Abstract
Macrophages are essential components of innate immunity, and apoptosis of these cells impairs mucosal defense to microbes. Helicobacter pylori is a gastric pathogen that infects half of the world population and causes peptic ulcer disease and gastric cancer. The host inflammatory response fails to eradicate the organism. We have reported that H. pylori induces apoptosis of macrophages by generation of polyamines from ornithine decarboxylase (ODC), which is dependent on c-Myc as a transcriptional enhancer. We have now demonstrated that expression of c-Myc requires phosphorylation and nuclear translocation of ERK, which results in phosphorylation of c-Fos and formation of a specific activator protein (AP)-1 complex. Electromobility shift assay and immunoprecipitation revealed a previously unrecognized complex of phospho-c-Fos (pc-Fos) and c-Jun in the nucleus. Fluorescence resonance energy transfer demonstrated the interaction of pc-Fos and c-Jun. The capacity of this AP-1 complex to bind to putative AP-1 sequences was demonstrated by oligonucleotide pulldown and fluorescence polarization. Binding of the pc-Fos·c-Jun complex to the c-Myc promoter was demonstrated by chromatin immunoprecipitation. A dominant-negative c-Fos inhibited H. pylori-induced expression of c-Myc and ODC and apoptosis. H. pylori infection of mice induced a rapid infiltration of macrophages into the stomach. Concomitant apoptosis depleted these cells, and this was associated with formation of a pc-Fos·c-Jun complex. Treatment of mice with an inhibitor of ERK phosphorylation attenuated phosphorylation of c-Fos, expression of ODC, and apoptosis in gastric macrophages. A unique AP-1 complex in gastric macrophages contributes to the immune escape of H. pylori.
Keywords: AP-1 Transcription Factor, Apoptosis, Innate Immunity, Macrophage, Polyamines
Introduction
In infection studies, apoptosis of macrophages has been demonstrated to play a pathogenic role in colonization of bacteria (1). Helicobacter pylori is a microaerophilic, Gram-negative bacterium that selectively colonizes the human stomach and infects half of the world population (2). Infected individuals exhibit chronic active gastritis and can develop peptic ulcer disease or gastric adenocarcinoma, the second leading cause of cancer deaths worldwide (3). The infection is usually acquired in childhood and persists for the life of the host despite eliciting a vigorous innate and adaptive immune response (2). Although H. pylori has generally been considered to be a noninvasive pathogen, strong evidence has emerged that H. pylori itself and its products can invade the mucosa and have direct contact with lamina propria immune cells (4–6). These findings suggest that the failure of the immune response could be directly related to the inability of effector cells, especially macrophages, to kill this bacterium. We have demonstrated that H. pylori induces apoptosis in macrophages in vitro by a polyamine-dependent mechanism (7–9). However, the signaling mechanisms involved in this process and their in vivo relevance remains to be elucidated.
H. pylori has been reported to activate mitogen-activated protein kinase (MAPK)3 enzymes (10). MAPKs belong to an important group of serine and threonine signaling kinases consisting of three family member proteins: JNK, p38 MAPK, and ERK1/2. These proteins mediate signal transduction in response to extracellular stimuli and affect diverse cellular functions such as proliferation, differentiation, and death (11, 12). In particular, ERK, which is activated upon phosphorylation by dual specificity MEK1 and MEK2 (13), can have biological effects by phosphorylating membrane or cytoskeletal proteins (14). Moreover, when phosphorylated ERK (pERK) translocates to the nucleus (15, 16), it can result in activation of transcription factors, including activator protein-1 (AP-1) (17).
AP-1 complexes most often consist of c-Fos and c-Jun, and other Fos and Jun family proteins can also form functional AP-1 (18). When these subfamily proteins form homodimers or heterodimers, they become active AP-1 complexes. Such complexes bind to AP-1 DNA recognition elements and activate transcription in stimulated cells (19). Fos proteins do not form homodimers, whereas c-Jun can form homodimers that have a low capacity to transactivate genes (20). When c-Fos heterodimerizes with c-Jun, this results in a more stable AP-1 complex that increases the capacity of c-Jun to transactivate target genes (21). JNK can phosphorylate c-Jun at Ser73 in the transactivation domain and thus potentiate its ability to induce transcription (22). Similarly, phosphorylation of c-Fos at Ser374 by ERK potentiates AP-1 transactivation capabilities and primes c-Fos for phosphorylation at Thr325; this stabilizes c-Fos heterodimers and enhances promoter transactivation by AP-1 complexes (23). Activation of AP-1 (18) can result in effects on cell proliferation (24), cell differentiation (25), and apoptosis (26).
Mutation of the AP-1 binding site inhibits IL-6 promoter activity in H. pylori-stimulated gastric epithelial cell lines, and all of the MAPKs, namely ERK, p38, and JNK were implicated in AP-1 binding and IL-6 expression, and other transcription factors were involved (27). Also nuclear extracts from H. pylori-infected gerbil stomach tissues bind to the AP-1 consensus binding sequence (28). However, in both of these studies the constituents of the AP-1 complex were not determined. In two other studies in gastric epithelial cells, increased expression of several potential AP-1 constituents (29) and induction of AP-1 DNA binding activity was demonstrated (29, 30), but phosphorylation of AP-1 members and the biological effects of AP-1 were not studied. In the case of THP-1 monocytes, H. pylori-stimulated IL-18 expression that was reduced by either ERK or p38 inhibitors and AP-1 binding to the IL-18 promoter were demonstrated (31), but the functional requirement for AP-1 in this process and the nature of the AP-1 complex was not examined. Inhibition of ERK and, to a greater degree, inhibition of p38 have been shown to reduce H. pylori-stimulated IL-8 expression in THP-1 cells, and although AP-1 was implicated in this process, it had less of an effect than NF-κB activation (32). In U937 monocytic cells, induction of IL-8 was shown to be largely p38-dependent and only partially AP-1-dependent (33). Taken together, these studies suggested that MAPKs and AP-1 may be involved in biological effects of H. pylori infection but that more investigation was warranted in macrophages.
Previously, we have shown that H. pylori induces c-Myc gene and protein expression and nuclear translocation in macrophages (9). This enhances expression of ornithine decarboxylase (ODC), the rate-limiting enzyme for polyamine synthesis, which causes apoptosis by a mechanism that involves oxidation of spermine (8). We now investigated the particular MAPK pathways activated in macrophages, the components of the AP-1 complex, and the role of these responses in the induction of apoptosis. Herein we show that activation of ERK, but not p38 or JNK, by H. pylori results in apoptosis through activation of c-Myc and ODC. This process occurs by ERK-dependent formation of a specific AP-1 complex that appears to be unique to H. pylori, consisting of a phospho(p)-c-Fos·c-Jun heterodimer. This AP-1 complex binds to the c-Myc promoter in macrophages and thus causes induction of c-Myc, ODC, and apoptosis. We have also demonstrated that in addition to these events occurring in a macrophage cell line, they also occur in gastric macrophages in H. pylori-infected gastric tissues and that the apoptosis of gastric macrophages in vivo that contributes to the loss of host defense can be abrogated by interruption of this pathway. The specificity of these events is demonstrated by our findings that two other enteric bacterial pathogens that cause mucosal inflammation that were tested, namely Campylobacter jejuni and Citrobacter rodentium, did not induce c-Fos phosphorylation, c-Myc, or ODC in macrophages despite activation of ERK and that H. pylori did not induce the pc-Fos-c-Myc-ODC pathway in gastric epithelial cells.
EXPERIMENTAL PROCEDURES
Reagents
All of the reagents used for cell culture and RNA extraction were purchased from Invitrogen. MAPK inhibitors PD98059, SB203580, SP600125, and U0126 were from Calbiochem. All other chemicals were from Sigma unless specified.
Bacteria, Cells, and Culture Conditions
H. pylori SS1 was grown, lysates were prepared using a French press, and the multiplicity of infection (MOI) was determined as described (34). The murine macrophage cell line RAW264.7 was maintained in complete Dulbecco's modified Eagle's medium. The cells were activated with H. pylori lysates at an MOI of 100. In some studies macrophages were also activated with live H. pylori SS1 at an MOI of 10 (2, 34). C. rodentium was grown, and lysates were prepared as described (35). C. jejuni was obtained from ATCC, and lysates were prepared as for H. pylori. RAW 264.7 cells were activated with C. rodentium and C. jejuni lysates at an MOI of 100. AGS gastric epithelial cells were activated with H. pylori strain 60190 at an MOI of 200.
Mouse Infections
H. pylori SS1 was grown in liquid culture in Brucella broth. C57BL/6 mice were inoculated by oral gavage with 5 × 108 colony-forming units of H. pylori in 0.1 ml of Brucella broth or broth vehicle control. For the colonization time points of 14, 60, and 120 days, the mice were gavaged every other day three times (7, 36), whereas for the time points of 1–7 days, the mice were inoculated once (35). In some studies, the mice were treated with PD98059 (5 mg/kg) or vehicle control (2% dimethyl sulfoxide) by intraperitoneal injection just prior to inoculation.
Cell Viability
The viability of macrophages was determined using an XTT assay as described (8, 36).
Apoptosis Detection by Annexin V Staining
The cells were stained with annexin V conjugated to fluorescein isothiocyanate (FITC) and either propidium iodide (PI) or 7-amino-actinomycin D (7-AAD; BD Biosciences, San Jose, CA). The cells were analyzed with a flow cytometer (FACSCalibur; BD Biosciences) using CellQuest software (BD Biosciences) for annexin V-FITC/PI-stained cells (8, 9) and FlowJo software (Tree Star, Inc., Ashland, OR) for the annexin V-FITC/7-AAD-stained cells (36).
RT-PCR and Real Time PCR
ODC and c-Myc mRNA expression was measured by real time PCR and RT-PCR as described (9). For c-Fos and c-Jun, the sense and antisense primer sequences and PCR product sizes were: c-Fos, 5′-GCCCAGTGAGGAATATCTGGA-3′ and 5′-ATCGCAGATGAAGCTCTGGT-3′, 187 bp; c-Jun, 5′-AGCCTACCAACGTGAGTGCT-3′ and 5′-AGAACGGTCCGTCACTTCAC-3′, 228 bp. For c-Fos and c-Jun, 1 μg of mRNA was reverse-transcribed with the iScriptTM cDNA synthesis kit (Bio-Rad). PCR was performed with thermal cycling conditions and methods to calculate the relative expression as described (2, 8, 9, 35).
Immunoprecipitation and Immunoblot Analysis
RAW 264.7 cells were lysed, and Western blotting for c-Myc and ODC was performed as described (9). Nuclear and cytoplasmic extracts were prepared using an extraction kit from Thermo Scientific (Rockford, IL). ERK1/2 was detected with a rabbit polyclonal antibody from BioSource (Camarillo, CA; 1:1000 dilution). For detection of pERK, a rabbit polyclonal antibody to phosphothreonine 185 and phosphotyrosine 187, pTh,pY185/187-ERK, was used (BioSource; 1:1000 dilution). For c-Fos, a mouse monoclonal antibody (EMD Chemicals Inc., Gibbstown, NJ; 1:2000 dilution) was used. Specific antibodies against the following c-Fos phosphorylation sites were used: serine 374 (pS374; mouse monoclonal antibody; EMD Chemicals Inc.; 1:1000 dilution), threonine 325 (pTh325; rabbit polyclonal antibody; Abcam, Cambridge, MA; 1:2000 dilution), and threonine 232 (pTh232; rabbit polyclonal; EMD Chemicals Inc; 1:2000 dilution). For c-Jun, a mouse monoclonal antibody (Cell Signaling, Boston, MA; 1:2000 dilution) was used. For pc-Jun, a rabbit polyclonal antibody to the phosphorylation site at serine 73 (pS73; Cell Signaling; 1:2000 dilution) was used. The proteins from cell lysates were immunoprecipitated with rabbit polyclonal anti-pTh325c-Fos, mouse monoclonal anti-c-Jun, or rabbit polyclonal anti-pS73c-Jun antibodies, followed by immunoblotting with rabbit polyclonal anti-pTh325c-Fos, mouse monoclonal anti-c-Jun, or rabbit polyclonal anti-pS73c-Jun antibodies.
ERK Enzyme Activity
ERK activity was measured with an ERK enzyme assay kit (Millipore, Temecula, CA) using myelin basic protein as the substrate, according to the manufacturer's instructions.
Immunofluorescence Staining
RAW 264.7 cells were plated on chamber slides (1 × 104 cells/well) and stimulated with H. pylori lysates for 0–120 min. The cells were fixed, permeabilized, and blocked as described (37) and then incubated with anti-pERK antibody (1:500 dilution) overnight at 4 °C. A rabbit anti-mouse secondary antibody conjugated to FITC (Jackson ImmunoResearch Laboratories Inc., Westgrove, PA; 1:1000 dilution) was used. The nuclei were visualized with PI, and staining was imaged as described (8, 37).
Electrophoretic Mobility Shift Assay (EMSA)
The nuclear extracts were prepared as above. 21-bp complimentary oligonucleotides (sense, 5′-CGCTTGATGAGTCAGCCGGAA-3′ and antisense, 5′-GCGAACTACTCAGTCGGCCTT-3′) containing the AP-1 consensus binding site were purchased from Promega (Madison, WI). Double-stranded oligonucleotides (50 ng) were labeled with [γ-32P]ATP using T4 polynucleotide kinase (Promega). Binding reactions were carried out with labeled oligonucleotides and 2 μg of nuclear protein. For competition experiments, a 200-fold excess of cold AP-1 oligonucleotides was used. The entire reaction was electrophoresed on 6% polyacrylamide gels. The gels were dried and phosphorimaged (9, 37).
Transient Transfection of Dominant-negative A-Fos Plasmid
RAW 264.7 cells were transfected with 1 μg of dominant-negative c-Fos (A-Fos) plasmid provided by C. Vinson from the National Cancer Institute (38) or control plasmid (pCMV), using Lipofectamine Plus (Invitrogen) as described (8).
Oligonucleotide Pulldown Assay
Both strands of the 21-bp oligonucleotides containing the AP-1 binding consensus sequence were labeled with biotin at the 3′ end using the Biotin 3′ end DNA labeling kit (Thermo Scientific). Biotin-labeled oligonucleotides were incubated with 100 μg of nuclear extracts in EMSA binding buffer (Promega). Biotinylated oligonucleotides bound to protein complexes were captured using immobilized streptavidin-agarose beads on a spin column (Thermo Scientific). After washing with Tris buffer three times, the slurry was resuspended in 100 μl of radioimmunoprecipitation assay buffer and centrifuged. The supernatants were collected and resolved on SDS-PAGE followed by Western blotting for pc-Fos (Thr325) and c-Jun.
Chromatin Immunoprecipitation
The cells were treated with H. pylori in the presence or absence of MAPK inhibitors and fixed with 1% formaldehyde to cross-link proteins to DNA. The cells were washed, lysed, sonicated on ice, and centrifuged at 14,000 × g for 10 min. Precleared supernatants were immunoprecipitated overnight at 4 °C using 2 μg of anti-pTh325c-Fos, anti-c-Jun, and anti-pS73c-Jun antibodies. The DNA·protein complex was eluted, and cross-linking was reversed. DNA was recovered and purified with a spin column. PCR was performed on the immunoprecipitated samples and inputs using the following conditions: 95 °C for 5 min; 35 cycles of 95 °C for 1 min, 58 °C for 1 min, and 72 °C for 1 min; and final elongation at 72 °C for 7 min. Primers were used that flanked three different AP-1 binding sites in the mouse c-Myc promoter. The sense and antisense primer sequences and PCR product sizes were: primer 1, flanking the −917 bp to −1136 bp region, 5′-TTGAGGCATGATGCTGAGAC-3′ and 5′-AGGCCTTAGACCATCCAGGT-3′, 219 bp; primer 2, flanking the −1896 bp to −2083 bp region, 5′-CTAATGCTCCTGCCTTGCAT-3′ and 5′-GTGGATGGGGAAAATGAATG-3′, 187 bp; and primer 3, flanking the −1921 bp to −2150 bp region, 5′-TCTGACTCCTTTTGCCCAGT-3′ and 5′-TTGCAAAGAGGGGGAGTAGA-3′, 235 bp. PCR products were analyzed on 1% agarose gels.
Gastric Macrophage Isolation and Quantification
C57BL/6 mice were infected with H. pylori SS1 for 1–120 days. The glandular stomach was digested, and isolation of gastric macrophages was performed as described using CD11b-positive selection (2, 39). Isolated cells were incubated with anti-F4/80 antibody conjugated with phycoerythrin (PE) (Invitrogen; 1:100 dilution). The cells were analyzed by flow cytometry to determine the percentage of F4/80-PE positive cells using a BD LSRII system (BD Biosciences). The number of macrophages was determined by multiplying the percentage of F4/80-positive cells by the number of CD11b-positive cells and then dividing by 100.
Analysis of Apoptosis in Gastric Macrophages
CD11b-positive cells from mouse stomachs were stained with anti-F4/80-PE, followed by staining with annexin V-FITC and PI (BD Biosciences). PE-positive cells were gated and analyzed for FITC and PI staining by flow cytometry (8, 9).
Isolation of RNA from Gastric Macrophages and Real Time PCR
Gastric macrophages were isolated from C57BL/6 mice by F4/80 positive selection. RNA was isolated, and real time PCR was performed as described (2, 35).
Analysis of Proteins in Gastric Macrophages
F4/80-positive macrophages were stained with mouse monoclonal anti-pERK antibody conjugated with FITC (BD Biosciences; 1:250 dilution), anti-pTh325c-Fos antibody (1:100 dilution), and goat polyclonal anti-ODC antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA; 1:200 dilution) and incubated on ice for 30 min. The cells were then incubated with goat polyclonal anti-rabbit secondary antibody conjugated with PE to detect pc-Fos and donkey polyclonal anti-goat secondary antibody conjugated with allophycocyanin (Jackson ImmunoResearch Laboratories Inc.) to detect ODC. Stained cells were analyzed with an LSRII flow cytometer.
Fluorescence Polarization
An Alexa 488-labeled 28-mer double-stranded oligonucleotide containing the AP-1 binding site from the c-Myc promoter was used. Alexa 488-labeled sense and antisense oligomers (Invitrogen) were annealed in at a final concentration of 100 nm by heating the mixture to 95 °C for 2.5 min. RAW 264.7 cells and gastric macrophages were lysed in radioimmune precipitation assay buffer. Detergents and salts were removed using a spin column. The cell lysates were resuspended in Tris buffer. The proteins were incubated with 20 nm of the labeled oligonucleotides for 20 min at room temperature in microtiter plates. Fluorescence polarization was measured on a Tecan Ultra plate reader (Durham, NC) with excitation at 485 nm and emission at 535 nm. For each condition, polarization was calculated using the formula (Ih − Iv)/(Ih + Iv), where Ih represents intensity measured with horizontally polarized excitation, and Iv represents intensity measured with vertically polarized excitation and expressed as millipolarization (40).
Fluorescence Resonance Energy Transfer (FRET) and Confocal Microscopy
RAW 267.4 cells or gastric macrophages were plated in chamber slides and fixed with 4% paraformaldehyde containing 0.1% Triton X-100. The cells were permeabilized with ice-cold 100% methanol with 0.1% Triton X-100. The cells were blocked with 5% normal goat serum containing 0.1% Triton X-100. Further, the cells were incubated with anti-pTh325c-Fos or anti-pS374c-Fos antibodies (both 1:250 dilution) overnight at 4 °C. The cells were washed and incubated at room temperature for 1 h with anti-rabbit secondary antibody conjugated with Cy3 (Jackson ImmunoResearch Laboratories Inc.; 50 μg/ml) as the donor. The cells were washed and incubated with anti-c-Jun antibody (1:250 dilution) for 2 h at room temperature. The cells were washed and incubated with anti-mouse secondary antibody conjugated with Cy5 (Jackson ImmunoResearch Laboratories Inc.; 25 μg/ml) as an acceptor for 2 h at room temperature.
Images of cells were captured with a Zeiss inverted LSM510 confocal microscope, using a cooled charge-coupled device camera and an IC300 digital imaging system. Cy3 and Cy5 intensity in cells was measured using MetaMorph 7.6 software (Molecular Devices, Sunnyvale, CA). To quantify FRET, Cy5 was photobleached for 1.5 min such that mean fluorescence for Cy5 decreased to background. Fluorescence of Cy3 prebleach (Cy3pre) and postbleach (Cy3post) was calculated in the region of interest in photobleached cells. Background Cy3 fluorescence (Cy3back) was subtracted from the prebleached fluorescence. FRET efficiency was calculated as [(Cy3pre − Cy3back) − Cy3post]/(Cy3pre − Cy3back) as described (41).
Statistical Analysis
The quantitative data are shown as the means ± S.E. Comparisons between multiple groups were made by using analysis of variance with the Student-Newman-Keul's post hoc multiple comparison test. When comparisons between only two groups were made, a paired t test was performed.
RESULTS
Inhibition of ERK Phosphorylation Increases Cell Viability and Attenuates Apoptosis
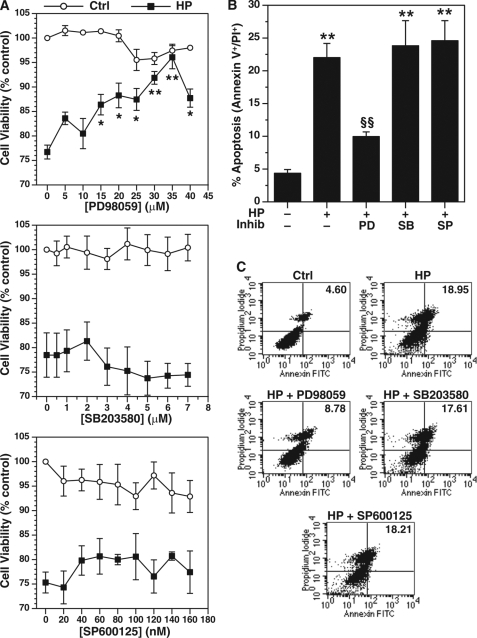
MAPK has been shown to modulate apoptosis in different cell types (11). We determined the effects of a dose range of MAPK inhibitors on cell death induced by H. pylori by performing XTT cell viability assays in RAW 264.7 cells. After 24 h of H. pylori stimulation, viability was decreased to 76.2 ± 2.3% of levels in untreated cells (Fig. 1A). Inhibition of ERK phosphorylation by pretreatment with PD98059 improved cell viability of H. pylori-stimulated macrophages in a concentration-dependent manner with a maximum effective dose of 35 μm (Fig. 1A, top panel). In addition, this dose had no significant effect on cell viability when added to unstimulated cells (Fig. 1A, top panel). Inhibition of phosphorylation of p38 MAPK with SB203580 or of JNK kinase with SP600125 had no beneficial effect on H. pylori-induced macrophage cell death and no affect on cell viability in unstimulated cells (Fig. 1A, middle and bottom panels).
FIGURE 1.
Effect of MAPK inhibitors on H. pylori-induced apoptosis in RAW 264.7 macrophages. The cells were pretreated with inhibitors of ERK phosphorylation (PD98059), p38 phosphorylation (SB203580), or JNK phosphorylation (SP600125) for 45 min before activation with H. pylori lysate (HP) of strain SS1 at an MOI of 100 for 24 h. A, cell viability was measured by XTT assay with different concentrations of PD98059, SB203580, and SP600125 in the presence or absence of H. pylori. B, apoptosis was measured by annexin V-FITC and PI staining and flow cytometry in macrophages pretreated with 35 μm of PD98059 (PD), 12.5 μm SB203580 (SB), or 50 nm SP600125 (SP). C, representative dot plots of annexin V-FITC versus PI. The values for the percentages of annexin V and PI double-positive cells (late apoptosis) in the upper right quadrants are shown. For A, *, p < 0.05; **, p < 0.01 versus HP alone without inhibitor; for B, **, p < 0.01 versus control (Ctrl); §§, p < 0.01 versus HP alone; Inhib, inhibitor. For A and B, n = 4.
We have reported that decreased macrophage cell viability with exposure to H. pylori correlates with apoptosis that peaks at 24 h poststimulation in RAW 264.7 cells (8). We have now observed that PD98059 inhibited the 5.3 ± 0.9-fold increase in H. pylori-stimulated apoptosis by 69.0 ± 2.1% (Fig. 1, B and C). In contrast, SB203850 and SP600125 did not inhibit the induction of apoptosis (Fig. 1, B and C).
ERK Phosphorylation Is Required for Induction of c-Myc and ODC Expression
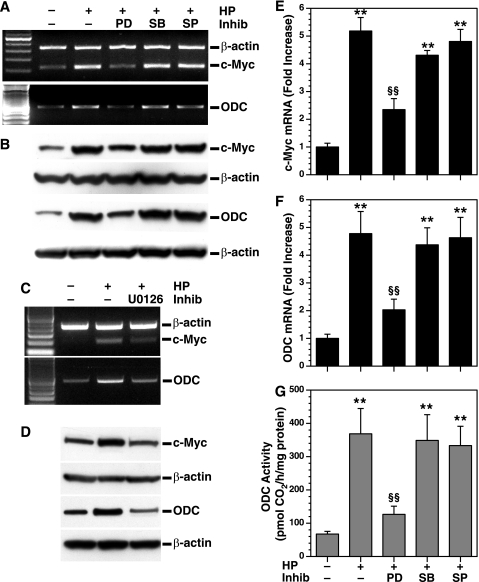
We have reported that H. pylori-induced apoptosis is dependent on the induction of ODC, which requires c-Myc binding to the ODC promoter (9). In the latter study, expression of c-Myc and ODC mRNA peaked at 2 and 6 h, respectively, and both were stable out to 12 h; protein expression peaked at 2–4 h for c-Myc and 6 h for ODC (9). We therefore tested the effects of MAPK inhibitors on c-Myc and ODC expression at 6 h after stimulation. PD98059 inhibited H. pylori-induced c-Myc and ODC mRNA and protein expression, whereas SB203580 or SP600125 had no such effect (Fig. 2, A and B). In addition, a second ERK inhibitor, U0126, also blocked H. pylori-induced c-Myc and ODC mRNA and protein expression (Fig. 2, C and D). To quantify these data further, we used real time PCR and found that H. pylori increased c-Myc and ODC mRNA expression by 5.1 ± 0.4- and 4.7 ± 0.7-fold, respectively; these increases were inhibited by 68.0 ± 10.4 and 73.2 ± 7.4%, respectively, with PD98059; SB203580 and SP600125 had no effect (Fig. 2, E and F). Similarly, the increase in ODC activity with H. pylori exposure was significantly reduced with PD98059 but not SB203580 or SP600125 (Fig. 2G).
FIGURE 2.
Effect of MAPK inhibitors on c-Myc and ODC mRNA and protein expression and ODC enzyme activity. RAW 264.7 macrophages were pretreated with PD98059 (PD), SB203580 (SB), or SP600125 (SP) and activated with HP, all as described for Fig. 1B. A, RT-PCR for c-Myc, ODC, and β-actin. B, Western blot analysis for c-Myc (64 kDa), ODC (53 kDa), and β-actin (42 kDa) proteins. C and D, RAW 264.7 cells were pretreated with U0126, an additional ERK phosphorylation inhibitor (Inhib), for 30 min prior to activation with HP and mRNA expression determined by RT-PCR (C), and the protein levels were assessed by Western blotting (D), for c-Myc, ODC, and β-actin. E and F, real time PCR analysis for c-Myc and ODC mRNA levels. G, ODC activity determined by radiochemical assay. For A, C, and E–G, the cells were activated with HP for 6 h, and for B and D, the cells were activated for 24 h with HP. For E–G, **, p < 0.01 versus unstimulated control macrophages; §§, p < 0.01 versus HP-stimulated macrophages. In A–D, the data are representative of three experiments. For E–G, each experiment was performed a minimum of three times, each in duplicate.
H. pylori Induces Phosphorylation of ERK and Its Nuclear Translocation
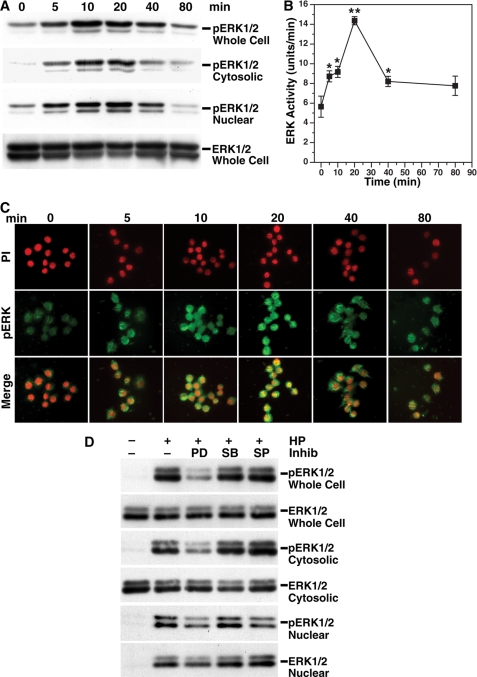
Because inhibition of ERK reduced expression of c-Myc and ODC and attenuated apoptosis, we assessed its phosphorylation status in H. pylori-stimulated macrophages. Levels of pERK in whole cell lysates, as well as cytosolic and nuclear fractions, were increased in a time-dependent manner (Fig. 3A). ERK phosphorylation was readily detected at 5 min and peaked at 10–20 min in both the cytosolic and nuclear fractions, whereas the ERK levels were not altered by H. pylori stimulation. ERK activity was also significantly increased by H. pylori and peaked at 20 min (Fig. 3B). Immunofluorescence staining demonstrated increased phosphorylation of ERK by 5 min after stimulation and rapid translocation of pERK to the nucleus that peaked at 20 min (Fig. 3C). Pretreatment of macrophages with PD98059 significantly attenuated phosphorylation of ERK in the whole cell extract, cytosol, and nucleus. Taken together, these data indicate that H. pylori rapidly induces nuclear translocation of pERK.
FIGURE 3.
H. pylori induces phosphorylation and translocation of ERK into the nucleus. RAW 264.7 macrophages were stimulated with HP for the times indicated. A, Western blot analysis for phosphorylated ERK (pERK1/2; 42 and 44 kDa) and ERK1/2 (42 and 44 kDa) in whole cell lysate, cytoplasmic, and nuclear fractions. B, ERK activity was measured in 1 μg of protein from whole cell lysates. *, p < 0.05; **, p < 0.01 versus unstimulated cells, n = 4. C, cells were cultured in chamber slides in the absence or presence of HP, followed by fixation and permeabilization, and staining with rabbit polyclonal antibody to pTh,pY185/187-ERK. pERK was detected with FITC-conjugated secondary antibody. The nuclei were labeled with PI. Translocation of pERK to nucleus is shown as yellow. All of the images shown were photographed at a magnification of 400×. D, macrophages were pretreated with PD98059 (PD), SB203580 (SB), or SP600125 (SP) at the concentrations used in Fig. 1B for 45 min before activation with HP for 40 min; Western blot analysis for pERK1/2 and ERK1/2 in whole cell lysate and in cytoplasmic and nuclear fractions. In A, C, and D, the data are representative of three experiments. Inhib, inhibitor.
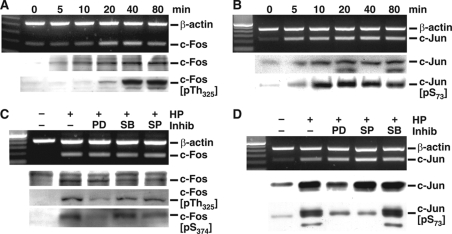
H. pylori Induces Expression of c-Fos and c-Jun
Because ERK phosphorylation has been shown to up-regulate AP-1 complex activity (19, 20, 42), we determined whether H. pylori activates AP-1 and whether this occurs in a pERK-dependent manner in macrophages. H. pylori induced c-Fos mRNA and protein expression in a time-dependent manner (Fig. 4A). c-Fos mRNA induction peaked at 20 min after stimulation, and protein levels peaked at 40 min (Fig. 4A). Phosphorylation of c-Fos at Thr325 was detected at 20 min, peaked at 40 min, and was sustained for 80 min (Fig. 4A). H. pylori induced c-Jun mRNA expression at 5 min after stimulation; c-Jun protein expression and phosphorylation at Ser73 peaked at 10 min and was sustained for 80 min (Fig. 4B).
FIGURE 4.
H. pylori-induces expression and phosphorylation of c-Fos and c-Jun in RAW 264. 7 cells. A, RT-PCR for c-Fos mRNA, and Western blotting for c-Fos (55 kDa) and pTh325c-Fos (58 kDa) in nuclear lysates. B, RT-PCR for c-Jun mRNA, and Western blotting for c-Jun (41 kDa) and pS73c-Jun (48 kDa) in nuclear lysates. In C and D, RAW 267.4 cells were pretreated with PD98059 (PD), SB203580 (SB), or SP600125 (SP) with the concentrations used in Fig. 1B for 45 min before activation with HP. C, RT-PCR for c-Fos mRNA, and Western blot analysis for c-Fos (55 kDa), pTh325c-Fos (58 kDa), and pS374c-Fos (55 kDa). D, RT-PCR for c-Jun and Western blot analysis for c-Jun (41 kDa) and pS73c-Jun (48 kDa). For Western blots, 30 μg of protein/lane was used. The data are representative of three independent experiments. Inhib, inhibitor.
H. pylori-stimulated c-Fos mRNA and protein expression was not affected by inhibition of any of the three MAPKs (Fig. 4C). However, PD98059 blocked phosphorylation of c-Fos at both Thr325 and Ser374. SB203580 or SP600125 had no such effect (Fig. 4C). PD98059 inhibited stimulated c-Jun protein expression and c-Jun phosphorylation at Ser73, without altering c-Jun mRNA levels (Fig. 4D). Inhibition of JNK by SP600125 inhibited c-Jun phosphorylation, whereas inhibition of p38 with SB203580 had no effect. These results suggest that H. pylori induces c-Fos and c-Jun and that the phosphorylation of c-Fos depends on the activation of ERK, whereas the phosphorylation of c-Jun depends on activation of both ERK and JNK.
H. pylori Induces Nuclear Protein Binding to AP-1 Consensus Sequences
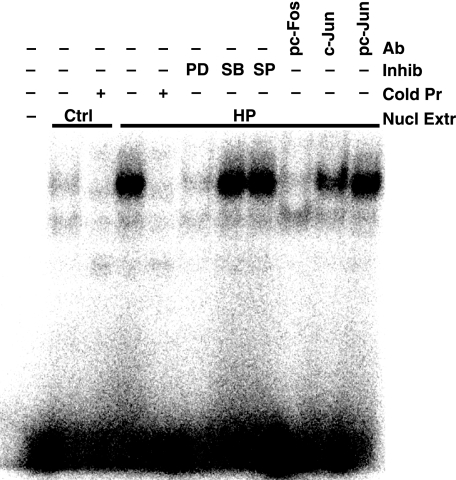
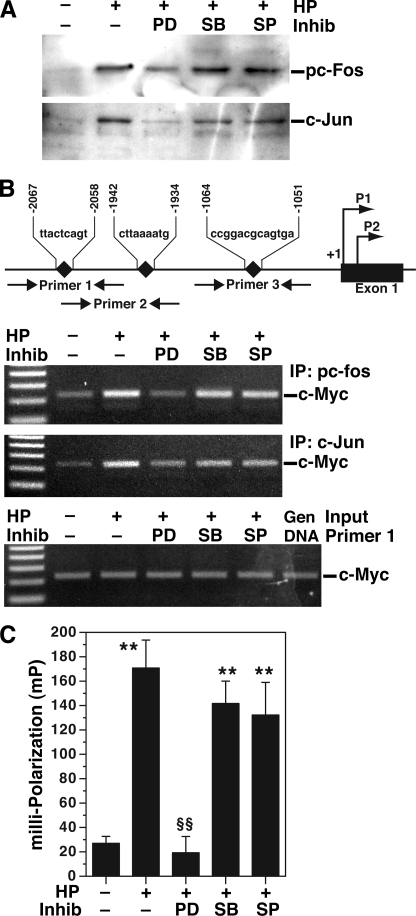
The c-Myc promoter has putative AP-1 consensus binding sequences, and its activity can be up-regulated by AP-1 complex proteins (43). EMSA using nuclear lysates from H. pylori-activated macrophages showed binding to AP-1 consensus binding sequences that was prevented by pretreatment of macrophages with PD98059 (Fig. 5). In contrast, nuclear lysates from macrophages pretreated with SB203580 or SP600125 did not exhibit any loss of binding to the AP-1 consensus sequences (Fig. 5). When nuclear extracts were immunodepleted with antibody to pc-Fos, there was complete inhibition of binding to the AP-1 consensus sequence; antibody to c-Jun showed a partial inhibitory effect, whereas antibody to pc-Jun had no effect (Fig. 5). Thus, H. pylori induces binding of proteins to AP-1 consensus binding sequences, and this involves ERK phosphorylation and the presence of pc-Fos and c-Jun proteins but not pc-Jun.
FIGURE 5.
Binding of pc-Fos·c-Jun to the AP-1 putative binding sequence as assessed by EMSA. RAW 264.7 cells were stimulated for 40 min with HP. Nuclear proteins were extracted, and 1 μg from each condition was incubated with a 32P-labeled probe specific for the consensus binding sequence for AP-1, electrophoresed, and phosphorimaged. First lane, free probe; second lane, nuclear lysates from unstimulated cells; fourth through eleventh lanes, nuclear lysates from HP stimulated cells. In the second and fifth lanes, excess unlabeled (cold) probes for AP-1 were used; the cold AP-1 competed away the binding of the labeled probe. In the sixth through eighth lanes, nuclear lysates were from cells pretreated with PD98059 (PD), SB203580 (SB), or SP600125 (SP) before activation with HP. In the ninth through eleventh lanes, nuclear lysates were immunodepleted with antibodies to pTh325c-Fos (2 μg), c-Jun (1 μg), or pS73c-Jun (2 μg), respectively. The data are representative of three independent experiments. Ab, antibody; Inhib, inhibitor; Cold pr, cold probes; Nucl extr, nuclear extract; Ctrl, control.
Inhibition of AP-1 Binding Attenuates c-Myc and ODC Expression and Apoptosis
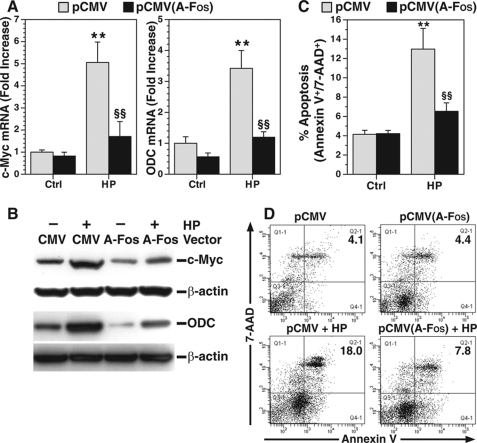
To assess the effect of H. pylori-induced AP-1 complex proteins on c-Myc and ODC expression, macrophages were transfected with A-Fos, dominant-negative c-Fos, which inhibits AP-1 complex occupancy to its consensus binding sequences (38). Transfection with A-Fos resulted in a nearly complete attenuation of H. pylori-induced expression of c-Myc and ODC mRNA (Fig. 6A) and protein (Fig. 6B) when compared with transfection with the cytomegalovirus empty vector. In parallel, A-Fos attenuated H. pylori-induced apoptosis (Fig. 6, C and D). These results suggest that binding of the AP-1 complex to its cognate sequence is a crucial step in inducing c-Myc and ODC expression and apoptosis in macrophages exposed to H. pylori.
FIGURE 6.
Effect of dominant-negative c-Fos on expression of c-Myc and ODC and apoptosis. RAW 264.7 cells were transfected with either empty vector (pCMV) or pCMV plasmid expressing A-Fos; transfected cells were activated with HP. A, real time PCR for c-Myc and ODC mRNA levels; transfected cells were activated for 6 h. B, Western blotting for c-Myc and ODC protein; transfected cells were activated for 24 h. C, apoptosis was measured after 24 h of activation using annexin V-FITC and 7-AAD staining. D, representative dot plots for apoptosis; percentages of cells positive for annexin V-FITC and 7-AAD in the upper right quadrant (late apoptosis) are shown. In A–C, **, p < 0.01 versus unstimulated control cells transfected with empty vector; §§, p < 0.01 versus HP-stimulated cells transfected with A-Fos vector. In A and C, each experiment was performed three times, each in duplicate. For B, the data are representative of three experiments. Ctrl, control.
Phosphorylation of ERK Is Essential for Heterodimerization of pc-Fos and c-Jun
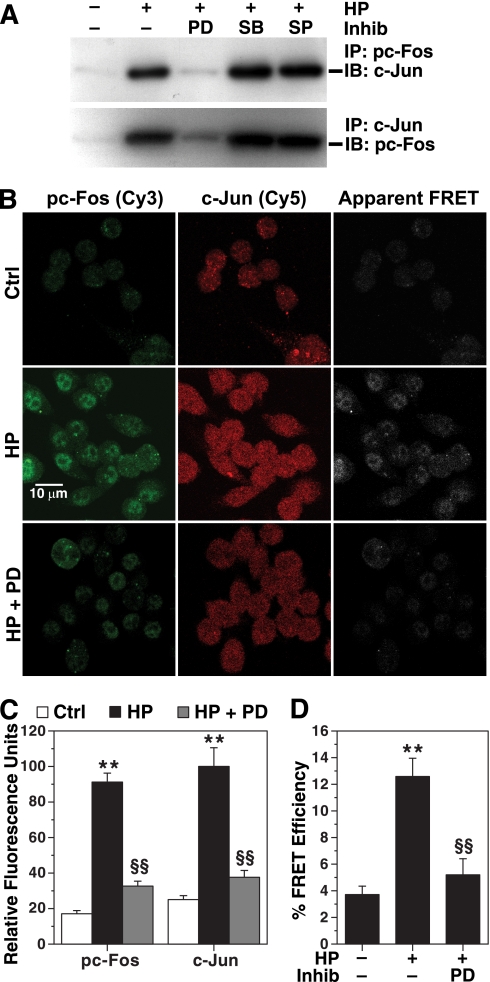
Because we demonstrated the importance of AP-1 in the causation of apoptosis, we investigated the major constituents of this complex in H. pylori-stimulated macrophages. To assess the direct interaction of pc-Fos with its potential partner protein, c-Jun, we performed immunoprecipitation with anti-pTh325c-Fos antibody. When immunoblotting was performed on this immunoprecipitate, c-Jun was abundant (Fig. 7A), whereas when it was probed with anti-pS73c-Jun antibody, no band was present (not shown). Cell lysates immunoprecipitated with anti-c-Jun also showed the presence of pTh325c-Fos (Fig. 7A). Pretreatment of cells with PD98059 prevented the H. pylori-stimulated interaction of c-Jun and pTh325c-Fos proteins (Fig. 7A).
FIGURE 7.
Heterodimerization of pc-Fos and c-Jun in H. pylori-activated macrophages. A, RAW 267.4 cells were pretreated with PD98059 (PD), SB203580 (SB), or SP600125 (SP) as described above and activated with HP for 40 min. Immunoprecipitation (IP) of nuclear proteins was performed with 2 μg of anti-pTh325c-Fos or anti-c-Jun antibodies, followed by immunoblotting (IB) for c-Jun and pTh325c-Fos proteins, respectively. B, cells were plated in chamber slides and treated with HP with or without PD98059 as above. The cells were fixed and permeabilized and stained for pTh325c-Fos and c-Jun proteins, followed by detection with secondary antibodies conjugated with Cy3 (green) and Cy5 (red), respectively. Using confocal microscopy, apparent FRET was imaged (white). C, Cy3 and Cy5 intensity was measured in 100 cells in five microscopic fields. D, Cy5 as an acceptor molecule was photo-bleached in 100 cells from five different fields, and FRET efficiency was measured. For C and D, **, p < 0.01 versus control; §§, p < 0.01 versus HP. For A and B, the data are representative of three experiments. For C and D, summary data from three separate experiments are shown. Inhib, inhibitor; Ctrl, control.
To confirm the direct physical interaction of these proteins, antibody-based confocal FRET microscopy was performed. H. pylori induced phosphorylation of c-Fos and expression of c-Jun that was attenuated by PD98059 (Fig. 7, B and C). When apparent FRET was measured, a positive signal was observed in H. pylori-activated cells that was attenuated in cells pretreated with PD98059 (Fig. 7B). H. pylori increased FRET efficiency by 3.5 ± 0.8-fold, indicating a substantial increase in physical proximity between pTh325c-Fos and c-Jun (Fig. 7D). PD98059 attenuated H. pylori-induced FRET efficiency by 80.2 ± 3.2%. These data indicate that H. pylori increases the interaction between pc-Fos and c-Jun, and this depends on phosphorylation of ERK.
The pc-Fos·c-Jun Complex and the Induction of c-Myc and ODC Are Specific to the Macrophage Response to H. pylori
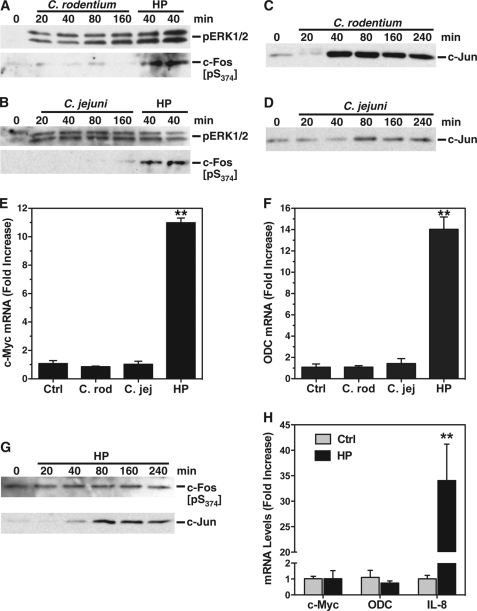
Because we demonstrated that phosphorylation of c-Fos was required for the formation of the AP-1 complex and induction of c-Myc and ODC in the response to H. pylori, we sought to determine whether this effect was specific or generalized in macrophages. We therefore tested the host response to two other relevant enteric Gram-negative bacterial pathogens, namely C. rodentium, which causes colitis in mice, and C. jejuni, which is phylogenetically related to H. pylori and causes bloody diarrhea in humans. Both C. rodentium (Fig. 8A) and C. jejuni (Fig. 8B) failed to induce an increase in the levels of pc-Fos when compared with H. pylori. In contrast, pERK was increased in the same nuclear lysates, and this was sustained at the times assessed from 20 to 160 min. Additionally, c-Jun was induced by both of these pathogens (Fig. 8, C and D). However, both agents failed to induce expression of either c-Myc (Fig. 8E) or ODC (Fig. 8F) when compared with H. pylori used as a positive control. We then added H. pylori strain 60190 to AGS gastric epithelial cells, using a defined model in which we have previously shown marked induction of apoptosis (44). However, even when assessed out to 240 min, there was no increase in pc-Fos levels when compared with unstimulated control cells, but there was an induction of c-Jun expression (Fig. 8G). Furthermore, H. pylori failed to induce any expression of either c-Myc or ODC in the gastric epithelial cells (Fig. 8H), in contrast to a 34-fold increase in IL-8, which was used as a positive control. Taken together, these data show that pc-Fos is very unlikely to be a constituent of an AP-1 complex either in macrophages activated with other prototype bacterial pathogens or in gastric epithelial cells stimulated with H. pylori and that in both of these cases c-Myc and ODC are not part of the host response.
FIGURE 8.
pc-Fos, c-Myc, and ODC are not induced in macrophages activated with C. rodentium or C. jejuni or in gastric epithelial cells activated with H. pylori. A–D, RAW 264.7 cells were activated with French-pressed lysates of C. rodentium (A and C) or C. jejuni (B and D) or H. pylori (A and B) at an MOI of 100 for the times indicated. Western blotting was performed for pERK1/2 and pS374c-Fos (A and B) and c-Jun (C and D) using nuclear lysates as in Fig. 4. E and F, real time PCR analysis for c-Myc and ODC mRNA levels in RAW 264.7 cells stimulated with lysates of C. rodentium (C. rod) or C. jejuni (C. jej) at an MOI of 100 or live H. pylori bacteria (HP) at an MOI of 10. G, Western blot analysis of pS374c-Fos and c-Jun in AGS gastric epithelial cells stimulated with live HP strain 60190 at an MOI of 100 for the times indicated. Note that because of the absence of substantial signal at standard exposure times, the pc-Fos blot shown represents a long enhanced chemiluminescence exposure time (10 min). H, real time PCR analysis for c-Myc, ODC, and IL-8 in AGS cells stimulated with live H. pylori 60190 for 6 h. In E, F, and H, **, p < 0.01 versus control. In A–H, the data are from a minimum of two experiments. Ctrl, control.
Phosphorylation of ERK Is Essential for Binding of pc-Fos·c-Jun to the c-Myc Promoter
Because of the apparently unique nature of the AP-1 complex and the induction of c-Myc in H. pylori-stimulated macrophages, we sought to determine whether the pc-Fos and c-Jun heterodimer that we had identified binds to the AP-1 consensus sequence. We therefore performed oligonucleotide pulldown assays with the biotinylated AP-1 consensus sequence and H. pylori-activated macrophage nuclear extracts (Fig. 9A). When the pulldown product was analyzed by Western blotting, we found that it contained pTh325c-Fos and c-Jun, but not pS73c-Jun. Pretreatment of cells with PD98059 prevented the pulldown of pTh325c-Fos and c-Jun (Fig. 9A). These findings indicate that H. pylori induces dimerization of pc-Fos and c-Jun, this complex binds to the AP-1 consensus sequence, and this process depends on phosphorylation of ERK.
FIGURE 9.
The pc-Fos·c-Jun dimer binds to the c-Myc promoter in H. pylori-activated macrophages. RAW 267.4 cells were stimulated with HP with or without MAPK inhibitors as above. A, oligonucleotide pulldown assay. Biotinylated 21-bp oligonucleotides containing the AP-1 binding consensus sequence were incubated with 100 μg of nuclear extracts. Captured proteins were immunoblotted for pTh325c-Fos and c-Jun. B, chromatin immunoprecipitation assay. Top panel, illustration of the three putative AP-1 binding sites in the c-Myc promoter. Two middle panels, immunoprecipitation (IP) was performed with anti-pTh325c-Fos or c-Jun antibodies on chromatin from cells treated as indicated. PCR for c-Myc was conducted using primers 1–3, followed by agarose gel electrophoresis. The data shown are for primer 1; PCR with primers 2 and 3 did not generate a product (not shown). Bottom panel, input for primer 1; PCR products from sheared DNA without immunoprecipitation are shown, indicating that an equal amount of DNA was used. Genomic (Gen) DNA was used as a positive control for PCR. C, fluorescence polarization, 20 nm of Alexa 488-labeled DNA probe was incubated with nuclear proteins (0.5 μg). In C, **, p < 0.01 versus control, §§, p < 0.01 versus HP; n = 3 separate experiments. For A and B, the data are representative of three experiments. Inhib, inhibitor; PD, PD98059; SB, SB203580; SP, SP600125.
Chromatin immunoprecipitation assay was performed to determine whether this unique complex binds to the c-Myc promoter in H. pylori-stimulated macrophages. As shown in Fig. 9B, c-Myc has three AP-1 putative binding sites in its promoter. When anti-pTh325c-Fos or anti-c-Jun antibodies were used to immunoprecipitate DNA for PCR, only primer 1 (flanking the AP-1 binding region between −2067 to −2058) showed amplified products, and treatment of cells with PD98059 inhibited the formation of these products (Fig. 9B). In contrast, anti-pS73c-Jun antibody did not reveal an amplified product (data not shown).
To verify binding of the AP-1 complex to this −2067 to −2058 region of the c-Myc promoter, fluorescence polarization was performed using an Alexa 488-labeled DNA fragment of this region. Cell lysates from macrophages activated with H. pylori exhibited increased fluorescence polarization of the labeled oligonucleotide, indicative of binding of the AP-1 complex to the DNA probe; this effect was completely blocked by PD98059 (Fig. 9C).
Apoptosis of Infiltrating Gastric Macrophages in H. pylori Infection
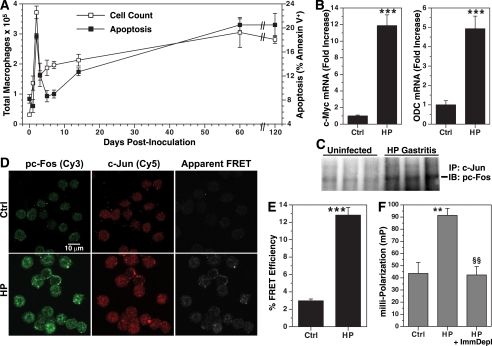
To assess the biological relevance of the AP-1 complex formation, we analyzed events in mice infected with H. pylori. Using flow cytometry we quantified the number of macrophages present in the stomach and the amount of apoptosis (Fig. 10A). In uninfected mice there were 0.3 ± 0.03 × 105 macrophages/stomach; there was an increase at 1 day postinoculation, and this peaked at day 2, with a 12-fold increase (Fig. 10A). The number of macrophages decreased by 62% at 3 days postinoculation. In parallel, there was a marked increase in apoptosis that peaked at 48 h postinoculation (p < 0.01 versus uninfected), suggesting that the depletion of macrophages was attributable to this apoptosis. The number of gastric macrophages gradually increased after day 3, but because of a concomitant increase in apoptosis at time points of chronic gastritis at 60 and 120 days postinoculation (p < 0.01 versus uninfected), the number of macrophages in the stomach remained below the peak level at 2 days postinoculation.
FIGURE 10.
H. pylori infection induces apoptosis and pc-Fos·c-Jun complex formation in gastric macrophages in vivo. A, macrophages from C57BL/6 mice sacrificed at the time points indicated after inoculation with H. pylori were positively selected with CD11b and stained with F4/80 as well as annexin V-FITC/7-AAD. The number of macrophages (F4/80+ cells, left y axis) and the percentage of macrophages that were apoptotic (annexin V+, total apoptosis, right y axis) are shown. B, real time PCR for c-Myc and ODC mRNA levels in F4/80+ cells (48 h postinoculation). C, pc-Fos·c-Jun AP-1 complex is present in gastric tissues from infected mice (120 days postinoculation). Tissue homogenates (100 μg of protein) were immunoprecipitated (IP) with anti-c-Jun antibody (2 μg) followed by immunoblotting (IB) for pTh325c-Fos. D–F, macrophages were positively selected with F4/80 from mouse stomach at 48 h postinoculation. FRET imaging (D) and efficiency analysis (E) were performed as in Fig. 7. For F, fluorescence polarization was measured in cell lysates with or without immunodepletion with anti-pTh325c-Fos antibody. In B and E, ***, p < 0.001 versus control; for F, **, p < 0.01 versus control, and §§, p < 0.01 versus HP without immunodepletion. For A–F, n = 3–6 mice in each group. Ctrl, control.
Induction of c-Myc and ODC Expression and Formation of an Active AP-1 Complex in vivo
Because H. pylori induced peak apoptosis at 2 days postinoculation, we measured c-Myc and ODC levels in gastric macrophages at this time point and found that mRNA expression of both genes was increased when compared with cells from uninfected mice (Fig. 10B). To assess for the presence of the AP-1 complex in vivo, whole gastric tissue lysates were immunoprecipitated with anti-c-Jun antibody, followed by immunoblotting with anti-pTh325c-Fos antibody (Fig. 10C). There was a marked increase in pc-Fos·c-Jun in tissues from H. pylori-infected mice compared with those from uninfected mice.
To assess for heterodimerization of pc-Fos and c-Jun within gastric macrophages, FRET analysis was performed. The cells isolated from infected mice showed an increase in pc-Fos and c-Jun staining and increased FRET events (Fig. 10D). There was a marked, 4.3 ± 0.3-fold increase in FRET efficiency (Fig. 10E). This interaction of pc-Fos and c-Jun was confirmed by fluorescence polarization analysis, with the specificity for this complex demonstrated by attenuation following immunodepletion with pc-Fos antibody (Fig. 10F).
Inhibition of ERK Decreases H. pylori-induced Phosphorylation of c-Fos and Apoptosis in Vivo
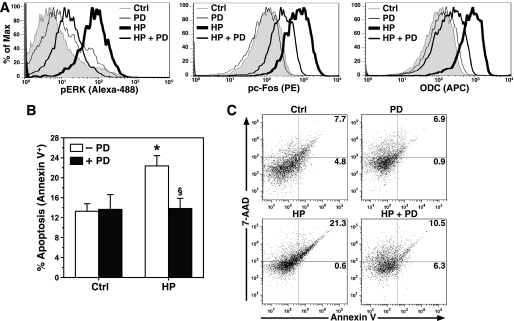
To investigate the importance of this AP-1 complex, the mice were treated with PD98059. We chose the peak time point of macrophage influx and apoptosis at 48 h postinoculation for these studies. Because of the limited numbers of isolated gastric macrophages, we used flow cytometry to evaluate proteins in cells positively selected with F4/80. H. pylori infection resulted in a significant increase in the levels of pERK, pc-Fos, and ODC in gastric macrophages, each of which was attenuated by injection of mice with PD98059 (Fig. 11A). There was a significant increase in macrophage apoptosis with H. pylori infection that was restored to the same level as uninfected mice by the PD98059 treatment (Fig. 11, B and C). Taken together, these data indicate that ERK phosphorylation plays a causal role in the apoptosis of infiltrating gastric macrophages in H. pylori infection through the effects on AP-1 complex formation that leads to ODC activation.
FIGURE 11.
Effect of ERK inhibition on levels of pc-Fos, ODC, and apoptosis in vivo. The mice were treated with PD98059 or vehicle by intraperitoneal injection prior to infection with H. pylori. A, flow cytometry histogram analysis for levels of pERK, pTh325c-Fos, and ODC proteins in macrophages positively selected with F4/80. The right shift in histograms represents increased protein expression. B, apoptosis was measured in gastric macrophages by flow cytometry as in Fig. 9. C, representative dot plots for percentages of cells positive for annexin V-FITC. In B, *, p < 0.05 for HP-infected versus control; §, p < 0.05 for HP with PD98059 (PD) versus HP alone. For A-B, n = 3–8 mice/group. Ctrl, control.
DISCUSSION
In the current report we demonstrate that macrophages exposed to H. pylori exhibit the formation of a unique AP-1 heterodimer consisting of pc-Fos·c-Jun, which was present with both in vitro and in vivo infection. The formation of this dimer was dependent on ERK phosphorylation and resulted in c-Myc and ODC expression and apoptosis. These findings are important because failure of the immune response to H. pylori contributes to its strong ability to colonize, as evidenced by prevalence rates approaching 100% in underdeveloped regions (45). Our work provides new insight into the molecular mechanism underlying the polyamine-mediated macrophage apoptosis that contributes to the immunopathogenesis of H. pylori infection.
We have observed that H. pylori-induced macrophage apoptosis requires ERK phosphorylation. Although ERK activation has been linked to anti-apoptotic and pro-survival signals, new evidence has emerged that this can also result in apoptosis (46, 47). Similarly, c-Myc promotes proliferation in the presence of growth factors but also causes apoptosis under conditions of stress (48). We show that H. pylori-induced phosphorylation of ERK mediates induction of c-Myc and ODC, with similar increases in mRNA and protein levels for both genes. Overexpression of constitutively active MEK1 in mammary epithelial cells has been reported to increase the levels of both c-Myc and ODC mRNA that was attenuated with ERK phosphorylation inhibitors (49). However, this study was microarray-based, without assessment of protein levels, and utilized ectopic expression rather than a biological stimulus, as we have used. It should be noted that pERK can enhance c-Myc protein levels without affecting mRNA expression by increasing protein stability through phosphorylation of c-Myc (50). However, we found that H. pylori stimulation of macrophages resulted in a consistent, parallel increase in c-Myc mRNA and protein levels, indicating that c-Myc transcriptional activation is sufficient to generate abundant c-Myc protein.
The duration of ERK activation has fundamentally different effects on the balance of apoptosis and proliferation. As an example, platelet-derived growth factor treatment in fibroblasts causes sustained phosphorylation of ERK, which promotes entry of cells into the S phase and proliferation, whereas epidermal growth factor causes transient activation of ERK that fails to induce entry into the S phase (51). We have observed a very rapid phosphorylation of ERK that occurs as early as 5 min after activation with H. pylori and returns to base-line levels by 80 min. Several groups have demonstrated that apoptosis that requires ERK activation is dependent on nuclear localization of pERK (52). In our study, pERK was found to translocate into the nucleus by 5 min after H. pylori stimulation, but this was transient, consistent with our findings of pERK-dependent induction of c-Fos phosphorylation and apoptosis.
ERK phosphorylation may lead to diverse biological outcomes, and one important determinant is the effect on downstream sensor molecules, such as c-Fos and Egr-1 (51). In the current study, we observed an increase in the expression of c-Fos and c-Jun and the phosphorylation of both proteins. The relative time kinetics for nuclear accumulation of pERK and phosphorylation of c-Fos have been shown to decide the biological outcome of stimuli (20). We have shown that accumulation of pERK in the nucleus precedes phosphorylation of c-Fos in response to H. pylori. These findings are consistent with a report that activated ERK accumulates in the nucleus and phosphorylates c-Fos at Ser374 and Thr325 in NIH3T3 fibroblasts activated with platelet-derived growth factor (53). This leads to the concept that phosphorylation of c-Fos can increase AP-1 transcriptional activity and, thus, biological outcomes (53). Expression of c-Jun has been shown to be dependent on ERK phosphorylation in neuronal cells (54). Phosphorylation at Ser73 stabilizes c-Jun protein and increases AP-1 transcriptional activity (18, 20). In our experiments, we found that ERK phosphorylation is required for c-Jun protein expression and phosphorylation at Ser73. Therefore, stabilization of c-Jun protein by its phosphorylation may be an important mechanism of transcriptional activation of c-Myc in H. pylori-stimulated cells.
The AP-1 complex is characterized by dimers of c-Fos and c-Jun family proteins (20). In our study, EMSA demonstrated that inhibition of ERK phosphorylation attenuated binding of nuclear proteins to AP-1 binding sequences. These data, coupled with our c-Fos and c-Jun expression and phosphorylation data, indicate that pc-Fos, rather than unphosphorylated c-Fos, is of primary importance in the host signaling response to H. pylori in macrophages. Moreover, the immunodepletion studies in the EMSA confirmed that pc-Fos is a component of the H. pylori-induced AP-1 complex and also showed that pc-Jun is not involved. In addition, our finding that immunodepletion of c-Jun from nuclear proteins caused a partial reduction in binding to the AP-1 DNA probe implicates c-Jun as a partner protein in the H. pylori-inducible AP-1 complex. It should be noted that higher concentrations of anti-c-Jun antibody had no additional effect on binding of nuclear proteins (data not shown), indicating that pc-Fos may bind to other c-Jun family proteins in the nucleus of H. pylori-stimulated macrophages.
Because the EMSA suggested that pc-Fos and c-Jun were both binding to the AP-1 DNA probe, it was essential to establish whether these proteins interact with each other in H. pylori-activated macrophages. The ability of pc-Fos to immunoprecipitate c-Jun and vice versa indicates that these proteins are principal components of the AP-1 complex. Our FRET data further demonstrated that these two proteins are in close physical proximity each other, because it has been reported that detectable FRET occurs when partner proteins are in the range of 10–100 Å (41). The attenuation of this FRET with the inhibition of ERK activity that we detected indicates that the close approximation of the pc-Fos·c-Jun dimer depends on phosphorylation of ERK. Several studies have assessed potential AP-1 constituents in the response to H. pylori (27–32), but in contrast to our study, they have not demonstrated the composition of the complex, the physical interaction of the components, the involvement of pc-Fos, or the role of AP-1 in apoptosis.
A major finding of our study was that AP-1 complex formation mediated apoptosis in cells activated with a prototype bacterial stimulus. In fact, AP-1 activation has been liked to apoptosis in a variety of cell types (20). For example ectopic overexpression of c-Fos or c-Jun causes apoptosis in fibroblasts (55) and colonic epithelial cells (42). Also knockdown of c-Fos and c-Jun by antisense oligonucleotides (56) or dominant-negative c-Jun (57) suppresses apoptosis induced by growth factor withdrawal in lymphoid cells and neurons, respectively. We have now directly demonstrated the role of AP-1 complex formation in H. pylori-stimulated macrophages, because dominant-negative c-Fos markedly attenuated apoptosis. Furthermore, this dominant-negative c-Fos inhibited c-Myc and ODC expression in parallel, consistent with our previous implication of these genes in H. pylori-stimulated macrophage apoptosis.
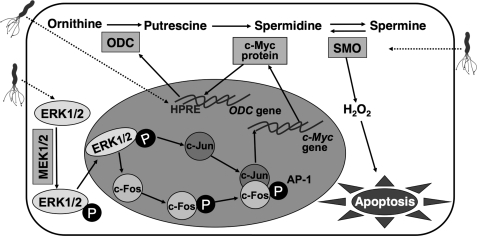
We have found that H. pylori activates ERK, which in turn phosphorylates c-Fos. To our knowledge, there are no reports of downstream targets of a pERK·pc-Fos pathway. However, there is an AP-1 response element in the promoter of the c-Myc gene (43). Consistent with this, we have shown that the c-Myc promoter can recruit the pc-Fos·c-Jun complex in macrophages exposed to H. pylori, leading to expression of c-Myc. These findings suggest the existence of a novel biochemical mechanism that links H. pylori-induced phosphorylation of ERK, c-Myc expression, and apoptosis through phosphorylation of c-Fos and activation of the AP-1 transcription factor (Fig. 12). This pathway also involves the coordinate action of an unknown H. pylori response element in the ODC promoter (Fig. 12), because we have found that deletion of the minimal ODC promoter upstream of the c-Myc binding site prevents promoter activation.4 Additionally, the induction of apoptosis also requires the oxidative metabolism of the polyamine spermine by the enzyme spermine oxidase that we have shown to generate sufficient H2O2 to cause mitochondrial membrane depolarization and thus apoptosis (8), as shown in Fig. 12.
FIGURE 12.
Pathway for induction of apoptosis in H. pylori-stimulated macrophages by an AP-1-dependent mechanism leading to polyamine-derived oxidative stress. As demonstrated in the current report, H. pylori causes activation of ERK1/2 phosphorylation by MEK1/2 that leads to nuclear translocation of pERK. This results in the induction of both c-Fos and c-Jun and the phosphorylation of c-Fos. pc-Fos and c-Jun form an AP-1 complex that binds to the c-Myc gene leading to expression of c-Myc mRNA and protein. This c-Myc protein in the cytoplasm translocates to the nucleus and enhances ODC transcription (9) in concert with activation of a purported H. pylori response element (HPRE). This induction of ODC leads to increased generation of the polyamines putrescine, spermidine, and spermine. In parallel, H. pylori up-regulates the expression and activity of the enzyme spermine oxidase (SMO), which back-converts spermine to spermidine, a process that generates H2O2, which causes apoptosis through mitochondrial membrane depolarization (8).
These conclusions led to the question of whether the pERK-pc-Fos·c-Jun-c-Myc-ODC pathway is canonical or potentially specific to H. pylori. To this end, we have shown in this report that other relevant bacterial stimuli that cause intestinal mucosal inflammation in mice (C. rodentium) and humans (C. jejuni) failed to cause phosphorylation of c-Fos and also failed to induce c-Myc or ODC expression in macrophages. Because both pathogens increased the expression of c-Jun, they likely induce a different AP-1 complex than the pc-Fos·c-Jun heterodimer that we have found to activate c-Myc transcription. In addition, our findings that both of these infectious agents induced accumulation of nuclear pERK, and yet there were not increases in the levels of pc-Fos, c-Myc, or ODC provide further evidence that the pathway in macrophages when stimulated with H. pylori is not canonical in nature. Moreover, the pERK activation was sustained with exposure to these other enteric pathogens compared with H. pylori, which was transient, again suggesting that the pERK-pc-Fos·c-Jun pathway is not the same under all stimulatory conditions. Additionally, our data in AGS gastric epithelial cells further suggest that the pathway in macrophages is noncanonical, because H. pylori did not induce phosphorylation of c-Fos and expression of c-Myc or ODC in these cells.
It should be noted that in monocytic cells, ERK and AP-1 have been studied in the response to H. pylori (31–33). However, in those studies, a role for pc-Fos has not been shown, and other pathways including p38 MAPK and NF-κB have been implicated in the cytokine responses (32, 33). In contrast, we found that inhibition of p38 had no effect and that interference with c-Fos phosphorylation with a dominant-negative plasmid completely attenuated the induction of apoptosis. Similarly, in studies in gastric epithelial cells (27–30), the role of pc-Fos in AP-1 complex formation has not been assessed, and p38 as well as JNK have been implicated in the cytokine response, in contrast to our results in macrophages. Further, the role of pERK or AP-1 has not been previously studied in the setting of H. pylori-induced expression of c-Myc, ODC, or apoptosis in either macrophages or epithelial cells.
Another goal of our study was to establish the involvement of the pERK·pc-Fos pathway in gastric macrophages during H. pylori infection in vivo. Rapid infiltration of inflammatory cells, such as macrophages and neutrophils (39), as well as dendritic cells (58) has been demonstrated in murine H. pylori infection, which is followed by a decrease in the number of these cells. We now show that the depletion of infiltrating macrophages is likely attributable to apoptosis. Furthermore, we have demonstrated that the pc-Fos·c-Jun dimer is present in the gastric macrophages from H. pylori-infected mice, and this correlates with c-Myc and ODC expression. This is the first report of the formation of this AP-1 complex in tissue-derived macrophages. Our findings also suggest that that this complex is functional in vivo, because inhibition of ERK phosphorylation prevented H. pylori-induced apoptosis in the gastric macrophages. It has been reported that inhibition of ERK with PD98059, as used, resulted in a reduction in brain injury in a mouse model of cerebral infarction (59), but apoptosis was not specifically assessed.
Our data have elucidated a novel aspect of immune dysregulation in H. pylori infection, namely the formation of a specific AP-1 complex that we have implicated in the causation of macrophage apoptosis. There is substantial evidence that enhancing the innate and adaptive immune response via vaccine strategies can limit or prevent colonization in mouse models (60, 61). Strategies that could lead to protection of infiltrating gastric macrophages from apoptosis could lead to 1) enhanced antimicrobial effector function of these cells and 2) potentiation of the adaptive immune response that depends on antigen-presenting and paracrine signaling functions from macrophages. Therefore, targeted treatments that would selectively block this unique pathway of c-Myc/ODC-mediated apoptosis could prove valuable in the management of H. pylori infection. Such strategies are potentially very important because of the large disease burden of H. pylori infection worldwide and issues of antibiotic resistance. α-Difluoromethylornithine is a known inhibitor of ODC but would inhibit ODC systemically, which could limit its broad utility for this purpose. Small molecule library screening has yielded candidate compounds that can inhibit AP-1 in cell-free assays (62). Assessment of such molecules in H. pylori-activated macrophages could prove to be a fruitful area of future investigation.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK053620 and R01AT004821 (to K. T. W.), P01CA116087 (to K. T. W.), P01CA028842 (to K. T. W.), F31GM083500 (to N. D. L.), T32CA009592 (to N. D. L.), and T32DK007673 (D. P. B.). This work was also supported by the Flow Cytometry Core and the Cell Imaging Core of the Vanderbilt University Digestive Disease Research Center, which was supported by National Institutes of Health Grant P30DK058404, a Merit Review Grant from the Office of Medical Research, Department of Veterans Affairs (to K. T. W.), and the Philippe Foundation (to T. d. S. and A. P. G.).
M. Asim, R. Chaturvedi, N. D. Lewis, K. Singh, D. P. Barry, T. de Sablet, A. P. Gobert, and K. T. Wilson, unpublished observations.
- MAPK
- mitogen-activated protein kinase
- 7-AAD
- 7-amino-actinomycin D
- AP-1
- activator protein-1
- EMSA
- electromobility shift assay
- ERK
- extracellular signal-regulated kinase
- FRET
- fluorescence resonance energy transfer
- FITC
- fluorescein isothiocyanate
- HP
- H. pylori lysate
- JNK
- c-Jun N-terminal kinase
- MOI
- multiplicity of infection
- ODC
- ornithine decarboxylase
- PE
- phycoerythrin
- PI
- propidium iodide
- RT
- reverse transcription
- pc-Fos
- phospho-c-Fos
- pc-Jun
- phospho-c-Jun
- MEK
- MAPK/ERK kinase
- pERK
- phosphorylated ERK
- IL
- interleukin
- XTT
- 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxyaniline disodium salt.
REFERENCES
- 1.Weinrauch Y., Zychlinsky A. (1999) Annu. Rev. Microbiol. 53, 155–187 [DOI] [PubMed] [Google Scholar]
- 2.Chaturvedi R., Asim M., Lewis N. D., Algood H. M., Cover T. L., Kim P. Y., Wilson K. T. (2007) Infect. Immun. 75, 4305–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peek R. M., Jr., Blaser M. J. (2002) Nat. Rev. Cancer 2, 28–37 [DOI] [PubMed] [Google Scholar]
- 4.Necchi V., Candusso M. E., Tava F., Luinetti O., Ventura U., Fiocca R., Ricci V., Solcia E. (2007) Gastroenterology 132, 1009–1023 [DOI] [PubMed] [Google Scholar]
- 5.Aspholm M., Olfat F. O., Nordén J., Sondén B., Lundberg C., Sjöström R., Altraja S., Odenbreit S., Haas R., Wadström T., Engstrand L., Semino-Mora C., Liu H., Dubois A., Teneberg S., Arnqvist A., Borén T. (2006) PLoS Pathog. 2, e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh J. D., Karam S. M., Gordon J. I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5186–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gobert A. P., Cheng Y., Wang J. Y., Boucher J. L., Iyer R. K., Cederbaum S. D., Casero R. A., Jr., Newton J. C., Wilson K. T. (2002) J. Immunol. 168, 4692–4700 [DOI] [PubMed] [Google Scholar]
- 8.Chaturvedi R., Cheng Y., Asim M., Bussière F. I., Xu H., Gobert A. P., Hacker A., Casero R. A., Jr., Wilson K. T. (2004) J. Biol. Chem. 279, 40161–40173 [DOI] [PubMed] [Google Scholar]
- 9.Cheng Y., Chaturvedi R., Asim M., Bussière F. I., Scholz A., Xu H., Casero R. A., Jr., Wilson K. T. (2005) J. Biol. Chem. 280, 22492–22496 [DOI] [PubMed] [Google Scholar]
- 10.Keates S., Keates A. C., Warny M., Peek R. M., Jr., Murray P. G., Kelly C. P. (1999) J. Immunol. 163, 5552–5559 [PubMed] [Google Scholar]
- 11.Kyriakis J. M. (2001) Nature 414, 265–266 [DOI] [PubMed] [Google Scholar]
- 12.Weston C. R., Davis R. J. (2001) Science 292, 2439–2440 [DOI] [PubMed] [Google Scholar]
- 13.Ferrell J. E., Jr., Bhatt R. R. (1997) J. Biol. Chem. 272, 19008–19016 [DOI] [PubMed] [Google Scholar]
- 14.Roberts P. J., Der C. J. (2007) Oncogene. 26, 3291–3310 [DOI] [PubMed] [Google Scholar]
- 15.Matsubayashi Y., Fukuda M., Nishida E. (2001) J. Biol. Chem. 276, 41755–41760 [DOI] [PubMed] [Google Scholar]
- 16.Chen R. H., Sarnecki C., Blenis J. (1992) Mol. Cell. Biol. 12, 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J., Yuan X., Frödin M., Grummt I. (2003) Mol. Cell 11, 405–413 [DOI] [PubMed] [Google Scholar]
- 18.Angel P., Karin M. (1991) Biochim. Biophys. Acta. 1072, 129–157 [DOI] [PubMed] [Google Scholar]
- 19.Karin M., Liu Z., Zandi E. (1997) Curr. Opin. Cell Biol. 9, 240–246 [DOI] [PubMed] [Google Scholar]
- 20.Shaulian E., Karin M. (2002) Nat. Cell Biol. 4, E131–E136 [DOI] [PubMed] [Google Scholar]
- 21.Deng T., Karin M. (1994) Nature 371, 171–175 [DOI] [PubMed] [Google Scholar]
- 22.Smeal T., Binetruy B., Mercola D., Grover-Bardwick A., Heidecker G., Rapp U. R., Karin M. (1992) Mol. Cell. Biol. 12, 3507–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen R. H., Juo P. C., Curran T., Blenis J. (1996) Oncogene 12, 1493–1502 [PubMed] [Google Scholar]
- 24.Holt J. T., Gopal T. V., Moulton A. D., Nienhuis A. W. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 4794–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szabo E., Preis L. H., Brown P. H., Birrer M. J. (1991) Cell Growth Differ. 2, 475–482 [PubMed] [Google Scholar]
- 26.Yamamura Y., Hua X., Bergelson S., Lodish H. F. (2000) J. Biol. Chem. 275, 36295–36302 [DOI] [PubMed] [Google Scholar]
- 27.Lu H., Wu J. Y., Kudo T., Ohno T., Graham D. Y., Yamaoka Y. (2005) Mol. Biol. Cell 16, 4954–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kudo T., Lu H., Wu J. Y., Ohno T., Wu M. J., Genta R. M., Graham D. Y., Yamaoka Y. (2007) Gastroenterology 132, 1024–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding S. Z., Olekhnovich I. N., Cover T. L., Peek R. M., Jr., Smith M. F., Jr., Goldberg J. B. (2008) FEMS Immunol. Med. Microbiol. 53, 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naumann M., Wessler S., Bartsch C., Wieland B., Covacci A., Haas R., Meyer T. F. (1999) J. Biol. Chem. 274, 31655–31662 [DOI] [PubMed] [Google Scholar]
- 31.Yamauchi K., Choi I. J., Lu H., Ogiwara H., Graham D. Y., Yamaoka Y. (2008) J. Immunol. 180, 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharyya A., Pathak S., Datta S., Chattopadhyay S., Basu J., Kundu M. (2002) Biochem. J. 368, 121–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hisatsune J., Nakayama M., Isomoto H., Kurazono H., Mukaida N., Mukhopadhyay A. K., Azuma T., Yamaoka Y., Sap J., Yamasaki E., Yahiro K., Moss J., Hirayama T. (2008) J. Immunol. 180, 5017–5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson K. T., Ramanujam K. S., Mobley H. L., Musselman R. F., James S. P., Meltzer S. J. (1996) Gastroenterology 111, 1524–1533 [DOI] [PubMed] [Google Scholar]
- 35.Lewis N. D., Asim M., Barry D. P., Singh K., de Sablet T., Boucher J. L., Gobert A. P., Chaturvedi R., Wilson K. T. (2010) J. Immunol. 184, 2572–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bussiere F. I., Chaturvedi R., Asim M., Hoek K. L., Cheng Y., Gainor J., Scholz A., Khan W. N., Wilson K. T. (2006) Cancer Res. 66, 6834–6842 [DOI] [PubMed] [Google Scholar]
- 37.Singh K., Chaturvedi R., Asim M., Barry D. P., Lewis N. D., Vitek M. P., Wilson K. T. (2008) J. Biol. Chem. 283, 16752–16761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olive M., Krylov D., Echlin D. R., Gardner K., Taparowsky E., Vinson C. (1997) J. Biol. Chem. 272, 18586–18594 [DOI] [PubMed] [Google Scholar]
- 39.Scott Algood H. M., Gallo-Romero J., Wilson K. T., Peek R. M., Jr., Cover T. L. (2007) FEMS Immunol. Med. Microbiol. 51, 577–586 [DOI] [PubMed] [Google Scholar]
- 40.Rusinova E., Tretyachenko-Ladokhina V., Vele O. E., Senear D. F., Alexander Ross J. B. (2002) Anal. Biochem. 308, 18–25 [DOI] [PubMed] [Google Scholar]
- 41.Sekar R. B., Periasamy A. (2003) J. Cell Biol. 160, 629–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preston G. A., Lyon T. T., Yin Y., Lang J. E., Solomon G., Annab L., Srinivasan D. G., Alcorta D. A., Barrett J. C. (1996) Mol. Cell. Biol. 16, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iavarone C., Catania A., Marinissen M. J., Visconti R., Acunzo M., Tarantino C., Carlomagno M. S., Bruni C. B., Gutkind J. S., Chiariello M. (2003) J. Biol. Chem. 278, 50024–50030 [DOI] [PubMed] [Google Scholar]
- 44.Xu H., Chaturvedi R., Cheng Y., Bussiere F. I., Asim M., Yao M. D., Potosky D., Meltzer S. J., Rhee J. G., Kim S. S., Moss S. F., Hacker A., Wang Y., Casero R. A., Jr., Wilson K. T. (2004) Cancer Res. 64, 8521–8525 [DOI] [PubMed] [Google Scholar]
- 45.Mera R., Fontham E. T., Bravo L. E., Bravo J. C., Piazuelo M. B., Camargo M. C., Correa P. (2005) Gut 54, 1536–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cagnol S., Chambard J. C. (2010) FEBS J. 277, 2–21 [DOI] [PubMed] [Google Scholar]
- 47.Bacus S. S., Gudkov A. V., Lowe M., Lyass L., Yung Y., Komarov A. P., Keyomarsi K., Yarden Y., Seger R. (2001) Oncogene 20, 147–155 [DOI] [PubMed] [Google Scholar]
- 48.Evan G. I., Wyllie A. H., Gilbert C. S., Littlewood T. D., Land H., Brooks M., Waters C. M., Penn L. Z., Hancock D. C. (1992) Cell 69, 119–128 [DOI] [PubMed] [Google Scholar]
- 49.Grill C., Gheyas F., Dayananth P., Jin W., Ding W., Qiu P., Wang L., Doll R. J., English J. M. (2004) Biochem. J. 381, 635–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niklinski J., Claassen G., Meyers C., Gregory M. A., Allegra C. J., Kaye F. J., Hann S. R., Zajac-Kaye M. (2000) Mol. Cell. Biol. 20, 5276–5284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy L. O., Smith S., Chen R. H., Fingar D. C., Blenis J. (2002) Nat. Cell Biol. 4, 556–564 [DOI] [PubMed] [Google Scholar]
- 52.Teixeiro E., Daniels M. A. (2010) FEBS J. 277, 30–38 [DOI] [PubMed] [Google Scholar]
- 53.Monje P., Marinissen M. J., Gutkind J. S. (2003) Mol. Cell. Biol. 23, 7030–7043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leppä S., Saffrich R., Ansorge W., Bohmann D. (1998) EMBO J. 17, 4404–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bossy-Wetzel E., Bakiri L., Yaniv M. (1997) EMBO J. 16, 1695–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Colotta F., Polentarutti N., Sironi M., Mantovani A. (1992) J. Biol. Chem. 267, 18278–18283 [PubMed] [Google Scholar]
- 57.Ham J., Babij C., Whitfield J., Pfarr C. M., Lallemand D., Yaniv M., Rubin L. L. (1995) Neuron 14, 927–939 [DOI] [PubMed] [Google Scholar]
- 58.Kao J. Y., Rathinavelu S., Eaton K. A., Bai L., Zavros Y., Takami M., Pierzchala A., Merchant J. L. (2006) Am. J. Physiol. Gastrointest. Liver Physiol. 291, G73–G81 [DOI] [PubMed] [Google Scholar]
- 59.Alessandrini A., Namura S., Moskowitz M. A., Bonventre J. V. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12866–12869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeLyria E. S., Redline R. W., Blanchard T. G. (2009) Gastroenterology 136, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson K. T., Crabtree J. E. (2007) Gastroenterology 133, 288–308 [DOI] [PubMed] [Google Scholar]
- 62.Rishi V., Potter T., Laudeman J., Reinhart R., Silvers T., Selby M., Stevenson T., Krosky P., Stephen A. G., Acharya A., Moll J., Oh W. J., Scudiero D., Shoemaker R. H., Vinson C. (2005) Anal. Biochem. 340, 259–271 [DOI] [PubMed] [Google Scholar]