Abstract
The Epstein-Barr virus basic leucine zipper transcriptional activator ZEBRA was shown recently to cross the outer membrane of live cells and to accumulate in the nucleus of lymphocytes. We investigated the potential application of the Epstein-Barr virus trans-activator ZEBRA as a transporter protein to facilitate transduction of cargo proteins. Analysis of different truncated forms of ZEBRA revealed that the minimal domain (MD) required for internalization spans residues 170–220. MD efficiently transported reporter proteins such as enhanced green fluorescent protein (EGFP) and β-galactosidase in several normal and tumor cell lines. Functionality of internalized cargo proteins was confirmed by β-galactosidase activity in transduced cells, and no MD-associated cell toxicity was detected. Translocation of MD through the cell membrane required binding to cell surface-associated heparan sulfate proteoglycans as shown by strong inhibition of protein uptake in the presence of heparin. We found that internalization was blocked at 4 °C, whereas no ATP was required as shown by an only 25% decreased uptake efficiency in energy-depleted cells. Common endocytotic inhibitors such as nystatin, chlorpromazine, and wortmannin had no significant impact on MD-EGFP uptake. Only methyl-β-cyclodextrin inhibited MD-EGFP uptake by 40%, implicating the lipid raft-mediated endocytotic pathway. These data suggest that MD-reporter protein transduction occurs mostly via direct translocation through the lipid bilayer and not by endocytosis. This mechanism of MD-mediated internalization is suitable for the efficient delivery of biologically active proteins and renders ZEBRA-MD a promising candidate for therapeutic protein delivery applications.
Keywords: Endocytosis, Heparan Sulfate, Multifunctional Protein, Protein-DNA Interaction, Protein Domains, Protein Translocation, Epstein-Barr Virus, Protein Transduction Domain, Protein Transduction, ZEBRA
Introduction
Recently, cell-penetrating peptides (CPPs)2 were discovered that have the ability to cross lipid bilayers of mammalian cells, which naturally constitute a tight biological barrier allowing only the uptake of non-polar molecules <500 Da into the cell (1). Coupling of CPPs to molecules such as small interfering RNA, peptide nucleic acids, and bioactive proteins facilitates delivery of therapeutic agents into live cells (2–5). The most intensively studied CPPs include the Tat peptide, which is derived from human immunodeficiency virus protein-1 (6, 7); the third helix of the Antennapedia homeodomain (also referred to as Penetratin) from Drosophila (8); and the VP22 peptide of the herpes simplex virus (9, 10).
The activity of therapeutic molecules depends on their availability and functionality after their delivery into cells, both of which are determined by the transduction mechanism of the particular CPP (11). Consequently, extensive research was directed to elucidate pathways underlying protein transduction. Because of its apparent temperature and ATP dependence, it is widely assumed that cell entry occurs via endocytosis after binding of the CPP to negatively charged heparan sulfate proteoglycans (HSPGs) located on the cell surface (12–14). Tat fusion proteins were shown to be internalized via macropinocytosis, whereas different endocytotic pathways such as clathrin-dependent or caveolin-dependent endocytosis were shown to be implicated in the transport of other CPPs (10, 12, 15–17). In addition, an alternative route of direct translocation of the small CPPs such as the SSHR-based peptide was reported (18). Currently, one of the major challenges for the application of CPPs is to increase endosomal escape of CPP cargoes to provide sufficient biologically active molecules at their intracellular destinations. Fusion of the CPP to pH-sensitive fusogenic peptides or co-administration of endosome-disrupting agents such as chloroquine, Ca2+, sucrose, and photosensitizers results in an enhanced release of the CPP cargoes from the endosomal compartments (19–22). However, these strategies come along with considerable medical and technical disadvantages, including the cytotoxicity of the disrupting agents such as chloroquine and the problematic application of photosensitizers for in vivo protein delivery. Therefore, there is a strong need for the identification of alternative CPPs that ideally bypass the endocytotic uptake route, thereby facilitating efficient delivery of therapeutic molecules to their intracellular destinations.
This study focuses on the characterization of the cell-penetrating properties of the major Epstein-Barr virus trans-activator, the ZEBRA protein (also referred to as Zta, Z, EB1, or BZLF1) (23, 24). Recently, it was shown that this protein not only binds DNA and exerts cell cycle control functions but also has the ability to penetrate lymphoid cells (25). The ZEBRA protein belongs to the basic leucine zipper (bZIP) family of transcription factors and is responsible for the initiation of the Epstein-Barr virus lytic cycle (24, 26). Furthermore, this multifunctional protein controls its own expression, virus replication, cell cycle arrest, and DNA damage response in the host cells (24, 27–30). The structure of the 245-amino acid ZEBRA protein has been resolved recently in its DNA-bound form: the protein is divided into an N-terminal transactivation region and a basic DNA-binding domain flanked by a coiled-coil dimerization region (zipper). The C-terminal domain interacts with the zipper region by forming intra- and intermolecular interactions, resulting in a hydrophobic pocket (31). These complex interactions are unique among the bZIP members and result in the stabilization of the ZEBRA dimer when bound to DNA.
In this work, we investigated the transduction properties of ZEBRA. We designed different truncated forms of ZEBRA to identify the minimal domain (MD) required for the delivery of different reporter proteins, including enhanced green fluorescence protein (EGFP) and β-galactosidase, in various cell lines. The 48-amino acid bZIP region of ZEBRA (ZEBRA-MD) was shown to be sufficient to facilitate reporter protein internalization. In addition, we demonstrate that ZEBRA-MD fused to EGFP requires interaction with cell-surface HSPGs and crosses the cell membrane in a pathway largely independent of endocytosis. Taken together, our results reveal the powerful transduction properties of ZEBRA-MD, rendering it a promising candidate for the development an efficient protein delivery vector.
EXPERIMENTAL PROCEDURES
Cloning of ZEBRA Protein and Fragments
The DNA fragments encoding the full-length protein and all truncated forms of ZEBRA were generated by PCR and ligated into the Escherichia coli expression vector pET15b (Novagen), which provided an N-terminal hexahistidine tag for subsequent purification. Sequences of primers used for generation of truncated ZEBRA fragments, EGFP, and LacZ have been described previously (32). The reporter genes were C-terminally fused to truncated ZEBRA fragments and cloned into the pET15b expression plasmid.
Expression and Purification of Recombinant Proteins
All recombinant fusion proteins were expressed in E. coli BL21(DE3) after induction with 0.5 mm isopropyl β-d-thiogalactopyranoside for 15 h at 16 °C. Cells were lysed by sonication in 20 mm Tris buffer (pH 6.8 or 8) containing 250 mm NaCl and 10% glycerol and subsequently treated with DNase I (Roche Applied Science) for nucleic acid removal. Purifications of His6-tagged proteins were performed by nickel affinity chromatography. Proteins were washed using a 0.5–1.5 m NaCl gradient and eluted in 500 mm imidazole, 20 mm Tris, 75 mm KCl, 0.5 m NaCl, and 10% glycerol. All purification steps were carried out at 4 °C and in the presence of protease inhibitors (pepstatin, E-64, aprotinin, Pefabloc, and complete protease inhibitor mixture; Roche Applied Science). Prior to transduction experiments, purified proteins were dialyzed against phosphate-buffered saline (PBS).
Electrophoretic Mobility Shift Assay (EMSA)
EMSA binding reactions were performed for all purified recombinant proteins. The AP-1 probe was made by annealing two oligonucleotides (5′-AGCACTGACTCATGAAGT-3′ and 5′-TACTTCATGAGTCAGTGCT-3′). The unlabeled probe was labeled with biotin and purified over a Microspin G-25 spin column (Active Motif). Up to 500 μg of full-length ZEBRA and truncated proteins were preincubated with 4× binding buffer B-1/2× stabilizing buffer (Active Motif) and 1 mm dithiothreitol for 15 min on ice. The biotin-labeled probe was mixed with 4× binding buffer C-1/2× stabilizing buffer (Active Motif) supplemented with 50 ng/μl poly(dI-dC) and added to protein-containing solution. After incubation of the reaction mixture for 15 min at 4 °C, the samples were separated on 4–8% nondenaturing polyacrylamide gels in 0.5× Tris borate/EDTA and transferred to Hybond N+ membrane (Amersham Biosciences). The LightShift chemiluminescent EMSA kit (Pierce) was used for detection according to the manufacturer's protocol.
Cell Culture and Transduction Experiments
HeLa cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen), and Saos2 cells were grown in McCoy's 5A medium (Invitrogen) supplemented with 10–20% heat-inactivated fetal bovine serum (Invitrogen), 50 units/ml penicillin, 50 μg/ml streptomycin, and 2 mm l-glutamine (Invitrogen). 7.5 × 105 cells/well were seeded on a 12-well plate 24 h before transduction experiments. For microscopic analysis, cells (2.5 × 105) were plated on 4-well chamber slides at least 24 h before treatment. Internalization experiments were performed at 60–80% confluence. Cells were rinsed twice with PBS before the addition of fresh serum-free culture medium containing the indicated amounts of protein. After 4 h, the culture medium was supplemented with 10% heat-inactivated fetal bovine serum for long-term incubation.
Drug Treatment
Heparin; endocytotic inhibitors such as wortmannin, nystatin, chlorpromazine hydrochloride, and methyl-β-cyclodextrin; 2-deoxy-d-glucose; and sodium azide was purchased from Sigma. Cells were incubated for 30 min prior to EGFP fusion protein addition in serum-free medium containing the indicated concentrations of individual drugs (20 μg/ml heparin, 100 nm wortmannin, 50 μg/ml nystatin, 30 μm chlorpromazine hydrochloride, and 5–10 mm methyl-β-cyclodextrin). Subsequently, cells were incubated for 3 h in the presence of inhibitors and proteins at 37 or 4 °C and trypsinized (0.5% trypsin/EDTA, Invitrogen) to remove surface-bound protein before analyzing fluorescence by flow cytometry. For depletion of the ATP pool, cells were preincubated for 1 h in PBS containing 6 mm 2-deoxy-d-glucose and 10 mm sodium azide.
Immunocytochemistry and Fluorescence Microscopy
When incubated with EGFP fusion proteins, cells were washed with PBS, followed by a mild trypsinization (0.5% trypsin/EDTA) and several washes with heparin (20 μg/ml). For immunolocalization studies, cells were washed two times for 5 min with heparin (20 μg/ml) in PBS and fixed for 10 min in 4% paraformaldehyde at room temperature. Cells were permeabilized and blocked with 0.25% Triton X-100 and 5% bovine serum albumin in PBS for 1 h at room temperature, followed by a 1-h incubation at room temperature with the corresponding primary antibodies in 0.1% Triton X-100 and 5% bovine serum albumin in PBS. To detect endogenous endosomal proteins, anti-EEA1 (4 μg/ml; Calbiochem), anti-Rab7 (5 μg/ml; Cell Signaling), or anti-clathrin or anti-caveolin-1 (5 μg/ml; Santa Cruz Biotechnology) antibody was applied. After three washes with PBS for 10 min, cells were incubated with the corresponding Alexa Fluor® 647 (Molecular Probes)-labeled anti-rabbit secondary antibody in 0.1% Triton X-100 and 5% bovine serum albumin in PBS. Cells were washed five times for 10 min with PBS, and nuclei were counterstained with Hoechst 33258 (Molecular Probes). Cell fluorescence on unfixed cells was visualized using a Nikon Eclipse TE2000-E inverted fluorescence microscope with a GFP filter (465–495 nm excitation and 515–555 nm emission). For localization studies, cells were analyzed by confocal microscopy (TCS-SP2, Leica, Mannheim, Germany). Images were acquired sequentially, with 488 nm excitation for MD-EGFP (fluorescence collection between 500 and 540 nm; displayed in green); 633 nm excitation for Alexa Fluor® 647-labeled EEA1, Rab7, clathrin, and caveolin-1 (fluorescence collection between 650 and 700 nm; displayed in red); and 405 nm excitation for Hoechst (fluorescence collection between 415 and 460 nm; displayed in blue).
β-Galactosidase Staining
After transduction experiments, cells were washed with 20 μg/ml heparin in PBS and carefully trypsinized for cell surface-bound protein removal. Fixation and staining were done according to the β-galactosidase staining kit protocol (Sigma). Briefly, cells were incubated for 10 min at room temperature with 1× fixation solution containing 2% formaldehyde and 0.2% glutaraldehyde. After three wash steps with PBS at room temperature, cells were stained with a solution containing 20 mm magnesium chloride, 40 mm potassium ferricyanide, 40 mm potassium ferrocyanide, and 2 mg/ml X-gal for 3 h at 37 °C. Images were taken with a Nikon Eclipse TE2000-E microscope.
Western Blot Analysis
After transduction experiments, cells were collected, and non-internalized proteins were removed by trypsinization. Whole cell extracts were prepared by lysing cells in ice-cold mammalian cell lysis buffer (Sigma), and separation in cytosolic and nucleic fractions was carried out using a cell compartment kit (Pierce). A total of 50–100 μg of protein were subjected to SDS-PAGE separation and transferred to nitrocellulose membranes. Western blotting was carried out as described previously (33). Mouse anti-ZEBRA Z125/Z130 monoclonal antibody (mouse anti-EB1/Zta Z125 antibody, 1:100 dilution, in Tween/Tris-buffered saline) and mouse anti-GFP monoclonal antibody (1:500 dilution, in Tween/Tris-buffered saline) (Euromedex) were used as primary antibodies. After incubation with peroxidase-labeled anti-mouse secondary antibody (1:5000 dilution, in Tween/Tris-buffered saline; Amersham Biosciences), the blots were washed again and analyzed using an enhanced chemiluminescence detection system (Amersham Biosciences).
Flow Cytometric Analysis
Internalization of EGFP fusion proteins alone or in the presence of inhibitors was measured by flow cytometry. Cells were treated with 0.5% trypsin and 20 μg/ml heparin for 10 min to remove surface-bound proteins before analyzing green fluorescence. Only live cells were assayed using 7-aminoactinomycin D exclusion (34). Flow cytometric analysis was carried out with a FACSCalibur (BD Biosciences).
Cytotoxicity Assay
Membrane integrity was measured using a cytotoxicity detection kit (Roche Applied Science). In brief, 1 × 104 HeLa or Saos2 cells were seeded in 96-well plates 24 h before treatment with ZEBRA-MD fusion proteins at the indicated concentrations in serum-free medium. After 24 h, lactate dehydrogenase assay was carried out according to the manufacturer's protocol (Roche Applied Science).
Synthetic Liposomes
Synthetic lipid vesicles were prepared as described previously (33). Liposomes were labeled with the lipophilic dye Nile red (10 μm), incubated at 37 °C in the presence of 0.2 μm MD-EGFP, and subsequently analyzed by confocal microscopy (TCS-SP2).
RESULTS
Identification of the MD Required for Protein Translocation
The full-length ZEBRA protein contains three major regions: a transactivation domain (residues 1–140), a highly basic DNA-binding domain (DBD; residues 175–195), and a dimerization domain (DIM domain; residues 195–220) (23, 26, 31). To identify the MD required for the translocation of ZEBRA into mammalian cells, nine different truncations of the full-length ZEBRA protein were constructed that covered the entire amino acid sequence of the native protein (Fig. 1A). Proteins extending from residues 1 to 195 covering the transactivation domain (Z1) and shorter fragments of the N-terminal part of ZEBRA (Z2 and Z3) were produced. Fragments Z4, Z6, and Z9 contained both the DBD and the DIM domain and varied with respect to the regions flanking those functional domains. Furthermore, three fragments bearing either the DBD (Z5 and Z7) or the DIM domain (Z8) were constructed (Fig. 1A). The full-length and truncated proteins had an N-terminal His6 tag to facilitate purification of the soluble protein fractions by nickel affinity chromatography. As shown in Fig. 1 (B and C), all truncated ZEBRA fragments and recombinant EGFP fusions could be successfully overexpressed and purified to near homogeneity. All N-terminal fragments (Z1, Z2, and Z3) and Z5 showed higher protein yields compared with C-terminal ZEBRA protein fragments (Fig. 1B). Furthermore, fusion of ZEBRA truncations to EGFP resulted in a higher yield of soluble purified protein compared with β-galactosidase fusions (data not shown).
FIGURE 1.
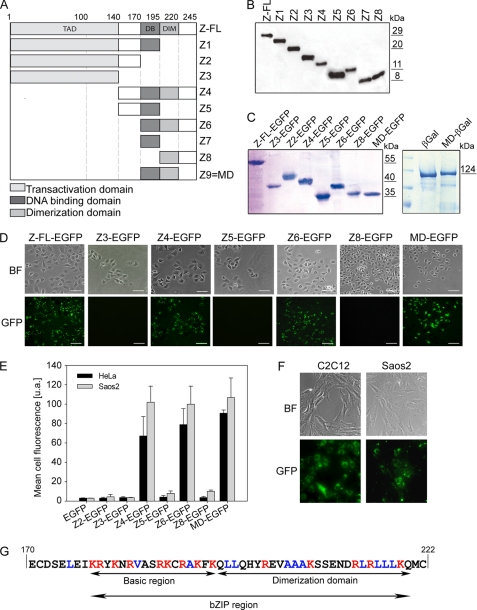
Identification of the MD required for protein translocation. A, shown is a scheme of full-length ZEBRA protein (Z-FL) and all corresponding protein truncations. The full-length ZEBRA protein contains a transactivation domain (TAD), a DBD (DB), and a DIM domain. Nine truncated forms of ZEBRA (Z1–Z9) were created to characterize the MD required for transduction. B, truncated ZEBRA proteins after expression in E. coli BL21(DE3) and purification by nickel affinity chromatography were separated by SDS-PAGE and detected by Western blotting using anti-His antibody. C, aliquots of ZEBRA-EGFP and MD-β-galactosidase (βGal) fusion proteins were loaded on a 13% SDS-polyacrylamide gel and visualized by Coomassie Blue staining after electrophoresis. D, shown are the results from analysis of intracellular fluorescence after a 15-h incubation with each ZEBRA-EGFP fusion protein (0.2 μm) in living HeLa cells by fluorescence microscopy. E, shown are the results from flow cytometric analysis of live HeLa and Saos2 cells after exposure to 0.2 μm ZEBRA-EGFP fusion proteins. Cells were treated with trypsin prior to flow fluorescence-activated cell sorter analysis. The mean ± S.D. of the cell fluorescence of three independent experiments is reported. F, shown is the uptake of MD-EGFP in C2C12 and Saos2 cells. 0.2 μm MD-EGFP was added to the serum-free culture medium. Intracellular fluorescence was visualized by fluorescence microscopy. G, shown is the amino acid sequence of the MD of the ZEBRA protein. Basic amino acids (seven lysines and seven arginines) are shown in red, whereas hydrophobic amino acids (one valine, five alanines, and seven leucines) are presented in blue. The presence of both the DBD and the DIM domain is absolutely required for the translocation property of ZEBRA-MD. BF, bright field.
To use ZEBRA as a protein carrier, we first evaluated the transduction properties of full-length ZEBRA and several different truncations fused to EGFP, including Z2-EGFP, Z3-EGFP, Z4-EGFP, Z5-EGFP, Z6-EGFP, Z8-EGFP, and Z9-EGFP. The fusion proteins were added in the culture medium of cervical cancer (HeLa) or osteosarcoma (Saos2) cells and incubated for 24 h, and cell fluorescence was monitored either by flow cytometry or by fluorescence microscopy of live unfixed cells (Fig. 1, D and E). We found that only the full-length ZEBRA-EGFP, Z4-EGFP, Z6-EGFP, and Z9-EGFP proteins were efficiently transduced into HeLa cells (Fig. 1D). No fluorescence signal was observed in association with the EGFP-fused truncations containing only the N-terminal part (Z3-EGFP) of the wild-type protein, an internal deletion in the zipper domain (Z5-EGFP), or a deletion of the basic domain (Z8-EGFP) (Fig. 1D). Transduction efficiency was further evaluated in HeLa and Saos2 cell lines by flow cytometry after a 15-h incubation with each protein at 0.2 μm. The flow cytometric analyses confirmed the efficient uptake of Z4-EGFP, Z6-EGFP, and Z9-EGFP in the two cell lines (Fig. 1E). In addition, internalization of Z9-EGFP (MD-EGFP) in mouse myoblasts (C2C12) and Saos2 cells (Fig. 1F) was demonstrated by fluorescence microscopy.
From these results, we concluded that the presence of both the DBD and the DIM domain is required and sufficient for internalization. The smallest tested truncated protein containing both domains was Z9 (MD), which facilitated the delivery of EGFP with an almost 100% efficiency (number of fluorescent cells over the total cell number) in various mammalian cell lines. Therefore, all translocation experiments described below were performed using the Z9 peptide, which is denoted below as the MD. This MD of ZEBRA is composed of 54 amino acid residues and combines two important domains of the ZEBRA protein: the DBD and the DIM domain, which together form the bZIP region (Fig. 1G). Additionally to 14 positively charged residues such as lysine and arginine, the MD provides 13 hydrophobic amino acid (leucine, alanine, and valine), which are located mainly in the DIM domain (Fig. 1G).
DNA Binding Activity
As ZEBRA is a transcription factor that binds DNA through its central basic region (DBD, residues 175–195), we investigated whether the DNA binding activity is preserved in different ZEBRA truncations. It was shown previously that ZEBRA recognizes the AP-1 consensus heptamer TGA(G/C)TCA (31). We used this heptamer as a probe to evaluate the DNA binding activity with EMSAs (Fig. 2). The truncations containing both the DBD and the DIM domain, Z4 (Fig. 2, seventh and eighth lanes) and Z6 (tenth, eleventh, and seventeenth lanes), bound the AP-1 probe with nearly the same efficiency as the full-length ZEBRA protein. In contrast, truncations carrying deletions in the DBD (Z8, thirteenth lane), the DIM domain (Z5, ninth lane; and Z7, twelfth lane), or both domains (Z2, fifth and sixth lanes) failed to bind the AP-1 probe. In agreement with the work of Petosa et al. (31), these data indicate that the presence of the DBD and the DIM domain is required and sufficient for DNA binding. However, residual DNA-protein complex formation was also detected for the Z1 truncation, which contained only the DBD but no DIM domain (Fig. 2).
FIGURE 2.
DNA binding activity. Shown are the results from EMSA of recombinant ZEBRA variants. Truncated ZEBRA proteins and their respective fusion proteins were analyzed for their ability to bind the AP-1 DNA probe. Signals were detected by a chemiluminescent assay (Pierce). ZEBRA-FL and Z-FL, full-length ZEBRA; βGal, β-galactosidase.
Because the truncated forms Z6 and Z9 (MD) were shown to transduce into cells, we further investigated the DNA binding activity of both peptides when fused N-terminally to EGFP or β-galactosidase reporter proteins. All fusion proteins failed to recognize the AP-1 probe as demonstrated by EMSA (Fig. 2). In contrast, fusion of EGFP to full-length ZEBRA or Z4 (data not shown) had no effect on the DNA binding activity (Fig. 2). These results suggested that conformational changes occurred after fusion of the reporter proteins with Z6 and MD, compromising their ability to bind DNA.
Kinetics of MD-EGFP Internalization and Cytotoxicity
The translocation of MD (Z9)-EGFP was monitored by the measurement of fluorescence in live cells by flow cytometric analysis. The addition of a low concentration of MD (Z9)-EGFP (0.2 μm) to the serum-free culture medium of HeLa or Saos2 cells resulted in a rapid intracellular accumulation of the fusion protein (Fig. 3A). This cellular uptake was detected after removal of cell surface-bound protein by extensive trypsin/heparin washes and remained stable for at least 24 h in the cells (Fig. 3A and supplemental Fig. 1). The increased fluorescence intensity in Saos2 cells after transduction is most likely due to their larger size compared with the smaller HeLa cells and is not caused by better translocation efficiency in the former.
FIGURE 3.
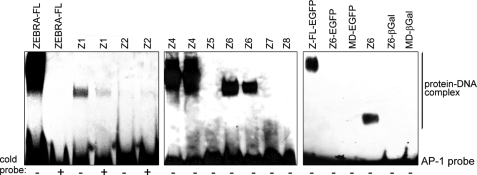
Kinetics of MD-EGFP internalization and cytotoxicity. A, time-dependent intracellular accumulation of MD-EGFP in HeLa and Saos2 cells. Both cell lines were incubated with 0.2 μm MD-EGFP at 37 °C for the times indicated and subsequently treated with trypsin for 10 min at 37 °C. The mean cell fluorescence was analyzed by flow cytometry. B, dose-dependent uptake of MD-EGFP in live HeLa and Saos2 cells. The indicated concentrations of MD-EGFP were added to the serum-free cell culture medium. After 4 h, cells were washed with PBS and trypsinized for 10 min at 37 °C, and cell fluorescence was measured by flow cytometry. All transduction experiments were performed in triplicates in two independent analyses, and the mean ± S.D. is indicated. C, MD-mediated protein delivery causes no cytotoxicity. HeLa and Saos2 cells were exposed to the indicated concentrations of MD-EGFP and MD-β-galactosidase (βGal) and incubated in growth medium at 37 °C for 24 h. Cytotoxicity after the uptake of both fusion proteins was determined by the leakage of lactate dehydrogenase into the culture medium. Each bar represents the mean ± S.D. of the viability of two independent experiments performed in triplicates. a.u., arbitrary units.
We also investigated the dose-dependent internalization of MD (Z9)-EGFP in these cell lines. Cells were incubated for 4 h with different concentrations of the fusion protein ranging from 10 to 200 nm. After extensive washing and trypsinization, transduction efficiency was analyzed by flow cytometry. Transduced EGFP was already detected in HeLa and Saos2 cells after incubation with low amounts (10 and 20 nm) of MD (Z9)-EGFP (Fig. 3B). About 50% of the cells were transduced in the presence of 100 nm MD (Z9)-EGFP (Fig. 3B). Incubation of cells with a higher concentration (200 nm) of MD (Z9)-EGFP resulted in a 100% transduced cell population (supplemental Movies 1 and 2). The fluorescence of internalized EGFP increased linearly with the concentration of MD (Z9)-EGFP in the culture medium and reached saturation at 200 nm (data not shown).
The toxicity of both MD (Z9)-EGFP and MD (Z9)-β-galactosidase fusion proteins was tested with a lactate dehydrogenase-based assay. The cytosolic enzyme lactate dehydrogenase could be detected in the cell culture medium after disruption of the cell membranes. Saos2 and HeLa cells were incubated with different concentrations of the fusion protein ranging from 0.1 to 3 μm. 24 h after the addition of the MD fusion proteins, no variation in cell viability was observed as shown by the absence of extracellular lactate dehydrogenase activity (Fig. 3C).
Mechanisms of Uptake
Several studies have demonstrated that cell-surface HSPGs play a key role in the cellular internalization of CPPs (14, 35). Mutational analysis of cation-rich sequences demonstrated that the cellular uptake properties of Tat depends mostly on the presence of positively charged residues that facilitate binding to negatively charged HSPGs at the cell surface (36, 37). To evaluate the role of HSPGs in the uptake of MD-EGFP, HeLa and Saos2 cells were incubated for 30 min with 20 μg/ml heparin prior to the addition of the fusion protein. Heparin is a structural homolog of HSPGs and may compete for binding of the latter to MD-EGFP. The uptake of MD-EGFP was significantly inhibited by the presence of heparin in the culture medium compared with the control condition without heparin (Fig. 4A). These data indicated that cellular internalization of MD-EGFP required interactions between negatively charged HSPGs and the basic amino acids in the sequence of MD (Z9) (Fig. 1G).
FIGURE 4.
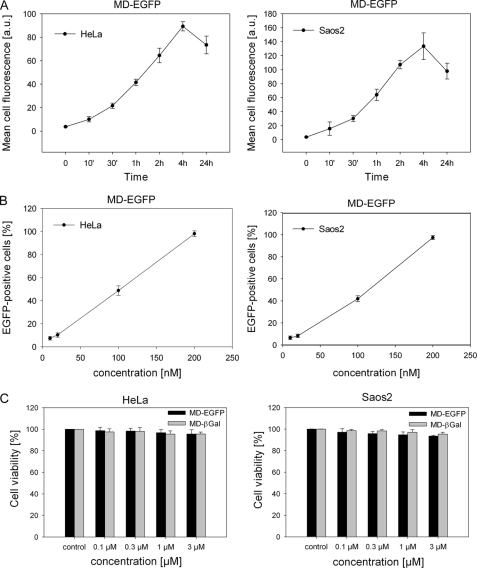
Mechanisms of uptake. A, HeLa and Saos2 cells were incubated with 20 μg/ml heparin for 30 min at 37 °C and afterward exposed to 0.2 μm MD-EGFP for 3 h at 37 °C. Cells were trypsinized for 10 min at 37 °C and washed prior to flow cytometric analysis. B, shown is the effect of low temperature and cellular ATP depletion on the internalization of MD-EGFP. Both cell lines were incubated with 0.2 μm MD-EGFP for 1 h at 4 °C. To deplete the cellular ATP pool, HeLa and Saos2 cells were incubated for 1 h with 6 mm 2-deoxy-d-glucose and 10 mm sodium azide and then exposed to 0.2 μm MD-EGFP for 1 h. After trypsinization, cells were analyzed by flow cytometry. C, shown is the effect of endocytotic inhibitors on MD-EGFP transduction. Both cell lines were treated with 30 μm chlorpromazine, 100 nm wortmannin, 50 μg/ml nystatin, or 5–10 mm methyl-β-cyclodextrin (MBCD) 30 min prior to the addition of 0.2 μm MD-EGFP. In general, the cellular uptake of pretreated cells was measured as mean fluorescence and normalized to the mean cell fluorescence of the untreated control. Each experiment was performed three times in triplicates, and the mean ± S.D. is indicated.
Recent studies on the uptake mechanisms of Tat and VP22 reported a significant contribution of endocytotic pathways to their cellular internalization (12, 15, 22, 38). Therefore, we first investigated the effect of low temperature and ATP depletion on the cellular uptake of MD-EGFP. As shown in Fig. 4B, the intracellular EGFP signal for HeLa and Saos2 cells was strongly reduced after incubation at 4 °C. The cellular ATP pool was depleted by treatment with sodium azide and 2-deoxy-d-glucose. We observed only a 20–30% decrease in cell fluorescence in both cell lines (Fig. 4B), indicating that the uptake of MD-EGFP is mostly ATP-independent.
To clarify whether MD-EGFP internalization involves endocytosis, we explored the effect of several drugs that specifically inhibit caveolin-, clathrin-, or lipid raft-dependent endocytosis. Nystatin is a known inhibitor of caveolin-dependent endocytosis (12). The internalization by a caveolar process is slow and occurs in general upon cell stimulation (39). HeLa and Saos2 cells were treated with 50 μg/ml nystatin prior to the addition of 0.2 μm MD-EGFP, and internalization was analyzed by flow cytometry. In both cell lines, the intracellular fluorescence signal for MD-EGFP in the presence of nystatin was identical to that under the control conditions (Fig. 4C). Thus, caveolin-dependent endocytosis is not involved in the uptake of MD-EGFP. Macropinocytosis is a rapid and nonspecific mechanism of internalization and has been described to be responsible for the cellular uptake of some CPPs (15). Macropinocytosis depends on the activity of phosphatidylinositol 3-kinase and is inhibited by wortmannin (40). We examined the impact of wortmannin on the internalization of MD-EGFP in HeLa and Saos2 cells. Pretreatment of both cell lines with 100 nm wortmannin did not alter the cellular uptake of MD-EGFP compared with untreated cells (Fig. 4C). Thus, MD-mediated protein translocation does not occur via macropinocytosis. To test whether MD-EGFP uptake involves clathrin-coated pit-mediated endocytosis, MD-EGFP internalization was measured in the presence of chlorpromazine. Saos2 and HeLa cells were incubated with 30 μm chlorpromazine. After a 30-min incubation with chlorpromazine, the MD-EGFP fusion protein was added. Interestingly, EGFP fluorescence was significantly reduced in Saos2 cells but not in HeLa cells (Fig. 4C). These results indicate that the internalization process of MD-EGFP may differ depending on the cell type. Finally, we investigated the lipid raft-mediated endocytotic pathway, which has been also reported to be implicated in the uptake of CPPs (15, 22). Cells were treated with methyl-β-cyclodextrin to deplete cell surface-associated cholesterol, resulting in the disruption of lipid rafts. As shown in Fig. 4C, an impaired uptake of MD-EGFP in both cell lines was observed. These data suggest that lipid raft-mediated endocytosis contributes to the uptake of MD-EGFP. However, 60% of MD-EGFP occurred through an alternative route of internalization. This observation is supported by the entry of MD-EGFP into synthetic liposomes (data not shown). The lipophilic dye Nile red (10 μm) was added to the synthetic liposomes preparation for fluorescence labeling. Immediately after the labeling reaction, liposomes were incubated for 30 min with MD-EGFP and analyzed by confocal microscopy. GFP fluorescence was detected around and inside the lipid vesicles, suggesting a direct translocation across the lipid layer (data not shown).
Intracellular Localization of MD-EGFP
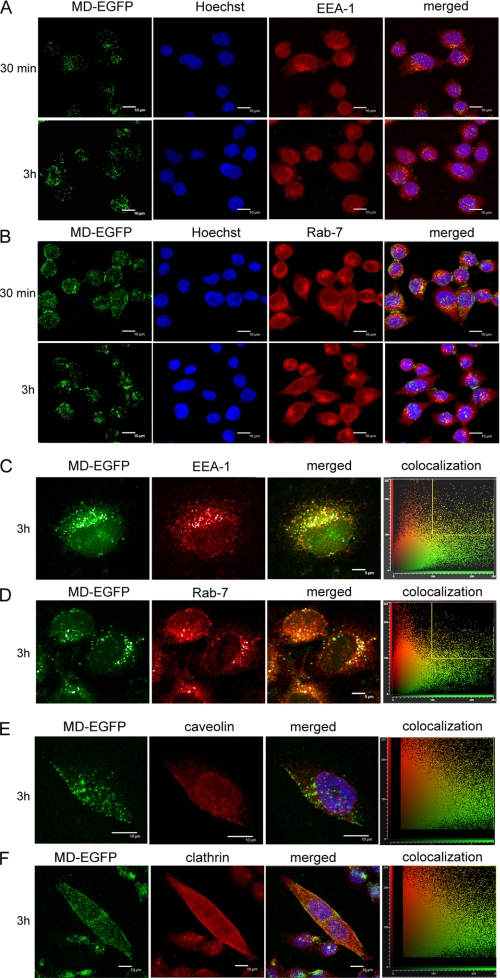
In the case of an endosome-dependent pathway of MD uptake, co-localization with early and late endosomes would be expected. We performed immunofluorescence microscopy to study the subcellular co-localization of internalized MD-EGFP with endosomal marker proteins such as EEA1 (early endosomal marker), Rab7 (endosomal marker) (Fig. 5, A–D; and supplemental Fig. 2), caveolin-1 (caveosome marker), and clathrin (marker for clathrin-coated pits) (Fig. 5, E and F) (41). HeLa cells were incubated for 30 min to 15 h with MD-EGFP at 37 °C, and protein internalization was analyzed by confocal microscopy. The uptake of MD-EGFP was confirmed by direct visualization of the intracellular fluorescence of EGFP or by antibody staining against EGFP.
FIGURE 5.
Intracellular localization of MD-EGFP in HeLa cells. HeLa cells were exposed to 0.2 μm MD-EGFP for 30 min to 3 h at 37 °C. Surface-bound proteins were removed by several heparin washes. Cells were then fixed, immunostained for the endosomal marker proteins EEA1 (A and C) and Rab7 (B and D), and detected with Alexa Fluor® 647-labeled secondary antibody, and nuclei were counterstained with Hoechst 33258. Cells were visualized by confocal microscopy (TCS-SP2). Images were acquired sequentially as described under “Experimental Procedures,” with MD-EGFP in green, EEA1 and Rab7 in red, and DNA in blue (A and B). EEA1 and Rab7 fluorescence is visualized in red and MD-EGFP in green (C and D). An estimation of co-localization was determined with the CF2D software from Leica. The diagrams represent the relative intensity of the pixels in the green and red images. Dots gated in the yellow square, corresponding to co-localization, are represented in white in the images. E and F, HeLa cells were incubated for 3 h with 0.2 μm MD-EGFP at 37 °C. Cells were washed with 20 μg/ml heparin in PBS, fixed, and incubated with anti-caveolin-1 (E) or anti-clathrin (F) antibody and the corresponding Alexa Fluor® 647-labeled anti-rabbit secondary antibody. Fluorescence was analyzed sequentially by confocal microscopy as described under “Experimental Procedures.” MD-EGFP signals are shown in green, the nucleus in blue, and clathrin and caveolin-1 markers in red.
The majority of the EGFP signals did not co-localize with EEA1 or Rab7 signals (Fig. 5, A–D). However, a portion of the EGFP signals overlapped with endosomal markers. Variation of the incubation times did change this only slightly. At earlier time points (30 min) after the addition of MD-EGFP to the cells, we observed minor co-localization of the internalized protein with the endosomal marker EEA1 and at later time points (3 h) with Rab7. Furthermore, MD-EGFP did not co-localize with caveolin or clathrin signals at any analyzed time points (Fig. 5, E and F).
Cell entry of MD-EGFP was characterized by live cell imaging in HeLa cells. The ZEBRA-EGFP fusion protein (0.3 μm) was added to the cells and directly visualized by fluorescence microscopy for 1 h (supplemental Fig. 3). A fast accumulation at the cell membrane level was observed within the first 15 min, followed by a rapid trafficking of EGFP signals inside the cell. Furthermore, internalized proteins were detected inside the cell after immunofluorescence analysis by examining different Z-sections of one cell (supplemental Fig. 3).
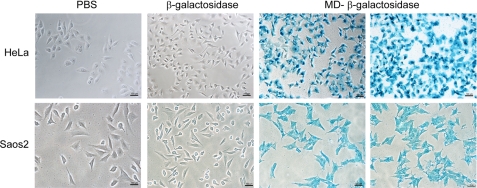
Delivery of β-Galactosidase into Cells
To use MD as a protein carrier, we tested the intracellular functionality of MD-delivered proteins using the 120-kDa enzyme β-galactosidase fused to MD as a model. The fusion protein was added to the serum-free culture medium of Saos2 or HeLa cells. Cells were fixed and stained according to the X-gal staining kit (Sigma). Fig. 6 shows the successful delivery of functional β-galactosidase into HeLa and Saos2 cells as proven by the blue cell staining. Analogous to the delivery of EGFP by ZEBRA-MD, we observed a 100% transduced cell population using MD-β-galactosidase. The reporter protein β-galactosidase alone was used as a negative control, and no β-galactosidase activity was detected in these control cells (Fig. 6). These results confirmed that β-galactosidase-positive staining did not occur through a fixation artifact.
FIGURE 6.
MD-mediated delivery of β-galactosidase into HeLa and Saos2 cells. After a 16-h incubation with 0.2 μm MD-β-galactosidase at 37 °C, cells were washed with 20 μg/ml heparin, fixed, and stained according to the β-galactosidase reporter gene staining kit (Sigma). β-Galactosidase staining was visualized by phase-contrast microscopy.
DISCUSSION
With the prospect of developing ZEBRA as a protein delivery tool, it was of interest to identify the minimal transduction domain required for internalization. For this reason, we engineered numerous protein truncations and characterized their transduction capacities when fused to the reporter protein EGFP. We identified three truncations of ZEBRA, namely Z4, Z6, and Z9 (MD), that had the ability to transfer EGFP into live cells. All truncations with protein translocation ability contained the basic positively charged DBD as well as the hydrophobic leucine-rich DIM domain of ZEBRA (26, 31), whereas truncated proteins missing either of the two domains failed to translocate EGFP into cells. The 48-amino acid peptide Z9 was the smallest ZEBRA truncation with transduction abilities. It was therefore denoted the MD and further characterized regarding its uptake mechanism, functionality of delivered proteins, and cell toxicity.
The uptake of MD-EGFP was temperature-dependent, presumably due to the fluidity loss of the lipid bilayer at low temperatures (18). Because the uptake of MD-EGFP into ATP-depleted cells decreased by only 20%, we concluded that the MD fusion protein is internalized by a largely ATP-independent process. Furthermore, several endocytotic inhibitors such as nystatin, wortmannin, and chlorpromazine did not interfere with MD-EGFP uptake, indicating no participation of receptor-mediated endocytosis (12, 15, 22, 38, 41). This finding was further supported by the lack of co-localization of MD-EGFP with caveolin-1 and clathrin. Furthermore, neither the early endosomal marker EEA1 nor the late endosomal marker Rab7, involved in the transport of cargo from early to late endosomes, showed a complete co-localization in microscopic images. Only incubation with methyl-β-cyclodextrin, a drug that inhibits lipid raft-mediated endocytosis (22), caused a 40% decreased uptake of MD-EGFP. Thus, the endocytotic pathway may have contributed in part to the cellular uptake of MD-EGFP under the applied conditions but did not account for the majority of the internalized fusion protein. In addition, uptake of MD-EGFP required the availability of cell-surface HSPGs because its internalization was clearly impaired in the presence of heparin. Similar to other CPPs that are taken up in a HSPG-dependent manner, the cell-surface binding of MD appears to be facilitated by the interaction of the highly positively charged DBD and the negatively charged HSPGs (14, 42, 43). However, as shown by the failure of the DBD-containing Z5 truncation to enable protein uptake, the presence of a positively charged domain alone is not sufficient for protein internalization. Only when the DBD was present together with the hydrophobic DIM domain was efficient protein uptake observed. It was established previously that the DBD and the DIM domain together form a continuous stretch of an α-helix (31), which is similar to other amphipathic CPPs, including MAP and transportan (43, 44). For those peptides, an increase in membrane permeability that correlates with the presence of amphiphilic α-helical structures has been observed (45). Accordingly, internalization of MD may require the α-helix formed by the DBD and the DIM domain. Furthermore, the DBD might mediate surface binding of the MD while the DIM domain subsequently facilitates translocation through the lipid bilayer by hydrophobic interactions in a similar way as reported for Penetratin (46). This CPP is internalized via a non-endocytotic and receptor- and transporter-independent pathway. Penetratin requires the hydrophobic tryptophan residues and crosses lipid bilayers without pore formation (47, 48). In line with these arguments, also the MD-mediated protein translocation did not cause disruption or pore formation in the plasma membrane as shown by the absence of both 7-aminoactinomycin D uptake and leakage of lactate dehydrogenase in transduced cells. Finally, we observed a direct translocation of MD-EGFP into synthetic liposomes (data not shown). Taken together, the energy-independent uptake of MD-EGFP, the decreased uptake of MD-EGFP into cells at low temperatures, the lack of exclusive co-localization with endosomal markers, and the incomplete inhibition of MD-EGFP uptake by endocytotic inhibitors suggest that MD-EGFP is translocated, at least in part, directly across the lipid bilayer. This energy-independent internalization property of ZEBRA-MD-EGFP is highly favorable because endosomal trapping can be circumvented, which seems to be the major bottleneck for most CPP, as, for example, TAT, which enters cell by macropinocytosis (15) and fails to enter into liposomes (43, 49). Further study to determine the amino acids from the MD responsible for the translocation properties will permit a better understanding of the molecular mechanism of ZEBRA cellular uptake.
In addition, we tested the ability of the truncated ZEBRA proteins to bind DNA. As we expected, constructs encompassing both the basic DBD and the DIM domain recognized and bound the AP-1 site in vitro. Interestingly, this specific binding was lost when these proteins were fused at the C terminus with either EGFP or β-galactosidase. As was shown in the crystal structure of ZEBRA bound to the AP-1 promoter element, DNA binding of the protein requires complex conformation of its C-terminal part, and mutations in the latter were suggested to inactivate the transcription factor activity of ZEBRA (31). Thus, the C-terminal fusion of proteins to truncated forms of ZEBRA may have compromised its ability to form DNA-protein complexes. The loss of its DNA binding activity when fused to proteins is certainly a tremendous advantage for the development of a new potential protein delivery system. However, a possible interaction between ZEBRA-MD and cellular proteins after its cellular uptake cannot be completely ruled out because it was not examined in this study. As the full-length ZEBRA protein can induce growth arrest independently of its ability to bind DNA in host cells through direct interplay with cellular proteins and its basic region (27, 28), possible interaction with cellular proteins and control of their activities need to be further analyzed.
In addition to the previously reported lymphocyte-specific uptake of ZEBRA (25), we observed a rapid internalization of MD-EGFP into a wide range of different cells. MD-delivered proteins remained functional when transduced into cells, as demonstrated by the enzymatic activity of internalized MD-β-galactosidase. In conclusion, the MD is a new carrier suitable for the efficient transport of proteins into live mammalian cells. Because its uptake is largely independent of known endocytotic pathways, ZEBRA-MD can carry proteins into cells without being trapped in endosomal compartments. Additionally, it does not show any toxic side effects and transduces a wide range of mammalian cells with 100% efficiency. Taken all together, these favorable biochemical properties of ZEBRA-MD indicate that ZEBRA represents a novel and powerful delivery system for therapeutic proteins.
Supplementary Material
Acknowledgments
We thank Kathrin Lang (Institute of Genetic, TU Dresden, Dresden, Germany) for providing the plasmid with the coding sequence of the β-galactosidase gene, Madiha Derouazi and Thomas Walther for intensive discussion and all helpful comments concerning the manuscript, and Barry Stidder for proofreading the manuscript.
This work was supported by Marie Curie Excellence Grant 014320.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Movies 1 and 2.
- CPP
- cell-penetrating peptide
- HSPG
- heparan sulfate proteoglycan
- bZIP
- basic leucine zipper
- MD
- minimal domain
- EGFP
- enhanced green fluorescence protein
- PBS
- phosphate-buffered saline
- EMSA
- electrophoretic mobility shift assay
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- DBD
- DNA-binding domain
- DIM
- dimerization.
REFERENCES
- 1.Murriel C. L., Dowdy S. F. (2006) Expert Opin. Drug Deliv. 3, 739–746 [DOI] [PubMed] [Google Scholar]
- 2.Endoh T., Sisido M., Ohtsuki T. (2008) Bioconjug. Chem. 19, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 3.Abes S., Turner J. J., Ivanova G. D., Owen D., Williams D., Arzumanov A., Clair P., Gait M. J., Lebleu B. (2007) Nucleic Acids Res. 35, 4495–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harada H., Hiraoka M., Kizaka-Kondoh S. (2002) Cancer Res. 62, 2013–2018 [PubMed] [Google Scholar]
- 5.Kilic U., Kilic E., Dietz G. P., Bähr M. (2003) Stroke 34, 1304–1310 [DOI] [PubMed] [Google Scholar]
- 6.Fawell S., Seery J., Daikh Y., Moore C., Chen L. L., Pepinsky B., Barsoum J. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vivès E., Brodin P., Lebleu B. (1997) J. Biol. Chem. 272, 16010–16017 [DOI] [PubMed] [Google Scholar]
- 8.Derossi D., Joliot A. H., Chassaing G., Prochiantz A. (1994) J. Biol. Chem. 269, 10444–10450 [PubMed] [Google Scholar]
- 9.Elliott G., O'Hare P. (1997) Cell 88, 223–233 [DOI] [PubMed] [Google Scholar]
- 10.Nishi K., Saigo K. (2007) J. Biol. Chem. 282, 27503–27517 [DOI] [PubMed] [Google Scholar]
- 11.Langel Ü. (ed) (2007) Handbook of Cell-penetrating Peptides, 2nd Ed., Taylor and Francis Group, Boca Raton, FL [Google Scholar]
- 12.Richard J. P., Melikov K., Brooks H., Prevot P., Lebleu B., Chernomordik L. V. (2005) J. Biol. Chem. 280, 15300–15306 [DOI] [PubMed] [Google Scholar]
- 13.Lundberg P., Langel U. (2003) J. Mol. Recognit. 16, 227–233 [DOI] [PubMed] [Google Scholar]
- 14.Ziegler A., Seelig J. (2004) Biophys. J. 86, 254–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan I. M., Wadia J. S., Dowdy S. F. (2005) J. Control Release 102, 247–253 [DOI] [PubMed] [Google Scholar]
- 16.Ferrari A., Pellegrini V., Arcangeli C., Fittipaldi A., Giacca M., Beltram F. (2003) Mol. Ther. 8, 284–294 [DOI] [PubMed] [Google Scholar]
- 17.Fittipaldi A., Ferrari A., Zoppé M., Arcangeli C., Pellegrini V., Beltram F., Giacca M. (2003) J. Biol. Chem. 278, 34141–34149 [DOI] [PubMed] [Google Scholar]
- 18.Veach R. A., Liu D., Yao S., Chen Y., Liu X. Y., Downs S., Hawiger J. (2004) J. Biol. Chem. 279, 11425–11431 [DOI] [PubMed] [Google Scholar]
- 19.Oliveira S., Høgset A., Storm G., Schiffelers R. M. (2008) Curr. Pharm. Des. 14, 3686–3697 [DOI] [PubMed] [Google Scholar]
- 20.Oliveira S., van Rooy I., Kranenburg O., Storm G., Schiffelers R. M. (2007) Int. J. Pharm. 331, 211–214 [DOI] [PubMed] [Google Scholar]
- 21.Shiraishi T., Nielsen P. E. (2006) Nat. Protoc. 1, 633–636 [DOI] [PubMed] [Google Scholar]
- 22.Wadia J. S., Stan R. V., Dowdy S. F. (2004) Nat. Med. 10, 310–315 [DOI] [PubMed] [Google Scholar]
- 23.Miller G., El-Guindy A., Countryman J., Ye J., Gradoville L. (2007) Adv. Cancer Res. 97, 81–109 [DOI] [PubMed] [Google Scholar]
- 24.Sinclair A. J. (2003) J. Gen. Virol. 84, 1941–1949 [DOI] [PubMed] [Google Scholar]
- 25.Mahot S., Fender P., Vivès R. R., Caron C., Perrissin M., Gruffat H., Sergeant A., Drouet E. (2005) Virus Res. 110, 187–193 [DOI] [PubMed] [Google Scholar]
- 26.Sinclair A. J. (2006) Trends Microbiol. 14, 289–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez A., Armstrong M., Dwyer D., Flemington E. (1999) J. Virol. 73, 9029–9038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez A., Jung E. J., Yin Q., Cayrol C., Flemington E. K. (2001) Virology 284, 159–169 [DOI] [PubMed] [Google Scholar]
- 29.Cayrol C., Flemington E. (1996) J. Biol. Chem. 271, 31799–31802 [DOI] [PubMed] [Google Scholar]
- 30.Cayrol C., Flemington E. K. (1996) EMBO J. 15, 2748–2759 [PMC free article] [PubMed] [Google Scholar]
- 31.Petosa C., Morand P., Baudin F., Moulin M., Artero J. B., Müller C. W. (2006) Mol. Cell 21, 565–572 [DOI] [PubMed] [Google Scholar]
- 32.Rothe R., Lenormand J. L. (2008) Curr. Protoc. Protein Sci. Chapter 18, Unit 18, 11 [DOI] [PubMed] [Google Scholar]
- 33.Liguori L., Marques B., Villegas-Mendez A., Rothe R., Lenormand J. L. (2008) J. Control Release 126, 217–227 [DOI] [PubMed] [Google Scholar]
- 34.Schmid I., Krall W. J., Uittenbogaart C. H., Braun J., Giorgi J. V. (1992) Cytometry 13, 204–208 [DOI] [PubMed] [Google Scholar]
- 35.Tyagi M., Rusnati M., Presta M., Giacca M. (2001) J. Biol. Chem. 276, 3254–3261 [DOI] [PubMed] [Google Scholar]
- 36.Ho A., Schwarze S. R., Mermelstein S. J., Waksman G., Dowdy S. F. (2001) Cancer Res. 61, 474–477 [PubMed] [Google Scholar]
- 37.Mukai Y., Sugita T., Yamato T., Yamanada N., Shibata H., Imai S., Abe Y., Nagano K., Nomura T., Tsutsumi Y., Kamada H., Nakagawa S., Tsunoda S. (2006) Biol. Pharm. Bull. 29, 1570–1574 [DOI] [PubMed] [Google Scholar]
- 38.Fuchs S. M., Raines R. T. (2004) Biochemistry 43, 2438–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson R. G. (1998) Annu. Rev. Biochem. 67, 199–225 [DOI] [PubMed] [Google Scholar]
- 40.Araki N., Johnson M. T., Swanson J. A. (1996) J. Cell Biol. 135, 1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watson P., Jones A. T., Stephens D. J. (2005) Adv. Drug Deliv. Rev. 57, 43–61 [DOI] [PubMed] [Google Scholar]
- 42.Fuchs S. M., Raines R. T. (2006) Cell. Mol. Life Sci. 63, 1819–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zorko M., Langel U. (2005) Adv. Drug Deliv. Rev. 57, 529–545 [DOI] [PubMed] [Google Scholar]
- 44.Lindgren M., Gallet X., Soomets U., Hällbrink M., Bråkenhielm E., Pooga M., Brasseur R., Langel U. (2000) Bioconjug. Chem. 11, 619–626 [DOI] [PubMed] [Google Scholar]
- 45.Dietz G. P., Bähr M. (2004) Mol. Cell. Neurosci. 27, 85–131 [DOI] [PubMed] [Google Scholar]
- 46.Joliot A., Prochiantz A. (2008) Adv. Drug Deliv. Rev. 60, 608–613 [DOI] [PubMed] [Google Scholar]
- 47.Thorén P. E., Persson D., Karlsson M., Nordén B. (2000) FEBS Lett. 482, 265–268 [DOI] [PubMed] [Google Scholar]
- 48.Terrone D., Sang S. L., Roudaia L., Silvius J. R. (2003) Biochemistry 42, 13787–13799 [DOI] [PubMed] [Google Scholar]
- 49.Krämer S. D., Wunderli-Allenspach H. (2003) Biochim. Biophys. Acta 1609, 161–169 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.