Abstract
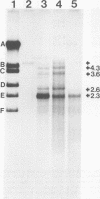
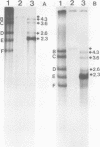
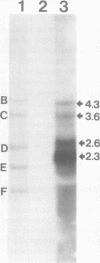
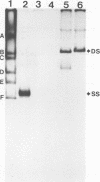
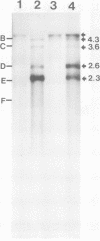
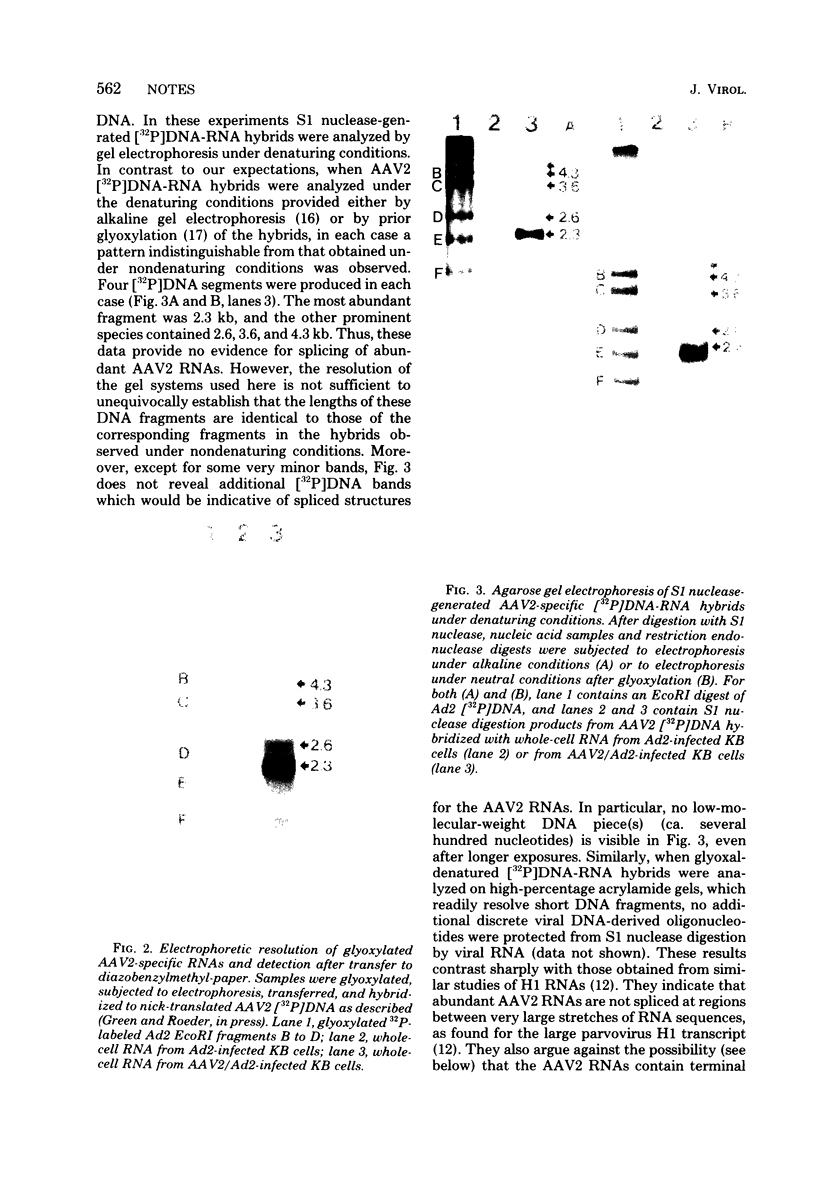
Adenovirus-associated virus type 2 synthesizes four prominent viral transcripts, containing 4.3, 3.6, 2.6, and 2.3 kilobases (kb), in productively infected human KB cells (coinfected with adenovirus type 2). All species are polyadenylated and present in both nuclear and whole-cell RNA preparations, but only the predominant 2.3-kb (and possibly the 2.6-kb) RNA species are found on polysomes. Electrophoretic analyses under denaturing conditions of S1 nuclease-generated and exonuclease VII-generated DNA-RNA hybrids revealed, in each case, four protected DNA fragments which are equal in length (within 50 to 100 nucleotides) to the four S1 nuclease-generated hybrids resolved by electrophoresis under nondenaturing conditions. These results suggest that in the infected cell, abundant adenovirus-associated virus type 2 transcripts are present predominantly (by mass) as unspliced RNAs or, alternatively, they are spliced but contain very short (less than or equal to 50 nucleotides) leader sequences. That the 2.3-kb RNA represents such a spliced transcript is suggested by exonuclease VII mapping experiments and our more detailed RNA mapping studies (M. R. Green and R. G. Roeder, J. Virol., in press).
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berget S. M., Sharp P. A. Structure of late adenovirus 2 heterogeneous nuclear RNA. J Mol Biol. 1979 Apr 25;129(4):547–565. doi: 10.1016/0022-2836(79)90468-6. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom M. E., Rose J. A. Transcription of adenovirus-associated virus RNA in isolated KB cell nuclei. Virology. 1978 Jan;84(1):118–126. doi: 10.1016/0042-6822(78)90223-4. [DOI] [PubMed] [Google Scholar]
- Carter B. J., Fife K. H., de la Maza L. M., Berns K. I. Genome localization of adeno-associated virus RNA. J Virol. 1976 Sep;19(3):1044–1053. doi: 10.1128/jvi.19.3.1044-1053.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B. J., Khoury G., Rose J. A. Adenovirus-associated virus multiplication. IX. Extent of transcription of the viral genome in vivo. J Virol. 1972 Dec;10(6):1118–1125. doi: 10.1128/jvi.10.6.1118-1125.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter B. J., Rose J. A. Transcription in vivo of a defective parvovirus: sedimentation and electrophoretic analysis of RNA synthesized by adenovirus-associated virus and its helper adenovirus. Virology. 1974 Sep;61(1):182–199. doi: 10.1016/0042-6822(74)90253-0. [DOI] [PubMed] [Google Scholar]
- Chase J. W., Richardson C. C. Exonuclease VII of Escherichia coli. Mechanism of action. J Biol Chem. 1974 Jul 25;249(14):4553–4561. [PubMed] [Google Scholar]
- Darnell J. E., Jr Implications of RNA-RNA splicing in evolution of eukaryotic cells. Science. 1978 Dec 22;202(4374):1257–1260. doi: 10.1126/science.364651. [DOI] [PubMed] [Google Scholar]
- Green M. R., Lebovitz R. M., Roeder R. G. Expression of the autonomous parvovirus H1 genome: evidence for a single transcriptional unit and multiple spliced polyadenylated transcripts. Cell. 1979 Aug;17(4):967–977. doi: 10.1016/0092-8674(79)90336-2. [DOI] [PubMed] [Google Scholar]
- Jay F. T., de la Maza L. M., Carter B. J. Parvovirus RNA transcripts containing sequences not present in mature mRNA: a method for isolation of putative mRNA precursor sequences. Proc Natl Acad Sci U S A. 1979 Feb;76(2):625–629. doi: 10.1073/pnas.76.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin C. A., Westphal H., Carter B. J. Spliced adenovirus-associated virus RNA. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5567–5571. doi: 10.1073/pnas.76.11.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey J. K., Brackmann K. H., Green M. R., Green M. Preparation and characterization of highly radioactive in vitro labeled adenovirus DNA and DNA restriction fragments. Biochemistry. 1977 Oct 4;16(20):4478–4483. doi: 10.1021/bi00639a023. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. A., Koczot F. Adenovirus-associated virus multiplication. VI. Base compostion of the deoxyribonucleic acid strand species and strand-specific in vivo transcription. J Virol. 1971 Nov;8(5):771–777. doi: 10.1128/jvi.8.5.771-777.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal J., Ron D., Tattersall P., Bratosin S., Aloni Y. About 30% of minute virus of mice RNA is spliced out following polyadenylation. Nature. 1979 Jun 14;279(5714):649–651. doi: 10.1038/279649a0. [DOI] [PubMed] [Google Scholar]
- Zieve G., Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976 May;8(1):19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]