Abstract
RIG-I (retinoic acid-inducible gene I) and TRIM25 (tripartite motif protein 25) have emerged as key regulatory factors to induce interferon (IFN)-mediated innate immune responses to limit viral replication. Upon recognition of viral RNA, TRIM25 E3 ligase binds the first caspase recruitment domain (CARD) of RIG-I and subsequently induces lysine 172 ubiquitination of the second CARD of RIG-I, which is essential for the interaction with downstream MAVS/IPS-1/CARDIF/VISA and, thereby, IFN-β mRNA production. Although ubiquitination has emerged as a major factor involved in RIG-I activation, the potential contribution of other post-translational modifications, such as phosphorylation, to the regulation of RIG-I activity has not been addressed. Here, we report the identification of serine 8 phosphorylation at the first CARD of RIG-I as a negative regulatory mechanism of RIG-I-mediated IFN-β production. Immunoblot analysis with a phosphospecific antibody showed that RIG-I serine 8 phosphorylation steady-state levels were decreased upon stimulation of cells with IFN-β or virus infection. Substitution of serine 8 in the CARD RIG-I functional domain with phosphomimetic aspartate or glutamate results in decreased TRIM25 binding, RIG-I ubiquitination, MAVS binding, and downstream signaling. Finally, sequence comparison reveals that only primate species carry serine 8, whereas other animal species carry an asparagine, indicating that serine 8 phosphorylation may represent a primate-specific regulation of RIG-I activation. Collectively, these data suggest that the phosphorylation of RIG-I serine 8 operates as a negative switch of RIG-I activation by suppressing TRIM25 interaction, further underscoring the importance of RIG-I and TRIM25 connection in type I IFN signal transduction.
Keywords: Innate Immunity, Interferon, RNA Viruses, Viral Immunology, Virus, RIG-I, Signal Transduction, Virus-host Interaction
Introduction
Viral infection of mammalian cells results in a series of events that culminate in the activation of innate immune responses, including the production of type I IFNs3 and other proinflammatory cytokines. Type I IFNs act in an autocrine and paracrine manner to protect cells from infection, and they do so by stimulating transcription of IFN-stimulated genes (ISGs) that create a cellular antiviral state, thus preventing the spread of the virus infection (1). Type I IFNs also modulate the immune response by NK and macrophage activation, antiviral cytotoxic T-cell response induction, and B-cell proliferation, in addition to promoting major histocompatibility complex antigen cross-presentation (2). Among type I IFNs, IFN-β is secreted by most cells upon viral infection, and IFN-β synthesis is triggered by two major sensor proteins of viral infection located in the cytoplasm, RIG-I and MDA5 (3). RIG-I has been reported to recognize 5′-triphosphate-containing double-stranded RNA (4, 5), a by-product of replication of several RNA viruses, whereas MDA5 has preference for longer double-stranded RNA molecules (6). Upon RNA recognition, both cytoplasmic sensors alter their conformation (7), allowing their N-terminal tandem CARD region to interact with the CARD domain of MAVS (mitochondrial antiviral signaling protein), also known as IPS-1, Cardif, or VISA, on the mitochondrial membrane (8–11). This event leads to the activation of a signaling complex on the surface of mitochondria, with a subsequent downstream signaling cascade resulting in the activation of the transcription factors AP1, IRF-3, and NF-κB (12). Binding of these transcription factors to the IFN-β enhancer region of the IFN-β gene in the nucleus mediates the formation of a pretranscriptional complex named IFN-β enhanceosome (13, 14). As a result, the remaining cellular factors required for the initiation of transcription are recruited to this enhanceosome, stimulating the synthesis of IFN-β mRNA by RNA polymerase II (15).
RIG-I is a DExH RNA helicase protein that belongs to the SF2 family of helicases. It is composed of three functional domains: the N-terminal RIG-I CARD functional domain, formed by two CARD-like domains (CARD1 and CARD2), the central part that contains a helicase domain, and the C-terminal end that contains a zinc-binding domain named the repressor domain or regulatory domain (RD) (16–18). It is believed that RIG-I remains in a dormant form in uninfected cells, where RD represses the active form of RIG-I by masking crucial residues of the CARD functional domain. Upon infection, activating RNA substrates are recognized by RIG-I in two ways; whereas the RD binds the 5′-triphosphate moiety, the helicase domain binds the double-stranded RNA (16, 19). Binding to activating RNA substrates induces a conformational change that exposes key residues in RIG-I CARDs, thereby allowing the interaction with the CARD domain of MAVS (19). This event is facilitated by Lys63-linked ubiquitination of the RIG-I CARD, which is mediated by the E3 ubiquitin ligases TRIM25 (also known as EFP or ZNF147) and REUL (also known as RNF135 or Riplet) (20, 21). Both E3 ligase proteins contain an N-terminal RING domain, a central coiled-coil domain, and a C-terminal PRY-SPRY domain. In the case of TRIM25, the SPRY domain binds to CARD1 of RIG-I, and the RING domain mediates the ubiquitination of the CARD2 of RIG-I (22).
Because phosphorylation is a critical post-translational modification affecting many signaling events, we investigated whether RIG-I phosphorylation plays a role in regulating RIG-I antiviral activity. In this report, we describe how human RIG-I undergoes phosphorylation at the Ser8 residue located within the CARD1 domain of RIG-I. Viral infection as well as treatment with IFN decreased the levels of RIG-I serine 8 phosphorylation. Substitution of serine 8 by phosphomimetic amino acids resulted in decreased binding of RIG-I to TRIM25, decreased RIG-I ubiquitination, and thereby decreased binding to the CARD domain of MAVS. As a consequence, S8D and S8E RIG-I mutants were compromised in their ability to induce IFN-β and to inhibit viral replication. Overall, our study suggests that RIG-I serine 8 phosphorylation in human and possibly primate RIG-I plays an important role in the regulation of RIG-I activity to induce IFN-mediated antiviral responses.
EXPERIMENTAL PROCEDURES
Plasmids
FLAG-tagged or HA-tagged versions of the proteins used in the experiments were generated by PCR amplification using RIG-I and MAVS clones described before (23) as templates. PCR products were inserted into the pCAGGS expression plasmid. Introduced mutations were confirmed by DNA sequence analysis. pCDNA3-HA-TRIM25 and GST-CARD2 plasmid constructs have been described previously (21). For the production of bacterial recombinant protein RIG-I, the RIG-I cDNA sequence was amplified by PCR and cloned into pGEX-6p1 (GE Healthcare). pGFP-IRF-3 has been described elsewhere (24). Renilla luciferase reporter vector pRL-TK was purchased from Promega.
Cell Culture and Viruses
293T and A549 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Hyclone) and 1% penicillin/streptomycin (Invitrogen). Stocks of influenza A/PR/8/34 ΔNS1, a recombinant PR8 virus lacking the NS1 gene, were grown in 7-day-old embryonated eggs (25). Vesicular stomatitis virus (VSV) expressing green fluorescent protein (VSV-GFP) virus stock was grown for 2 days in Vero cells. Sendai virus (Cantell strain) was grown for 2 days in 10-day-old embryonated eggs. The cells were infected at 90% confluence for 1 h with the corresponding viruses diluted in Opti-MEM. After 1 h, the cells were washed in PBS, and Dulbecco's modified Eagle's medium supplemented with 0.3% bovine albumin was added to cells until harvest. When indicated, IFN treatment was carried out by adding 1000 units/ml universal type I IFN (PBL) to the medium. RIG-I−/− MEFs have been described elsewhere (22). RIG-I−/− MEFs stably complemented with RIG-I WT or its mutants were constructed by retroviral transduction. Briefly, DNA encoding RIG-I wild type and its mutants S8D and S8E were cloned into the pBabe-puro vector. Each plasmid was transfected into EcoPack2-293 cells (Clontech) to produce pseudotyped retroviruses. For the control, an empty pBabe-puro vector was transfected. The RIG-I−/− MEF cells were infected with the pseudotyped retroviruses encoding each construct and selected with 0.8 μg/ml puromycin (Sigma).
Protein Purification
GST-CARD2 purification has been described previously (21). Endogenous RIG-I was purified from A549 cells treated for 24 h with the indicated stimuli. Cells were harvested, washed, and lysed in a hypotonic based buffer (25 mm Tris-HCl (pH 7.6), 25 mm NaCl, 1% Nonidet P-40 supplemented with the protease inhibitor mixture Complete (Roche Applied Science) and with a phosphatase inhibition mixture (Calbiochem) following the manufacturer's instructions. After 15 min, the hypotonic lysate was equilibrated with NaCl to 200 mm (isotonic conditions) and incubated on ice for an additional 30 min. Total cell lysates were precleared for 1 h at 4 °C using protein G-agarose beads (Roche Applied Science) and further clarified by centrifugation at 17,000 rpm on a Beckman SW28 rotor for 45 min. The resulting supernatants were used for immunoprecipitation using 1C3 anti RIG-I monoclonal antibodies (0.01 mg/ml cellular lysate). After 12 h in slow rotation at 4 °C, lysates were incubated for 2 h at 4 °C with protein G-agarose beads. Precipitated beads were washed extensively with 25 mm Tris-HCl (pH 7.6), 200 mm NaCl, and 1% Nonidet P-40. 2× Laemmli SDS buffer was used to elute the proteins. Proteins were separated electrophoretically using a 7% SDS-PAGE gel. RIG-I corresponding bands were excised and stored at −80 °C until analysis.
Recombinant RIG-I was expressed in BL21 pLys bacteria. Bacteria containing pGEX-6p-1 full-length RIG-I plasmid were maintained in 2XYT medium and induced for 24 h at 18 °C with 100 μm isopropyl 1-thio-β-d-galactopyranoside (Sigma). Bacterial cell pellet was resuspended in lysis buffer (25 mm Tris-HCl (pH 8.0), 1 m NaCl, 0.1% Nonidet P-40, 1 mm tris(2-carboxyethyl)phosphine), sonicated, and centrifuged at 17,000 rpm on a Sorvall RC-34 rotor for 40 min. The lysate was passed through a filter and loaded onto GE Healthcare glutathione-Sepharose 4B beads. The column was washed extensively with 25 mm Tris-HCl (pH 8.0), 1 m NaCl, 0.1% Nonidet P-40. Recombinant protein was eluted with 25 mm Tris-HCl (pH 8.0), 200 mm NaCl, and 5% glycerol containing 10 mm glutathione. GST was cleaved from the GST-RIG-I fusion protein by digestion with PreScission protease (Amersham Biosciences) for 10 h at 4 °C. Recombinant RIG-I protein was further purified by ion exchange using a Q-Sepharose resin (GE Healthcare), followed by a gel filtration step using a Superdex 200 (Amersham Biosciences).
Antibodies
1C3 monoclonal antibody was generated by injecting mice with incomplete Freund's adjuvant together with 100 μg of recombinant RIG-I CARD (amino acids 1–211) protein purified from bacteria. Three weeks after immunization, animals were boosted with incomplete Freund's adjuvant and 50 μg of protein and euthanized 10 days later. Splenocytes were isolated to generate the fusion hybridomas. Monoclonal antibodies from different hybridoma clones were screened by an enzyme-linked immunosorbent assay using the purified recombinant protein. Monoclonal antibodies were also used to detect overexpressed or endogenous RIG-I in 293T cells. The phosphospecific serine 8-RIG-I antibody was generated by immunizing rabbits with the phosphopeptide Cys-MTTEQRRpSLQAFQDYIRKTLDPTYILSY covalently coupled to keyhole limpet hemocyanin. For purification purposes, a second group of rabbits was immunized with the non-phospho-Cys-MTTEQRRSLQAFQDYIRKTLDPTYILSY peptide. The presence of phosphospecific immunoreactivity was detected by an enzyme-linked immunosorbent assay using both phosphorylated and non-phosphorylated peptides. Phosphopeptide-specific antibodies were purified by first passing the antibodies over immobilized non-phosphorylated peptide to remove antibodies that are reactive to non-phosphorylated epitopes. The non-absorbed fraction was then passed over a column of immobilized phosphopeptide. After extensive washing, the retained immunoglobulins were eluted, dialyzed, and concentrated. Anti-FLAG M2 and anti HA monoclonal antibody were purchased from Sigma. Anti-α-tubulin was purchased from Abcam. Anti-GST antibody was purchased from Sigma.
Mass Spectrometry
The detailed procedures of the in-gel digestion have been described previously (26). Briefly, bands corresponding to RIG-I protein were cut to small pieces, destained and washed using a mixture of 50 mm ammonium bicarbonate, and followed by acetonitrile and 50 mm ammonium bicarbonate. Then 10 mm dithiothreitol and 55 mm iodoacetamide were added to reduce and alkylate the protein sequentially. The protein was then digested with trypsin (12.5 ng/μl in 25 mm ammonium bicarbonate) at 37 °C overnight. The resulting tryptic peptides were extracted from the gel pieces with 5% formic acid in 50% acetonitrile and evaporated in a vacuum centrifuge. Trypsin (sequencing grade) was purchased from Promega.
MALDI Time-of-flight Analysis
Dried peptides were resuspended in 0.1% trifluoroacetic acid in 5% acetonitrile. 1 μl of the peptide solution was mixed with 1 μl of matrix solution (saturated α-cyano-4-hydroxycinnamic acid in a 1:4:4 (v/v/v) mixture of formic acid/water/isopropyl alcohol solution). A small amount of mixture (around 1 μl) was spotted to a sample plate. A Voyager-DE STR time-of-flight mass spectrometer (Applied Biosystems) was used to collect spectra in linear and reflector mode. The spectra were smoothed, calibrated, and analyzed using a program called M-over-Z. All chemical compounds were provided by Sigma.
Transfection and Reporter Assays
Transient transfection of 293T cells was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were transfected with 100 ng of the IFN-β promoter-driven firefly luciferase reporter vector pGL4-IFNβ-firefly luciferase (described previously (27)) and 100 ng of the constitutive expression plasmid Renilla luciferase reporter vector pRL-TK together with the indicated RIG-I constructs. 12–18 h after transfection, the cells were harvested and lysed in dual reporter lysis buffer (Promega). IFN reporter firefly luciferase values were normalized using Renilla luciferase values. Protein expression levels were determined by Western blot using specific antibodies against the indicated proteins. When indicated, cells were infected for 1 h with influenza PR8 ΔNS1 at a multiplicity of infection (MOI) of 2, Sendai Cantell at 1000 HA units/ml, or VSV-GFP at an MOI of 0.05 for 1 h, followed by incubation of the cells in Dulbecco's modified Eagle's medium supplemented with 0.3% bovine albumin or Opti-MEM for 24 h.
IRF-3 Translocation
2 million 293T cells were co-transfected with 200 ng of the different 2CARD plasmid constructs together with 500 ng of GFP-IRF-3 plasmid. 12 h after transfection pictures of living cells were taken, and the GFP translocation percentage was calculated.
Immunofluorescence
Cells were fixed with cold methanol for 10 min at 4 °C, washed three times in 1× PBS for 10 min, permeabilized with 0.25% Triton X-100 for 60 min, and blocked with 10% fetal bovine serum at 37 °C for 30 min. Cells were incubated for 2 h with the primary antibody (M2 FLAG monoclonal antibody, 1:500 dilution) and anti-whole PR8 virion polyclonal serum (1:1000) diluted in blocking buffer. Samples were washed three times with 0.1% Tween 20, 1× PBS for 30 min and finally incubated with the secondary antibodies (donkey anti-mouse Texas Red from Jackson ImmunoResearch and goat anti-rabbit Alexa Fluor 350 from Invitrogen) at a 1:3000 dilution in 0.1% Triton X-100 1× PBS for 2 h at room temperature. Cells were finally washed three times with 0.1% Tween 20, 1× PBS for 30 min before fluorescence analysis using a Zeiss LSM 510 Meta confocal microscope.
Immunoprecipitation
Transfected cells were washed, collected, and resuspended in PBS. Cells were pelleted by centrifugation and lysed in immunoprecipitation buffer (Tris-HCl, pH 7.6, 150–400 mm NaCl, and 1% Nonidet P-40, 1 mm EDTA, and Complete protease inhibitor (Roche Applied Science)). The lysates were cleared by centrifugation and incubated with anti-FLAG M2-agarose beads (Sigma) at 4 °C for several h. The beads were then washed several times with the immunoprecipitation buffer. 2× SDS Laemmli buffer was added to the beads, and the beads were boiled to elute the proteins. After centrifugation, the immunoprecipitated eluted proteins from the beads were subjected to SDS-PAGE and Western blotting.
Western Blotting
Proteins were separated by SDS-PAGE (Bio-Rad) and then transferred onto polyvinylidene difluoride membranes (Millipore) following the manufacturer's instructions. After blocking with 5% milk, the membranes were incubated with the indicated antibodies. After being washed with phosphate-buffered saline containing 0.5% Tween 20, the membranes were incubated with a horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin secondary antibody (Amersham Biosciences). The horseradish peroxidase immunocomplexes were detected using an enhanced chemiluminescence kit from PerkinElmer Life Sciences (catalogue no. NEL101).
Structural Modeling
The automated protein structure homology-modeling server Phyre (28) was used to generate the structural model of CARD1 using as a template the best fitting prototype CARD domain that corresponds to the CARD domain of human caspase 9 protein (Protein Data Bank code 3ygs). PyMOL (Delano Scientific) (29) was used to generate the final figures for the structural model.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism software (version 4.0). The significance between the luciferase activities of CARD and full-length constructs was determined by paired two-sample Student's t test.
RESULTS
Phosphorylation of RIG-I Serine 8
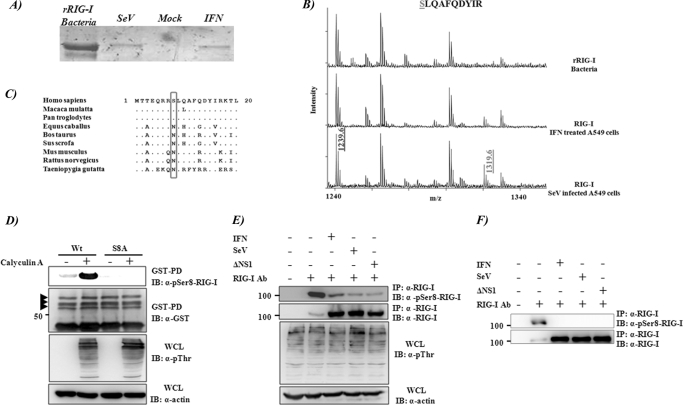
To identify additional post-translational modifications of RIG-I, endogenous human RIG-I was purified from A459 lung carcinoma cells using monoclonal antibody 1C3 (Fig. 1A). Because the endogenous protein levels of RIG-I are low in unstimulated cells, we stimulated RIG-I expression by treating cells with recombinant universal type I IFN or by infecting with Sendai virus. In both cases, RIG-I protein was highly induced and could be isolated in enough amounts to be analyzed by mass spectrophotometry. MALDI analysis of peptides detected a phosphorylated species of the peptide SLQAFQDYIR corresponding to amino acid residues 8–17 within RIG-I CARD1. This species was not present in recombinant RIG-I purified from bacteria (Fig. 1B), suggesting that RIG-I undergoes phosphorylation at the serine 8 residue. To confirm the in vivo phosphorylation of RIG-I serine 8, we generated a polyclonal antibody specific for the serine 8-phosphorylated peptide. This antibody specifically recognizes the overexpressed GST-2CARD WT form of RIG-I but not a RIG-I CARD mutant in which serine 8 was mutated to alanine (S8A) (Fig. 1D). The signal for serine 8 phosphorylation was further enhanced when 293T cells were treated with calyculin A, a strong serine/threonine protein phosphatase inhibitor (Fig. 1D, first two lanes). We next compared the serine 8 phosphorylation levels of endogenous RIG-I under different conditions by using the same serine 8 phosphospecific serum. 293T cells were left untreated or were treated with different stimuli shown to induce RIG-I protein expression, such as IFN treatment or Sendai Cantell or influenza A/PR/8/34 ΔNS1 virus infections. Cells were treated again with calyculin A for 45 min in order to block serine/threonine phosphatases or left untreated. Interestingly, whereas a high level of serine 8 phosphorylation was detected in untreated cells, this phosphorylation was strongly reduced upon IFN or virus stimulation (Fig. 1, E and F). Sequence comparison among RIG-I proteins from different species revealed that serine 8 is present in RIG-I proteins derived from primate species, whereas other animal species carry an asparagine (Asn8) (Fig. 1C). This indicates that serine 8 phosphorylation could mediate a primate-specific regulation of RIG-I activation.
FIGURE 1.
RIG-I serine 8 phosphorylation. A, RIG-I was expressed and purified from bacteria, or endogenous RIG-I was purified by immunoprecipitation from human lung carcinoma epithelial A549 cells (10 million cells/sample) that were untreated or treated with 1000 units/ml of universal type I IFN (PBL) or infected with Sendai Cantell virus (50 HA units/ml) for 24 h. Recombinant protein purification details are described under “Experimental Procedures.” An SDS-polyacrylamide gel shows purified RIG-I from these samples. Bands are detected by Coomassie staining. B, MALDI spectra of peptides from trypsin-digested RIG-I proteins from A (from top to bottom, recombinant RIG-I expressed in bacteria and endogenous RIG-I from A549 treated with type I universal IFN or infected with Sendai virus, respectively). The peak labeled 1239.6 (m/z) corresponds to unphosphorylated peptide SLQAFQDTYIR, and peak 1319.6 (m/z) represents the phosphorylated peptide. C, alignment of the first 20 amino acids of RIG-I from different species. Position 8 is boxed. D–F, detection of serine 8 phosphorylation of RIG-I in vivo by using a phospho-Ser8-RIG-I antibody. D, serine 8 phosphorylation of RIG-I 2CARD. At 48 h after transfection with GST-RIG-I 2CARD WT or S8A mutant, HEK293T cells were mock-treated or treated with a 100 nm concentration of the phosphatase inhibitor calyculin A (Invitrogen) for 45 min. WCLs were subjected to GST-PD, followed by immunoblot (IB) with anti-Ser(P)8 (pSer8)-RIG-I or anti-GST antibody. WCLs were further used for immunoblotting with anti-Thr(P) (α-pThr) and anti-actin antibodies. The arrows indicate ubiquitinated RIG-I bands, as described previously (21). The phosphorylated serine 8 band corresponds to the lower GST-CARD band (unubiquitinated). E and F, serine 8 phosphorylation of endogenous RIG-I. HEK293T cells were non-treated, treated with IFN-β (1000 units/ml) for 12 h, or infected with either SeV (50 HA units/ml) or ΔNS1 A/PR8/34 influenza (MOI 3) for 16 h. Before harvesting, cells were either treated (E) or non-treated (F) with 100 nm calyculin A for 45 min. WCLs were subjected to immunoprecipitation (IP) with an anti-RIG-I antibody, followed by immunoblot with anti-Ser(P)8-RIG-I or anti-RIG-I antibody. WCLs were also used for immunoblotting with anti-Thr(P) (as an indication of calyculin activity) or anti-actin antibody.
Functional Effect of RIG-I Serine 8 Substitution on IFN-β Induction
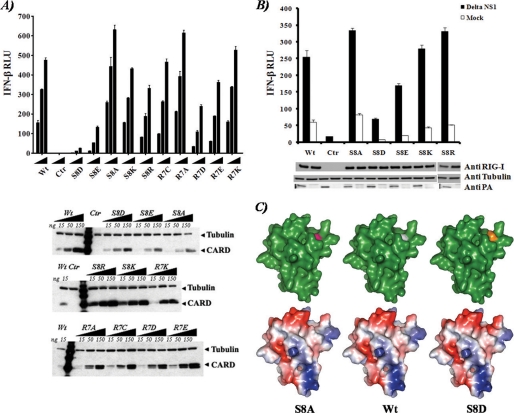
In order to investigate the effect of serine 8 phosphorylation on RIG-I function, serine 8 was mutated to the phosphomimetic amino acids aspartate (S8D) and glutamate (S8E) as well as to the non-phosphorylatable residues alanine (S8A), lysine (S8K), and arginine (S8R). To test the site specificity of this residue, we generated equivalent mutations at the proximal amino acid arginine 7 (R7D, R7E, R7A, R7K, and R7C). We then tested the ability of the 2CARD region of RIG-I to induce IFN-β in 293T cells by co-transfecting the aforementioned constructs along with reporter plasmids containing the IFN-β enhancer-promoter sequence fused to firefly luciferase reporter gene (Fig. 2A) and the pRL-TK plasmid to normalize IFN-β-driven firefly luciferase levels. Phosphomimetic amino acid substitutions of RIG-I serine 8 reduced IFN-β promoter activation, with S8D resulting in the strongest reduction. In contrast, the substitution S8A resulted in a slightly increased activation (p < 0.01) of the IFN-β promoter as compared with the WT CARD. The S8K and S8R mutations had only minor effects on the activation of the promoter. By contrast, mutations at arginine 7 had only a moderate impact in IFN-β promoter activation (Fig. 2A). Collectively, these results suggest that specific phosphorylation of RIG-I at serine 8 reduces the ability of RIG-I to induce IFN-β.
FIGURE 2.
Functional effect of RIG-I serine 8 substitutions on IFN-β induction. A, top, IFN-β reporter assay of RIG-I CARD WT or mutants in 293T cells. Experiments were performed in triplicate, and error bars indicate the S.D. for each sample. Bottom, Western blot analysis of the different samples. B, IFN-β reporter assay and Western blot analysis of the different overexpressed full-length RIG-I in 293T cells. 12 h after transfection, the cells were mock-infected or infected for 24 h with influenza A/PR/8/34 ΔNS1 virus (Delta NS1) at an MOI of 2. Experiments were performed in triplicate, and error bars indicate the S.D. for each sample. C, structural modeling of RIG-I CARD1. The Phyre modeling server was used to model the structure of the first CARD-like domain of RIG-I. Top (from left to right), surface representation of S8A, WT, and S8D CARD1 (green). Amino acid residue 8 is highlighted in different colors. Bottom, charge surface representation of the upper models. Red, negatively charged areas; blue, positively charged areas. Statistical analysis for the experiments was performed using GraphPad software. RLU, relative luciferase units.
We next introduced the S8D, S8E, S8A, S8K, and S8R mutations into full-length human RIG-I (Fig. 2B). In order to test the effect of these RIG-I mutants on IFN-β induction in the context of viral infection, we infected the cells overexpressing the indicated mutants with influenza A/PR/8/34 ΔNS1 virus. Influenza virus infection has been reported to be recognized only by RIG-I in order to stimulate the induction of IFN-β production (30). Influenza ΔNS1 virus is attenuated in its ability to interfere in the production of IFN-β (31) and can therefore be used to induce IFN. As compared with WT RIG-I, RIG-I S8D mutant exhibited a reduction (∼5-fold) in its ability to induce IFN-β-driven reporter protein in response to influenza A virus infection despite similar RIG-I expression levels, whereas S8E had activity that was slightly reduced and S8K had activity that was similar to that of WT. In contrast, S8A and S8R showed a significant (p < 0.01) but modest enhancement of the IFN-β reporter induction compared with WT.
In order to visualize the relative position of the residue Ser8 in the first CARD-like domain of RIG-I, we generated a molecular model using the structure of the homologous CARD domain of caspase 9 as a template (Fig. 2C). The modeled structures of the WT, S8A, and S8D CARD1-like domains show no major structural changes. Nevertheless, the amino acid residue serine 8 is exposed, and a mutation of such residue to aspartate affects the overall surface charge of the surrounding area. The model also predicts that the serine 8 is going to be in close proximity to the C-terminal end of the CARD1 domain and may bring it in close proximity to the CARD2 domain. Together, these results indicate that the RIG-I serine 8 is a sensitive position whose charge may have a negative effect on the activation of RIG-I and the IFN-β pathway, raising the possibility that the serine 8 phosphorylation could be used to inhibit the induction of IFN-β synthesis.
Functional Effect of RIG-I Serine 8 Substitutions on the Localization of IRF-3
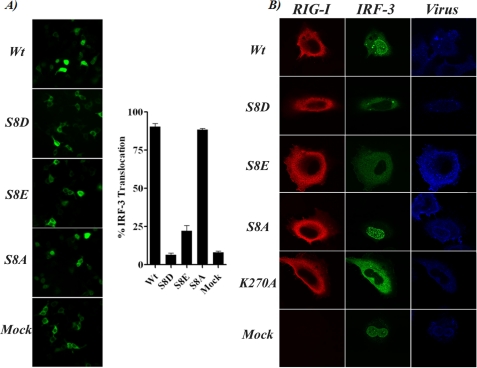
IRF-3 translocation from the cytoplasm to the nucleus is an essential step for the induction of IFN-β. Therefore, we tested the effect of mutating RIG-I serine 8 on the induction of the nuclear translocation of IRF-3. For this, 293T cells were co-transfected with plasmids expressing the different 2CARD serine 8 mutants together with GFP-IRF-3, and the localization of the live GFP signal was analyzed and quantified (Fig. 3A). Cells expressing RIG-I 2CARD WT and S8A mutant showed a strong pattern of nuclear GFP-IRF-3, whereas S8D and S8E CARD mutants were defective in inducing translocation of this transcription factor. S8E presented a less attenuated phenotype than the S8D CARD, which correlates with the reporter assay data in Fig. 2A. We further tested the effect of mutating RIG-I serine 8 on the cytoplasmic-nuclear translocation of IRF-3 in the context of virus infection. For this, HeLa cells were transfected with plasmids expressing full-length RIG-I WT, S8D, S8E, S8A, or K270A mutant together with GFP-IRF-3 followed by infection with influenza virus A/PR/8/34 ΔNS1. K270A RIG-I, known to be non-functional (17), was included as control. RIG-I and IRF-3 localization was analyzed by immunofluorescence using confocal microscopy. All RIG-I constructs showed a similar cytoplasmic localization (Fig. 3B). Although IRF-3 translocated to the cell nucleus in the presence of WT RIG-I upon virus infection, it remained mainly cytoplasmic in the presence of the RIG-I S8D and S8E mutants, in agreement with the impairment of S8D and S8E RIG-I to induce IFN-β (Fig. 2B).
FIGURE 3.
Functional effect of RIG-I serine 8 substitutions on IRF-3 localization. A, GFP-IRF-3 localization in 293T cells co-transfected with RIG-I CARD WT or with the indicated serine 8 mutants. Images of live cells were taken 12 h after transfection, and translocation percentage was calculated by dividing the amount of cells with nuclear GFP-IRF3 by the total number of GFP-positive cells. Left, live image of GFP fluorescence in co-transfected cells. Right, quantification of nucleus/cytoplasm ratio (translocation) of GFP-IRF-3. Three individual transfection experiments for each CARD construct and four fields for each transfection experiment were used for the analysis. Error bars represent the S.D. for the different measurements. B, immunofluorescence for overexpressed full-length RIG-I WT or mutants (red) or GFP-IRF-3 (green) in HeLa cells infected with influenza A/PR/8/34 ΔNS1 virus (blue).
Functional Effect of RIG-I Serine 8 Substitutions on Interaction with TRIM25 and MAVS
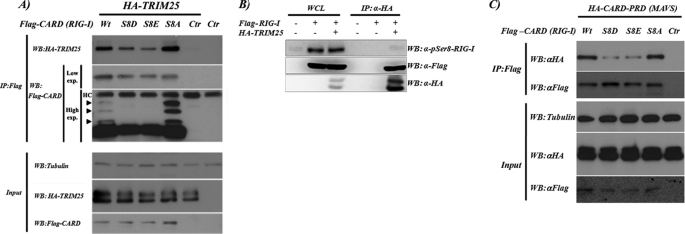
Because the CARD region of RIG-I interacts with TRIM25 and MAVS, we were intrigued by the possibility that serine 8 phosphorylation may affect these interactions. We compared the ability of WT CARD and S8D, S8E, and S8A mutant constructs to interact with TRIM25 by co-transfecting 293T cells with plasmids expressing full-length TRIM25 and the different RIG-I CARD constructs. In co-immunoprecipitation experiments, TRIM25 interacted with all of the CARD constructs, but the phosphomimetic mutants S8D and S8E showed a markedly decreased binding ability for TRIM25 compared with WT CARD. In contrast, the S8A mutant showed a stronger binding to TRIM25 than WT CARD (Fig. 4A). These results indicate that RIG-I serine 8 phosphorylation could have an inhibitory effect on the TRIM25 binding to the RIG-I first CARD. Immunoprecipitated CARD WT presented three additional bands of higher molecular weight that have been reported to be CARD ubiquitinated forms (21). The S8A CARD construct showed a stronger signal for those higher molecular weight bands, whereas in the S8D and S8E CARD mutants, the post-translational modified bands disappeared. Following these observations, we tried to confirm a higher affinity of dephosphorylated serine 8 RIG-I for TRIM25. 293T cells were transfected with plasmids expressing FLAG-RIG-I and HA-TRIM25. We compared the amount of phosphorylated serine 8 RIG-I present in the whole cell lysate (WCL) with the one present in RIG-I co-immunoprecipitated with TRIM25 for a similar amount of total RIG-I (Fig. 4B). We observed that TRIM25 preferentially binds dephosphorylated RIG-I because the amount of phosphorylated serine 8 RIG-I is undetectable in the RIG-I co-immunoprecipitated with TRIM25. In addition, this observation strengthens the results obtained with the phosphomimetic mutants, indicating that S8D and S8E mimic RIG-I serine 8 phosphorylation.
FIGURE 4.
RIG-I CARD interaction with TRIM25 and MAVS CARD-PRD. A, S8D and S8E RIG-I 2CARD mutants exhibit a decreased TRIM25 binding ability. Top, 293T cells (2 million cells) were co-transfected with the indicated plasmids. 24 h after transfection, the interactions between TRIM25 and RIG-I 2CARD WT or mutants were analyzed by immunoprecipitation (IP), followed by Western blot (WB). Bottom, Western blot showing the expression levels of the transfected proteins and tubulin. B, RIG-I phosphorylation at serine 8 decreases its ability to bind TRIM25. At 48 h after transfection with vector or HA-TRIM25 together with RIG-I-FLAG, HEK293T cells were treated with 100 nm calyculin A (Invitrogen) for 45 min. WCLs were subjected to immunoprecipitation with α-HA. Immunoprecipitates and WCLs were then immunoblotted with anti-Ser(P)8 (pSer8)-RIG-I, anti-FLAG, or anti-HA antibodies. C, S8D and S8E RIG-I 2CARD mutants have a decreased ability for MAVS binding. Top, 293T cells (2 million cells) were co-transfected with the indicated plasmids. 24 h after transfection, CARD interaction with the MAVS CARD-PRD region was analyzed by immunoprecipitation followed by Western blot. Bottom, Western blot showing the expression levels of the different transfected proteins and tubulin.
The ubiquitination of the CARD region of RIG-I is required for optimal MAVS binding and efficient downstream signaling. We next analyzed by immunoprecipitation the ability of RIG-I 2CARD WT, S8D, S8E, and S8A mutants to interact with MAVS. 293T cells were co-transfected with the indicated CARD RIG-I-expressing plasmids together with the cytoplasmic CARD-PRD region of MAVS. In line with their decreased ubiquitination level, the S8E and S8D mutants showed a decreased ability to bind MAVS (Fig. 4C) compared with WT RIG-I CARD and S8A mutant.
RIG-I S8D and S8E Mutants Are Defective in Inducing Antiviral Responses
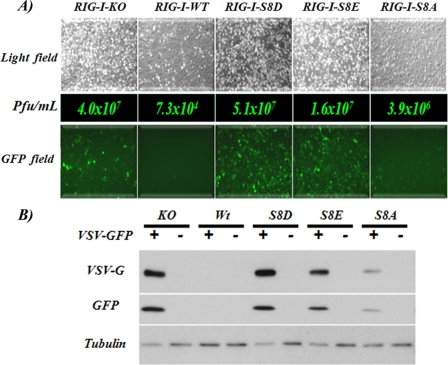
To explore the potential effects of the RIG-I serine 8 phosphorylation on RIG-I-mediated antiviral responses, RIG-I KO MEFs were transduced with constructs expressing WT, S8D, S8E, and S8A RIG-I. We then infected these cells with VSV-GFP at low MOI (0.05). 24 h after the initial infection, RIG-I−/− MEFs showed remarkably increased levels of VSV-GFP-positive cells and increased VSV yields (over 100-fold) compared with RIG-I WT MEF, as reported previously (32). By contrast, RIG-I S8D and S8E MEF behaved as RIG-I−/− MEF. RIG-I S8A expression also resulted in inhibition of replication of VSV as compared with RIG-I KO MEF. However, the degree of inhibition did not reach that of MEFs complemented with RIG-I WT. This reduced inhibition of the RIG-I S8A mutant cannot be accounted for by different expression levels of WT and S8A RIG-I (data not shown). The inability of the S8A mutant RIG-I to provide as good inhibition of VSV replication as WT RIG-I might reflect the fact that in this system, human RIG-I is expressed in mouse cells. Nevertheless, it is clear that the S8A mutant is functionally more active than the S8E and S8D mutants.
Levels of VSV-induced cell death correlated with virus titers (Fig. 5A, top). Analysis of protein expression level (Fig. 5B) showed increased levels of GFP and VSV G protein in MEF RIG-I−/− cells as well as S8D and S8E RIG-I cell lines, as compared with the RIG-I WT and S8A cells. Collectively, these results indicate that RIG-I phosphomimetics aspartate and glutamate at position 8 suppress RIG-I antiviral activity, suggesting that RIG-I phosphorylation at the serine 8 residue is critical for the regulation of RIG-I-mediated antiviral IFN response.
FIGURE 5.
RIG-I phosphomimetic mutants are impaired in inducing an antiviral response. A, RIG-I−/− MEFs (RIG-I-KO) and RIG-I−/− MEFs expressing full-length RIG-I WT or different mutants were infected with VSV-GFP at an MOI of 0.05. 24 h after infection, images of living cells were taken in order to compare the virus cytopathic effect (top; bright field) and the degree of virus replication (bottom; GFP field). Middle, cell supernatants were analyzed for the presence of virus. VSV-GFP titers from supernatants are indicated in plaque-forming units (Pfu)/ml. B, infected cells from A or mock-infected cells were collected and lysed, and the protein levels of VSV-G, GFP, and tubulin were analyzed by Western blot.
DISCUSSION
Here we provide data showing that RIG-I serine 8 is phosphorylated in human cell lines and that this phosphorylation is related to a repressive mechanism that affects the ability of RIG-I to induce IFN-β synthesis. Although phosphorylation is commonly associated with activation mechanisms, there are many examples where phosphorylation of a protein results in its functional repression. Phosphorylation events in the microtubule plus-end tracking proteins inhibit its interaction with EB1, thereby affecting the targeting of plus-end tracking proteins to growing microtubules (33). NF-κB regulation can also be negatively regulated by phosphorylation of NEMO. Specifically, phosphorylation of serine 68 in the IκB kinase-binding domain of NEMO interferes with the structure of the IκB kinase complex and the tumor necrosis factor-α-induced NF-κB activity (34).
By combining mass spectrometry analysis with biochemical and cellular biology experiments, we provide evidence of a new mechanism that may control the IFN-β induction pathway. We show that a negative charge at serine 8 in RIG-I has a negative impact on the ability of TRIM25 to bind the RIG-I CARD functional domain and that it hinders RIG-I CARD ubiquitination and thereby its ability to form a complex with MAVS (Fig. 4). Structural modeling analysis (Fig. 2C) suggests that serine 8 is an exposed residue and that this residue is in close proximity to the 5-amino acid linker between the two RIG-I CARD-like domains. Because TRIM25 binds to the CARD1 region of RIG-I, we propose that serine 8 phosphorylation regulates the activation of RIG-I by two possible but not mutually exclusive mechanisms. First, serine 8 phosphorylation may affect the ability of the TRIM25 SPRY to bind RIG-I CARD1 and thereby decreases RIG-I CARD ubiquitination. This is consistent with the decrease in the ability of serine 8 phosphomimetic mutants to bind to TRIM25. Second, the close proximity of the phosphorylation site to the C-terminal portion of the CARD1 domain puts this residue in close proximity to the CARD2 of RIG-I, whose lysines 154, 164, and 172 are predicted by structural modeling (data not shown) to be in close proximity to the N terminus of CARD2. These lysines are critical ubiquitination sites for TRIM25 and REUL (20, 21), and their ubiquitination is required for an optimal RIG-I-mediated IFN-β mRNA induction. The closeness of CARD1 serine 8 to Lys154, Lys164, and Lys172 in CARD2 could have a negative impact on the catalytic ubiquitination reaction of these lysines. Thus, unphosphorylated serine 8 in CARD1 may allow both optimal TRIM25 binding and TRIM25-mediated ubiquitination.
A portion of endogenous RIG-I purified from infected and IFN-treated cells was phosphorylated at serine 8, but we found that IFN induction and viral infection lead to lower levels of phosphorylated serine 8 (Fig. 1). Thus, although RIG-I serine 8 phosphorylation may help to maintain an inactive state under non-inducing conditions, stimulation with IFN results not only in increased levels of RIG-I but also in decreased serine 8 phosphorylation, which is consistent with the known positive feedback mechanism of IFN in priming cells to induce higher amounts of IFN when stimulated by virus infection (35). The S8A RIG-I mutant showed a modest but consistently higher capacity to induce the IFN-β promoter as compared with wild type RIG-I. This increased potential could be an indication of the contribution of the serine 8 phosphorylation to the partial repression of RIG-I. The presence of a serine at position 8 is exclusive to primates, with the other species containing an asparagine at this position (Fig. 1C). Thus, RIG-I serine 8 phosphorylation appears to be a primate-specific negative regulatory mechanism for the control of RIG-I activity. It remains to be determined whether a different residue is phosphorylated in RIG-I from non-primate species and if that contributes to the negative regulation of IFN in these species.
Overexpressed GST-RIG-I 2CARD contained phosphorylated serine 8 in both ubiquitinated and non-ubiquitinated species, although the ratio between phosphorylated and non-phosphorylated peptides at serine 8 was higher in the non-ubiquitinated CARDs (data not shown). This indicates that RIG-I serine 8 phosphorylation could occur independently of its ubiquitination status, but more experiments will be needed to evaluate whether this is the case.
We do not know at present which mechanism triggers RIG-I serine 8 phosphorylation or dephosphorylation and which cellular kinases and phosphatases are responsible for regulating this phosphorylation. RIG-I serine 8 phosphorylation may also be regulated differentially in different cell types or by yet unknown stimuli, such as cellular stress, hormone environment, or cytokine and chemokine stimulation. We have also found that threonine 170 in the CARD2 domain of RIG-I undergoes phosphorylation and, similar to serine 8 phosphorylation, it has a negative impact on RIG-I ubiquitination and activation (36). It will then be interesting to know whether RIG-I serine 8 and threonine 170 phosphorylations are controlled by the same or a different set of cellular kinases/phosphatases. Our observations of the impact of phosphorylation on RIG-I activity open a new aspect of RIG-I regulation that will require further research in this area.
In addition to RIG-I phosphorylation described here, ubiquitination and ISGylation have been shown to be involved in the regulation of RIG-I activity (20, 21, 37, 38). Lys63 polyubiquitination of the RIG-I CARD region has been shown to be essential for the activation of RIG-I. TRIM25 and REUL E3 are the ubiquitin ligases that mediate this ubiquitination, but the mechanism by which these ligases are activated or repressed remains unknown. It has been suggested that the key amino acids in RIG-I CARD for its ubiquitination are exposed following the recognition of viral RNA by RIG-I (7, 18, 22), but the sequence of these associated events and their molecular regulation are unclear. It is only now becoming clearer that although RIG-I is able to bind several types of RNA, only 5′-triphosphate moieties of RNA (39) that contain double-stranded regions (4, 5) are able to trigger RIG-I-mediated induction of IFN-β. In addition, although RIG-I activation is linked to cellular processes triggered by virus infection (40–42), the actual activator template of RIG-I in virus-infected cells remains unknown, and it remains unknown whether this activator has to be accompanied by other co-stimuli that may deactivate the RIG-I suppressive mechanisms. In this context, protein phosphatases are likely to play an important role in controlling RIG-I serine 8 phosphorylation. Together, the data we present here on RIG-I repression by serine 8 phosphorylation open a new aspect of the regulation in the IFN pathway that may play an important role in pathological processes related to viral infection and/or autoimmunity.
Acknowledgments
We thank Alina Baum, Mirco Schmolke, Ben Hale, Alissa Pham, Benjamin TenOever, and Pablo DeIoannes for reagents and valuable discussions.
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grants RR00168 (to M. U. G.); CA082057, U19AI083025, and AI083355 (to J. U. J. and M. U. G.); AI041706 (to A. K. A.); and U19AI083025 and R01AI46954 and NIH, NIAID, Contract HHSN266200700010C (to A. G.-S.). This work was also supported by grants from the Fletcher Jones Foundation and Hastings Foundation (to J. U. J. and M. U. G.).
- IFN
- interferon
- ISG
- IFN-stimulated gene
- CARD
- caspase recruitment domain
- RD
- repressor domain or regulatory domain
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline
- MEF
- mouse embryo fibroblast
- GST
- glutathione S-transferase
- MALDI
- matrix-assisted laser desorption ionization
- WCL
- whole cell lysate
- VSV
- vesicular stomatitis virus
- GFP
- green fluorescent protein
- MOI
- multiplicity of infection.
REFERENCES
- 1.Stark G. R., Kerr I. M., Williams B. R., Silverman R. H., Schreiber R. D. (1998) Annu. Rev. Biochem. 67, 227–264 [DOI] [PubMed] [Google Scholar]
- 2.Stetson D. B., Medzhitov R. (2006) Immunity 25, 373–381 [DOI] [PubMed] [Google Scholar]
- 3.Kawai T., Akira S. (2008) Ann. N.Y. Acad. Sci. 1143, 1–20 [DOI] [PubMed] [Google Scholar]
- 4.Schlee M., Roth A., Hornung V., Hagmann C. A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G., Juranek S., Kato H., Kawai T., Poeck H., Fitzgerald K. A., Takeuchi O., Akira S., Tuschl T., Latz E., Ludwig J., Hartmann G. (2009) Immunity 31, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt A., Schwerd T., Hamm W., Hellmuth J. C., Cui S., Wenzel M., Hoffmann F. S., Michallet M. C., Besch R., Hopfner K. P., Endres S., Rothenfusser S. (2009) Proc. Natl. Acad. Sci. U. S. A. 106, 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T. S., Fujita T., Akira S. (2008) J. Exp. Med. 205, 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M., Jr., Inagaki F., Fujita T. (2008) Mol. Cell 29, 428–440 [DOI] [PubMed] [Google Scholar]
- 8.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 9.Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 10.Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. (2005) Mol. Cell 19, 727–740 [DOI] [PubMed] [Google Scholar]
- 11.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. (2005) Nature 437, 1167–1172 [DOI] [PubMed] [Google Scholar]
- 12.McWhirter S. M., Tenoever B. R., Maniatis T. (2005) Cell 122, 645–647 [DOI] [PubMed] [Google Scholar]
- 13.Wathelet M. G., Lin C. H., Parekh B. S., Ronco L. V., Howley P. M., Maniatis T. (1998) Mol. Cell 1, 507–518 [DOI] [PubMed] [Google Scholar]
- 14.Thanos D., Maniatis T. (1995) Cell 83, 1091–1100 [DOI] [PubMed] [Google Scholar]
- 15.Agalioti T., Lomvardas S., Parekh B., Yie J., Maniatis T., Thanos D. (2000) Cell 103, 667–678 [DOI] [PubMed] [Google Scholar]
- 16.Saito T., Hirai R., Loo Y. M., Owen D., Johnson C. L., Sinha S. C., Akira S., Fujita T., Gale M., Jr. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. (2004) Nat. Immunol. 5, 730–737 [DOI] [PubMed] [Google Scholar]
- 18.Cui S., Eisenächer K., Kirchhofer A., Brzózka K., Lammens A., Lammens K., Fujita T., Conzelmann K. K., Krug A., Hopfner K. P. (2008) Mol. Cell 29, 169–179 [DOI] [PubMed] [Google Scholar]
- 19.Yoneyama M., Fujita T. (2008) Immunity 29, 178–181 [DOI] [PubMed] [Google Scholar]
- 20.Gao D., Yang Y. K., Wang R. P., Zhou X., Diao F. C., Li M. D., Zhai Z. H., Jiang Z. F., Chen D. Y. (2009) PLoS One 4, e5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gack M. U., Shin Y. C., Joo C. H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., Jung J. U. (2007) Nature 446, 916–920 [DOI] [PubMed] [Google Scholar]
- 22.Gack M. U., Kirchhofer A., Shin Y. C., Inn K. S., Liang C., Cui S., Myong S., Ha T., Hopfner K. P., Jung J. U. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16743–16748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mibayashi M., Martínez-Sobrido L., Loo Y. M., Cárdenas W. B., Gale M., Jr., García-Sastre A. (2007) J. Virol. 81, 514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Escalante C. R., Nistal-Villán E., Shen L., García-Sastre A., Aggarwal A. K. (2007) Mol. Cell 26, 703–716 [DOI] [PubMed] [Google Scholar]
- 25.Talon J., Salvatore M., O'Neill R. E., Nakaya Y., Zheng H., Muster T., García-Sastre A., Palese P. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4309–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steen H., Pandey A., Andersen J. S., Mann M. (2002) Sci. STKE 2002, pl16. [DOI] [PubMed] [Google Scholar]
- 27.Kochs G., García-Sastre A., Martínez-Sobrido L. (2007) J. Virol. 81, 7011–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 29.DeLano W. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 30.Loo Y. M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martínez-Sobrido L., Akira S., Gill M. A., García-Sastre A., Katze M. G., Gale M., Jr. (2008) J. Virol. 82, 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X., Li M., Zheng H., Muster T., Palese P., Beg A. A., García-Sastre A. (2000) J. Virol. 74, 11566–11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato H., Sato S., Yoneyama M., Yamamoto M., Uematsu S., Matsui K., Tsujimura T., Takeda K., Fujita T., Takeuchi O., Akira S. (2005) Immunity 23, 19–28 [DOI] [PubMed] [Google Scholar]
- 33.Honnappa S., Gouveia S. M., Weisbrich A., Damberger F. F., Bhavesh N. S., Jawhari H., Grigoriev I., van Rijssel F. J., Buey R. M., Lawera A., Jelesarov I., Winkler F. K., Wüthrich K., Akhmanova A., Steinmetz M. O. (2009) Cell 138, 366–376 [DOI] [PubMed] [Google Scholar]
- 34.Palkowitsch L., Leidner J., Ghosh S., Marienfeld R. B. (2008) J. Biol. Chem. 283, 76–86 [DOI] [PubMed] [Google Scholar]
- 35.Levy D. E. (2002) J. Exp. Med. 195, F15–F18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gack M. U., Nistal-Villán E., Inn K. S., García-Sastre A., Jung J. U.J. Virol. 84, 3220–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oshiumi H., Matsumoto M., Hatakeyama S., Seya T. (2009) J. Biol. Chem. 284, 807–817 [DOI] [PubMed] [Google Scholar]
- 38.Kim M. J., Hwang S. Y., Imaizumi T., Yoo J. Y. (2008) J. Virol. 82, 1474–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pichlmair A., Schulz O., Tan C. P., Näslund T. I., Liljeström P., Weber F., Reis e Sousa C. (2006) Science 314, 997–1001 [DOI] [PubMed] [Google Scholar]
- 40.Chiu Y. H., Macmillan J. B., Chen Z. J. (2009) Cell 138, 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ablasser A., Bauernfeind F., Hartmann G., Latz E., Fitzgerald K. A., Hornung V. (2009) Nat. Immunol. 10, 1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malathi K., Dong B., Gale M., Jr., Silverman R. H. (2007) Nature 448, 816–819 [DOI] [PMC free article] [PubMed] [Google Scholar]