Abstract
E1 ubiquitin-activating enzymes (UBAs) are large multidomain proteins that catalyze formation of a thioester bond between the terminal carboxylate of a ubiquitin or ubiquitin-like modifier (UBL) and a conserved cysteine in an E2 protein, producing reactive ubiquityl units for subsequent ligation to substrate lysines. Two important E1 reaction intermediates have been identified: a ubiquityl-adenylate phosphoester and a ubiquityl-enzyme thioester. However, the mechanism of thioester bond formation and its subsequent transfer to an E2 enzyme remains poorly understood. We have determined the crystal structure of the human UFM1 (ubiquitin-fold modifier 1) E1-activating enzyme UBA5, bound to ATP, revealing a structure that shares similarities with both large canonical E1 enzymes and smaller ancestral E1-like enzymes. In contrast to other E1 active site cysteines, which are in a variably sized domain that is separate and flexible relative to the adenylation domain, the catalytic cysteine of UBA5 (Cys250) is part of the adenylation domain in an α-helical motif. The novel position of the UBA5 catalytic cysteine and conformational changes associated with ATP binding provides insight into the possible mechanisms through which the ubiquityl-enzyme thioester is formed. These studies reveal structural features that further our understanding of the UBA5 enzyme reaction mechanism and provide insight into the evolution of ubiquitin activation.
Keywords: Crystal Structure, Crystallography, Enzyme Structure, Ubiquitin, Ubiquitination, E1, UBA5, UFM1, Ubiquitin-activating Enzyme, Thioester
Introduction
Protein modification by covalent attachment of ubiquitin or ubiquitin-like molecules (the acronym UBL is used in this article to refer to both ubiquitin and ubiquitin-like modifiers) plays an essential role in the function and regulation of molecular processes in all eukaryotic organisms (1, 2). UBL conjugation has been shown to have a multitude of effects, including protein degradation, cellular trafficking, cell division, and antiviral responses (3–12). UBL modification reactions are initiated by E1 enzymes (also called UBAs or ubiquitin-activating enzymes), which catalyze UBL activation via two intermediates; an adenylated UBL that forms a noncovalent complex with E1 (A-site binding), and a covalently bound UBL that is attached via a thioester bond to the E1 catalytic cysteine (T-site binding). The thioester bound UBL is subsequently transferred to a conserved cysteine of an E2 enzyme through a trans-thioesterification reaction. Ligation of the activated UBLs to substrates is mediated by E3 ligases, resulting in an isopeptide bond between the ϵ-amino group of the target protein lysine and the terminal carboxyl group of the UBL (1).
Canonical E1-activating enzymes are characterized by the presence of two conserved adenylation domains (active and inactive adenylation domains) with similar Rossman-like folds that are believed to have evolved from bacterial enzymes such as ThiF or MoeB (13). E1 homologues differ from these ancestral proteins by means of additional domains that are inserted or appended to the adenylation domain. These domains include two catalytic cysteine half-domains known as first and second catalytic cysteine half-domains (FCCH3 and SCCH, respectively), and a ubiquitin fold domain that has been shown to be required for E2 recruitment and to facilitate thioester bond transfer (14–17). All E1 enzymes also conserve a loop region called the crossover loop that separates the active adenylation domains from the SCCH.
UFM1 is one of only a handful of UBLs with a dedicated E1 (UBA5), E2 cofactor (UFC1), and E3 ligase (UFL1) (18–20). UBA5 has been shown to also be reactive, however weakly, with SUMO2, and the catalytic cysteine (Cys250) has been identified (18, 21). Although a protein target of UFMylation, C20orf116, was recently identified, the biological role of UFM1 modification reactions is currently unknown (20). UBA5 is the least characterized of all human E1 enzymes, is comparatively smaller than the other E1 enzymes (∼400 residues compared with >1000 residues), and is only present in multicellular organisms. Sequence analyses revealed that UBA5 does not appear to contain FCCH or SCCH domains but is instead composed of only an adenylation domain followed by a short C-terminal domain (CTD). The smaller size of UBA5 compared with other E1 enzymes thus raises many questions as to how this enzyme catalyzes adenylation and transthiolation reactions, in addition to mediating protein-protein interactions, which are generally catalyzed by larger multidomain E1 enzymes.
To further characterize the mechanism of UBA5 catalysis, we have targeted the human enzyme for structural and biochemical analyses. The results presented here reveal insight into how this minimalistic E1 enzyme performs its multiple biochemical activities.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
Human UBA5 (residues 57–329) was cloned from a Mammalian Gene Collection cDNA template (AT9-F4) into the pET28a-LIC vector (GenBankTM EF442785) using the In-Fusion CF Dry-Down PCR cloning kit (Clontech, catalog no. 639605). Competent BL21 (DE3) cells (Invitrogen, catalog no. C6000-03) were transformed and grown using the LEX system (HarbingerBiotech) at 37 °C in 2-liter bottles (VWR, catalog no. 89000-242) containing 1800 ml of Terrific Broth (Sigma, catalog no. T0918) supplemented with 150 mm glycerol, 100 μm kanamycin, and 600 μl anti-foam 204 (Sigma, catalog no. A-8311). When an A600 of ∼6 was reached, the temperature was reduced to 15 °C, and 1 h later, protein expression was induced with 100 μm isopropyl 1-thio-β-d-galactopyranoside (BioShop catalog no. IPT001), and the culture was incubated overnight (16 h) at 15 °C. Cell pellets were collected by centrifugation (12,227 × g for 20 min) and frozen. Cell pellets were thawed and resuspended in 40 ml lysis buffer (20 mm Tris, pH 7.5, 500 mm NaCl, 2 mm imidazole), and lysis was accomplished by sonication. Four milliliters of TALON metal affinity resin (BD Bioscience) was mixed for 2 h at 4 °C with 150 ml of lysate, centrifuged for 3 min (SX4750 rotor, Allegra X-12R, Beckman Coulter), and decanted. Beads were transferred into a 25 ml Econo-Column (Bio-Rad, catalog no. 732-1010) and washed with 3 × 15 ml wash buffer (20 mm Tris, pH 7.5, 500 mm NaCl, 10 mm imidazole). Sample was eluted with 3 column volumes of elution buffer (20 mm Tris, pH 7.5, 50 mm NaCl, 1 mm Tris(2-carboxyethyl) phosphine, 2 mm ATP, 5 mm MgCl2, 200 mm imidazole). Sample was then gel-filtered (XK 16 × 65 packed with HighLoad Superdex 200 resin, GE Healthcare) using an AKTAxpress (GE Healthcare, catalog no. 18-6645-05) in crystallization buffer (20 mm Tris, pH 7.5, 50 mm NaCl, 1 mm Tris(2-carboxyethyl) phosphine, 2 mm ATP, 5 mm MgCl2). Fractions containing protein were pooled and centrifuged through concentrators with a 10,000 kDa cut-off (Amicon Ultra-15, UFC900524, Millipore) for 30 min at 3750 rpm.
Crystallization and Structure Determination
Crystals of UBA5 were grown at 287 K using the hanging drop method by mixing equal volumes of 1 m lithium sulfate, 0.3 m ammonium sulfate, 0.1 m sodium citrate, pH 6.2, and 20 mg/ml protein solution in crystallization buffer. The crystals were cryoprotected by dragging the crystal through a drop containing cryoprotectant solution (9% sucrose (w/v), 2% glucose (w/v), 8% glycerol (v/v), 8% ethylene glycol (v/v)) and reservoir buffer. Diffraction data were collected at the Advanced Photon Source (24-ID-C) and processed using HKL2000 (22). Phase estimates were obtained by molecular replacement using Phaser (search model: Protein Data Bank code 1ZFN) (23, 24), followed by iterative model building and refinement using COOT and REFMAC (25, 26). The coordinates and structure factors of the UBA5 structure have been deposited in the Protein Data Bank with the code 3H8V.
Biochemical Assays
UFM1 loading and transfer activity of UBA5 was analyzed by incubating indicated concentrations of UBA5, UFC1, and UFM1 (Boston Biochem) in a 10-μl reaction buffer containing 50 mm Tris-HCl, pH 7.0, and 5 mm MgCl2, in the presence and absence of 1 μm ATP or 5 mm dithiothreitol at room temperature for 90 min. Products were resolved by nonreducing 10–20% gradient SDS-PAGE followed by immunoblotting using anti-UFM1 primary rabbit polyclonal antibody (Boston Biochem) and anti-rabbit secondary antibodies (Thermo Scientific) followed by enhanced chemiluminescent detection (GE Healthcare).
Thermal Denaturation Studies
An amount of 0.1 mg/ml of UBA5 in a buffer consisting of 100 mm HEPES, pH 7.5, and 150 mm NaCl was screened in the presence and absence of 2 mm ATP using an reverse transcription-PCR instrument (Stratagene) in the presence of the fluorophore, SYPRO Orange (Invitrogen). The reaction mixtures were screened in a 25-μl reaction volume in a 96-well PCR microplate (ABGene, Surrey, UK). Plots of fluorescence intensity versus temperature were fitted from the inflection point of the curves with the Boltzmann equation to obtain the temperature at which 50% of the protein was unfolded, as described in Ref. 27.
RESULTS
Overall Structure and Function
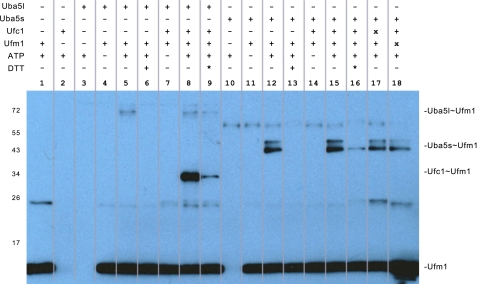
UBA5 is expressed in humans as two distinct isoforms that result from differential splicing of the full-length transcript (1–404 and 57–404) (18, 21). The role of the additional N-terminal residues in the longer splice variant is unknown; however, they are not strongly conserved (supplemental Fig. S1) and are not required for UFM1 activation (21). To obtain high resolution crystals of UBA5, the N-terminal region of the long isoform (residues 1–56) was deleted and residues 330–404 of the CTD were also removed. This UBA5 protein preparation was shown to catalyze ATP- and dithiothreitol-dependent formation of the UBA5-UFM1 thioester intermediate (Fig. 1). The removal of the CTD thus does not abrogate formation of the UBA5-UFM1 thioester intermediate, demonstrating that the UBA5 CTD is not required for adenylation or thioester transfer of UFM1 to the UBA5 catalytic cysteine. The UBA5ΔCTD construct was unable to conjugate UFM1 to UFC1, confirming the requirement of the CTD for UFC1 recruitment and thioester bond transfer (Fig. 1). This finding is in agreement with biochemical analyses of other E1 enzymes, which maintain a C-terminal ubiquitin fold domain that is required for E2 trans-thioesterification reactions (14–17) The CTD of UBA5 thus likely serves an analogous role to the ubiquitin fold domain found in other E1 enzymes.
FIGURE 1.
UFM1 activation and loading activity of UBA5. 0.5 μm UBA5, 1 μm UFC1, and 1 μm UFM1 were incubated in a 10-μl reaction buffer containing 50 mm Tris-HCl, pH 7, 5 mm MgCl2, in the presence and absence of 1 μm ATP or 5 mm dithiothreitol at room temperature for 90 min. Products were resolved by nonreducing 10–20% gradient SDS-PAGE and immunoblotted with anti-UFM1 rabbit polyclonal primary (Boston Biochem) and anti-rabbit secondary antibodies (Thermo Scientific) followed by enhanced chemiluminescent detection (GE Healthcare). *, 5 mm dithiothreitol added to assay mix following initial reaction incubation. x, 10 μm UFC1 or UFM1. Full-length (Uba5l) and truncated (Uba5s) reaction products are indicated. C, C-terminal, N, N-terminal.
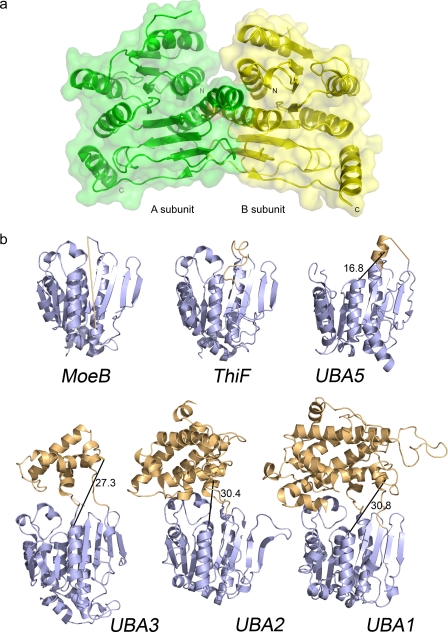
The UBA5 structure was solved to 2.0 Å resolution as a noncrystallographic homodimer using molecular replacement with a ThiF structure as the search model (1ZFN) (23) in space group P3221 (Table 1). The homodimer interaction buries 3020 Å2 surface area per UBA5 monomer (Fig. 2a). A crystallographic tetramer, which buries a dimer-dimer surface area of 2930 Å2, was also observed. However, the full-length enzyme was shown by using analytical ultracentrifugation analysis to form primarily dimers (1.85:1 dimers to monomers ratio), which indicates the enzyme is likely to be active as a dimer (supplemental Fig. S2).
TABLE 1.
UBA5 data collection and refinement statistics
PDB, Protein Data Bank; r.m.s.d., root mean square deviation; APS, Advanced Photon Source.
| Data collection | |
| PDB code | 3H8V |
| Space group | P3221 |
| Unit cell (Å) | a = b = 78.0, c = 207.01 |
| Beamline | APS (24-ID-C) |
| Wavelength (Å) | 0.97944 |
| Resolution | 50.0-2.0 |
| Unique reflections | 45,414 |
| Data redundancy | 3.9 (3.8) |
| Completeness (%) | 90.1 (93.3) |
| I/σI | 16.75 (1.65) |
| Rsym | 0.08 (0.859) |
| Rp.i.m.a | 0.045 (0.475) |
| Refinement | |
| Resolution | 25.53-2.0 |
| Reflections used | 42,909 |
| All atoms, including solvent | 3615 (159) |
| Rwork/Rfree (%) | 19.1/22.4 |
| r.m.s.d. bond length (Å) | 0.011 |
| r.m.s.d. bond angle | 1.251° |
| Mean B value (Å2) | 68.49 |
| Ramachandran analysis (%) | |
| Favored | 99.1 |
| Allowed | 100 |
| Disallowed | 0 |
a R-factor precision-indicating merging.
FIGURE 2.
Ribbon representation of the UBA5 and related crystal structures. a, molecular surface representation of the UBA5 dimer. Catalytic cysteine is shown in stick format, and N and C termini are labeled. b, architecture of E1 and E1-like enzymes. Conserved adenylation domains (shown in blue) and SCCH domains (or structurally analogous regions; shown in orange) of the following are shown: MoeB (1JWA) (13), ThiF (1ZFN) (23), UBA5, UBA3 (1YOV) (28), UBA2 (1Y8Q) (15), UBA1 (3CMM) (30). Only one of the subunits of the heterodimeric E1s are shown. Distances between catalytic cysteines and A-site regions are shown with a black line. The flexible region in UBA5 between the crossover loop and the catalytic cysteine and the analogous flexible loop region in MoeB are indicated as straight light-orange lines. Catalytic cysteines are shown in stick format. All structure figures were prepared with PyMOL.
The UBA5 structure shows similarities to both E1 and E1-like enzymes and is composed of an ATP-binding domain that consists of an eight-stranded β-sheet surrounded by seven α-helices (Fig. 2b). Similar to the adenylation domain of other E1 and E1-like structures, UBA5 maintains a zinc-binding site that is coordinated by four cysteines with tetrahedral geometry. Several residues in two loop regions (β2–α3 loop and β6–α6 loop) could not be modeled in the UBA5 model, suggesting structural flexibility for these regions, and a few residues at the N- and C-terminal ends were also disordered. The β6–α6 loop is structurally analogous to crossover loops in E1-like structures, and it is also partially disordered in most other MoeB and ThiF crystal structures (Fig. 2b). This loop precedes the catalytic cysteine in E1 structures and forms a link between the active adenylation domains and SCCH domains. In contrast to other E1 structures, which have their catalytic cysteine within the SCCH domain, the UBA5 catalytic cysteine (Cys250) is near the N terminus of the long α6-helix in the adenylation domain. The UBA5 crossover loop is thus shorter compared with other E1 structures and more closely resembles crossover loops of bacterial E1-like structures. Finally, electron density for the glycine-rich, ATP-binding active sites of UBA5 is stronger in the A subunit, and density supporting the binding of ATP is also only visible in this subunit.
ATP-binding Site
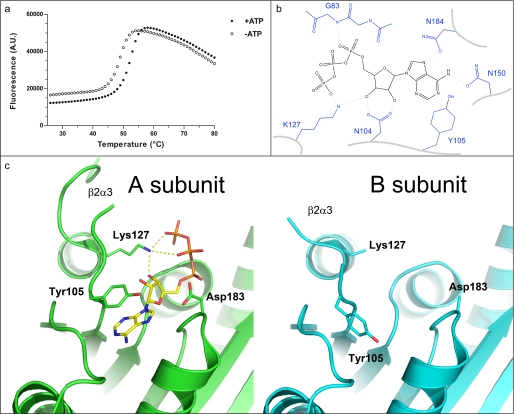
The E1 adenylation reaction step is thought to occur through a penta-coordinated α-phosphate transition state (13). This reaction is catalyzed in a glycine-rich active site; a conserved binding pocket that includes a hydrophobic patch and several charged residues that promote binding and catalysis. To characterize interactions in this pocket, UBA5 was co-crystallized in the presence of ATP. ATP was also shown by fluorescence-based thermal denaturation studies to stabilize the enzyme (Fig. 3a). Despite the presence of 2 mm ATP in the crystallization sample, ATP was observed in only the A subunit of the UBA5 homodimer and was observed to be stabilized by at least six hydrogen bonds in this monomer (Fig. 3b). The adenyl moiety of ATP is also protected from the solvent by Tyr105 in the A subunit, while in the B subunit Tyr105 is buried in the adenine pocket (Fig. 3c). Movement of Tyr105 may also affect the stability of the β2–α3 loop, which immediately follows Tyr105, because several more residues showed clear electron density and could be modeled in this loop in the ATP-bound A chain. Another residue that immediately follows the disordered β2–α3 loop, Lys127, also comes in to close proximity to the terminal phosphates and ribose of ATP (Fig. 3c), suggesting this residue plays a direct role in ATP binding (or catalysis), which could also affect the stability of the β2–α3 loop.
FIGURE 3.
ATP binding pocket of UBA5. a, thermal stabilization by ATP. Solid and empty circles represent UBA5 protein in the presence and absence of 2 mm ATP, respectively. An increase in fluorescence is indicative of protein denaturation. Plots of fluorescence intensity versus temperature were fitted from the inflection point of the curves to interpolate the temperature at which 50% of the protein was unfolded. This transition temperature was increased by 3.2 °C in the presence of ATP. b, schematic diagram of hydrogen bonding network around ATP. ATP is shown in black, labeled side chains of UBA5 are shown in blue, and hydrogen bonds are shown as dashed lines. c, the ATP-binding active site of the A (green) and B (cyan) subunits are shown from the same perspective, side by side. Side chains that show structural variation in the two subunits and ATP are shown in stick format. Distances between Lys127 and ATP γ- and β-phosphates and ribose are 2.7, 3.5, and 2.7 Å, respectively. A. U., absorbance units.
Mutagenesis studies have demonstrated the requirement for at least one positively charged residue in the vicinity of the β/γ-phosphate moiety of ATP for catalysis (13, 28). In addition to Lys127, two other positively charged UBA5 residues that could potentially act in catalysis are not visible in the electron density (Arg61 and Arg115). Arg61 is near the N-terminal region of the construct and is not visible, whereas Arg115 is in the disordered region of the β2–α3 loop. Presumably not required for ATP binding, the absence of Arg61 and Arg115 in our structure does not preclude a role for these residues in catalysis.
A strictly conserved aspartate residue, Asp183, shown to be required for the adenylation reactions through its interaction with magnesium (13), also appears to be well ordered in the UBA5 structure A subunit but does not point toward the bound ATP (Fig. 3c). In addition, there is no electron density to support the presence of magnesium (despite 5 mm present in the crystallization buffer). This observation suggests that magnesium coordination is required only during the adenylation reaction to alleviate electrostatic repulsion between the C terminus of UFM1 and the α-phosphate of ATP (13).
Catalytic Cysteine
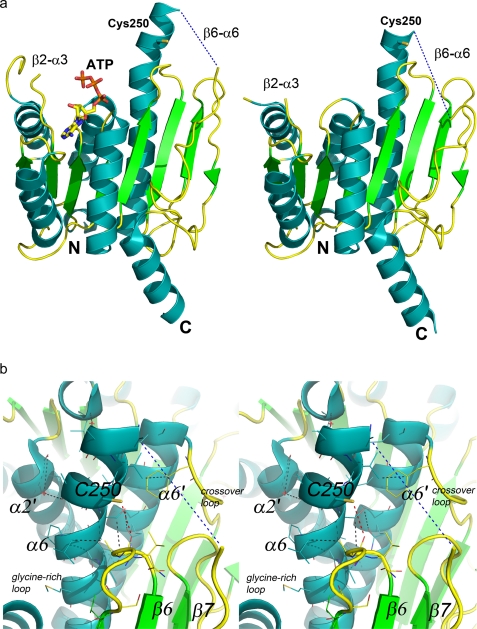
The UBA5 structure shows that both E1 catalytic cysteine half-domains are absent and that the catalytic cysteine is, in contrast to all other known E1 enzymes, part of the adenylation domain in the α6-helix (Fig. 4a). Although partially solvent exposed, this residue is adjacent to the β5–β6 loop and is within hydrogen-bonding distance of the main chain carbonyl of Asn210 (Fig. 4b). Numerous other hydrogen-bonding and van der Waals interactions are made between the helix containing the catalytic cysteine and the rest of the molecule, including the other subunit (Fig. 4b). Therefore, the helical region following the catalytic cysteine is not likely to undergo significant conformational changes during thioester bond formation. However, structural differences between the A and B subunits of UBA5 place the catalytic cysteine near a kink in the helix that is preceded by either four (A subunit) or one (B subunit) helical residues (Fig. 4a). The presence of disordered residues at the N terminus of α6 combined with the structural disorder of the β6–α6 loop that also directly precedes the catalytic region indicate some structural mobility that may allow the catalytic cysteine to move at least partially toward the adenylated UFM1 molecule to form the thioester bond.
FIGURE 4.
Ribbon representations of the A and B subunits of UBA5. a, catalytic cysteine (Cys250), ATP, and boundaries of partially disordered loop regions (β2–α3 and β6–α6) are labeled. Helices, strands, and loops are colored cyan, green, and yellow, respectively. b, stereoscopic representation of the catalytic cysteine and its environs. Van der Waals interactions are shown as dashed black lines. The disordered region of the crossover loop is shown as a dashed blue line, and the hydrogen bond between the catalytic cysteine sulfur and the Asn210 main chain carbonyl is shown as a dashed red line. Secondary structures are labeled; B subunit labels are primed. Water molecules are shown as red stars. N, N-terminal; C, C-terminal.
Model of UFM1 Binding to Adenylation Site
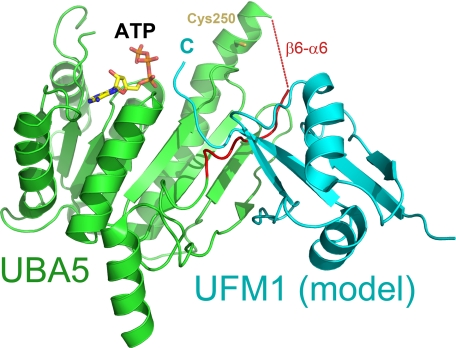
Several structures of E1 and E1-like ligases have revealed that the binding of UBLs to their cognate E1 A-site is a conserved structural interaction, and models of UBLs in complex with their respective E1s can be confidently performed by structural superposition (28). Attempts to co-crystallize UBA5 with UFM1 were unsuccessful, and therefore, a model of UFM1 binding was generated by superposition of UBA5 to the MoeB-MoaD adenylate complex (13) (DaliLite root mean square deviation, 2.1 Å; Z-score, 24.1) and then superimposing the UFM1 NMR model (29) into the site of MoaD (DaliLite root mean square deviation, 3.6 Å; Z-score, 3.1) (Fig. 5). A similar approach was initially used for the APPB1-UBA3-NEDD8 A-site complex structure (28). Although the C-terminal region of UFM1 clashes with the crossover loop in our model, because all other UBLs, including NEDD8, SUMO, and yeast ubiquitin, bind under their respective crossover loops during adenylation, the UFM1 C-terminal tail is also hypothesized to bind under this loop prior to thioester bond formation.
FIGURE 5.
Predicted model of the UBA5-UFM1 complex. UBA5 (green) and UFM1 (cyan) are shown as a schematic. The region encompassing the crossover loop is colored red. ATP and catalytic cysteine are shown in stick format and labeled. Position of the UFM1 C terminus (C) is also labeled.
Most of the interactions between E1 ligases and their A-site bound UBLs are localized near four strands of the β-sheet that are structurally conserved in the ligases (13, 15, 23, 30, 31). However for UBA5, the presence of a conserved proline, Pro293 (supplemental Figs. S1 and S3), results in the loss of a hydrogen bond in the two final β-strands of the sheet, suggesting greater flexibility for this region in UBA5.
DISCUSSION
Structural and biochemical analysis of UBA5 has revealed insight into a structurally minimalistic E1 that does not contain canonical FCCH or SCCH domains. E1 enzymes are thought to perform their multiple activities and interactions through the dynamic action of several modular domains, which are connected by flexible linkers that can undergo extensive structural rearrangements (1, 14, 30). Similar to other E1 enzymes, the results from the binding assay demonstrate that the CTD of UBA5 also appears to be required for interactions with its E2 enzyme, UFC1. The E1 C-terminal UBL domain is conserved in all E1 enzymes that transfer UBLs to E2-conjugating proteins, and our binding studies of UBA5 also show a dependence on the CTD for thioester transfer of UFM1 to UFC1.
UBL binding specificity and stabilization has been shown to be mediated by a conserved β-sheet in the adenylation domain and also through polar interactions with the FCCH domain (28, 30). Because native UBA5 does not contain the FCCH domain, we looked for other structural factors that may contribute to UBL substrate specificity or stabilization. In this context, it is interesting to note the presence of the conserved Pro293 residue, which results in the loss of a hydrogen bond between the two final β-strands of the β-sheet that forms part of the UBL-binding surface of the adenylation domain. Further experiments will be required to delineate residues that play important roles in stabilizing interactions with UFM1, while also acting to discriminate against other UBL molecules.
The SCCH domain contains the catalytic cysteine in canonical E1 enzymes and varies in size from ∼80 residues in UBA3, ∼220 residues in UBA2, and ∼265 residues in UBA1. The evolution of the SCCH domain in E1 enzymes has been proposed to be an adaptation to promote reactions with UBLs and E2 enzymes or to allow binding of two UBL molecules simultaneously (A-site and T-site) (14, 16). For UBA5, the position of the catalytic cysteine in the long α6-helix places it in a position that projects away from the rest of the molecule and would thus likely also allow the simultaneous binding of a UBL to both the A-site and T-site.
Formation of the thioester bond between E1 enzymes and their respective UBL proceeds through the nucleophilic attack of the E1 active site cysteine on the adenylated UBL, with AMP acting as a leaving group. Although it is generally accepted that the E1 catalytic cysteine must be in a deprotonated state to form the thioester bond, there has been little evidence of factors that enhance the cysteine nucleophilicity (30, 32). A conserved threonine residue that is found in several E1 structures immediately following the catalytic cysteine may contribute to cysteine reactivity and mutation of the residue to an alanine has been shown to greatly reduce thioester bond formation without affecting adenylation of NEDD8 (28). This threonine is not conserved in UBA5, and the equivalent residue is in fact an alanine for this enzyme. Mutation of residues Arg223 and His227 in UBA3 have also been shown to reduce the efficiency of thioester bond formation for this enzyme (14). However, because these residues are in the SCCH domain of UBA3, a structurally analogous mechanism is not likely for UBA5. Catalytic cysteines are often found at or near the N terminus of α-helices, and the positive charge dipole in this region of various other enzymes has been shown to contribute to a decrease in cysteine pKa (33, 34). For example, active site cysteines of the thioredoxin fold family of proteins are commonly found at the N termini of α-helices, and this at least partially accounts for the dramatically lower reported pKa of these cysteines to as low as 3.0 (35). Other studies on the influence of helix dipole effects have shown that the position of cysteines at or near the N terminus of α-helices can dramatically affect cysteine pKa (33, 34). For example, cysteines introduced into the H helix of myoglobin demonstrated that helix dipole effects alone could account for the pKa reduction of cysteines at (−ΔpKa 2.1) or near the N-terminal position (−ΔpKa 0.5) in the helix (34). Another study with helical peptides showed that cysteines two or three residues from the N terminus demonstrated decreases in pKa of 1.6 and 1.5 pH units, respectively, whereas a cysteine at the N-terminal position demonstrated a pKa decrease of 1.1 pH units (33). The unique location of UBA5 Cys250 near the N terminus of the long α6-helix thus also suggests that helix dipole effects at least partially account for the nucleophilic character of this residue. While other E1 enzymes likely employ different mechanisms to enhance cysteine nucleophilicity, the potential need for UBA5 to utilize helix dipole effects may be a consequence of the smaller size of this enzyme when compared with other E1 enzymes.
In all known E1 structures, the catalytic cysteine thiols are >15 Å away from the terminal glycine of their respective A-site bound UBLs (Fig. 2b). This suggests that juxtaposition of the catalytic cysteine and the C terminus of the UBL would require either a major conformational change by the E1 enzyme, displacement of the reaction intermediates, or a combination of both mechanisms. Although it is possible that the adenylated UFM1 C terminus undergoes some displacement between the phosphoester transition state and the thioester transition state, it is also plausible that the Cys250-containing helical segment partially remodels toward this intermediate. Accordingly, the catalytic cysteine of UBA5 is located near a kink in the α6-helix, which is likely to affect the strength of the helix dipole, but could also afford the residue some structural mobility. With this knowledge, structural rearrangements that affect the pKa of the active site cysteine could be used as a mechanism to modulate nucleophilicity to enhance thioester bond formation and transfer reactions.
CONCLUSIONS
Structural analysis of UBA5 has revealed insight into a structurally minimalistic E1 that does not contain canonical FCCH or SCCH domains. The novel position of the UBA5 catalytic cysteine in the long α6-helix of the adenylation domain and structural rearrangements associated with the binding of ATP are proposed to be adaptations that allow UBA5 to catalyze specific thioester bond formation and transfer reactions. These studies reveal insight into the evolution of E1 enzymes and provide the foundations for a mechanistic model of the multiple reaction steps catalyzed by UBA5.
Supplementary Material
Acknowledgments
We thank Yanjun Li, Lianet Lopez, Patrick J. Finerty, Jr., and Taras Makhnevych for technical assistance. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357.
The Structural Genomics Consortium is a registered charity (1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.
The atomic coordinates and structure factors (code 3H8V) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- FCCH
- first catalytic cysteine half-domain
- SCCH
- second catalytic cysteine half-domain
- CTD
- C-terminal domain
- UFC
- ubiquitin fold modifier conjugating enzyme
- UFM
- ubiquitin fold modifier
- SUMO
- small ubiquitin-like modifier.
REFERENCES
- 1.Schulman B. A., Harper J. W. (2009) Nat. Rev. Mol. Cell Biol. 10, 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hochstrasser M. (2000) Nat Cell Biol. 2, E153–7 [DOI] [PubMed] [Google Scholar]
- 3.Ciechanover A. (1998) EMBO J. 17, 7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnell J. D., Hicke L. (2003) J. Biol. Chem. 278, 35857–35860 [DOI] [PubMed] [Google Scholar]
- 5.Tang Z., Hecker C. M., Scheschonka A., Betz H. (2008) FEBS. J. 275, 3003–3015 [DOI] [PubMed] [Google Scholar]
- 6.Geoffroy M. C., Hay R. T. (2009) Nat. Rev. Mol. Cell Biol. 10, 564–568 [DOI] [PubMed] [Google Scholar]
- 7.Gong L., Yeh E. T. (1999) J. Biol. Chem. 274, 12036–12042 [DOI] [PubMed] [Google Scholar]
- 8.Chiba T., Tanaka K. (2004) Curr. Protein Pept. Sci. 5, 177–184 [DOI] [PubMed] [Google Scholar]
- 9.Haas A. L., Ahrens P., Bright P. M., Ankel H. (1987) J. Biol. Chem. 262, 11315–11323 [PubMed] [Google Scholar]
- 10.Zhao C., Denison C., Huibregtse J. M., Gygi S., Krug R. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10200–10205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M. D., Klionsky D. J., Ohsumi M., Ohsumi Y. (1998) Nature 395, 395–398 [DOI] [PubMed] [Google Scholar]
- 12.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. (2000) Nature 408, 488–492 [DOI] [PubMed] [Google Scholar]
- 13.Lake M. W., Wuebbens M. M., Rajagopalan K. V., Schindelin H. (2001) Nature 414, 325–329 [DOI] [PubMed] [Google Scholar]
- 14.Huang D. T., Hunt H. W., Zhuang M., Ohi M. D., Holton J. M., Schulman B. A. (2007) Nature 445, 394–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lois L. M., Lima C. D. (2005) EMBO J. 24, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J., Hu W., Cai S., Lee B., Song J., Chen Y. (2007) Mol. Cell 27, 228–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J., Lee B., Cai S., Fukui L., Hu W., Chen Y. (2009) J. Biol. Chem. 284, 20340–20348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komatsu M., Chiba T., Tatsumi K., Iemura S., Tanida I., Okazaki N., Ueno T., Kominami E., Natsume T., Tanaka K. (2004) EMBO J. 23, 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dou T., Gu S., Liu J., Chen F., Zeng L., Guo L., Xie Y., Mao Y. (2005) Mol. Biol. Rep. 32, 265–271 [DOI] [PubMed] [Google Scholar]
- 20.Tatsumi K., Sou Y. S., Tada N., Nakamura E., Iemura S., Natsume T., Kang S. H., Chung C. H., Kasahara M., Kominami E., Yamamoto M., Tanaka K., Komatsu M. (2010) J. Biol. Chem. 285, 5417–5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng M., Gu X., Zheng D., Yang Z., Li F., Zhao J., Xie Y., Ji C., Mao Y. (2008) J. Cell. Biochem. 104, 2324–2334 [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z., Borek D., Majewski W., Minor W. (2003) Acta Crystallogr. A. 59, 228–234 [DOI] [PubMed] [Google Scholar]
- 23.Duda D. M., Walden H., Sfondouris J., Schulman B. A. (2005) J. Mol. Biol. 349, 774–786 [DOI] [PubMed] [Google Scholar]
- 24.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 27.Senisterra G. A., Finerty P. J., Jr. (2009) Mol. Biosyst. 5, 217–223 [DOI] [PubMed] [Google Scholar]
- 28.Walden H., Podgorski M. S., Schulman B. A. (2003) Nature 422, 330–334 [DOI] [PubMed] [Google Scholar]
- 29.Sasakawa H., Sakata E., Yamaguchi Y., Komatsu M., Tatsumi K., Kominami E., Tanaka K., Kato K. (2006) Biochem. Biophys. Res. Commun. 343, 21–26 [DOI] [PubMed] [Google Scholar]
- 30.Lee I., Schindelin H. (2008) Cell 134, 268–278 [DOI] [PubMed] [Google Scholar]
- 31.Walden H., Podgorski M. S., Huang D. T., Miller D. W., Howard R. J., Minor D. L., Jr., Holton J. M., Schulman B. A. (2003) Mol. Cell 12, 1427–1437 [DOI] [PubMed] [Google Scholar]
- 32.Szczepanowski R. H., Filipek R., Bochtler M. (2005) J. Biol. Chem. 280, 22006–22011 [DOI] [PubMed] [Google Scholar]
- 33.Kortemme T., Creighton T. E. (1995) J. Mol. Biol. 253, 799–812 [DOI] [PubMed] [Google Scholar]
- 34.Miranda J. J. (2003) Protein Sci. 12, 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charbonnier J. B., Belin P., Moutiez M., Stura E. A., Quéméneur E. (1999) Protein Sci. 8, 96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.