Abstract
Integrase (IN) is the catalytic component of the preintegration complex, a large nucleoprotein assembly critical for the integration of the retroviral genome into a host chromosome. Although partial crystal structures of human immunodeficiency virus IN alone and its complex with the integrase binding domain of the host factor PSIP1/lens epithelium-derived growth factor (LEDGF)/p75 are available, many questions remain regarding the properties and structures of LEDGF-bound IN oligomers. Using analytical ultracentrifugation, multiangle light scattering, and small angle x-ray scattering, we have established the oligomeric state, stoichiometry, and molecular shapes of IN·LEDGF complexes in solution. Analyses of intact IN tetramers bound to two different LEDGF truncations allow for placement of the integrase binding domain by difference analysis. Modeling of the small angle x-ray scattering envelopes using existing structural data suggests domain arrangements in the IN oligomers that support and extend existing biochemical data for IN·LEDGF complexes and lend new insights into the quaternary structure of LEDGF-bound IN tetramers. These IN oligomers may be involved in stages of the viral life cycle other than integration, including assembly, budding, and early replication.
Keywords: Growth Factors, Integrase, Protein Structure, Viral Protein, X-ray Scattering
Introduction
After a retrovirus such as HIV3 binds to and enters a sensitive cell, the viral RNA is reverse-transcribed to yield a cDNA copy of the genome. A large nucleoprotein assembly called the preintegration complex is assembled on this DNA. The preintegration complex includes an assortment of virus- and host-derived proteins and carries out integration of viral cDNA into a host chromosome. The primary catalytic component of this assembly is the viral integrase enzyme (IN). IN catalyzes the concerted integration of the viral DNA ends via two distinct chemical reactions as follows: a cleavage reaction that exposes 3′-ends of the viral DNA and a trans-esterification reaction where the 3′-ends attack the host DNA (1). The reaction is completed by host DNA repair enzymes that connect the remaining unjoined strands (2).
The host protein LEDGF/p75 is a component of the preintegration complex for lentiviruses, playing a key role in the viral life cycle. LEDGF binds tightly to HIV IN via a well characterized IN binding domain (IBD, Fig. 1A), and this interaction is required for proviral integration and for viral fitness (3–9). LEDGF functions as a target site-selection factor, directing integration to sites of high transcriptional activity in the host chromosome (10–13).
FIGURE 1.
A, IN and LEDGF/p75 proteins. IN is composed of three structural domains as follows: an N-terminal domain (green), a catalytic core domain (red), and a C-terminal domain (yellow). The integrase binding domain of LEDGF/p75 is shown in magenta. B, representative data for sedimentation equilibrium analysis of IN(tetra)·LEDGF(IBD). Global fits were performed at three concentrations at three rotor speeds. Top panels are radial absorbance data (symbols) and model fits (lines). Bottom panels are residuals from the fits. Experiments were carried out at 12,000, 16,000, and 20,000 rpm. C, representative SEC-MALS analyses of IN·LEDGF complexes. The top panel shows the elution profile of IN(tetra)·LEDGF(IBD) (red line) from a TSK3000 analytical column. The middle panel and lower panels show the elution profiles for wild-type IN·LEDGF(Cterm) (green) and IN(tetra)(D116N)·LEDGF(Cterm) (cyan) from a TSK4000 column. Wild-type IN preparations showed evidence of higher order species that are greatly reduced in IN(tetra) preparations. D, representative sedimentation velocity analyses. c(S) distributions for IN(tetra)·LEDGF(IBD) (red line, top panel), IN·LEDGF(Cterm) (green, middle panel), and IN(tetra)·LEDGF(Cterm) (cyan, lower panel) are shown.
HIV IN is a 32-kDa protein with three distinct structural domains (Fig. 1A) as follows: an N-terminal zinc binding domain (NTD), a catalytic core domain (CCD), and a C-terminal DNA binding domain (CTD) (14–18). The NTD contains a conserved zinc-binding motif and affects both oligomerization and catalytic function. The CCD contains an RNase H fold that is well conserved among the retroviral integrases, including the conserved DDE active site residues (Asp-64, Asp-116, and Glu-152) common to a superfamily of polynucleotidyltransferases (16). The CTD features an Src homology 3-like fold that is involved in DNA binding (19) and tetramerization (20) but is poorly conserved among the retroviral integrases.
The individual domains of IN form dimers, both in solution and in their respective crystal lattices, although intact IN has been shown to exist in an equilibrium between dimeric, tetrameric, and higher order oligomeric forms. LEDGF(IBD) stimulates the tetramerization of intact IN (21, 22). Structural models are available for the LEDGF(IBD), both alone (23) and in complex with IN(CCD) for HIV-1 (24). Crystallographic models are also available for NTD-CCD fragments from HIV-2 and maedi-visna virus in complex with LEDGF(IBD) (25, 26). The IN·IBD structures have revealed interactions at both the CCD dimerization interface and with the NTD that have been shown to be key to high affinity binding. Although each IN CCD dimer has the capacity to bind two LEDGF(IBD)s, IN:LEDGF stoichiometries ranging from 4:4 to 2:1 have been observed, both in solution experiments and related experimental structures (22, 24–27).
Although there is general agreement that four IN molecules are required for concerted integration of viral long terminal repeats (LTRs) into the host chromosome, there are differing views on the precise oligomeric state and arrangement of IN in the relevant protein-DNA assemblies. Crystal structures of IN fragments and in vitro analyses have implicated a tetramer as the catalytically relevant arrangement of IN in this complex (18, 25, 28), although other studies have implicated IN dimers in the 3′-processing steps and in assembly of the higher order nucleoprotein complex (29, 30). A recent crystal structure of the prototype foamy virus IN in complex with viral DNA reveals a quaternary arrangement that relies on a domain swap between the NTDs of two IN dimers to create a tetrameric arrangement (31). However, this related retroviral IN does not bind LEDGF, and it remains unclear whether a similar domain arrangement is utilized by HIV IN, which lacks some of the extended linkers found within prototype foamy virus IN. Recent work also suggests that the synaptic complex formed between HIV IN and viral LTR DNA is biochemically distinct from the strand transfer complex formed between IN, viral LTRs, and host target DNA (30, 32).
HIV IN is the target of the therapeutics raltegravir and eltegravir (33). Therefore, structural models of IN complexes would not only be useful in understanding the biological mechanism of retroviral integration but also in the optimization of pharmacological agents. Several studies have led to proposed structural models of the IN tetramer and its complex with DNA, based in large part on the crystallographic packing interactions observed in crystal structures of IN fragments (18, 25, 26, 34–39). Most recently, a model for the HIV IN·LEDGF tetramer was proposed based on cryo-electron microscopy (cryo-EM) studies of a reconstituted IN·LEDGF complex containing full-length and wild-type proteins (27). However, in this model, the IN dimers do not form a substantial protein-protein interface, and the tetrameric arrangement is inconsistent with existing data regarding the IN·LEDGF interaction and the properties of intact LEDGF (24–26). The lack of agreement among proposed models for IN oligomers indicates that additional characterization of the properties of IN complexes in solution is needed and that alternative experimental approaches might be useful.
To address these questions, we have analyzed IN·LEDGF complexes in solution in the absence of detergents or high salt conditions. We have established the oligomeric state, stoichiometry, and hydrodynamic properties of IN dimers and IN tetramers bound to two different LEDGF constructs and have combined the results with small angle x-ray scattering (SAXS) analysis to derive the shape of each complex in solution. Using existing crystal structures of IN dimers and IN·IBD complexes as the building blocks, we have generated models for the dimeric IN(NTD-CCD)·IBD and tetrameric IN·IBD complexes that are consistent with the measured biophysical parameters and with existing biochemical data. The model for the LEDGF-bound IN tetramer most consistent with our data builds on previous observations but contains a unique asymmetric arrangement of domains that differs from those previously discussed. These findings will be useful both in the interpretation of existing biochemical data for IN complexes in the absence of DNA and in considering mechanistic models for IN assemblies during the HIV life cycle.
EXPERIMENTAL PROCEDURES
Expression and Purification
IN variants and LEDGF(Cterm) constructs alone were expressed and purified as described previously (10, 40). IN and LEDGF constructs were co-expressed from pETDuet (Novagen) in BL21(DE3) cells at 37 °C. LEDGF was inserted into the vector in-frame with a C-terminal Mxe intein (New England Biolabs) containing the chitin binding domain and hexahistidine affinity tags. Proteins were purified using nickel-nitrilotriacetic acid (Qiagen) and chitin (New England Biolabs) resins. Fusion proteins were released by intein cleavage in 50 mm dithiothreitol overnight at 4 °C. Preparations of IN alone were further purified using SP-Sepharose chromatography (GE Healthcare). Proteins were concentrated at 4 °C in YM-3 Centricon (Millipore), and aliquots were flash-frozen in liquid nitrogen for storage at −80 °C. All preparations were stored and analyzed in 20 mm HEPES, pH 7.5, 150–450 mm NaCl, 0.1 mm EDTA, 10 μm ZnOAc2, and 1–10 mm dithiothreitol. The viscosity of this buffer at 4 and 20 °C was determined using a glass viscometer. Solvent density was determined gravimetrically. Prior to SAXS analyses, proteins were purified by size-exclusion chromatography (SEC) on a S200 10/300GL column, concentrated, and dialyzed. All purified proteins were analyzed by mass spectrometry (matrix-assisted laser desorption ionization time-of-flight) to verify the correct molecular weights of the individual components.
Sedimentation Equilibrium
Sedimentation equilibrium ultracentrifugation (SE) experiments were performed at either 4 or 20 °C with an XL-A analytical ultracentrifuge (Beckman Coulter) and a TiAn60 rotor with six-channel charcoal-filled Epon centerpieces and quartz windows. Radial absorption scan data at 280 nm for three protein concentrations were measured at 16 and 18 h for each of three different rotor speeds (12,000, 16,000, and 20,000 rpm). Comparison of radial absorption scans verified that equilibrium had been reached. Data were analyzed using the programs SEDFIT (41) and SEDPHAT (42). Single species or multimer equilibrium models were selected based on the smallest goodness of fit observed, with low and randomly distributed residual errors. An estimated error for the equilibrium constant was determined from a 1,000-iteration Monte Carlo simulation, as implemented in SEDPHAT.
Sedimentation Velocity
Sedimentation velocity ultracentrifugation experiments were performed at either 4 or 20 °C with an XL-A analytical ultracentrifuge (Beckman) and a TiAn60 rotor with two-channel charcoal-filled Epon centerpieces and quartz windows. Complete sedimentation velocity profiles were collected every 30 s at 45,000 rpm followed by data analysis using the program SEDFIT.
Size-exclusion Chromatography and Multiangle Light Scattering (SEC-MALS)
For determination of the Stokes radius (RS), SEC experiments were performed with a Superdex 200 10/300 GL column (GE Healthcare) at 0.4 ml/min at 20 °C in buffer containing 20 mm HEPES-NaOH, pH 7.5, 150 mm NaCl, 5 mm dithiothreitol, 0.1 mm EDTA, and 10 μm Zn(OAc)2. The column was calibrated using the following proteins (Bio-Rad): thyroglobulin (670 kDa, RS = 85 Å), γ-globulin (158 kDa, RS = 52.2 Å), ovalbumin (44 kDa, RS = 30.5 Å), myoglobin (17 kDa, RS = 20.8 Å), and vitamin B12 (1,350 Da). Blue dextran (Sigma) was used to define the void volume of the column.
Absolute molecular weights of LEDGF and IN·LEDGF heteromers were determined using MALS coupled with a TSK3000 or TSK4000 analytical SEC column (TosoHaas, Montgomeryville, PA). The columns were calibrated as described above. The scattered light intensity of the column eluant was recorded at 16 different angles using a DAWN-HELEOS MALS detector (Wyatt Technology Corp.) operating at 658 nm after calibration with RNase A. Protein concentration of the eluant was determined using an in-line Optilab DSP interferometric refractometer (Wyatt Technology Corp.). The weight-averaged molecular weight of species within defined chromatographic peaks was calculated using the ASTRA software version 5.2 (Wyatt Technology Corp.), by construction of Debye plots (KC/Rθ versus sin2[θ/2]) at 1-s data intervals. The weight-averaged molecular weight was then calculated at each point of the chromatographic trace from the Debye plot intercept, and an overall average molecular weight was calculated by averaging across the peak.
Small-angle X-ray Scattering
X-ray scattering data were measured at three different synchrotron sources as follows: beam line G1 at Cornell University High Energy Synchrotron Source (Ithaca, NY), beam line X21 at the National Synchrotron Light Source (Upton, NY), and beam line BL4-2 at the Stanford Synchrotron Radiation Light Source (Menlo Park, CA). Specific details of the experimental setup and procedures specific to each location are provided in the supplemental material. In all cases, the forward scattering from the samples studied was recorded on a CCD detector and circularly averaged to yield one-dimensional intensity profiles as a function of Q (Q = 4πsinθ/λ, where 2θ is the scattering angle). Samples were centrifuged at 10,000 × g for 10 min at 4 °C before 1–20-s exposures were taken at 20 °C. Scattering from a matching buffer solution was subtracted from the data and corrected for the incident intensity of x-rays. Replicate exposures were examined carefully for evidence of radiation damage by Guinier analysis and Kratky plot analysis. Silver behenate powder was used to locate the beam center and to calibrate the sample-to-detector distance.
SAXS Data Analysis
All of the preparations analyzed were monodisperse, as evidenced by linearity in the Guinier region of the scattering data and agreement of the I(0) and Rg values determined with inverse Fourier transform analysis by the programs GNOM (43) and AUTOGNOM (44). Molecular mass as derived from I(0) measurements, using the forward scatter from either bovine serum albumin or ovalbumin as a control, was consistent with the molecular masses determined by centrifugation and SEC-MALS. When fitting manually, the maximum diameter of the particle (Dmax) was adjusted in 10-Å increments in GNOM to maximize the goodness-of-fit parameter. This analysis also yielded determinations of Rg and I(0). The theoretical SAXS profiles for heteromer models of IN and LEDGF were created using the CRYSOL program (45).
Ab Initio Shape Reconstruction from SAXS Data
Low resolution shapes were determined from solution scattering data using the programs DAMMIF (46) and GASBOR (47). With GASBOR, the number of dummy residues used in shape reconstruction is prescribed by the user, requiring an understanding of the composition of the particle being modeled. Ten independent calculations were performed for each data set using default parameters. Initially, no symmetry constraints were applied. Calculations were then repeated assuming 2-fold symmetry, if justified by the apparent shape of the particle and improvement in the final χ and normalized spatial discrepancy (NSD) criterion. The models resulting from the independent runs were superimposed by the program SUPCOMB based on the NSD criterion (48). The 10 independent reconstructions were then averaged and filtered to a final consensus model using the DAMAVER suite of programs (49). Consensus models obtained by DAMMIF and GASBOR approaches yielded similar results, unless otherwise noted. Bead models were visualized in PyMOL (50) or converted to meshed envelopes using SITUS (51) and visualized using Chimera (52).
Modeling of LEDGF-bound IN Heteromers
Models of IN(NTD-CCD)2·LEDGF(IBD)2 and IN(tetra)4·LEDGF(IBD)2 were created by superposition of available crystal structures (Protein Data Bank codes 1K6Y, 2B4J, 3F9K, and 1EX4) in the program PyMOL, followed by energy minimization in the program CNS (53) to relieve bad side-chain contacts. Residues 270–288 from IN and 430–471 from LEDGF are all predicted to be disordered and are missing in our structural models. The program HYDROPRO (54) was used to determine theoretical hydrodynamic properties of these models and of SAXS bead ensembles from their respective atomic coordinates. Sculptor was used to rigid body dock structural models into low resolution envelopes using a feature-based docking algorithm (55).
RESULTS
Production of Soluble IN·LEDGF Complexes
Biophysical and crystallographic efforts to study HIV IN have been hindered by the poor solubility and yield of recombinant preparations of the full-length protein. Combinations of IN surface mutations and additives such as high salt, glycerol, and the detergent CHAPS have been used extensively to improve the stability and solubility of IN for in vitro experiments (56, 57). We found that by co-expressing truncated and full-length IN variants with LEDGF(IBD) in Escherichia coli, a number of IN:LEDGF(IBD) complexes could be co-purified in the absence of CHAPS or other additives, allowing the protein assemblies to be studied using more physiological buffer conditions.
We evaluated purification of IN complexes containing several of the previously described surface mutations, including IN(F185H), IN(F185K), and IN(tetra) (“tetra” refers to the C56S, F139D, F185H, C280S-substituted protein). IN(tetra) retains the binding and catalytic activities of wild-type IN (58).4 Substitutions at Phe-139, and Phe-185 improve solubility (15, 20), whereas the serine substitutions at positions Cys-56 and Cys-280 reduce the formation of oxidative side products that form during purification and storage (20, 59). The proteins and protein-protein complexes were purified using nickel-nitrilotriacetic acid and chitin chromatography, based on a modified self-cleaving intein fused to the C terminus of LEDGF. Only those complexes that were stable to repeated column washes with 300 mm NaCl (used for both resins) were recovered by co-purification. Table 1 summarizes the IN·LEDGF complex constructs that we have attempted to purify using this system.
TABLE 1.
Co-expression of IN truncations with LEDGF(IBD)
| Integrase | Mutations | Co-purification with LEDGF(IBD) | Co-purification with LEDGF(Cterm) |
|---|---|---|---|
| IN(CCD) | None | No | NAa |
| F185H | No | NA | |
| IN(NTD-CCD) | F185H | Yes | NA |
| F185K | Yes | NA | |
| C56S, F139D,F185H | Yes | NA | |
| IN | None | NA | Yes |
| F185H | Yes | Yes | |
| C56S, F139D, F185H | Yes | NA | |
| C56S, F139D, F185H, C280S (tetra) | Yes | Yes |
a NA means not attempted.
The smallest IN·LEDGF complex that we considered is the IN(CCD)·LEDGF(IBD) heterotetramer that was successfully crystallized (24). Although both components are produced during co-expression in E. coli, we found that the complex is not sufficiently stable to survive co-purification. Consistent with previous observations (8, 26), when the IN expression construct was extended to include both the NTD and CCD (Fig. 1A), we found that a stable complex could be obtained that was soluble in 0.3 m NaCl, defining a minimal three-domain composition for a stable IN·LEDGF complex under our conditions. This complex was stable to washes with 1 m NaCl and was not disrupted by 10 mm tetraphenylarsonium chloride (data not shown), a compound previously found to bind at the CCD-IBD interface (60). We were also able to obtain soluble complexes of LEDGF(IBD) with several full-length IN variants (Table 1). Interestingly, when we extended the LEDGF construct to include residues 325–530 (Cterm, Fig. 1A), the resulting IN·LEDGF complexes were obtained in higher yield and were highly soluble in 150 mm NaCl. We were not able to co-purify stable complexes of full-length LEDGF with IN for biophysical studies due to their limited solubility.
Stoichiometry and Oligomeric State of IN:LEDGF Complexes
We used several independent and complementary techniques to characterize the IN·LEDGF assemblies that we obtained from co-expression and co-purification. Our initial goal was to establish the oligomeric state and the protein stoichiometries of the complexes, both of which are crucial to interpretation of small angle x-ray scattering experiments. First we analyzed the complexes using sedimentation equilibrium (SE) ultracentrifugation. In all cases, we observed either a single species for which we were able to determine the molecular weight or we were able to fit the radial distribution curves to a simple monomer-dimer or dimer-tetramer association model. For the association models, we obtained estimates of the molecular weight and the dissociation constant. The results of the SE experiments are summarized in Table 2.
TABLE 2.
Oligomeric state of IN and IN·LEDGF complexes
| Construct | Modela | Concentrations | Molecular mass | Kd |
|---|---|---|---|---|
| μm | Da | μm | ||
| IN(F185H)b,c | 2 IN ⇆ IN2 | 6.6, 8.8, 17.6 | (32,283)d | 5.5 ± 0.004 |
| IN(tetra)b,c,e | 2 IN ⇆ IN2 | 8.8, 17.6 | (32,164)d | 9.3 ± 0.013 |
| LEDGF(Cterm)f | Single species | 34.1 | 29,786 ± 887 (23,452)d | |
| IN(NTD-CCD)(F185K)·LEDGF(IBD)f | IN2LEDGF2 | 4.0, 12.0, 20.0 | 75,634 ± 1,316 (75,947)d | |
| IN(tetra)·LEDGF(IBD)e,f | IN2LEDGF1 ⇆ (IN2LEDGF1)2 | 6.0, 9.6, 15.4 | (78,861)d | 8.9 ± 0.006 |
| IN(tetra)·LEDGF(Cterm)e,f | IN4LEDGF4 | 6.7, 11.1, 15.6 | 240,007 ± 4,996 (221,251)d |
a Data are as modeled by SEDPHAT. Experiments were carried out at 12,000, 16,000, and 20,000 rpm.
b Shown in the presence of 10 mm CHAPS.
c Measurements were made at 4 °C.
d Theoretical molecular mass is shown in daltons.
e Tetra corresponds to the mutational background of C56S, F139D, F185H, and C280S.
f Measurements were made at 20 °C.
In a second set of experiments, we used SEC coupled with MALS detector to determine the Stokes radius (RS) of the complex and to obtain an independent measurement of the complex mass. Finally, we used sedimentation velocity (SV) ultracentrifugation to determine sedimentation coefficients for several of the species studied. The measured Stokes radius and sedimentation coefficients then provided a third estimate of the complex mass via the Siegel and Monty equation (61). The results of these experiments for IN, LEDGF, and co-purified IN·LEDGF complexes are summarized in Tables 2 and 3, with representative examples of SE, SEC-MALS, and SV experiments shown in Fig. 1, B–D, respectively. Additional SE and SEC-MALS data are shown in supplemental Figs. 1 and 2, respectively.
TABLE 3.
Biophysical properties of co-expressed IN·LEDGF heteromers
NA means not attempted.
| Protein | SECa | MALSb | IN:LEDGFd | Sedimentation velocity | f/f0 | Siegel and Monty | DLSb |
|---|---|---|---|---|---|---|---|
| Rs (Å) | Molar massc | s20,w | Molar massc | Rh (Å) | |||
| LEDGF(Cterm) | 35.4 ± 0.1 | 29,390 ± 1,910 (23,452) | NA | 1.7 | 1.8 | 24,298 | NA |
| IN(NTD-CCD)(F185K) LEDGF(IBD) | 38.5 ± 1.2 | 73,213 ± 4,838e (75,947) | 2:2 | 4.3 | 1.8 | 80,802 | NA |
| IN(tetra)-LEDGF(IBD) | 54.9 ± 1.8 | 159,720 ± 7,456 (157,721) | 4:2 | 7.2 | 1.8 | 160,165 | NA |
| IN-LEDGF(Cterm) | 73.7 ± 2.1 | 217,080 ± 16,194 (221,251) | 4:4 | 7.4 | 1.6 | 229,170 | 58.0 ± 3.8 |
| IN(tetra) LEDGF(Cterm) | 76.7 ± 1.6 | NA | NA | 7.7 | 1.6 | 243,655 | NA |
| IN(tetra)(D166N) LEDGF(Cterm) | NA | 209,720 ± 9049 (221,251) | 4:4 | NA | NA | NA | 57.1 ± 2.8 |
a The results presented are the average of 2–4 replicates. SEC measurements were made at room temperature.
b The results presented are the average of 3–5 replicates. DLS measurements were performed at 4 °C.
c Molar masses are presented in units of daltons. In parentheses, theoretical values as computed from primary sequence using SEDNTERP are presented.
d Stoichiometry was inferred from experimentally determined molecular mass; theoretical values are in parentheses.
e This preparation shows evidence of dissociation on silica-based resins not seen in other gel filtration media. In contrast to the other SEC-MALS analysis, this sample was analyzed using a Superdex 200 column in-line with MALS, using the theoretical extinction coefficient in lieu of its refractive index for concentration determination.
Solution Structure of IN(NTD-CCD) Bound to LEDGF(IBD)
The IN(NTD-CCD)·LEDGF(IBD) complex exists as a single species in solution, with a molecular weight consistent with a 2:2 complex (Tables 2 and 3). Thus, two LEDGF(IBD) molecules bind to a dimer of IN(NTD-CCD). This result is consistent with the IN(CCD)·LEDGF(IBD) crystal structure, which reveals two IBDs bound to each IN CCD dimer (24), as well as that observed with the maedi-visna virus IN(NTD-CCD)·LEDGF(IBD) crystal structure (26). However, the 2:2 stoichiometry is not consistent with the crystal structure of HIV-2 IN(NTD-CCD)·LEDGF(IBD), which shows one IBD bound to each IN dimer (26).
In addition to the question of stoichiometry, the arrangement of NTDs in the IN dimer has not been unambiguously demonstrated based on structural data. In the HIV-2 IN(NTD-CCD) structure, two IN dimers are observed in the asymmetric unit, but the linkers connecting the NTDs to the CCDs are disordered (26). Two symmetric NTD arrangements can be considered based on the observed crystal packing (Fig. 2). In one configuration, two NTDs interact with one another and to a CCD dimer surface that is orthogonal to that formed by the IBD interface. In this “apical” arrangement, the 2-fold symmetry axis of the CCD dimer passes through the NTD dimer. In the alternative “lateral” configuration, each NTD interacts with one CCD adjacent to where the IBD binds. This arrangement is similar to the domain organization observed in one-half of the HIV-2 IN(NTD-CCD)·LEDGF(IBD) crystal structure, where the CCD-NTD linker is well ordered. In the other half of that IN dimer, the NTD adopts a different position, and there is no IBD bound. This same lateral arrangement is also observed in the maedi-visna virus IN(NTD-CCD)·LEDGF(IBD) crystal structure (25). The apical arrangement of NTDs has been proposed to occur in the context of a LEDGF-bound IN tetramer (37). The apical and lateral positions for the NTDs are compared in Fig. 2.
FIGURE 2.
SAXS analysis of IN(NTD-CCD)(C56S,F139D,F185H)·LEDGF(IBD). A, small angle x-ray scattering data from IN(NTD-CCD)(C56S,F139D,F185H)· LEDGF(IBD). Shown in black squares is the recorded intensity as a function of Q. Plotted against the data is the fit derived from GASBOR analysis (red line), with a χ value of 1.52. B, P(r) shape function (solid red line). Shown in dotted and solid lines are the theoretical shape functions for the lateral (blue) and apical (green) arrangement of the NTDs with respect to IN CCD. C and D, rigid body docking of structural models of IN(NTD-CCD)2·LEDGF(IBD)2 in both lateral (C) and apical (D) configurations into the SAXS envelope.
To address the question of NTD positions, we analyzed the IN(NTD-CCD)·LEDGF(IBD) complex using SAXS. We measured scattering for three different IN(NTD-CCD) mutants and at several concentrations and obtained similar results in each case (supplemental Table 1). The results of these experiments for IN(C56S, F139D, and F185H) are summarized in Fig. 2. Assuming a 2:2 stoichiometry, we constructed the symmetric IN(NTD-CCD)2IBD2 models discussed above and shown in Fig. 2, C and D. The models have similar maximum dimensions (Dmax) that correlate well to the Dmax of 114 Å experimentally determined from the scattering data.
We generated theoretical scattering profiles for both models and compared them to the experimental data. The model with NTDs in lateral positions shows better agreement with experimental data (χ2 = 1.75) than the apical model (χ2 = 2.32) (supplemental Fig. 3). This correlation is also mirrored in comparison with P(r) shape functions (Fig. 2B). The P(r) curve is a distribution of inter-atomic distances present in the molecule, so differently shaped molecules give rise to different features in the P(r) function. Because 80 amino acids in the constructs studied here are missing from the crystal structures used to derive the two models, an exact match between calculated and experimental P(r) functions for any model is unlikely. However, the general shape of the function should be similar for a correct model. As shown in Fig. 2B, the shape function associated with the lateral model is most similar to the experimental P(r) but compressed toward smaller interatomic distances due to the smaller size of the complex.
To generate shape reconstructions from the scattering data, we used the DAMMIF and GASBOR programs (see “Experimental Procedures”), which use different but complementary algorithms to generate shape envelopes of the scattering molecule. With a 2-fold symmetry constraint for the overall shape, both approaches reproducibly yielded envelopes with good correlations between experimental and calculated scattering data (sqrt(χ) ∼1.5). The ensemble of envelopes generated from multiple iterations of the reconstruction process also agreed well with one another, with NSD values of ranging from 0.7 to 0.8 for DAMMIF and 1.0 to 1.3 for GASBOR (where a value of unity corresponds to identity between two structures, and values below 1 indicate a high degree of overlap). We obtained similar solution parameters and SAXS results for constructs containing the F185H and F185K solubility mutations (data not shown).
As shown in Fig. 2 and in supplemental Fig. S4, the averaged envelope is an elongated prolate ellipsoid. The hydrodynamic properties calculated for this shape closely resemble those determined experimentally (Table 3). For example, the calculated sedimentation coefficient of 4.6 and the extended ellipsoidal shape of the envelope agree well with the values derived from SV analysis (s20,w = 4.3), and the calculated RS of 40 Å agrees well with results derived from SEC (38.5 Å) and DLS (40.8 Å) measurements (data not shown). From a comparison of the two alternative models docked into the SAXS envelope, it is clear that the lateral model (Fig. 2C) captures the overall shape of the IN(NTD-CCD)·IBD complex in solution much better than the apical model (Fig. 2D). These results strongly support the model shown in Fig. 2C, where each NTD of IN interacts with the CCD and IBD, but the NTDs do not interact with one another and do not contribute directly to the dimer interface. The model is also consistent with mutagenesis data that support the IBD·NTD interface observed in the IN(NTD-CCD)·IBD crystal structure (26).
Shape of an IN·LEDGF Tetramer in Solution
We next examined a series of full-length IN complexes with the LEDGF(IBD). The IN(tetra)·LEDGF(IBD) complex is soluble and monodisperse in buffers containing 300 mm salt, but it differs in two important ways with respect to the truncated IN(NTD-CCD) complexes described above. First, the full-length complex forms tetramers of IN, with a dimer-tetramer Kd ∼9 μm (Tables 2 and 3). Thus, the LEDGF(IBD) is alone capable of stabilizing the tetrameric form of IN, although the comparison to IN oligomerization in the absence of bound IBD is to a protein solution containing 10 mm CHAPS at a lower temperature (Table 2). The second difference is that the IN:IBD stoichiometry is 4:2 in the tetramer, where each CCD dimer binds only one IBD. In this case, experimental determination of the mass of the IN·IBD complex comes from a dimer-tetramer association model fit to the SE data, plus direct MALS analysis of the tetramer species observed on the SEC column (Fig. 1C). The IN(tetra)·IBD complex is stable and is not disrupted by 1 m NaCl (data not shown).
Our interpretation of the 4:2 IN·IBD stoichiometry is that IN tetramerization leads to formation of one pair of high affinity binding sites for the IBD and one pair of low affinity sites, a concept previously proposed based on modeling of available crystallographic structures (37). The IBDs bound to low affinity sites are presumably removed from the IN complex during purification. This conceptual model of the tetramer requires that the complex is at most 2-fold symmetric. To further understand this complex, we used SAXS to determine the shape of the particle in solution.
The IN(tetra)·LEDGF(IBD) heteromer is a well suited for SAXS analysis and interpretation, because it is very soluble and not prone to aggregation, and the component protein domains are represented by available crystal structures. We tested IN(tetra)·LEDGF(IBD) scattering over a range of concentrations well in excess of the dimer-tetramer Kd value to fully populate the tetramer state. Across the range of concentrations examined, the mass of the particle was constant (based on I(0) extrapolation and comparison with scattered bovine serum albumin standards) and consistent with the mass determined by biophysical measurements. The shape parameters (Rg and Dmax) derived from x-ray scattering at different concentrations are also consistent, with only a small increasing trend toward higher concentrations (supplemental Table 1).
Shape reconstructions using both the DAMMIF and GASBOR approaches yielded prolate ellipsoids of similar volume and dimension and are consistent with the hydrodynamic properties determined by SV analysis (Fig. 3 and Table 3). Independent reconstructions using DAMMIF showed good agreement, with sqrt(χ) values ranging from 1.8 to 1.9 and NSD values ranging from 0.78 to 0.87, indicating a very stable structural solution (Fig. 3B and supplemental Fig. 4). Although the reconstructions using GASBOR featured similar sqrt(χ) values (1.7 to 1.8), the NSD values were higher, ranging from 1.8 to 2.2. Examination of the individual models showed similar core ellipsoidal shapes, with the most apparent deviations located on the distal ends of each particle, perhaps suggesting inherent disorder within these regions (supplemental Fig. 4). However, the averaged envelope shares the same conserved features observed in all the reconstructions and recapitulates the shape observed in the averaged DAMMIF envelope. The averaged shape obtained from GASBOR also correlates well with measured hydrodynamic properties as follows: calculated versus measured RS values are 53.3 and 54.9 Å, respectively, and the calculated versus measured sedimentation coefficients are 7.3 and 7.4, respectively.
FIGURE 3.
SAXS analysis of IN(tetra)·LEDGF(IBD). A, small angle x-ray scattering data for IN(tetra)·LEDGF(IBD). Shown in black squares is the recorded intensity as a function of Q. Plotted against these data is the fit derived from GASBOR analysis (red lines). B, final averaged shape reconstructions for IN(tetra)·LEDGF(IBD) from GASBOR (light blue) and DAMMIF (dark blue). The bead radius shown is 1.9 Å.
Structural Models of the IN:LEDGF(IBD) Tetramer
There are currently no crystal structures of IN tetramers available to provide a basis for modeling the shape of the IN·LEDGF(IBD) complex in solution. However, contacts between neighboring molecules in the crystal structures of IN(NTD-CCD) and IN(CCD-CTD) reveal a number of interactions that could be relevant in the context of an IN tetramer (15, 18). We constructed a series of models based on these contacts, the determined 4:2 IN·IBD stoichiometry, the known binding position of the LEDGF(IBD), and two alternative positions for the NTD. Because the envelope reconstructions and stoichiometry strongly indicate that the complex is at most 2-fold symmetric in solution, we only considered 2-fold symmetric arrangements of the IN and LEDGF domains.
Four models that fit these criteria are shown in Fig. 4A. Models 1 and 2 are derived from crystallographic packing observed in the IN(CCD-CTD) crystal structure (15), where CTD-CCD and CTD-CTD interactions account for nearly all of the contacts between dimers. Models 3 and 4 are derived from the crystallographic packing observed in the IN(NTD-CCD) structure (18), where dimer-dimer contacts are mediated primarily by NTD-CCD interactions. As was the case in the IN(NTD-CCD) crystal structure, there are multiple ways to connect the NTDs to the CCDs in models 3 and 4. In models 1 and 2, however, the domain connectivity is unambiguous, but the complex is distinctly asymmetric with respect to the NTD arrangement.
FIGURE 4.
Modeling the LEDGF-bound IN tetramer. A, quaternary models of LEDGF-bound IN tetramer evaluated in this study. The models were derived from packing interactions observed in IN fragment crystal structures. B, comparison of theoretical shape functions for the four structural models to the experimental data. Theoretical data are shown as dotted purple lines, with experimental data for IN(tetra)-LEDGF(IBD) rendered as solid red lines. C, orthogonal views of the rigid body docking results for each of the four models into the experimental envelope derived from GASBOR analysis.
The differences between models 1 and 2 and between models 3 and 4 involve the location of the bound IBDs. In models 1 and 3, each IBD interacts with one NTD and one CCD of IN, as observed in the IN(NTD-CCD)·LEDGF(IBD) structure (26). In models 2 and 4, the IBD is moved to the alternative location on the CCD dimer, where it binds in the absence of NTD interactions. We attempted to construct models in which both the NTD and the IBD were moved to the alternative surface, but the resulting structures were rejected because distance constraints prevented connection of all four NTDs to a nearby CCD. Models 1 and 3 therefore have IBDs bound in high affinity sites, the most likely arrangement in a 4:2 IN·IBD complex. Models 2 and 4 have IBDs bound at low affinity sites, which as discussed later are likely to be occupied when in complex with the longer LEDGF(Cterm) construct.
To determine whether one or more of the models shown in Fig. 4A is consistent with the SAXS data for the IN(tetra)·LEDGF(IBD) complex, we computed the expected hydrodynamic properties and scattering profiles for each model and compared them with the experimental data (Table 4). Theoretical scattering data for the models agreed reasonably well with the observed I(q) curves (χ2 of 2.10, 2.33, 2.08, and 4.41, for models 1–4, respectively, see supplemental Fig. 3). As expected from the ellipsoidal shape of the reconstructed envelopes, the calculated molecular dimensions of model 1 showed the best overall agreement with the parameters derived from SAXS. The most discriminating comparison comes from a P(r) curve analysis (Fig. 4B). Of the four models, the calculated P(r) for model 1 showed the best correlation with the experimental shape function.
TABLE 4.
Comparison of IN(tetra)·LEDGF(IBD) shape reconstruction to theoretical models
| SAXS | Model 1 | Model 2 | Model 3 | Model 4 | EMa | |
|---|---|---|---|---|---|---|
| χ2b | 1.89 | 1.44 | 1.54 | 1.99 | 1.22 | |
| Rgc | 50 Å | 44 Å | 42 Å | 35 Å | 44 Å | 45 Å |
| Rsd | 55 Å | 49 Å | 50 Å | 47 Å | 52 Å | 51 Å |
| ϕ (Rg /Rs)e | 0.91 | 0.898 | 0.840 | 0.745 | 0.808 | 0.881 |
| Dmax | 147 Å | 160 Å | 130 Å | 142 Å | 149 Å | 147 Å |
| Ellipsoidal character | Prolate | Prolate | Oblate | Oblate | Prolate | Oblate |
|---|---|---|---|---|---|---|
| Dimensionsf | 66.11 × 153.00 × 85.00 Å | 69.90 × 137.43 × 108.90 Å | 102.14 × 115.24 × 109.08 Å | 84.07 × 117.86 × 90.19 Å | 86.10 × 131.63 × 111.78 Å | 106.87 × 115.13 × 107.57 Å |
| ccg | 0.76 | 0.67 | 0.55 | 0.67 | 0.61 |
a Parameters for HIV IN-LEDGF tetramer model were derived from cryo-electron microscopy (27).
b Statistical agreement between experimental data and structural model, as determined by CRYSOL, is shown.
c Rg values for models 1–4 were calculated using the program CRYSOL.
d Data were determined by HYDROPRO.
e Defined in Ref. 75.
f Dimensions were determined using the pdb2vol extension of the program SITUS.
g Real space correlation coefficient between rigid-body docking solution of model versus experimental SAXS envelope, as measured in the program Sculptor.
To compare more directly the models to the SAXS-derived molecular shape, each of the four candidate models was optimally docked as rigid bodies into the experimental envelope (Fig. 4C). Of the four models, only model 1 (correlation coefficient (cc) = 0.76) provided a solution that properly accounts for the particle volume, with all structural domains contained within the envelope. Despite being derived from the same IN tetramer, model 2 (cc = 0.67) fails to account for the longest dimension of the particle due to the alternative placement of the IBDs, which protrude out of the envelope. Models 3 and 4 (cc = 0.55 and 0.67, respectively) have a more compact, oblate ellipsoidal character in the core tetramer and lead to substantially poorer agreement with the molecular envelope.
Within the constraints of the relatively simple modeling performed here, we conclude that the IN tetramer represented by model 1 best accounts for the experimental solution scattering data. It is important to note, however, that models can be readily scored as unlikely based on their overall three-dimensional shapes, but it is difficult to conclude that any given model is correct based on agreement with the SAXS data alone. Alternative models with similar elongated shapes as that shown for model 1 may also exist.
Properties of the IN·LEDGF(Cterm) Complex
We next examined complexes containing the LEDGF “Cterm” fragment, which includes the entire C-terminal domain of LEDGF and is therefore larger than the IBD construct by an additional 80 amino acids (Fig. 1A). When we examined complexes containing full-length IN (both wild-type and the tetra mutant) bound to the longer LEDGF(Cterm) construct, we obtained a surprising result. The IN dimer-tetramer equilibrium strongly favors tetramer formation (i.e. the Kd is lower) and the IN:LEDGF stoichiometry is 4:4 (Tables 2 and 3). The additional sequences flanking the IBD in the LEDGF(Cterm) construct apparently enhance binding to IN, allowing all four LEDGF-binding sites in the tetramer to be stably occupied. A comparison of the SV and SEC profiles for these complexes is shown in Fig. 1, C and D. The IN(tetra)·LEDGF(Cterm) complex is exceptionally well behaved, with no evidence for formation of higher order species or aggregates.
The hydrodynamic properties of LEDGF(Cterm) alone indicate that LEDGF(Cterm) exists as a monomeric elongated particle in solution (Tables 2 and 3). The regions flanking the IBD in the LEDGF(Cterm) construct have eluded direct structural analysis (23) and are predicted to be unstructured. Our ability to model the IN(tetra)·LEDGF(Cterm) envelope is therefore limited by the lack of structural information for these additional regions. Nonetheless, we decided to analyze the IN(tetra)-LEDGF(Cterm) tetramer by SAXS. Not only is this complex likely to be more representative of the LEDGF·IN complexes that form in vivo, but a comparison of the molecular shape to that obtained for the IN·LEDGF(IBD) complex could provide information about the location of the additional LEDGF sequences present in the longer construct. Because this complex forms a stable tetramer at micromolar concentrations, we were able to perform the SAXS experiments under conditions where only tetramers exist in solution.
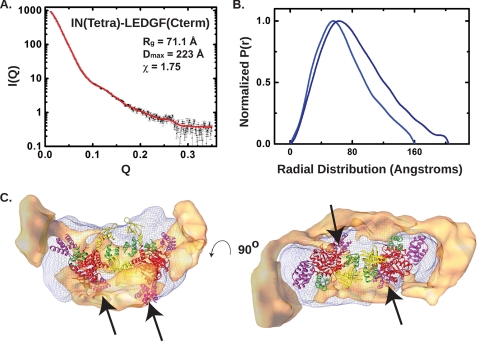
As shown in Fig. 5B and supplemental Table 1, both Rg and Dmax values are significantly larger for IN(tetra)· LEDGF(Cterm) than for the same IN tetramer with two bound IBDs. Shape reconstruction using DAMMIF yielded prolate ellipsoids similar to those obtained with IN(tetra)· LEDGF(IBD), but with a larger dimension along the longest axis (223 versus 147 Å). The NSDs of the reconstructions are small (∼0.6), indicating a tight clustering of nearly identical shapes (supplemental Fig. 4). Similar results were obtained with IN(F185H)·LEDGF(Cterm) (data not shown).
FIGURE 5.
SAXS analysis of IN(tetra)·LEDGF(Cterm). A, small angle scattering data from IN(tetra)·LEDGF(Cterm). Shown in black squares is the recorded intensity as a function of Q. Plotted against these data is the fit derived from Gasbor analysis (red lines). B, experimental P(r) shape functions for IN(tetra)·LEDGF(IBD) (light blue) and IN(tetra)·LEDGF(Cterm) (dark blue). C, volume difference between the DAMMIF-derived envelopes for IN(tetra)·LEDGF(IBD) and IN(tetra)·LEDGF(Cterm), rendered as a tan solid in orthogonal views. Two additional IBDs were added to model 1, and the resulting 4:4 IN:LEDGF complex was docked into the envelope exactly as shown in Fig. 4. The difference density is located near bound LEDGF molecules and could account for the flanking residues in LEDGF(Cterm) not present in IN(tetra)·LEDGF(IBD).
A difference envelope generated from the IN(tetra)· LEDGF(IBD) and IN(tetra)·LEDGF(Cterm) shapes computed by DAMMIF at similar nominal resolutions (Qmax = 0.25) is shown in Fig. 5C. This envelope shows regions of protein present in the IN(tetra)·LEDGF(Cterm) structure that are not present in IN(tetra)·LEDGF(IBD). We assume that these difference densities correspond primarily to the two additional molecules of bound LEDGF and the additional residues flanking the LEDGF(IBD). Based on this result, we can infer the locations of the four bound LEDGF molecules.
To construct a model for the IN(tetra)·LEDGF(Cterm) tetramer, we added two additional IBDs to model 1, creating a composite of models 1 and 2. The new IBDs were placed in the “low affinity” sites that do not include binding contributions from an adjacent NTD. Because the sequences flanking the IBDs are missing in this model, the protein inventory is not complete, but the model does serve to identify a plausible arrangement of domains in a 4:4 complex. We then docked the 4:4 IN·IBD model into the IN(tetra)·IBD envelope that we had determined for the 4:2 complex (Fig. 5C). As expected from consideration of model 2, the additional IBDs project out of the envelope. When we superimposed the difference density envelope, we found two distinct lobes of density located at opposite ends of the envelope, adjacent to the high affinity IBDs. Thus, the extension of the maximal dimension of the 4:4 particle as a result of increasing the size of the LEDGF construct can be readily understood in terms of adding additional residues to the IBDs, based on the domain arrangement in model 1. Two additional patches of difference density are located near the central region of the envelope. These regions correspond to the second pair of IBDs that were placed in the lower affinity sites of model 1.
DISCUSSION
The primary goal of these studies has been to probe the solution properties of IN tetramers, as this oligomeric state has been frequently implicated in the synaptic and strand transfer complexes formed between IN and DNA. Here, we have employed SAXS to consider models for the arrangements of domains in a series of complexes formed between HIV IN and human LEDGF. Although the resolution of SAXS is not sufficient to reveal specific inter-domain interactions, this method can rule out models of quaternary arrangements that are inconsistent with the determined shape properties and with hydrodynamic properties derived from complementary methods. The different IN and LEDGF truncations examined here have allowed us to consider specific questions about the relative positioning of domains such as the NTD and the IBD.
There are currently no crystal structures available containing full-length HIV IN. Thus, the intermolecular contacts observed in two-domain crystal structures have been used to construct plausible models of higher oligomeric forms of the IN protein (18, 25, 26, 34, 35, 37–39, 62). Indeed, the models we consider here are based on similar logic. We note that our use of the term “tetramer” is only intended to indicate a stoichiometry of an IN oligomer that involves four subunits and does not imply an equivalence or particular symmetry relationship between the protein domains. As noted earlier, the solution properties of the complexes we have studied here are largely consistent with tetrameric assemblies that could be best thought of as a dimer of dimers rather than a symmetric tetramer, a view that is very much in line with current thinking about the functional forms of IN (18, 21, 25, 28, 32, 63).
The IN tetramer model most consistent with our experimental data (Fig. 4, model 1) is based on elements from three separate crystal structures (15, 18, 26). The dimer-dimer interface of this model is formed from two NTDs and two CTDs of IN that interact with one another and with the CCDs. The IN CTDs contribute extensively to this interface, consistent with the previously established importance of this domain to oligomerization (20) and the associated impact of mutations at this interface (L241A, L242A) (64, 65). Interestingly, the remaining two CTDs do not contribute to the dimer-dimer interface in this model and are largely solvent-exposed. A more extensive kink in the helix linking the CCD and the CTD or an entirely different linker conformation could allow these additional domains to participate in the dimer-dimer interface, but we restricted our modeling to rigid-body placement of structural fragments available from existing crystal structures and did not employ any type of flexible fitting.
Model 1 also features an asymmetric positioning of the NTDs. One NTD is largely buried in the dimer-dimer interface, where it occupies an apical position with respect to the CCD dimer. The other NTD occupies a lateral position, where it is engaged in binding to LEDGF and is not directly involved in tetramer formation. Due largely to its extended form, this model provides the best agreement with the SAXS envelope and therefore best represents the shape of the IN:LEDGF tetramer in solution.
The second type of model we considered (Fig. 4, model 3) is based on a different dimer-dimer interface. In this case, the CTDs are not directly involved, and the interface is mediated entirely by interactions between the four NTDs in the tetramer, as well as between the NTDs and the CCDs. This dimer-dimer interface is based on crystal packing between two IN(NTD-CCD) dimers in the crystal structure (18), where a dimer of NTDs in apical positions is associated with each CCD dimer, and two of these NTDs are shared with the opposing CCD dimer where they occupy lateral positions and engage the LEDGF(IBD)s. The protein-protein interface in this model is more extensive in terms of buried surface area compared with model 1; however, the more compact globular shape is less consistent with the elongated nature of the SAXS envelope.
In addition to models 1 and 3, we considered the alternatives modes of IBD binding to CCD, where they would bind in the absence of interactions involving the NTD. These IBD positions (models 2 and 4) are unlikely for the 4:2 IN·IBD complex, but would be expected to be occupied in the 4:4 IN·LEDGF(Cterm) complex. Indeed, the SAXS data for this larger complex supports the placement of LEDGF molecules in the context of models 1 and 2.
If the IN·IBD tetramer represented by model 1 is in fact closely related to the oligomeric form that exists in vivo, then we should be able to explain previous observations based on the proposed domain organization. For example, the zwitterionic detergent CHAPS has a reported dissociative effect on IN tetramerization (66, 67). In the IN(CCD-CTD) crystal structure, two CHAPS molecules are bound to each CTD, and one of these binding sites would most likely be excluded by the dimer-dimer interface in model 1. At high concentrations, CHAPS would therefore be expected to compete for this binding site and disrupt formation of the IN tetramer.
A tetramer model should also be able to explain the observation that LEDGF binding lowers the dimer-tetramer Kd value, thereby stabilizing the tetrameric state (this study and see Refs. 21, 22). Because the IBD-binding sites are well separated from the surfaces used for oligomerization in all of our candidate models, the effects of IBD binding on tetramer formation are expected to be indirect. We propose that this stabilizing effect involves the positioning of NTDs in the dimeric versus tetrameric forms of IN. Upon tetramerization, model 1 requires that one NTD remain in the apical position and one move to the lateral position. This is likely to be true even in the absence of LEDGF, because there does not appear to be room in the dimer-dimer interface for both NTDs without making significant adjustments to the CTD positions. Binding of LEDGF to IN could therefore promote tetramer formation by stabilizing the lateral configuration of two of the NTDs. This mechanism of tetramerization also explains why two types of IBD-binding sites are created in the tetramer, resulting in a stable 4:2 IN·LEDGF complex with the IBD but a 4:4 complex with the higher affinity Cterm construct. Because we do not yet know the nature of the interactions between IN and the LEDGF sequences flanking the IBD, we cannot rule out additional effects where LEDGF plays a more direct role in facilitating tetramerization via these additional elements.
A recent cryo-EM study of wild-type IN bound to full-length LEDGF (∼60 kDa) both in the absence and presence of U5 DNA described an IN:LEDGF stoichiometry of 4:2 based on mass spectral analysis of a chemically cross-linked complex. This work proposed an IN·LEDGF tetramer structure in the absence of DNA that differs considerably from the models described here (27). We calculated geometric properties for an IN·IBD complex based on the EM tetramer and docked this model into our SAXS envelope. The results are summarized in Table 4 and in supplemental Fig. 5. Although the dimensions of the EM model result in favorable Dmax and Rg values, the shape of the model results in a poor fit to the SAXS envelope (cc = 0.61), with several regions protruding outside the envelope and parts of the envelope unaccounted for by model. The calculated P(r) distribution is also in poor agreement with that derived from the experimental data. It is thus difficult to reconcile the EM protein-only model with the hydrodynamic properties and shapes of the IN·LEDGF structures described here. Further biophysical characterization of the full-length IN·LEDGF complex in solution, perhaps coupled with EM studies of the minimal IN·LEDGF complexes described here, will no doubt be required to fully understand the differences.
The Kd values of dimerization and tetramerization reported here for IN and IN·LEDGF complexes are in the micromolar range, supporting the idea that IN may switch among oligomeric forms during viral replication (20, 22, 66–68). Indeed, viral DNA has been reported to dissociate IN tetramers (67), and recent studies suggest that distinct IN arrangements are formed on DNA during the various steps on the integration pathway (29, 32), with dimers first binding to each viral LTR (30). Thus, there are likely to be distinct quaternary structures of dimeric and tetrameric forms of IN that form during the integration reaction when bound to DNA. Similarly, the domain organization of IN oligomers may change upon binding viral DNA, an observation that has been made in a number of nucleic acid-binding proteins (69). Indeed, the distances between IN active sites in our tetramer models (77 Å for model 1) are far greater than the ∼18 Å required for catalysis of concerted integration (18, 26, 67), and recent studies suggest that distinct IN arrangements are formed on DNA during the various steps on the integration pathway (29, 32), with dimers binding to each viral LTR (30), indicating that they cannot represent the form of IN required for the final stage of the integration process.
LEDGF stimulates tetramerization of lentiviral INs. Although the IN·LEDGF interaction appears to be most important for viral integration, the capacity for IN alone to multimerize into tetramers could be important for several additional stages of the viral life cycle. Many amino acid substitutions in IN are known to affect assembly and morphology of viral particles (70), and recent work has demonstrated a role for IN in the uncoating of the viral core (71). Additionally, mutations that disrupt IN tetramerization affect its ability to interact with Gemin2 and assemble with reverse transcriptase on viral RNA (65).
In viral producer cells, IN is synthesized as a part of the Gag-Pol polyprotein precursor, which contains the myristoylated matrix (MA) protein at the N terminus, structural proteins, including capsid (CA), and enzyme precursors, including reverse transcriptase (RT) as intervening components, and IN at the C terminus. HIV is thought to contain ∼2000 gag molecules (composed of MA, CA, and nucleocapsid) and ∼100 Gag-Pol molecules (composed of Gag plus protease, RT, and IN) (72, 73). Thus, the great majority of IN monomers synthesized will not contribute to an IN tetramer involved in catalysis of integration but, judging from mutational studies, may well participate in some aspects of assembly. LEDGF does not appear to play an essential role in these potential nonintegration activities of IN (8, 74), but because LEDGF binds tightly to IN in infected cells, the properties of IN tetramers described here are likely to represent much of the IN present during viral assembly and maturation.
Our ultimate goal in this work is to develop structural models for IN assemblies that play a role in the virus life cycle and to understand the role of host factors in the formation and function of these assemblies. Here, we have presented initial studies that aimed to develop a biophysical basis for understanding the oligomeric states, stoichiometries, and solution shapes of IN·LEDGF complexes. The next step will be to carry out similar studies on IN assemblies bound to DNA, again considering different combinations of protein variants and substrate forms. For these macromolecular complexes, neutron scattering offers the additional advantage of providing contrast between the protein and DNA components.
Supplementary Material
Acknowledgments
We are grateful to Marc Ruff for supplying the model coordinates derived from the cryo-EM studies by his group; Richard Gillilan (Cornell University High Energy Synchrotron Source), Hiro Tsuruta (Stanford Synchrotron Radiation Light Source), and Lin Yang (National Synchrotron Light Source) for their technical expertise and support; and G. V. and F. B. laboratory members for helpful discussions. Cornell University High Energy Synchrotron Source is supported by National Science Foundation Grant DMR 0225180, and the MacCHESS facility is supported by National Institutes of Health Grant RR-01646. Financial support for the National Synchrotron Light Source comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the United States Department of Energy and from the National Center for Research Resources, National Institutes of Health. The Stanford Synchrotron Radiation Light Source is a national user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences.
This work was supported, in whole or in part, by National Institutes of Health Grants AI52845 and AI082020. This work was also supported by the University of Pennsylvania Center for AIDS Research, the Penn Genome Frontiers Institute, and a grant from the Pennsylvania Department of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Table S1.
T. Diamond, Y. Hwang, and F. Bushman, unpublished results.
- HIV
- human immunodeficiency virus
- IN
- integrase
- LEDGF
- lens epithelium-derived growth factor
- IBD
- integrase binding domain
- SAXS
- small angle x-ray scattering
- NTD
- N-terminal domain
- CCD
- catalytic core domain
- CTD
- C-terminal domain
- LTR
- long terminal repeat
- SE
- sedimentation equilibrium
- SV
- sedimentation velocity
- SEC-MALS
- size exclusion chromatography in-line with multiangle light scattering
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate
- NSD
- normalized spatial discrepancy.
REFERENCES
- 1.Craigie R. (2002) in Mobile DNA II (Craig N. L., Craigie R., Gellert M., Lambowitz A. M. eds) pp. 613–630, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 2.Yoder K. E., Bushman F. D. (2000) J. Virol. 74, 11191–11200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busschots K., Vercammen J., Emiliani S., Benarous R., Engelborghs Y., Christ F., Debyser Z. (2005) J. Biol. Chem. 280, 17841–17847 [DOI] [PubMed] [Google Scholar]
- 4.Cherepanov P., Devroe E., Silver P. A., Engelman A. (2004) J. Biol. Chem. 279, 48883–48892 [DOI] [PubMed] [Google Scholar]
- 5.Emiliani S., Mousnier A., Busschots K., Maroun M., Van Maele B., Tempé D., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., Christ F., Rain J. C., Dargemont C., Debyser Z., Benarous R. (2005) J. Biol. Chem. 280, 25517–25523 [DOI] [PubMed] [Google Scholar]
- 6.Llano M., Delgado S., Vanegas M., Poeschla E. M. (2004) J. Biol. Chem. 279, 55570–55577 [DOI] [PubMed] [Google Scholar]
- 7.Llano M., Saenz D. T., Meehan A., Wongthida P., Peretz M., Walker W. H., Teo W., Poeschla E. M. (2006) Science 314, 461–464 [DOI] [PubMed] [Google Scholar]
- 8.Maertens G., Cherepanov P., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y. (2003) J. Biol. Chem. 278, 33528–33539 [DOI] [PubMed] [Google Scholar]
- 9.Turlure F., Devroe E., Silver P. A., Engelman A. (2004) Front. Biosci. 9, 3187–3208 [DOI] [PubMed] [Google Scholar]
- 10.Ciuffi A., Diamond T. L., Hwang Y., Marshall H. M., Bushman F. D. (2006) Hum. Gene Ther. 17, 960–967 [DOI] [PubMed] [Google Scholar]
- 11.Ciuffi A., Llano M., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J. R., Bushman F. (2005) Nat. Med. 11, 1287–1289 [DOI] [PubMed] [Google Scholar]
- 12.Marshall H. M., Ronen K., Berry C., Llano M., Sutherland H., Saenz D., Bickmore W., Poeschla E., Bushman F. D. (2007) PLoS One 2, e1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shun M. C., Raghavendra N. K., Vandegraaff N., Daigle J. E., Hughes S., Kellam P., Cherepanov P., Engelman A. (2007) Genes Dev. 21, 1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai M., Zheng R., Caffrey M., Craigie R., Clore G. M., Gronenborn A. M. (1997) Nat. Struct. Biol. 4, 567–577 [DOI] [PubMed] [Google Scholar]
- 15.Chen J. C., Krucinski J., Miercke L. J., Finer-Moore J. S., Tang A. H., Leavitt A. D., Stroud R. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyda F., Hickman A. B., Jenkins T. M., Engelman A., Craigie R., Davies D. R. (1994) Science 266, 1981–1986 [DOI] [PubMed] [Google Scholar]
- 17.Lodi P. J., Ernst J. A., Kuszewski J., Hickman A. B., Engelman A., Craigie R., Clore G. M., Gronenborn A. M. (1995) Biochemistry 34, 9826–9833 [DOI] [PubMed] [Google Scholar]
- 18.Wang J. Y., Ling H., Yang W., Craigie R. (2001) EMBO J. 20, 7333–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woerner A. M., Marcus-Sekura C. J. (1993) Nucleic Acids Res. 21, 3507–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkins T. M., Engelman A., Ghirlando R., Craigie R. (1996) J. Biol. Chem. 271, 7712–7718 [DOI] [PubMed] [Google Scholar]
- 21.Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z. (2003) J. Biol. Chem. 278, 372–381 [DOI] [PubMed] [Google Scholar]
- 22.Hayouka Z., Rosenbluh J., Levin A., Loya S., Lebendiker M., Veprintsev D., Kotler M., Hizi A., Loyter A., Friedler A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8316–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cherepanov P., Sun Z. Y., Rahman S., Maertens G., Wagner G., Engelman A. (2005) Nat. Struct. Mol. Biol. 12, 526–532 [DOI] [PubMed] [Google Scholar]
- 24.Cherepanov P., Ambrosio A. L., Rahman S., Ellenberger T., Engelman A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17308–17313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hare S., Di Nunzio F., Labeja A., Wang J., Engelman A., Cherepanov P. (2009) PLoS Pathog. 5, e1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hare S., Shun M. C., Gupta S. S., Valkov E., Engelman A., Cherepanov P. (2009) PLoS Pathog. 5, e1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michel F., Crucifix C., Granger F., Eiler S., Mouscadet J. F., Korolev S., Agapkina J., Ziganshin R., Gottikh M., Nazabal A., Emiliani S., Benarous R., Moras D., Schultz P., Ruff M. (2009) EMBO J. 28, 980–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faure A., Calmels C., Desjobert C., Castroviejo M., Caumont-Sarcos A., Tarrago-Litvak L., Litvak S., Parissi V. (2005) Nucleic Acids Res. 33, 977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bera S., Pandey K. K., Vora A. C., Grandgenett D. P. (2009) J. Mol. Biol. 389, 183–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guiot E., Carayon K., Delelis O., Simon F., Tauc P., Zubin E., Gottikh M., Mouscadet J. F., Brochon J. C., Deprez E. (2006) J. Biol. Chem. 281, 22707–22719 [DOI] [PubMed] [Google Scholar]
- 31.Hare S., Gupta S. S., Valkov E., Engelman A., Cherepanov P. (2010) Nature 464, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M., Mizuuchi M., Burke T. R., Jr., Craigie R. (2006) EMBO J. 25, 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Havlir D. V. (2008) N. Engl. J. Med. 359, 416–418 [DOI] [PubMed] [Google Scholar]
- 34.Berthoux L., Sebastian S., Muesing M. A., Luban J. (2007) Virology 364, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bosserman M. A., O'Quinn D. F., Wong I. (2007) Biochemistry 46, 11231–11239 [DOI] [PubMed] [Google Scholar]
- 36.Chen A., Weber I. T., Harrison R. W., Leis J. (2006) J. Biol. Chem. 281, 4173–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKee C. J., Kessl J. J., Shkriabai N., Dar M. J., Engelman A., Kvaratskhelia M. (2008) J. Biol. Chem. 283, 31802–31812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L. D., Liu C. L., Chen W. Z., Wang C. X. (2005) Biochem. Biophys. Res. Commun. 337, 313–319 [DOI] [PubMed] [Google Scholar]
- 39.Yang Z. N., Mueser T. C., Bushman F. D., Hyde C. C. (2000) J. Mol. Biol. 296, 535–548 [DOI] [PubMed] [Google Scholar]
- 40.Diamond T. L., Bushman F. D. (2005) J. Virol. 79, 15376–15387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vistica J., Dam J., Balbo A., Yikilmaz E., Mariuzza R. A., Rouault T. A., Schuck P. (2004) Anal. Biochem. 326, 234–256 [DOI] [PubMed] [Google Scholar]
- 43.Semenyuk A. V., Svergun D. I. (1991) J. Appl. Crystallogr. 24, 537–540 [Google Scholar]
- 44.Petoukhov M. V., Konarev P. V., Kikhney A. G., Svergun D. I. (2007) J. Appl. Crystallogr. 40, S223–S228 [Google Scholar]
- 45.Svergun D., Barberato C., Koch M. H. (1995) J. Appl. Crystallgor. 28, 768–773 [Google Scholar]
- 46.Franke D., Svergun D. I. (2009) J. Appl. Crystallgor. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svergun D. I., Petoukhov M. V., Koch M. H. (2001) Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozin M. B., Svergun D. I. (2001) J. Appl. Crystallgor. 34, 33–41 [Google Scholar]
- 49.Volkov V. V., Svergun D. I. (2003) J. Appl. Crystallgor. 36, 860–864 [Google Scholar]
- 50.Delano W. (2002) The PYMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 51.Wriggers W., Milligan R. A., McCammon J. A. (1999) J. Struct. Biol. 125, 185–195 [DOI] [PubMed] [Google Scholar]
- 52.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comp. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 53.Brunger A. T. (2007) Nat. Protocols 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 54.García De La Torre J., Huertas M. L., Carrasco B. (2000) Biophys. J. 78, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Birmanns S., Wriggers W. (2007) J. Struct. Biol. 157, 271–280 [DOI] [PubMed] [Google Scholar]
- 56.Carteau S., Mouscadet J. F., Goulaouic H., Subra F., Auclair C. (1993) Arch. Biochem. Biophys. 300, 756–760 [DOI] [PubMed] [Google Scholar]
- 57.Sherman P. A., Fyfe J. A. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5119–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alian A., Griner S. L., Chiang V., Tsiang M., Jones G., Birkus G., Geleziunas R., Leavitt A. D., Stroud R. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8192–8197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao K., Butler S. L., Bushman F. D. (2001) EMBO J. 20, 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Molteni V., Greenwald J., Rhodes D., Hwang Y., Kwiatkowski W., Bushman F. D., Siegel J. S., Choe S. (2001) Acta Crystallogr D. Biol. Crystallogr. 57, 536–544 [DOI] [PubMed] [Google Scholar]
- 61.Siegel L., Monty K. (1966) 112, 346–362 [DOI] [PubMed] [Google Scholar]
- 62.Dolan J., Chen A., Weber I. T., Harrison R. W., Leis J. (2009) J. Mol. Biol. 385, 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bao K. K., Wang H., Miller J. K., Erie D. A., Skalka A. M., Wong I. (2003) J. Biol. Chem. 278, 1323–1327 [DOI] [PubMed] [Google Scholar]
- 64.Lutzke R. A., Plasterk R. H. (1998) J. Virol. 72, 4841–4848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nishitsuji H., Hayashi T., Takahashi T., Miyano M., Kannagi M., Masuda T. (2009) PLoS One 4, e7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deprez E., Tauc P., Leh H., Mouscadet J. F., Auclair C., Brochon J. C. (2000) Biochemistry 39, 9275–9284 [DOI] [PubMed] [Google Scholar]
- 67.Deprez E., Tauc P., Leh H., Mouscadet J. F., Auclair C., Hawkins M. E., Brochon J. C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10090–10095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baranova S., Tuzikov F. V., Zakharova O. D., Tuzikova N. A., Calmels C., Litvak S., Tarrago-Litvak L., Parissi V., Nevinsky G. A. (2007) Nucleic Acids Res. 35, 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Duyne G. D. (2008) in Protein-Nucleic Acid Interactions: Structural Biology (Rice P. A., Correll C. C. eds) pp. 303–328, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 70.Engelman A. (1999) Adv. Virus Res. 52, 411–426 [DOI] [PubMed] [Google Scholar]
- 71.Briones M. S., Dobard C. W., Chow S. A. (2010) J. Virol. 84, 5181–5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vogt V. M. (1997) in Retroviruses (Coffin J. M., Hughes S. H., Varmus H. E. eds) pp. 263–334, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 73.Jacks T., Power M. D., Masiarz F. R., Luciw P. A., Barr P. J., Varmus H. E. (1988) Nature 331, 280–283 [DOI] [PubMed] [Google Scholar]
- 74.Llano M., Vanegas M., Fregoso O., Saenz D., Chung S., Peretz M., Poeschla E. M. (2004) J. Virol. 78, 9524–9537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Damaschun G., Damaschun H., Gast K., Gerlach D., Misselwitz R., Welfle H., Zirwer D. (1992) Eur. Biophys. J. 20, 355–361 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.