Abstract
Fibrillin microfibrils are polymeric structures present in connective tissues. The importance of fibrillin microfibrils to connective tissue function has been demonstrated by the multiple genetic disorders caused by mutations in fibrillins and in microfibril-associated molecules. However, knowledge of microfibril structure is limited, largely due to their insolubility. Most previous studies have focused on how fibrillin-1 is organized within microfibril polymers. In this study, an immunochemical approach was used to circumvent the insolubility of microfibrils to determine the role of fibrillin-2 in postnatal microfibril structure. Results obtained from studies of wild type and fibrillin-1 null tissues, using monoclonal and polyclonal antibodies with defined epitopes, demonstrated that N-terminal fibrillin-2 epitopes are masked in postnatal microfibrils and can be revealed by enzymatic digestion or by genetic ablation of Fbn1. From these studies, we conclude that fetal fibrillin polymers form an inner core within postnatal microfibrils and that microfibril structure evolves as growth and development proceed into the postnatal period. Furthermore, documentation of a novel cryptic site present in EGF4 in fibrillin-1 underscores the molecular complexity and tissue-specific differences in microfibril structure.
Keywords: Connective Tissue, Extracellular Matrix, Fibrillin, Mouse Genetics, Protein Structure, Congenital Contractural Arachnodactyly, Marfan Syndrome, Microfibril
Introduction
Fibrillins are ubiquitous extracellular matrix molecules that assemble into microfibrils (1–3). Fibrillin-1, detected at gastrulation in the developing avian embryo (4), is present throughout development and postnatal life. In contrast, expression of Fbn2 and Fbn3 is largely limited to fetal development (2, 3, 5). In the mouse, there are only two Fbn genes, Fbn1 and Fbn2, because Fbn3 was inactivated during chromosomal evolution (3). Gene targeting experiments have shown that mice without Fbn2 are viable and fertile but are born with syndactyly and joint contractures (6), whereas mice without Fbn1 die in the early postnatal period (∼P14) from aortic rupture (7). Thus, it is thought that fibrillin-1 can compensate for the most part for the absence of fibrillin-2 and that fibrillin-2 can compensate for fibrillin-1 during development but not after P14, when expression levels of Fbn2 are very low.
Fibrillin-containing microfibrils are polymeric structures that have been best defined by their ultrastructural appearance, described as the small diameter nonstriated fibrils associated with elastic fibers or with basement membranes (8) and as beaded strings, after liberation from tissues and rotary shadowing electron microscopy (9). Most structural studies of fibrillin-containing microfibrils have focused on the contributions of fibrillin-1 (10–14), leaving the role of fibrillin-2 largely undetermined. However, some progress has been made using an immunochemical approach. Antibodies specific for fibrillin-1 and -2 showed that fetal microfibrils are heteropolymeric, containing both fibrillin-1 and -2 in tissues and in the same beaded strings (15). Because little is known about fibrillin-2 in microfibrils, it is possible that the structure of microfibrils may be more complex than the models based on fibrillin-1 as the major structural component.
Two conclusions can be drawn from published results. First, during the assembly of heteropolymeric microfibrils, regions of fibrillin-2 are masked. This conclusion is based on findings from epitope-mapped monoclonal antibodies (mAbs)2 specific for human fibrillin-2. These mAbs previously showed that the first 8-cysteine domain in fibrillin-2 is exposed and available for antibody binding within microfibril polymers. However, a domain adjacent to the first 8-cysteine domain and domains flanking the second hybrid domain in fibrillin-2 were found to be cryptic in polymerized microfibrils (15).
Second, functional roles for fibrillin-2 in postnatal tissues are not well understood. Both expression of Fbn2 (5) and immunostaining of fibrillin-2 protein (15) are minimal in postnatal tissues, suggesting that fibrillin-2 may not perform important functions after the early postnatal period. However, genetic evidence in humans and in mice indicates that mutations in fibrillin-2 affect postnatal skeletal growth. Mutations in FBN2 cause congenital contractural arachnodactyly in humans (OMIM 121050), and Fbn2 null mice demonstrate bone and tendon defects (17).
To better determine the contribution of fibrillin-2 to microfibril structure and to provide a better basis for elucidating mechanisms by which mutant fibrillin-2 contributes to skeletal abnormalities, we have performed additional immunochemical investigations of fibrillin-2 in postnatal microfibrils. Using this approach, we report results that demonstrate that fibrillin-2 is present, but masked, in postnatal tissues. Moreover, results obtained using Fbn1 null mice suggest a new model of microfibril structure in which fibrillin-2 forms a core within the microfibril, a core that is overlaid at least by fibrillin-1. In addition, we show that masked epitopes in fibrillin-1 and in fibrillin-2 are differentially masked or available, depending on the type of tissue. These are the first results that demonstrate tissue-specific differences in microfibril structure. From these data, new concepts of microfibril structure have emerged.
EXPERIMENTAL PROCEDURES
Antibodies, Recombinant Fibrillin Polypeptides, and Microfibril Preparations
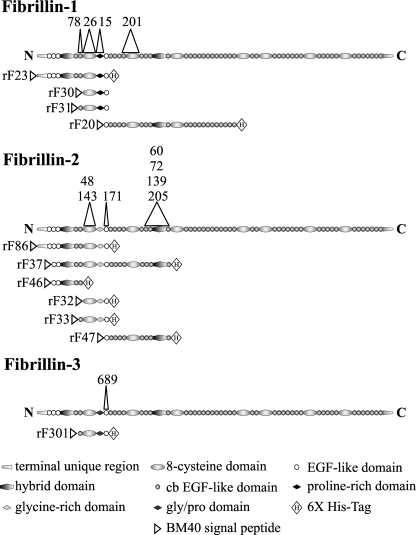
Polyclonal antibodies (pAbs) as well as mAbs specific for fibrillin-1, fibrillin-2, and fibrillin-3 have been described previously (1, 3, 15, 16). Characterization of a new mAb, mAb 689, is shown here. Construction and purification of recombinant fibrillin-1, fibrillin-2, and fibrillin-3 polypeptides used in this study have also been described (3, 9, 11, 15, 18). For easy reference, the peptides used and the mapped epitopes recognized by mAbs are depicted in Fig. 1. Extraction and purification of microfibrils from human amnion, using either guanidine or crude bacterial collagenase (Sigma), were performed according to described protocols (16).
FIGURE 1.
Domain structures of the full-length fibrillins and the recombinant polypeptides used in this study. Domain modules contained within each peptide are depicted. mAb clone numbers are shown above the domains that contain their epitopes. EGF, epidermal growth factor-like.
Mice
Heterozygous mice engineered with a targeted disruption in the Fbn1 gene (mgN) (7) were bred to yield wild type, heterozygous, and homozygous mgN offspring. Heterozygous mice with a targeted disruption in the Fbn2 gene (6) were bred to yield all genotypes as well. Wild type and heterozygous littermates were used as controls for the homozygous null animals. All experiments were conducted according to protocols approved by the Oregon Health and Science Institutional Animal Care and Use Committee.
Enzyme-linked Immunosorbent Assay (ELISA)
Various recombinant peptides (2 μg/ml in 15 mm Na2CO3, 35 mm NaHCO3, pH 9.2; 100 μl/well) were used to coat ELISA plates. After overnight incubation at 4 °C, wells were washed with Tris-buffered saline containing 0.025% Tween 20 (TBS-T) (Bio-Rad) and blocked with 100 μl of 4% powdered milk in TBS-T at room temperature for 1 h. Wells were washed again with TBS-T and incubated at room temperature with primary antibody diluted in milk/TBS-T. Dilutions started at 1:50 of a 1 mg/ml solution and continued 2-fold to 1:52,600. Plates were incubated at room temperature on a nutating platform for 3 h, washed with TBS-T, and incubated with secondary GAM-HRP (Sigma), diluted 1:1000 in milk/TBS-T for 2 h at room temperature on a nutating platform. After washing, wells were incubated with TMB substrate (Sigma) for 5 min. Color reaction was stopped with 2 n H2SO4, and absorbance was determined at 450 nm using a Molecular Devices Emax microplate reader. ELISAs were also performed using microfibrils extracted from human amnion. Conditions used were as described previously (16).
Whole-mount Immunolocalization and Confocal Microscopy
Autopods were removed from 6-day-old avian embryos. Whole-mount staining was performed using mAb 201 (specific for fibrillin-1) (1), JB3 (specific for fibrillin-2) (Developmental Studies Hybridoma Bank at the University of Iowa), and mAb 129 (3). Images were captured using a Leica confocal microscope.
Immunohistochemistry
Supranummary digits were fixed in 10% neutral buffered formalin after surgical removal from children (17 and 21 months of age). Standard immunohistochemical studies were performed on slides from paraffin-embedded tissues using a modified avidin-biotin (ABC) immunoperoxidase method. After clearing the slides with xylene and alcohols, endogenous peroxidase was blocked with 0.5% hydrogen peroxide in methanol. Pretreatment to enhance antigen retrieval is listed in Table 1 along with the concentrations and dilutions of the primary antibodies. Primary antibodies for fibrillin-1–3 were all previously described mouse mAbs (1, 3, 15). Secondary horse anti-mouse antibodies were utilized, and the tertiary antibody 3′,3′-diaminobenzidine chromagen complex in the Vectastain ABC kit for Elite PK-6100 (Vector Laboratories, Burlingame, CA) was employed. Control slides using phosphate-buffered saline in place of primary antibody were processed concurrently with the slides stained with primary antibodies. Immunostaining results were evaluated using a Zeiss Axiophot microscope, and digital photographs were taken of representative areas.
TABLE 1.
Conditions for epitope retrieval of formalin-fixed, paraffin-embedded sections
Pronase was obtained from Calbiochem.
| Antibody target | Clone no. | Concentration | Working dilution | Pretreatment |
|---|---|---|---|---|
| mg/ml | ||||
| Fibrillin-1 | mAb 15 | 1 | 1:5000 | 15-Min microwave retrieval in citrate (10 mm) buffer |
| Fibrillin-2 | mAb 48 | 1 | 1:200 | 10-Min Pronase digestion and 10-min microwave retrieval in citrate (10 mm) buffer |
| Fibrillin-3 | mAb 129 | 1.49 | 1:200 | 10-Min Pronase digestion and 10-min microwave retrieval citrate in (10 mm) buffer |
Enzyme Digestion Experiments
Tissues were snap-frozen, and 4-μm sections were processed for immunofluorescence microscopy (19). Prior to incubation with primary antibodies, sections were treated for 20 min with titrated solutions (0.05, 0.1, 0.2, and 0.4 mg/ml) of crude collagenase (Sigma), dissolved in 50 mm Tris, 0.4 m NaCl, 5 mm CaCl2, pH 7.5, or with buffer without collagenase. Sections were washed and then incubated with primary antibodies, followed by washing and incubation with secondary antibodies. Affinity-purified pAb 9543 (anti-fibrillin-1), affinity-purified pAb 0868 (anti-fibrillin-2), and indicated mAbs were used as primary antibodies. Secondary antibodies were Alexa 488 anti-rabbit, Alexa 488 anti-mouse, and Alexa 568 anti-rabbit (Invitrogen). Double immunofluorescence was performed using pAb 9543 and fibrillin-2 mAbs. Images were captured using a Zeiss Axiovert 200 microscope equipped with AxioVision software (version 4.5) and Apotome optical sectioning.
Electron Microscopy
Section-surface and en bloc diffusion immunolabeling experiments were performed as described previously (20).
RESULTS
Immunolocalization of Fibrillins in Digit Tissues
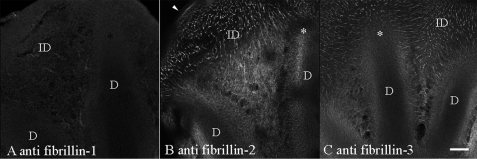
Confocal microscopy and whole-mount antibody staining of 6-day-old embryonic avian autopods revealed abundant fibrillin-2 and -3 staining that was primarily associated with the interdigital connective tissue space. Fibrillin-2 and -3 fibrils looked like guide wires, extending from the ectoderm (arrowhead) and connecting the interdigital space to the leading edges of the forming digit cartilages (asterisk) (Fig. 2, B and C). In contrast, fibrillin-1 staining yielded only very limited, if any, signals that were restricted to areas immediately adjacent to the developing digit cartilage (Fig. 2A), and the guide wires were not stained. The interdigital connective tissue is the first known example of a tissue that is structured with little to no apparent contribution from fibrillin-1.
FIGURE 2.
Whole-mount immunostaining of 6-day-old avian autopods. After immunostaining with mAb 201, specific for fibrillin-1 (A), mAb JB3, for fibrillin-2 (B), and mAb 129, for fibrillin-3 (C), confocal microscopy revealed abundant fibrillin-2 and -3 in the interdigital spaces and very little fibrillin-1 that was limited to the cartilage boundary. Discrete fibrils of fibrillin-2 and -3 appeared to connect the ectoderm (marked with an arrowhead) to the developing digit cartilages. Digit tips are marked with an asterisk. D, digit; ID, interdigital space. Scale bar, 80 μm.
To investigate the contributions of fibrillins in postnatal digits, conditions were first established for immunohistochemical staining of formalin-fixed paraffin-embedded samples. Previously, immunofluorescence experiments using postnatal tissues were complicated by the autofluorescence of elastin, which made documentation of fibrillin immunostaining difficult in elastin-rich tissues like the perichondrium. Therefore, an alternative staining method (immunoperoxidase) was employed. Because fetal tissues are known to contain all three fibrillins, sections of human fetal tissues were analyzed first. Positive immunostainings (data not shown) for fibrillin-specific mAbs were achieved using the conditions shown in Table 1. These conditions were then used to stain sections obtained from surgically removed extra digits.
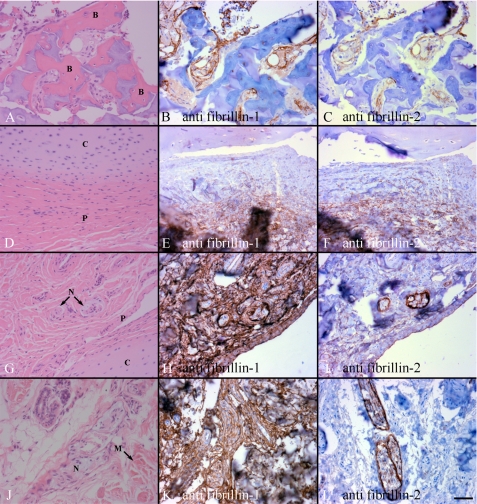
Immunohistochemical analyses of postnatal digit tissues revealed new information. First, both fibrillin-1 and -2 are present in bone marrow stromal matrix as well as in areas around woven bone (Fig. 3, A–C). Second, both fibrillin-1 and -2 are present in the fibrous perichondrium (Fig. 3, D–F). Third, although all of the juxta-articular soft tissues surrounding the cartilage, including joint capsule, demonstrated abundant fibrillin-1 staining (Fig. 3, H and K), fibrillin-2 staining was pronounced only around peripheral nerves, with fainter staining apparent in the surrounding perichondrium and skeletal muscle in the 17-month-old sections (Fig. 3I). However, this faint staining of surrounding tissues appeared to be lost by 21 months of age (Fig. 3L). Fibrillin-3 staining of postnatal digits was negative, even though conditions were used that resulted in clearly positive immunostaining of similar fetal tissues (data not shown).
FIGURE 3.
Immunohistochemical staining of human postnatal supranummary digits. Sections were stained with mAb 15, specific for fibrillin-1 (B, E, H, and K), and with mAb 48, specific for fibrillin-2 (C, F, I, and L). Corresponding hematoxylin and eosin-stained sections are shown in A, D, G, and J. B and C, fibrillin-1 and -2 were found in the periosteum and marrow matrix around ossifying woven bone (labeled B in A) from a 17-month extra digit. E and F, staining for fibrillin-1 and -2 was found in the fibrous perichondrium next to the cartilage (labeled P and C in D) of a 17-month extra digit. H and I, fibrillin-1 was abundant, whereas fibrillin-2 appeared to be much less abundant, in all joint capsule tissues of a 21-month extra digit. Peripheral nerve (N), perichondrium (P), and cartilage (C) of the joint capsule are shown in G. K and L of 21-month digit, abundant fibrillin-1 staining was found around peripheral nerves and muscle (labeled N and M in J), whereas fibrillin-2 staining was abundant only around the nerves. Scale bar, 50 μm.
Unmasking Fibrillin-2 Epitopes within Microfibrils
A drawing of fibrillin-2 domain structure with mapped epitopes for previously characterized mAbs (15) is shown in Fig. 1. We previously found that all fibrillin-2 mAbs recognized monomeric fibrillin-2 secreted by cells into the culture medium and analyzed by Western blotting. However, we found that only the epitopes recognized by mAb 48 and mAb 143 were available for binding in microfibrils present in fetal tissues or in cell culture extracellular matrix. All other epitopes were cryptic. Based on these data, we concluded that certain regions of fibrillin-2 are masked in matrix microfibrils (15).
Furthermore, we showed that microfibrils in all fetal connective tissues examined were well labeled with mAb 48 and mAb 143 and were likely to be heteropolymers of fibrillin-1 and -2 (15). In contrast, postnatal tissues, with the exception of peripheral nerves, were nonreactive or poorly immunofluorescent with mAb 48 or mAb 143 (15). In Fig. 3, we confirmed and extended previous observations in postnatal digit tissues using immunoperoxidase staining rather than immunofluorescence to exclude complications due to autofluorescence. To explain the difference between ubiquitous fibrillin-2 mAb 48 and mAb 143 immunoreactivity in fetal tissues and limited immunoreactivity in postnatal tissues, we hypothesized that fetal heteropolymeric microfibrils are masked in postnatal tissues by proteins that further elaborate microfibril structure. To test this hypothesis, we performed enzymatic digestions of tissue sections to unmask fibrillin-2 epitopes.
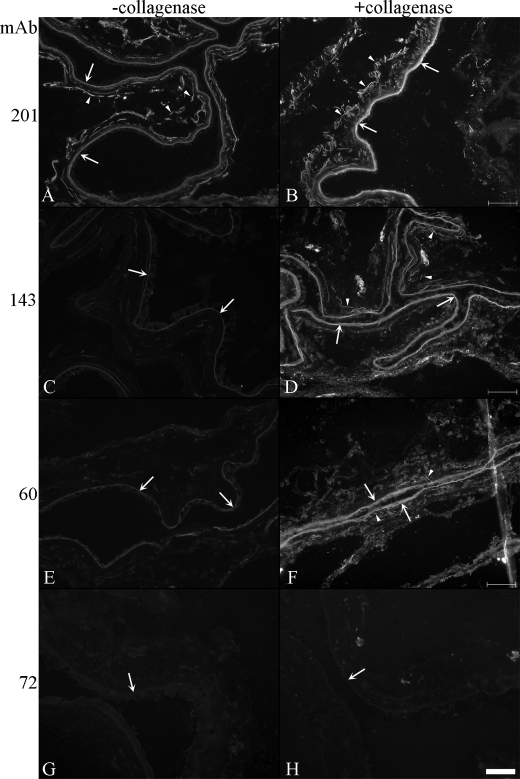
Human amnion was stained with mAbs specific for fibrillin-1, fibrillin-2, and elastin. Elastin staining was negative, and electron microscopic examination of amnion showed no accumulation of amorphous elastin (data not shown). Microfibrils in the amnion were stained with antibodies to fibrillin-1 (Fig. 4A). However, fibrillin-2 appeared to be absent in the microfibril bundles and present only in limited amounts in the amniotic basement membrane, after staining with mAb 143 (Fig. 4C). When amnion sections were treated with crude collagenase, mAb 143 stained the microfibril bundles as well as the amniotic basement membrane (Fig. 4D). Similarly, other fibrillin-2 epitopes, for example the one recognized by mAb 60, were revealed after enzymatic treatment (Fig. 4, F compared with E), whereas others remained masked (Fig. 4, G and H). Fibrillin-2 epitopes recognized by mAbs 48, 143, 60, 139, and 171 were revealed in these experiments, whereas epitopes recognized by mAbs 72 and 205 remained masked (Fig. 4 and data not shown).
FIGURE 4.
Unmasking fibrillin-2 immunostaining in human amnion. Tissue sections in A, C, E, and G were not digested with crude collagenase, whereas those in B, D, F, and H were treated with enzyme. mAb 201, specific for fibrillin-1, stained microfibril bundles as well as the amniotic basement membrane in untreated sections (A) as well as collagenase-digested sections (B). mAb 143, one of two antibodies that recognize fibrillin-2 in fetal tissues, displayed only limited staining of the amniotic basement membrane in untreated sections (C). Digestion of tissue sections with crude collagenase unmasked the epitope recognized by mAb 143 in the microfibril bundles (D). mAb 60, which recognizes a cryptic epitope in fibrillin-2, gave similar results as mAb 143 (E and F). In contrast, mAb 72, a fibrillin-specific antibody, failed to recognize fibrillin-2 in both untreated (G) and treated (H) sections. This result was also obtained using mAb 205 (data not shown). Arrows point to the amniotic basement membrane; arrowheads point to microfibril bundles. Scale bar, 50 μm.
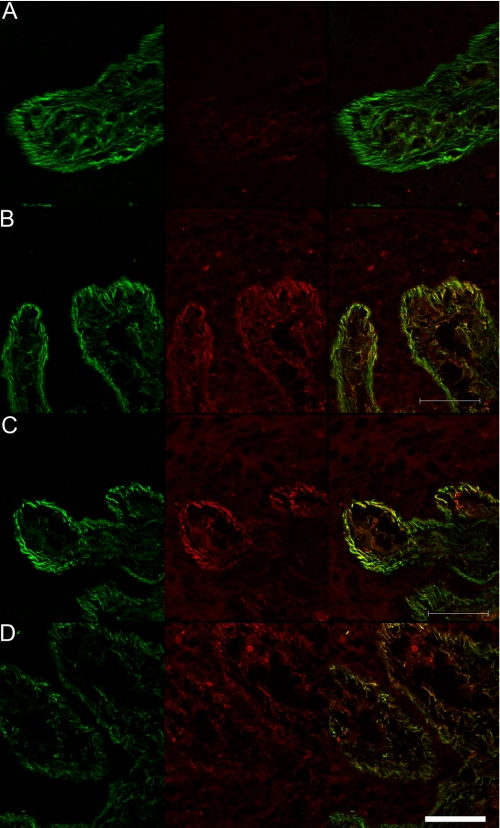
Similar results were obtained when a 20-month supranummary digit was stained after digestion with crude collagenase. In the skin, fibrillin-2 epitopes were masked (Fig. 5A) and could be revealed differentially using increasing concentrations of crude collagenase. Cryptic mAb 60 (data not shown), mAb 139 (Fig. 5B), and mAb 171 (Fig. 5C) were revealed with 0.2 mg/ml crude collagenase, although staining with cryptic mAbs 72 and 205 remained negative (data not shown). A higher concentration of crude collagenase (0.4 mg/ml) was required to reveal mAb 72 (Fig. 5D) and mAb 205 (data not shown). Double immunofluorescence labeling with fibrillin-2 mAbs in combination with fibrillin-1 pAb 9543 documented the presence of fibrillin-2 and fibrillin-1 in the same microfibril bundles (Fig. 5). In addition, a decrease in fibrillin-1 immunostaining was observed at the highest concentrations of crude collagenase (Fig. 5D).
FIGURE 5.
Colocalization of fibrillin-1 and unmasked fibrillin-2 in human skin. Tissue sections of 20-month extra digit were either untreated (A) or treated (B) with 0.2 mg/ml collagenase and stained with anti-fibrillin-1 (pAb 9543) and with mAb 139, which recognizes a cryptic epitope in fibrillin-2. No red immunofluorescence was seen in untreated tissue sections with mAb 139 (A) or with mAbs 60, 171, 72, or 205. Red fibrillin-2 immunofluorescence was revealed in sections treated with 0.2 mg/ml collagenase and stained with mAb 139 (B) and with mAb 171 (C). The cryptic epitope recognized by mAb 72 was not revealed with 0.2 mg/ml collagenase treatment (as in Fig. 4, G and H). Sections treated with 0.4 mg/ml collagenase showed red fibrillin-2 staining with mAb 72 (D), but at this concentration of collagenase green fibrillin-1 staining appeared reduced (D). Merged images show colocalization of green fibrillin-1 fibrils with unmasked red fibrillin-2 fibrils (B–D). Scale bar, 50 μm.
To determine whether fibrillin-2 epitopes could be revealed by removal of noncovalently bound microfibril components, we tested microfibrils after extraction from human amnion with 6 m guanidine (16). Guanidine-extracted microfibrils were adsorbed to ELISA plates. Fibrillin-2 mAbs 48, 143, 60, 72, 139, 171, and 205 were titrated and tested for reactivity. Fibrillin-1 mAb 201 was titrated and used as a control. Guanidine-extracted microfibrils reacted well with both mAb 201 and mAb 48 and weakly with mAb 143, whereas other fibrillin-2 mAbs were unreactive (Fig. 6). These data suggest that certain fibrillin-2 epitopes masked in amnion tissue (mAb 48 and mAb 143) may be unmasked by removing noncovalently bound microfibril-associated proteins and/or by altering the quaternary structure of the microfibril with denaturants. However, all other fibrillin-2 epitopes remained masked in the presence of 6 m guanidine. Because multiple fibrillin-1 epitopes were previously shown to be retained in guanidine-extracted microfibrils (16), we interpret these results to indicate that the cryptic fibrillin-2 epitopes are masked by the structure of the microfibril.
FIGURE 6.
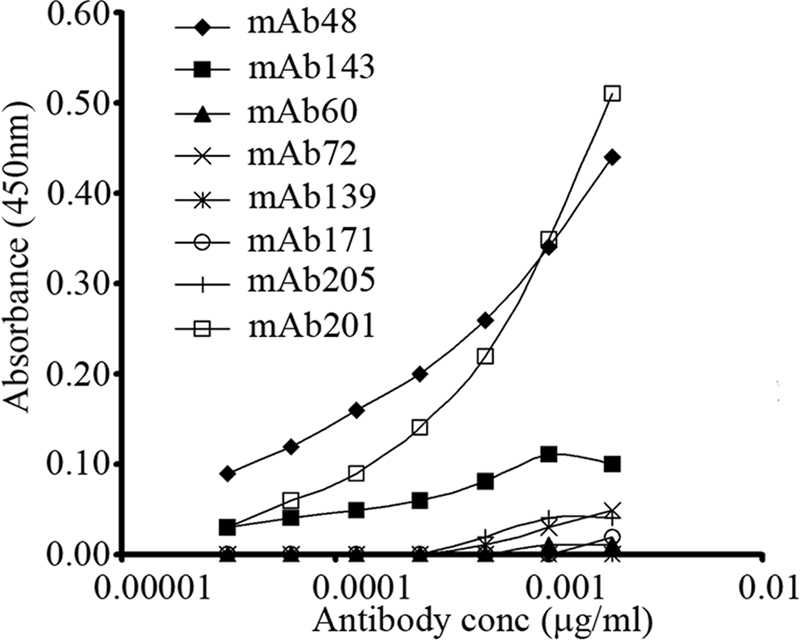
Detection of fibrillin-2 epitopes in microfibrils extracted from human amnion. Guanidine-extracted microfibrils were used to coat ELISA plates. Titrations of fibrillin-2 mAbs 48, 143, 60, 72, 139, 171, and 205 and fibrillin-1 mAb 201 were tested. Fibrillin-2 epitopes recognized by mAb 48, and possibly also by mAb 143, were revealed in guanidine-extracted microfibrils. However, cryptic epitopes recognized by mAbs 60, 72, 139, 171, and 205 remained cryptic even after treatment with 6 m guanidine. mAb 201 served as a control in these experiments.
Homopolymeric Fibrillin-2 Microfibrils Are Exposed in Early Postnatal Fbn1 Null Tissues
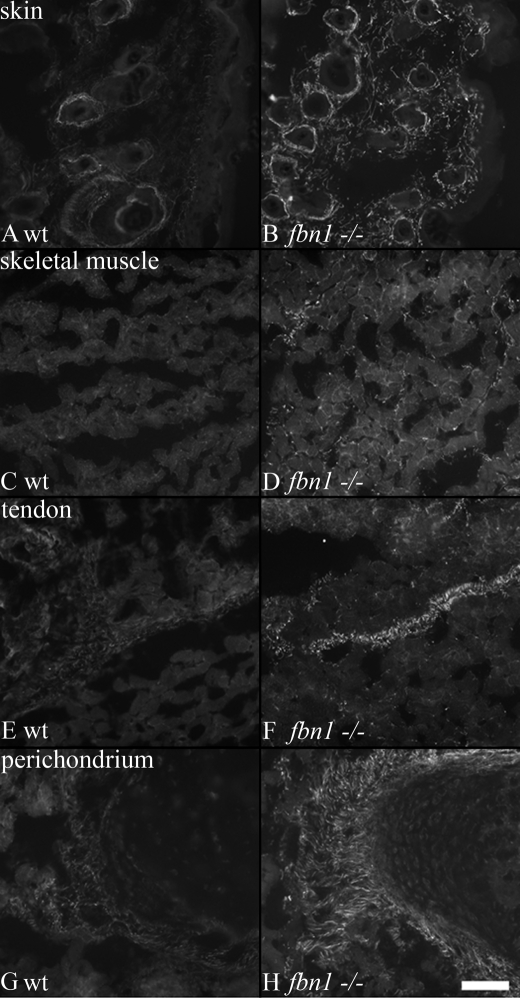
Microfibrils in postnatal wild type mouse tissues, as in human tissues, were not well stained with pAb 0868, specific for fibrillin-2 domains contained in rF37 (Fig. 1). In a second approach to determine the postnatal fate of fibrillin-2, we examined Fbn1 null mouse tissues. In contrast to wild type littermate mouse tissues, abundant pAb 0868 immunoreactive fibrillin-2 microfibrils were observed in all newborn and early postnatal Fbn1 null tissues. Fig. 7 shows that fibrillin-2 microfibril staining is revealed in the skin, skeletal muscle, tendon, and perichondrium of P8 Fbn1 null mice, compared with wild type littermate control sections. These results indicate that fibrillin-1 molecules or fibrillin-1-associated molecules mask fibrillin-2 epitopes in postnatal tissues.
FIGURE 7.
Fibrillin-2 microfibrils are unmasked in mouse Fbn1 null tissues. P8 wild type (A, C, E, and G) and Fbn1−/− (B, D, F, and H) littermate sections were stained with pAb 0868, specific for fibrillin-2. Fibrillin-2 immunostaining was mostly negative in wild type sections of skin (A) and skeletal muscle (C) at this time; there was weak fibrillin-2 immunostaining in wild type sections of tendon (E) and perichondrium (F). However, in Fbn1−/− sections, fibrillin-2 immunostaining was revealed in typical tissue-specific fibrillin microfibril patterns (B, D, F, and H). Scale bar, 50 μm.
mAb 689 Binds to an Epitope in the Fourth Epidermal Growth Factor-like Domain in All Three Fibrillins
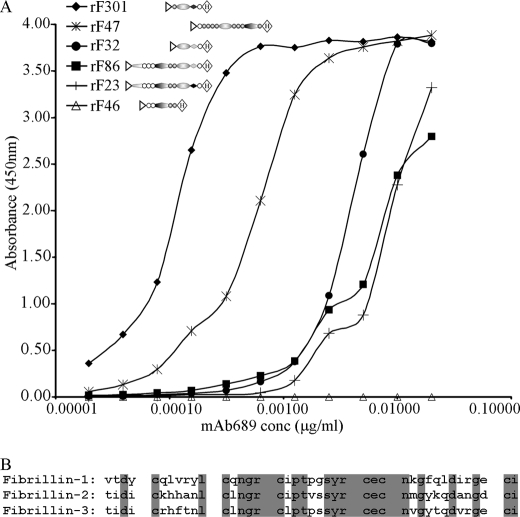
mAb 689 was generated using rF301 (depicted in Fig. 1), a recombinant human fibrillin-3 polypeptide that spans cbEGF1, the first 8-cysteine domain, the proline/glycine-rich region, and EGF4 (3). mAb 689 recognizes rF301 as well as polypeptides containing the homologous region from fibrillin-1 and -2. These include rF23 and rF30 (fibrillin-1) and rF86 and rF32 (fibrillin-2) (Figs. 1 and 8A, and data not shown). Using additional polypeptides (depicted in Fig. 1) as coated substrates, we performed ELISAs to map more precisely the epitope recognized by mAb 689. Fig. 8A shows that mAb 689 binds to peptides containing the fourth epidermal growth factor-like domain, but not to peptides that lack this domain. mAb 689 reacted well with the fibrillin-1 peptide rF20 (data not shown) and the fibrillin-2 peptide rF47 (Fig. 8A), which contain EGF4 but lack the preceding domains. Based on these data, we conclude that mAb 689 is a pan-fibrillin antibody that binds to an epitope contained largely in EGF4.
FIGURE 8.
mAb 689 binds to an epitope in EGF4 in all three fibrillins. When tested in ELISA (A), mAb 689 reacted with rF301, the recombinant fibrillin-3 peptide used to generate this mAb. mAb 689 also reacted with recombinant fibrillin-1 and -2 peptides containing EGF4 but not with peptides missing EGF4. Of the four domains contained in rF301, rF47 contains only EGF4, and mAb 689 reacted well with rF47. Similarly, mAb 689 reacted well with rF20, a fibrillin-1 recombinant peptide whose N-terminal domain is EGF4 (data not shown). B, identical residues in the amino acid sequences of EGF4 in fibrillin-1, -2, and -3 are shaded.
In addition to human fibrillin, mAb 689 recognizes mouse fibrillin (Figs. 9 and 10). The primary structure of human and mouse fibrillin-1 EGF4 is identical, as is the primary structure of human and mouse fibrillin-2 EGF4. However, only 25 of the 41 residues are identical in fibrillin-1 and -3 EGF4, whereas 31 of the 41 residues are identical in fibrillin-2 and -3. 22 residues are identical in all three fibrillins. Fig. 8B shows the region of identity in the EGF4 sequence. This region of identity clusters around the residues predicted to form the second strand of an anti-parallel β-sheet, which likely forms the core structure of EGF4. The mAb 689 epitope is stabilized by disulfide bonds, because reduction abolishes immunoreactivity (data not shown).
FIGURE 9.
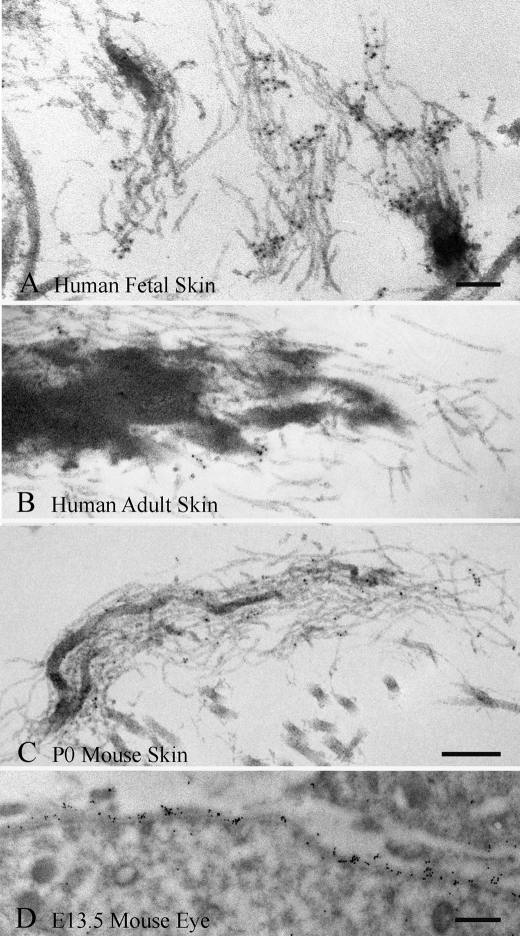
Immunolocalization of mAb 689 to fibrillin microfibrils and to basement membranes. Immunogold labeling was observed on microfibrils in 16-week human fetal digit skin (A), but little to no labeling was found when 21-year-old human skin was tested (B). Similarly, P0 wild type mouse skin demonstrated relatively little labeling of microfibrils (C), whereas the retinal pigment epithelial cell basement membrane of E13.5 wild type mouse eye was well labeled (D). Scale bars, 100 nm for A and B; 200 nm for C; 500 nm for D.
FIGURE 10.
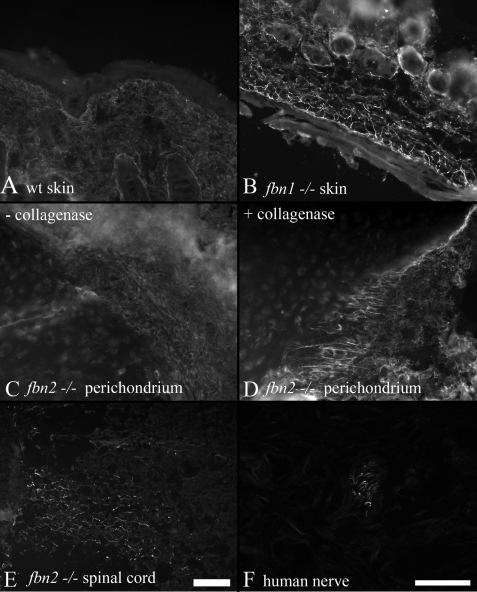
Tissue-specific differences in the availability of the mAb 689 epitope. Postnatal wild type mouse tissues showed relatively little immunostaining when mAb 689 was used. Dermal microfibrils in P8 wild type (wt) skin were very weakly stained with mAb 689 (A). In contrast, mAb 689 staining of fibrillin-2 microfibrils in Fbn1−/− skin demonstrated typical fibrillin microfibril patterns in the dermis and in the dermal-epidermal junction (B). Fibrillin-1 microfibrils in P9 Fbn2−/− perichondrium were relatively unstained (C), indicating that the mAb 689 epitope is cryptic in fibrillin-1 microfibrils. However, when Fbn2−/− tissue sections were digested with crude collagenase, mAb 689 stained fibrillin-1 microfibrils in the perichondrium of Fbn2−/− tissue sections (D). Undigested sections of P9 Fbn2−/−spinal cord displayed abundant microfibril bundles that stained with mAb 689 (E). Fibrillin-2 mAb 171 stained peripheral nerves in undigested tissue sections of 20-month human extra digit (F). Scale bars, 50 μm.
Because fibrillin-1, fibrillin-2, and fibrillin-3 antibodies have been immunolocalized to fetal microfibrils (3, 15), we expected that mAb 689 would label heteropolymeric fetal microfibrils. Microfibrils (Fig. 9A) and basement membranes (Fig. 9D) were well labeled by mAb 689 in fetal tissues. However, mAb 689 failed to label microfibrils in adult human skin (Fig. 9B), and labeling of P0 wild type mouse skin was sparse, even though microfibrils were abundant (Fig. 9C). Poor reactivity with adult or early postnatal microfibrils was unexpected, because mAb 689 binds well to fibrillin-1 in ELISA (Fig. 8A).
mAb 689 Epitope Is Cryptic in Postnatal Wild Type and Some Fbn2 Null Tissues
When tested by immunofluorescence on frozen sections, mAb 689 failed to yield classical fibrillin fibril staining patterns in postnatal wild type mouse tissues (Fig. 10A). However, mAb 689 stained all early postnatal Fbn1 null tissues, yielding classic fibrillin fibril patterns and demonstrating that mAb 689 reacts well with polymeric fibrillin-2 (Fig. 10B and data not shown). Therefore, in postnatal tissues, mAb 689 staining is similar to pAb 0868 fibrillin-2 antibody staining (Fig. 7).
Because mAb 689 reacts well with monomeric fibrillin-1 in ELISA (Fig. 8A), polymeric fibrillin-1 microfibrils in Fbn2 null tissues were tested. mAb 689 failed to stain most Fbn2 null tissues (Fig. 10C and data not shown), indicating that the mAb 689 epitope in EGF4 is unavailable in the polymerized fibrillin-1 microfibrils in most tissues. Digestion of Fbn2 null tissue sections with crude collagenase uncovered the mAb 689 epitope in the perichondrium (Fig. 10D) and in the dermis (data not shown). Because the mAb 689 epitope is accessible in Fbn1 null tissue, fibrillin-2 EGF4 is exposed in the absence of fibrillin-1 (Fig. 10B). In contrast, in the absence of fibrillin-2, fibrillin-1 EGF4 remains masked. We also found that polymerized fibrillin-1 microfibrils in the Fbn2 null spinal cord were available for binding by mAb 689, without enzymatic unmasking (Fig. 10E). Therefore, EGF4 in fibrillin-1 is variably exposed or masked in tissues, demonstrating that tissue-specific modulation of microfibril structure can make this domain accessible.
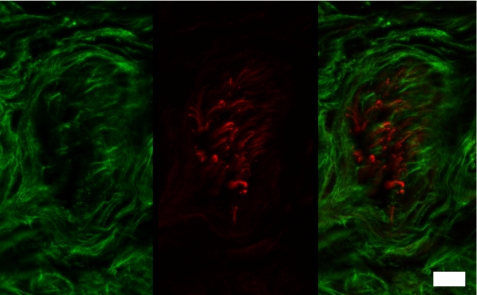
To further investigate microfibrils in neuronal tissues, human 20-month postnatal digit was examined. Consistent with results showing that the cryptic mAb 689 epitope in fibrillin-1 is exposed in spinal cord (Fig. 10E), cryptic fibrillin-2 epitopes recognized by mAbs 60 (data not shown), 139 (data not shown), and 171 (Fig. 10F) were available for immunostaining of untreated peripheral nerves in the human digit. Epitopes recognized by mAbs 72 and 205 were largely masked in peripheral nerve (data not shown). Double immunofluorescence clearly showed that fibrillin-1 and -2 microfibrils in the peripheral nerve are distinct. No colocalization was observed between the green fibrillin-1 immunofluorescence and the red fibrillin-2 immunofluorescence (Fig. 11).
FIGURE 11.
Double immunofluorescence of fibrillin-1 and -2 in human peripheral nerve. Undigested sections of 20-month human extra digit were incubated with antibodies specific for fibrillin-1 (pAb 9543) and fibrillin-2 (mAb 171). Fibrillin-1 antibodies (green, left panel) and fibrillin-2 antibodies (red, middle panel) stain discrete fibrils (merged images, right panel). Scale bar, 10 μm.
DISCUSSION
Little is known about the contributions of fibrillin-2 to microfibril structure. However, genetic evidence in humans and mice has shown that fibrillin-2 performs important functions in bone growth, joint function, and limb patterning. In humans, mutations in FBN2 cause congenital contractures, arachnodactyly, and crumpled ears, hallmark features of congenital contractural arachnodactyly (OMIM 121050). Fbn2 null mice demonstrate congenital contractures, syndactyly, tendon defects, and shorter, rather than longer, bones (6, 17). We show that both fibrillin-1 and -2 are ubiquitously distributed in digit tissues surrounding joints (perichondrium, skeletal muscle, tendon, and nerves). However, two differences were found as follows: fibrillin-2, but not fibrillin-1, was present in the interdigital tissues of the developing autopod, and fibrillin-2 immunostaining in digit tissues was lost in the early postnatal period. The differential distribution of fibrillins in the developing autopod could explain why fibrillin-1 is unable to compensate for the loss of fibrillin-2 in the interdigital space, with resulting syndactyly, in the Fbn2 null mouse. However, the apparent loss of fibrillin-2 in postnatal limb tissues confounds an understanding of why mutant fibrillin-2 in human postnatal limb tissues results in arachnodactyly, whereas loss of Fbn2 in mice results in decreased bone growth.
Results presented here reveal that fibrillin-2 is present, but masked, in postnatal tissues. Enzymatic digestion of human tissue sections with crude collagenase uncovered fibrillin-2 epitopes. Since we previously showed that crude collagenase contains enzymes that cleave fibrillin-1 (16), we hypothesized that fibrillin-1 molecules overlay fibrillin-2 molecules in the structure of postnatal microfibrils and mask fibrillin-2 epitopes. To test this hypothesis, immunodetection of fibrillin-2 epitopes present in rF37, the N-terminal third of fibrillin-2 (Fig. 1), was assayed in wild type and Fbn1 null tissues. Although N-terminal epitopes in fibrillin-2 were largely masked in postnatal wild type tissues, these fibrillin-2 epitopes were clearly available in Fbn1 null tissues. These data indicate that fibrillin-1 molecules themselves, or fibrillin-1 molecules in association with other ligands, mask fibrillin-2. With this new information, it is now reasonable to hypothesize that mutations in FBN2 may cause congenital contractural arachnodactyly by perturbing the core structure of microfibrils, leading to alterations in the successive structural and functional elaboration of postnatal microfibrils by fibrillin-1 and associated proteins.
Previous immunochemical studies have contributed significantly to current concepts of fibrillin microfibril structure. Biochemical approaches to identification of structural components of microfibrils have been hampered by the inability to solubilize and extract microfibril proteins from tissues. In fact, fibrillin was first identified as a major component of microfibrils, using an immunochemical approach (1). Asymmetric labeling of a single fibrillin-1 mAb just to one side of each bead in the microfibril established an antiparallel arrangement of linear fibrillin-1 molecules within the microfibril (10). Additional antibody labeling studies using epitope-mapped mAbs have been used to support various arrangements of fibrillin-1 within the microfibril (11, 12, 16). Antibodies specific for microfibril-associated glycoprotein-1 and -2 (MAGP-1 and -2) have also been used to demonstrate the presence of microfibril-associated glycoproteins near or on the globular beads in the microfibril (21). Double labeling with antibodies specific for fibrillin-1 and -2 showed that fetal microfibrils are composed of both fibrillins (15). However, current models of microfibril structure do not include a specific role for fibrillin-2.
In the studies reported here, mAbs with mapped epitopes were used to obtain additional information about the structure of microfibrils. First, the epitopes recognized by mAbs 48 and 143 demonstrated that the first 8-cysteine domain in fibrillin-2 is exposed in fetal microfibrils and masked in postnatal microfibrils. These results were identical to results obtained with pAb 0868, which recognizes epitopes throughout rF37 (Fig. 1). Therefore, we conclude that at least portions of the N-terminal region of fibrillin-2, corresponding to rF37, are masked in postnatal tissues, likely through the continual elaboration of fibrillin-1 overlaying a core structure that includes fibrillin-2. Second, cryptic epitopes described previously in fibrillin-2 (15) were shown to be differentially masked. EGF4 and some regions close to the second hybrid domain were easily unmasked or available in peripheral nerves, whereas other regions (containing epitopes for mAb 72 and mAb 205) could be revealed only with the highest concentrations of crude collagenase. Third, a cryptic epitope in fibrillin-1 was identified.
Our study also demonstrated tissue-specific differences in microfibril structure. We found that fibrillin-2 cryptic epitopes recognized by mAbs 60, 139, and 171 and fibrillin-1 cryptic epitope recognized by mAb 689 were available in neuronal tissues but unavailable in all other tissues examined. In contrast to colocalization of fibrillin-1 and unmasked fibrillin-2 in skin (Fig. 5), double immunofluorescence of untreated peripheral nerve discerned discrete fibrillin-1 and -2 microfibril bundles (Fig. 11). These results, together with studies of the interdigital space (Fig. 2), indicate that microfibrils in specific tissues can be composed of fibrillin-2, without overlaying fibrillin-1 molecules.
Taken all together, results show for the first time that microfibril structure evolves as growth and development proceed into the postnatal period. We propose that fibrillin-2 is masked in microfibrils in postnatal tissues (other than neuronal tissues) by the continued elaboration of microfibril structure. During fetal development, epitopes for both fibrillins are available in heteropolymeric microfibrils. As growth and development proceed, fibrillin-1 continues to be synthesized, whereas fibrillin-2 synthesis is turned off, resulting in an outer layer of fibrillin-1 polymers. This outer layer of fibrillin-1 molecules is mainly accessible in postnatal microfibrils. Together with associated molecules, this outer layer of fibrillin-1 masks fibrillin-2 epitopes. In addition, a site in fibrillin-1 EGF4 is also made cryptic, either by interaction with an associated molecule or by microfibril tertiary or quaternary structure.
Structural studies of relevant fibrillin-1 recombinant peptides have been performed (13, 22). These studies do not, however, clarify whether the cryptic epitopes that we have identified are made cryptic by the structure of polymerized fibrillin. An epitope in EGF4 that is common in all three fibrillins (recognized by mAb 689) as well as an epitope present only in fibrillin-2 EGF4 (recognized by mAb 171) were masked in most of the postnatal tissues examined. EGF4 is the domain adjacent to the proline/glycine-rich region, which is presumed to be flexible in fibrillin-1 (13). Adjacent domains in fibrillin-1 (recognized by mAbs 78, 26, and 15) and in fibrillin-2 (recognized by mAbs 48 and 143) are accessible in microfibril polymers, so a flexible region may allow EGF4 to be buried underneath these adjacent accessible domains. Tissue-specific factors could modulate this basic microfibril structure to allow EGF4 domains in both fibrillins to become accessible in neuronal tissues.
Structural studies of the region surrounding the second hybrid domain (recognized by mAbs 60, 72, 139, and 205) have produced contradictory conclusions. Solution structures of recombinant peptides containing the second hybrid domain of fibrillin-1 indicated that this region may be V-shaped, with the hybrid domain forming the “V” (13), but a crystallographic analysis of the second hybrid domain and flanking regions suggested, instead, a near linear arrangement of these domains (22). Therefore, it is unclear from these studies whether cryptic epitopes near the second hybrid domain might be hidden by fibrillin tertiary structure.
It is possible that fibrillin-2 cryptic epitopes may be masked by microfibril-associated proteins in addition to fibrillin-1. MAGP-1, for example, has been reported to bind to the third 8-cysteine domain in recombinant fibrillin-2 (23). The third 8-cysteine domain is just adjacent to the region containing cryptic epitopes recognized by mAbs 60, 72, 139, and 205, so it is possible that MAGP-1 may mask some of these epitopes. Studies similar to the ones performed here, using MAGP-1 null mice (24) and fibrillin-2 antibodies directed specifically toward these cryptic epitopes, could test this possibility.
There are important implications from the data presented here. First, cell culture models for microfibril structure, which have been used previously to indicate structural differences between normal and Marfan microfibrils (25, 26), may have only limited relevance to the status of microfibrils in vivo. In addition, cell culture models that are used to test mechanisms of microfibril assembly (27) may similarly have only limited relevance to in vivo mechanisms of assembly, which as we have shown result in both developmental and tissue-specific differences in microfibril structure.
Finally, an important implication of the new structural information presented here is that useful biomarkers for microfibril damage or degradation may include unmasking of fibrillin-2 epitopes in postnatal tissues or isolation of fibrillin-2 peptide fragments. Our current investigations are directed toward testing fibrillin-2 biomarkers in the pathogenesis of the Marfan syndrome, a disorder characterized by elastic fiber and microfibril fragmentation (28).
Acknowledgments
We are grateful to Dr. Juan Hurle for showing us how to perform whole-mount antibody staining using avian embryos. Sara Tufa and Valerie Carlberg provided excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants P01 AR049698 (to F. R., D. B. R., H. C. D., and L. Y. S.) and AR042044 (to F. R.). This work was also supported by Shriners Hospitals for Children (to D. R. K. and to L. Y. S.).
- mAb
- monoclonal antibody
- ELISA
- enzyme-linked immunosorbent assay
- pAb
- polyclonal antibody
- rF
- recombinant fibrillin.
REFERENCES
- 1.Sakai L. Y., Keene D. R., Engvall E. (1986) J. Cell Biol. 103, 2499–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H., Apfelroth S. D., Hu W., Davis E. C., Sanguineti C., Bonadio J., Mecham R. P., Ramirez F. (1994) J. Cell Biol. 124, 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corson G. M., Charbonneau N. L., Keene D. R., Sakai L. Y. (2004) Genomics 83, 461–472 [DOI] [PubMed] [Google Scholar]
- 4.Gallagher B. C., Sakai L. Y., Little C. D. (1993) Dev. Dyn. 196, 70–78 [DOI] [PubMed] [Google Scholar]
- 5.Zhang H., Hu W., Ramirez F. (1995) J. Cell Biol. 129, 1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arteaga-Solis E., Gayraud B., Lee S. Y., Shum L., Sakai L., Ramirez F. (2001) J. Cell Biol. 154, 275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carta L., Pereira L., Arteaga-Solis E., Lee-Arteaga S. Y., Lenart B., Starcher B., Merkel C. A., Sukoyan M., Kerkis A., Hazeki N., Keene D. R., Sakai L. Y., Ramirez F. (2006) J. Biol. Chem. 281, 8016–8023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Low F. N. (1962) Anat. Rec. 142, 131–137 [DOI] [PubMed] [Google Scholar]
- 9.Keene D. R., Maddox B. K., Kuo H. J., Sakai L. Y., Glanville R. W. (1991) J. Histochem. Cytochem. 39, 441–449 [DOI] [PubMed] [Google Scholar]
- 10.Sakai L. Y., Keene D. R., Glanville R. W., Bächinger H. P. (1991) J. Biol. Chem. 266, 14763–14770 [PubMed] [Google Scholar]
- 11.Reinhardt D. P., Keene D. R., Corson G. M., Pöschl E., Bächinger H. P., Gambee J. E., Sakai L. Y. (1996) J. Mol. Biol. 258, 104–116 [DOI] [PubMed] [Google Scholar]
- 12.Baldock C., Koster A. J., Ziese U., Rock M. J., Sherratt M. J., Kadler K. E., Shuttleworth C. A., Kielty C. M. (2001) J. Cell Biol. 152, 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baldock C., Siegler V., Bax D. V., Cain S. A., Mellody K. T., Marson A., Haston J. L., Berry R., Wang M. C., Grossmann J. G., Roessle M., Kielty C. M., Wess T. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11922–11927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S. S., Knott V., Jovanoviæ J., Harlos K., Grimes J. M., Choulier L., Mardon H. J., Stuart D. I., Handford P. A. (2004) Structure 12, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charbonneau N. L., Dzamba B. J., Ono R. N., Keene D. R., Corson G. M., Reinhardt D. P., Sakai L. Y. (2003) J. Biol. Chem. 278, 2740–2749 [DOI] [PubMed] [Google Scholar]
- 16.Kuo C. L., Isogai Z., Keene D. R., Hazeki N., Ono R. N., Sengle G., Bächinger H. P., Sakai L. Y. (2007) J. Biol. Chem. 282, 4007–4020 [DOI] [PubMed] [Google Scholar]
- 17.Boregowda R., Paul E., White J., Ritty T. M. (2008) Matrix Biol. 27, 661–666 [DOI] [PubMed] [Google Scholar]
- 18.Sengle G., Charbonneau N. L., Ono R. N., Sasaki T., Alvarez J., Keene D. R., Bächinger H. P., Sakai L. Y. (2008) J. Biol. Chem. 283, 13874–13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono R. N., Sengle G., Charbonneau N. L., Carlberg V., Bächinger H. P., Sasaki T., Lee-Arteaga S., Zilberberg L., Rifkin D. B., Ramirez F., Chu M. L., Sakai L. Y. (2009) J. Biol. Chem. 284, 16872–16881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakai L. Y., Keene D. R. (1994) Methods Enzymol. 245, 29–52 [DOI] [PubMed] [Google Scholar]
- 21.Hanssen E., Hew F. H., Moore E., Gibson M. A. (2004) J. Biol. Chem. 279, 29185–29194 [DOI] [PubMed] [Google Scholar]
- 22.Jensen S. A., Iqbal S., Lowe E. D., Redfield C., Handford P. A. (2009) Structure 17, 759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Werneck C. C., Trask B. C., Broekelmann T. J., Trask T. M., Ritty T. M., Segade F., Mecham R. P. (2004) J. Biol. Chem. 279, 23045–23051 [DOI] [PubMed] [Google Scholar]
- 24.Weinbaum J. S., Broekelmann T. J., Pierce R. A., Werneck C. C., Segade F., Craft C. S., Knutsen R. H., Mecham R. P. (2008) J. Biol. Chem. 283, 25533–25543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kielty C. M., Shuttleworth C. A. (1994) J. Cell Biol. 124, 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kielty C. M., Rantamäki T., Child A. H., Shuttleworth C. A., Peltonen L. (1995) J. Cell Sci. 108, 1317–1323 [DOI] [PubMed] [Google Scholar]
- 27.Hirai M., Horiguchi M., Ohbayashi T., Kita T., Chien K. R., Nakamura T. (2007) EMBO J. 26, 3283–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollister D. W., Godfrey M., Sakai L. Y., Pyeritz R. E. (1990) N. Engl. J. Med. 323, 152–159 [DOI] [PubMed] [Google Scholar]