues important for function in VopS Fic domain. A, shown is superposition of conserved residues of the Fic motif. Colored in green are VopS Fic secondary structural elements and carbon atoms. Superimposed onto VopS are Fic motif residues from H. pylori (white secondary structure and carbons; PDB ID 2F6S) and BepA from Bartonella henselae (yellow secondary structure and carbons; PDB ID 2JK8). Pyrophosphate and magnesium (black sphere) are complexed with BepA. Black dashed lines represent hydrogen bonds in the MgPPi BepA complex. B, in vitro AMPylation assay incubated with 100 μm His6-Cdc42-(1–179)-Q61L and [α-32P]ATP at 25 °C only or with 5 nm of wild-type GST-VopS-(31–387) or mutants is shown. The assay was stopped at 90 s with loading buffer. WT, wild type.
Structural superpositions of VopS with H. pylori and with BepA Fic domains show a conserved positioning of this loop and the corresponding side chains that define the motif (Fig. 2A). The VopS Phe-350 side chain stacks on His-287 of helix a2, whereas its main-chain carbonyl oxygen and amide nitrogen form hydrogen bonds with the main chain of another Fic motif residue, Asn-354 (Fig. 2A).
The structure of BepA complexed with magnesium pyrophosphate has magnesium coordinated by the acidic glutamate residue of the Fic motif (Fig. 2A). This observation supports a role of the corresponding VopS residue Asp-352 in magnesium coordination, as acidic residues commonly play a role in binding positively charged divalent metal ions. The BepA complex structure reveals that the conserved asparagine of the Fic motif interacts with pyrophosphate. This observation suggests that Asn-354 of VopS has dual functions; that is, interaction with the β-phosphate of ATP and positioning the active site loop through main-chain interactions. One notable difference between the VopS Fic motif and those of other Fic proteins is the relative positioning of the imidizolium group of the His-348 ring. In VopS His-348 is rotated by ∼90° relative to the position observed in other structures (Fig. 2A).
Arg-356 appears to have a role in protein stability because of its electrostatic interactions with Asp-259 from the neighboring helix a1 (Fig. 2A). Arg-356 also lines a shallow pocket that includes the other active site residues, such as Asp-352 and Asn-354 and, thus, may be involved in ATP binding. Accordingly, the corresponding Fic motif arginine residue in BepA is positioned near a phosphate moiety of the pyrophosphate.
Distal in sequence but conserved among Fic domains is Arg-299, which points downward into the active site and is positioned above His-348. Within the same hairpin loop, Leu-308 is the nearest residue to His-348 at 3.5 Å (Fig. 2A).
The VopS Fic motif residues appear to form a conserved active site proximal to the hairpin loop. Conserved active site residues were individually mutated to alanine, purified, and assayed for AMPylation activity using Cdc42 as protein substrate and [α-32P]ATP. VopS mutant analysis shows that the Fic motif residues (His-348, Phe-350, Asp-352, Asn-354, and Arg-356) and the distal but evolutionarily conserved residue, Arg-299, are critical for full catalytic activity (Fig. 2B). Mutation of the hairpin element residue proximal to the active site (Leu-308) resulted in a slight reduction of activity.
Steady-state Substrate Measurements
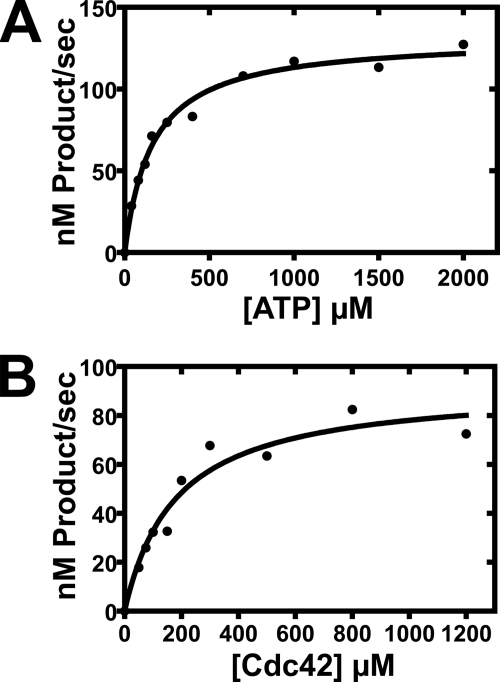
VopS utilizes both ATP and Rho family GTPases as substrates to catalyze the transfer of AMP to an invariant threonine residue located on the switch 1 region of Rho GTPases. The switch 1 region is important for binding both nucleotide and downstream effectors. We determined the apparent steady-state kinetic constants for ATP using a dominant active form His6-Cdc42-(1–179)-Q61L, where glutamine 61 was mutated to leucine to disrupt intrinsic GTPase activity and lock it into a GTP-bound state. The protein substrate had 12 amino acids deleted from its C terminus to eliminate variable C-terminal proteolysis of purified substrate and produce a single species substrate. The concentrations of VopS (5 nm) and His6-Cdc42-(1–179)-Q61L (600 μm) were kept constant while varying ATP concentrations from 50–2000 μm. The apparent Km for ATP was 160 ± 18 μm with a kcat of 26 s−1 ± 1.0 (Fig. 3, A and B; Table 1). The catalytic efficiency for ATP was 1.60 × 105 s−1 m−1. The kinetic constants for His6-Cdc42-(1–179)-Q61L were then determined with constant enzyme (5 nm) and ATP (2000 μm) concentrations. Varying Cdc42 concentrations from 50 to 1200 μm were used to determine apparent Km values. The kcat was 18 ± 1.5 s−1, and the Km was 180 ± 42 μm with a catalytic efficiency of 1.0 × 105 s−1 m−1 (Fig. 3B; Table 2).
FIGURE 3.
Apparent steady-state kinetic measurements for ATP and Cdc42. A, initial-velocity measurements for ATP were obtained using a constant concentration of His6-Cdc42-(1–179)-Q61L at 600 μm while varying ATP concentrations (40, 80, 120, 160, 250, 400, 700, 1000, 1500, 2000 μm). B, initial velocity measurements for Cdc42 were obtained with a constant concentration of ATP at 2 mm while varying the concentration of His6-Cdc42-(1–179)-Q61L (50, 75, 100, 200, 500 μm). Assays were preformed in triplicate with VopS (31–387) at 5 nm. In both parts the individual data points are depicted as circles, and the line represents the fit to these data using the Michaelis-Menten equation (Equation 1).
TABLE 1.
Apparent kinetic constant measurements for ATP
| kcata | Kma | kcat/Km | |
|---|---|---|---|
| s−1 | μm | s−1/m−1 | |
| Wild type | 26 ± 1.0 | 160 ± 18 | 1.6 × 105 |
| H348Ab | NDc | ND | ≤3.9 × 10−4 |
| F350A | 3.0 ± 0.32 | 1600 ± 290 | 1.9 × 103 |
| D352A | 0.098 ± 0.0070 | 580 ± 100 | 1.7 × 102 |
| N354A | 0.85 ± 0.11 | 2600 ± 480 | 3.3 × 102 |
| R356A | 0.035 ± 0.0039 | 1700 ± 330 | 2.0 × 101 |
| R299A | 0.036 ± 0.0019 | 220 ± 40 | 1.6 × 102 |
| L308A | 2.8 ± 0.28 | 590 ± 150 | 4.8 × 103 |
a Mean of three assays ± S.E.
b H348A VopS mutant was deemed severely inactive. H348A kcat/Km (s−1/m−1) was calculated from an endpoint measurement (performed in triplicate) at 10 μm enzyme, 2 mm ATP, 600 μm Cdc42.
c ND, not determined.
TABLE 2.
Apparent kinetic constant measurements for Cdc42
| kcata | Kma | kcat/Km | |
|---|---|---|---|
| s−1 | μm | s−1/m−1 | |
| Wild type | 18 ± 1.5 | 180 ± 40 | 1.0 × 105 |
| R299A | 0.027 ± 0.0036 | 660 ± 170 | 4.1 × 101 |
a Mean of three assays ± S.E.
The Fic motif (HPFX(D/E)GN(G/K)R) and the distal amino acid residue Arg-299 were demonstrated to be important for full catalytic activity (Fig. 2B). To address the question of which residue(s) of the Fic domain is critical for catalysis and to understand the function of each residue in protein AMPylation, we set out to determine the steady-state kinetic parameters for the small molecule substrate ATP for different VopS mutants. The most deleterious mutation was H348A; VopS harboring this mutation had a catalytic efficiency of less than 4.0 × 10−4 s−1 m−1, which is 9 orders of magnitude lower than wild-type protein and 5 orders of magnitude lower than the second worst mutation R356A (Table 1). Although the Michaelis-Menten constant Km is not strictly equivalent to KD, the value still provides a useful means to determine relative affinity when comparing the wild-type enzyme to its mutant form. The N354A mutation decreased the apparent kcat and had the most drastic increase in apparent Km for ATP: more than 2.5 mm, which is about 16-fold higher than that of the wild-type enzyme. This result further supports the hypothesis that Asn-354 plays a role in ATP binding.
VopS in which the hairpin residue Arg-299 had been mutated to an alanine had a low kcat value (0.036 s−1), but its apparent Km for ATP was statistically indistinguishable from the wild-type enzyme (220 μm). This phenomenon suggests that although catalysis is compromised, ATP binding is unaffected in the R299A mutant. To ascertain if the R299A mutant displays impaired binding of protein substrate, the Km with respect to Cdc42 was determined. The apparent Km was 3-fold higher than that of the wild-type enzyme (Table 2), implying that Arg-299 plays a minor role in protein substrate binding. Mutation of Arg-299 to a lysine was not sufficient to rescue activity in vitro (data not shown).
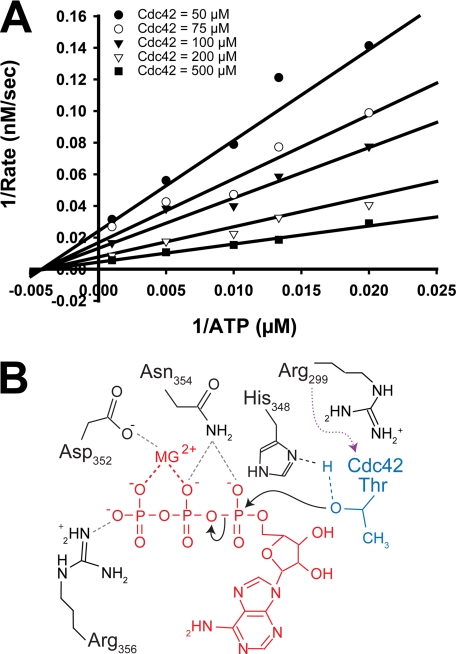
Kinetic Analysis of VopS Supports a Sequential Mechanism
The kinetic mechanisms of bisubstrate enzymes such as VopS can be distinguished with initial velocity studies analyzed using double-reciprocal plots. In such analyses a set of intersecting lines supports a sequential mechanism wherein the enzyme and both substrates form a ternary complex during the reaction. In a sequential (ternary complex) mechanism, VopS would interact with both ATP and the GTPase to allow transfer of the AMP group to the threonine residue. In contrast, a set of parallel lines supports a ping-pong reaction mechanism in which the enzyme forms a high energy covalent intermediate that leads to subsequent modification of the second substrate. In a ping-pong mechanism, VopS could potentially form a covalent phosphoramidate intermediate where the AMP group of ATP is transferred to the conserved His-348 of the Fic motif followed by AMP transfer to the GTPase. Initial velocity studies with VopS and its substrates, Cdc42 and ATP, reveal a set of intersecting lines that supports a sequential reaction mechanism. Kinetic analysis comparing a random (r2 = 98.6) to an ordered (r2 = 97.3) rapid-equilibrium model were statistically indistinguishable, where r2 is the square of the correlation coefficient (Fig. 4A, supplemental Fig. 4, A and B). The support for a ternary complex mechanism for VopS-mediated AMPylation and the kinetic results that the H348A mutant is essentially an inactive enzyme suggests histidine 348 functions as a general base during catalysis. The pKa of histidine is 6.0, and so the predicted optimal activity for the VopS enzyme would be at a pH greater than 6.0 where the histidine would be more basic. Consistent with our kinetic results, the pH profile of VopS WT activity demonstrate the enzyme is less active at lower pH conditions (supplemental Fig. 5).
FIGURE 4.
Ternary complex is required for catalysis. A, shown is a double-reciprocal plot of 1/rate versus 1/[ATP]. Assays were performed at fixed concentrations of His6-Cdc42-(1–179)-Q61L 50 μm (●), 75 μm (○), 100 μm (▾), 200 μm (▿), and 500 μm (■) while varying ATP concentrations (50, 75, 100, 200, 1000 μm). The data were fitted to a rapid-equilibrium random model (Equation 2) using Sigma Plot. The best-fit lines are shown. The square of the correlation coefficient (r2) for the global fit data is 98.6. Kinetic constants from the global fit have kcat = 89 ± 13 s−1, KATP = 280 ± 90 μm, KCdc42 = 490 ± 120 μm. B, shown is a proposed model of catalytic mechanism. His-348 functions as a general base to abstract a proton from the hydroxyl of the bound Rho GTPase threonine. The activated threonine of Cdc42 performs a nucleophilic attack on the α-phosphate of ATP. Asn-354 interacts with the β phosphate of ATP. The magnesium, coordinated by Asp-352, interacts with the β- and γ-phosphate of ATP. Arg-356 coordinates the γ phosphate of ATP. Arg-299 positioned in the hairpin has a role in protein-substrate binding. Dashed lines indicate proposed hydrogen bonds.
DISCUSSION
VopS, an AMPylator
The bacterial virulence factor VopS utilizes a Fic domain to posttranslationally modify GTPases with AMP. Our structural studies of the first characterized AMPylator revealed a predominantly α-helical enzyme with a novel N-terminal subdomain stabilizing a C-terminal Fic domain (Fig. 1A). Although the VopS structure is devoid of ATP and Cdc42, we can infer the position of the active site from kinetic characterization of VopS mutants (Table 1). The active site is formed by the Fic motif conserved residues (His-348, Asp-352, Asn-354, and Arg-356), which line a shallow pocket in the Fic domain. This pocket is completed by residues from the hairpin loop (Arg-299 and Leu-308). Bisubstrate kinetic analysis fitted best to a random, rapid-equilibrium model, suggesting random substrate binding. Future kinetic inhibition studies will be required to provide evidence of a random or possibly ordered mechanism for substrate binding, Attempted experiments were inconclusive due to solubility issues with potential inhibitors (not shown). Nevertheless, our kinetic studies support a mechanism whereby a ternary complex is formed between VopS, ATP, and Cdc42. In such a complex, VopS would position the threonine residue of the Rho GTPase near the imidazole side chain of the catalytic histidine (His-348) (Fig. 4B). The proposed catalytic mechanism is described in Fig. 4B, and the roles of the active site residues are discussed below.
Structural Mechanism of Substrate Interaction
Superposition of VopS with BepA structures and a representative Fic domain from H. pylori highlights the structure differences in the hairpin loop and illustrates movement within this structural element (Fig. 5A). The superposition reveals four distinct positions of the hairpin element. The VopS hairpin loop adopts a position that is intermediate to the others. The BepA hairpin loop appears to be in an open conformation, whereas the H. pylori Fic hairpin is in a relatively closed position that is lowered toward the active site. The BepA apo- and MgPPi-bound structures have two different hairpin loop conformations, supporting the notion that ATP binding induces structural changes in the hairpin element of the enzyme.
FIGURE 5.
Structural mechanism for protein substrate binding. A, structural comparison of β-hairpins and hairpin loops of Fic domains from VopS (green), BepA apo (cyan, PDB ID 2JK8), BepA MgPPi complex (yellow, PDB ID 2JK8), and H. pylori (white, PDB ID 2F6S) reveal four distinct structural conformations. The BepA structures used for superposition had two molecules per asymmetric unit, where one had MgPPi bound, and the other BepA molecule was unbound. The hairpin loop of VopS is in an intermediate conformation. VopS residues Arg-299 and His-348 have altered orientations relative to other Fic structures. B, the β-hairpin (green) of S. oneidensis Fic (PDB ID 3EQX) forms a three-stranded β sheet interaction with the N terminus (red) of a neighboring crystal mate. The conserved histidine of the Fic motif is depicted in blue. C, AvrB (PDB ID 2NUD) forms a structurally analogous anti-parallel β-sheet (green) formed via edge-on strand interactions with a high affinity peptide. AvrB lacks the Fic sequence motif; highlighted in blue spheres is the Met-262 residue that takes the place of the conserved histidine of the Fic motif.
Evidence for the β-hairpin mediating protein-substrate interaction is seen in the crystal structure of Shewanella oneidensis (PDB ID 3EQX) from the Joint Center for Structural Genomics (Fig. 5B). The N terminus of a crystallographic symmetry-related Fic domain binds edge-on to the β-hairpin to form a three-stranded β-sheet. AvrB (avirulence protein B) (PDB ID 2NUD) from Pseudomonas syringae possesses a structurally similar Fic topology that includes a similar β interaction with a bound peptide (Fig. 5C) (8). In support of this binding mechanism, a complex structure of Cdc42 with its downstream substrate, p21-activated kinase, has revealed β-strand interactions with the Cdc42 switch 1 region (24). Analogous to the interaction with p21-activated kinase, the Rho GTPase switch 1 region could interact with an induced Fic domain β-hairpin in VopS. Such an interaction would require a similar movement of the VopS hairpin loop illustrated in Fig. 5A. A conservative mutation of the VopS hairpin loop residue Leu-308 to alanine impairs catalytic activity, supporting a role for the hairpin loop in protein substrate binding.
The BepA MgPPi complex indicates an approximate binding location for the β- and γ-phosphates of ATP bound to Fic domains (Fig. 2A). The VopS R356A mutant displayed an increased apparent Km for ATP (10-fold) and a substantially decreased kcat (1000-fold). This residue is positioned to stabilize the increasing negative charge developing on the pyrophosphate during catalysis, thereby stabilizing the transition state. A similarly positioned positive charge in the molybdenum cofactor biosynthetic enzyme MoeB stabilizes the developing negative charge on pyrophosphate in forming a transient acyladenylate (25). The BepA MgPPi structure appears to be a product complex, with the corresponding Fic motif arginine guanidinium pointing away from the PPi and forming a salt bridge with a residue from helix a1, similar to the salt bridge formed by VopS Arg-356. An alternative side-chain conformation for the BepA (PDB ID 2VZA) arginine is seen in a structure with two bound sulfates replacing the PPi. In this structure, the guanidinium swings toward the active site, forming a salt bridge with the sulfate. A similar repositioning of VopS Arg-356 may occur during catalysis, allowing a stabilizing interaction with the PPi of the transition state but not with the product.
A decrease in apparent kcat and an increase in the apparent Km for ATP is observed in the F350A mutant, indicating that this residue side chain may stack on the adenine ring of the ATP. The nucleotide would be required to adopt a significantly bent conformation with the hinge at the α-phosphate to accommodate simultaneous interactions with Asp-352, Asn-354, Arg-356, and Phe-350. A similar conformation for bound ATP has been observed in yeast aspartyl-tRNA synthetase, which hydrolyzes ATP and transfers AMP to aspartic acid as the first step of the enzymatic reaction (25). This bent conformation would position the α-phosphate near the catalytic histidine and expose the phosphate bond for efficient nucleophilic attack by the Rho GTPase threonine hydroxyl.
VopS has three glycines within the catalytic loop positioned between helix a4 and a5, two of which are conserved in the Fic motif. These glycines may provide the flexibility necessary for efficient ATP binding and catalysis. Attempts to model ATP in the postulated active site of the known Fic domain structure while retaining interactions with conserved residues results in steric clashes with the main chain of the catalytic Fic motif loop and, for VopS, the β-hairpin loop. Substantial rearrangement of both loops in VopS may be required during the catalytic cycle.
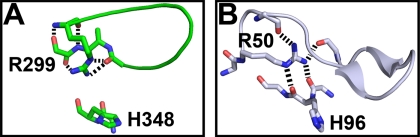
The Arg-299 residue of VopS adopts a different side-chain rotamer relative to arginines in other Fic domains (Fig. 5A). Kinetic studies with the R299A mutant show an increased Km for Rho GTPase and not for ATP, supporting a functional role for this residue in protein-substrate binding. Comparison of VopS Arg-299 and the analogous arginines of several Fic domain structures, such as the Fic protein of H. pylori, reveal differences in polar contacts. R299A resides in a loop just N-terminal to the VopS β-hairpin element (Fig. 6A). The Arg-299 guanidinium group forms hydrogen bonds to a backbone carbonyl oxygen atom at the C terminus of the hairpin loop, thus anchoring the loop at both ends. VopS Arg-299 anchors the hairpin loop by forming hydrogen bonds (main chain and side chain) with its C terminus. The analogous conserved arginine, Arg-50, of H. pylori interacts with its β hairpin through hydrogen bonds between the guanidinium group and main-chain carbonyl oxygen atoms at its N terminus and C terminus (Fig. 6B). However, the H. pylori R50 guanidinium group also forms hydrogen bonds to main-chain carbonyl oxygen atoms of the active site histidine. The evolutionarily conserved Arg-299 may function as an anchor that coordinates the hairpin loop toward the active site in addition to properly positioning the catalytic loop for efficient catalysis. The very low kcat value for Arg-299 supports a catalytic function. Similar to the arginine finger observed in GTPases, Arg-299 may be involved in neutralizing the negative charge of the α-phosphate in the transition state (26).
FIGURE 6.
The molecular anchor; possible role of arginine 299. A, VopS R299A interactions are mainly within the hairpin. B, in H. pylori Fic the analogous arginine, Arg-50, forms main-chain-side-chain interactions that function as an anchor that links and holds the hairpin loop in an appropriate conformation and anchors the catalytic histidine in the active site. Black dashes represent hydrogen bonds.
Protein AMPylation Is a Novel Mechanism of Cell Regulation
Posttranslational modification of proteins is a regulatory mechanism that enables molecules to control and diversify their function in a cell. Conserved from bacteria to humans, protein phosphorylation, acetylation, and methylation are just a few posttranslational mechanisms used to control and regulate complex signaling processes. Recently, protein AMPylation has joined this list. Similar to phosphorylation, VopS uses an abundant high energy substrate, ATP, to modify its substrates (11). Comparison of AMPylation with turnover rates from other posttranslational modifications, such as phosphorylation, acetylation, and methylation, suggests that VopS is a relatively fast enzyme with the apparent kcat = 18 s−1 (Table 1). Phosphorylation by mitogen-activated protein kinase p38 has a kcat of 22.6 s−1. Histone acetyltransferase GCN5 has a kcat of 1.7 s−1, and the G9a histone methyltransferase has a kcat of 0.0012 s−1 (27–29). The protein AMPylation turnover value is well within the range of other, established posttranslational modifications. Therefore, AMPylation is a potential regulatory mechanism that could mediate eukaryotic signaling processes.
Fic domains are present in proteins of higher eukaryotes, although genome analysis shows that they are less abundant when compared with the prevalence of Fic domains in bacteria. Nonetheless, activities that are functionally homologous to those of prokaryotic Fic domains may exist in higher eukaryotes. AMP, when covalently attached to a target protein, is a bulky moiety that has been demonstrated to inhibit cellular signaling through AMPylation. Moreover, this same type of AMP modification on a tyrosine in glutamate synthetase results in an increased stability of the active enzyme, thus providing an example of positive regulation for AMPylation in a signaling process (30). Covalently attached AMP could potentially promote protein complex assembly where the AMP moiety can serve as a recognition site. Additionally, AMPylation is not restricted to threonine. An AMPylated tyrosine was initially observed on glutamine synthetase and subsequently for a Fic domain containing protein from Histophilus somni, a respiratory pathogen (13). Protein AMPylation was also demonstrated in eukaryotes from huntington yeast-interacting protein (HYPE) and the Drosophila Fic domain protein FicD (10, 13).
The conservation of Fic domains from bacteria to worms, flies, and humans suggests a role for protein AMPylation as a regulatory mechanism in diverse signaling pathways. The nature of protein AMPylation as a common mechanism utilized by macromolecular molecules to regulate signal transduction is poised for future exploration.
Supplementary Material
Acknowledgments
We appreciate the generosity and support of our colleagues at the University of Texas Southwestern Medical Center at Dallas. We thank Margaret Phillips, Radha Akella, Carlos Huerta, and Melanie Yarbrough for helpful discussions and comments. We thank Yan Li and Hayden Ball for mass spectrometry analysis. We thank all of the members of the Orth laboratory for helpful discussions and support. Results in this report are derived from work at the Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the United States Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-AI056404 (NIAID, to K. O. and P. L.). This work was also funded in part by Welch Research Foundation Grant I-1561 (to K. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 2–5 and Table 1.
The atomic coordinates and structure factors (code 3LET) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
D. Palanivelu, M. Meury, C. Dehio, and T. Schirmer, unpublished information.
- T3SS
- type III secretion system
- GST
- glutathione S-transferase
- DTT
- dithiothreitol
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Broberg C. A., Orth K. (2010) Curr. Opin. Microbiol. 1, 34–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galán J. E., Wolf-Watz H. (2006) Nature 444, 567–573 [DOI] [PubMed] [Google Scholar]
- 3.Makino K., Oshima K., Kurokawa K., Yokoyama K., Uda T., Tagomori K., Iijima Y., Najima M., Nakano M., Yamashita A., Kubota Y., Kimura S., Yasunaga T., Honda T., Shinagawa H., Hattori M., Iida T. (2003) Lancet 361, 743–749 [DOI] [PubMed] [Google Scholar]
- 4.Nair G. B., Ramamurthy T., Bhattacharya S. K., Dutta B., Takeda Y., Sack D. A. (2007) Clin. Microbiol. Rev. 20, 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ono T., Park K. S., Ueta M., Iida T., Honda T. (2006) Infect. Immun. 74, 1032–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park K. S., Ono T., Rokuda M., Jang M. H., Iida T., Honda T. (2004) Microbiol. Immunol. 48, 313–318 [DOI] [PubMed] [Google Scholar]
- 7.Burdette D. L., Yarbrough M. L., Orvedahl A., Gilpin C. J., Orth K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12497–12502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yarbrough M. L., Li Y., Kinch L. N., Grishin N. V., Ball H. L., Orth K. (2009) Science 323, 269–272 [DOI] [PubMed] [Google Scholar]
- 9.Burdette D. L., Seeman J., Orth K. (2009) Mol. Microbiol, 73, 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinch L. N., Yarbrough M. L., Orth K., Grishin N. V. (2009) PLoS ONE 4, e5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yarbrough M. L., Orth K. (2009) Nat. Chem. Biol. 5, 378–379 [DOI] [PubMed] [Google Scholar]
- 12.Casselli T., Lynch T., Southward C. M., Jones B. W., DeVinney R. (2008) Infect. Immun. 76, 2202–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Worby C. A., Mattoo S., Kruger R. P., Corbeil L. B., Koller A., Mendez J. C., Zekarias B., Lazar C., Dixon J. E. (2009) Mol. Cell 34, 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 15.Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 16.Otwinowski Z. (1991) in Isomorphous Scattering and Anomalous Replacement (Wolf W., Evans P.R., Leslie A. G. W., eds) pp. 80–86, Science & Engineering Research Council, Cambridge, UK [Google Scholar]
- 17.Cowtan K., Main P. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 487–493 [DOI] [PubMed] [Google Scholar]
- 18.Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 19.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 20.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 21.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 23.Das D., Krishna S. S., McMullan D., Miller M. D., Xu Q., Abdubek P., Acosta C., Astakhova T., Axelrod H. L., Burra P., Carlton D., Chiu H. J., Clayton T., Deller M. C., Duan L., Elias Y., Elsliger M. A., Ernst D., Feuerhelm J., Grzechnik A., Grzechnik S. K., Hale J., Han G. W., Jaroszewski L., Jin K. K., Klock H. E., Knuth M. W., Kozbial P., Kumar A., Marciano D., Morse A. T., Murphy K. D., Nigoghossian E., Okach L., Oommachen S., Paulsen J., Reyes R., Rife C. L., Sefcovic N., Tien H., Trame C. B., Trout C. V., van den Bedem H., Weekes D., White A., Hodgson K. O., Wooley J., Deacon A. M., Godzik A., Lesley S. A., Wilson I. A. (2009) Proteins 75, 264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morreale A., Venkatesan M., Mott H. R., Owen D., Nietlispach D., Lowe P. N., Laue E. D. (2000) Nat. Struct. Biol. 7, 384–388 [DOI] [PubMed] [Google Scholar]
- 25.Lake M. W., Wuebbens M. M., Rajagopalan K. V., Schindelin H. (2001) Nature 414, 325–329 [DOI] [PubMed] [Google Scholar]
- 26.Ahmadian M. R., Stege P., Scheffzek K., Wittinghofer A. (1997) Nat. Struct. Biol. 4, 686–689 [DOI] [PubMed] [Google Scholar]
- 27.Chen G., Porter M. D., Bristol J. R., Fitzgibbon M. J., Pazhanisamy S. (2000) Biochemistry 39, 2079–2087 [DOI] [PubMed] [Google Scholar]
- 28.Tanner K. G., Langer M. R., Kim Y., Denu J. M. (2000) J. Biol. Chem. 275, 22048–22055 [DOI] [PubMed] [Google Scholar]
- 29.Patnaik D., Chin H. G., Estève P. O., Benner J., Jacobsen S. E., Pradhan S. (2004) J. Biol. Chem. 279, 53248–53258 [DOI] [PubMed] [Google Scholar]
- 30.Gill H. S., Pfluegl G. M., Eisenberg D. (2002) Biochemistry 41, 9863–9872 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.