Abstract
During the development of the sympathetic nervous system, the p75 neurotrophin receptor (p75NTR) has a dual function: promoting survival together with TrkA in response to NGF, but inducing cell death upon binding pro or mature brain-derived neurotrophic factor (BDNF). Apoptotic signaling through p75NTR requires activation of the stress kinase, JNK. However, the receptor also undergoes regulated proteolysis, first by a metalloprotease, and then by γ-secretase, in response to pro-apoptotic ligands and this is necessary for receptor mediated neuronal death (Kenchappa, R. S., Zampieri, N., Chao, M. V., Barker, P. A., Teng, H. K., Hempstead, B. L., and Carter, B. D. (2006) Neuron 50, 219–232). Hence, the relationship between JNK activation and receptor proteolysis remains to be defined. Here, we report that JNK3 activation is necessary for p75NTR cleavage; however, following release of the intracellular domain, there is a secondary activation of JNK3 that is cleavage dependent. Receptor proteolysis and apoptosis were prevented in sympathetic neurons from jnk3−/− mice, while activation of JNK by ectopic expression of MEKK1 induced p75NTR cleavage and cell death. Proteolysis of the receptor was not detected until 6 h after BDNF treatment, suggesting that JNK3 promotes cleavage through a transcriptional mechanism. In support of this hypothesis, BDNF up-regulated tumor necrosis factor-α-converting enzyme (TACE)/ADAM17 mRNA and protein in wild-type, but not jnk3−/− sympathetic neurons. Down-regulation of TACE by RNA interference blocked BDNF-induced p75NTR cleavage and apoptosis, indicating that this metalloprotease is responsible for the initial processing of the receptor. Together, these results demonstrate that p75NTR-mediated activation of JNK3 is required for up-regulation of TACE, which promotes receptor proteolysis, leading to prolonged activation of JNK3 and subsequent apoptosis in sympathetic neurons.
Keywords: Apoptosis, Brain, JNK, Neurodevelopment, Neurotrophin, γ-Secretase, Metalloprotease, Neurotrophin Receptors, Neurotrophins, Pro-neurotrophins
Introduction
During early stages of sympathetic nervous system development, an excess of neurons is produced; however, the neuronal population is eventually decreased by apoptosis. The final number of neurons is finely tuned through a combination of neuronal competition for limited amounts of trophic factors produced by the target organs and the activation of pro-apoptotic receptors (1, 2). The elimination of sympathetic neurons during development by apoptosis depends, in part, on the p75 neurotrophin receptor (p75NTR).2 The trophic factor produced in sympathetic neuron target organs is nerve growth factor (NGF) (3–5), which binds to a high affinity complex of p75NTR and the tyrosine kinase receptor TrkA (6). The interaction of p75NTR with TrkA not only enhances the binding affinity for NGF, it also increases the efficacy of TrkA-mediated signaling to promote survival (7–9).
Paradoxically, in addition to its pro-survival effects in conjunction with TrkA, p75NTR can also initiate independent signals that lead to axonal degeneration (10) and apoptotic cell death in sympathetic neurons (11). p75NTR-mediated apoptosis contributes to the normal developmental elimination of these neurons in vivo, as indicated by the fact that p75ntr−/− mice exhibit excess neurons in the superior cervical ganglia (SCG) during the apoptotic period (11, 12). It is unclear which p75NTR ligand is responsible for sympathetic neuron apoptosis in vivo; however, BDNF has been suggested as a candidate, because this neurotrophin can kill sympathetic neurons in vitro by binding to p75NTR, and BDNF null mice have increased numbers of sympathetic neurons (11). Recently, Ginty and colleagues demonstrated that BDNF is synthesized by SCG neurons upon NGF exposure, resulting in BDNF binding to p75NTR on neighboring neurons to eliminate them (2).
How p75NTR mediates such divergent responses is poorly understood. However, activation of the stress kinase, c-Jun N-terminal kinase (JNK) is an essential component of the receptor's apoptotic signal (13–15). Recently, p75NTR was shown to undergo regulated intramembrane proteolysis by the γ-secretase complex (16, 17), and this cleavage could be initiated in sympathetic neurons in response to BDNF or pro-BDNF (18). The release of the intracellular domain of p75NTR allowed nuclear translocation of the DNA-binding protein Neurotrophin receptor interacting factor (NRIF), which was necessary for receptor-mediated apoptosis (18, 19). The mechanism by which p75NTR proteolysis is regulated and how it relates to JNK activation in sympathetic neurons remains unknown. Here, we report that BDNF binding to p75NTR in sympathetic neurons activates JNK3, which is necessary and sufficient to up-regulate the metalloprotease TACE/ADAM17, resulting in receptor cleavage by this enzyme and, subsequently, by the γ-secretase complex. Release of the intracellular domain (ICD) of p75NTR then triggers further activation of JNK3, ultimately leading to cell death.
EXPERIMENTAL PROCEDURES
Cell Culture
All experiments using animals were approved by the Animal Care and Use Committee at Vanderbilt University. SCG were isolated from postnatal day 4 (P4) Sprague-Dawley rats, wild-type, traf6−/−, and jnk3−/− mice, and sympathetic neurons were cultured as previously described (20). Briefly, SCGs were first dissociated using 0.25% trypsin (Worthington, Freehold, NJ) and 0.3% collagenase (Sigma), then the neurons were cultured on poly-l-ornithine and laminin (Nalge Nunc, Naperville, IL) in Ultraculture medium (BioWhittaker, Walkersville, MD) containing 3% fetal bovine serum, 2 mm l-glutamine,100 units/ml penicillin, 100 μg/ml streptomycin, 1 μg/ml gentamicin, and 20 ng/ml NGF (Harlan, Indianapolis, IN). Neurons were maintained in the presence of NGF before using for p75NTR activation. HEK293 cells were cultured and maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum, 2 mm l-glutamine 100 units/ml penicillin, 100 μg/ml streptomycin, and were transfected using the standard calcium phosphate method.
p75NTR Activation and Western Blotting
Sympathetic neurons from P4 rats, wild-type, jnk3−/−, and traf6−/− P4 mice were cultured and maintained in NGF for 48 h, then NGF was removed, cells were rinsed, and switched to medium containing anti-NGF (0.1 μg/ml, Chemicon, Temecula, CA) together with 12.5 mm KCl, to promote survival, with or without the addition of 200 ng/ml BDNF (generously provided by Regeneron Inc.) for p75NTR activation for various times, as indicated. In some experiments, the neurons were pre-treated for different times with 10 μm of the JNK inhibitor SP600125 (catalogue no. 420119, Calbiochem), 100 nm of the γ-secretase inhibitor Compound E (kindly provided by Dr. Todd Golde, Mayo Clinic, Jacksonville, FL and Dr. Graham Carpenter, Vanderbilt University, Nashville, TN), 10 μm of the metalloprotease inhibitor TAPI-1 (catalogue no. 579051, Calbiochem) or 100 ng/ml the protein synthesis inhibitor cycloheximide (Sigma), in the presence and absence of 200 ng/ml BDNF. The cells were harvested and lysed in Nonidet P-40 lysis buffer (10% glycerol, 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 2 μg/ml leupeptin and aprotinin). Lysates were subjected to Western blot analysis using antibodies to p-JNK (1:500, catalogue no. 9251, Cell Signaling Technology), total-JNK (1:500, catalogue no. SC-474, Santa Cruz Biotechnology, Santa Cruz, CA), TACE/ADAM-17 (1:500, catalogue no. PC491, Calbiochem), Presenilin1 (1:1000, catalogue no. SC-7860, Santa Cruz Biotechnology), HA (1:500, clone 12CA5, Roche Applied Science), TRAF6 (1:1000, catalogue no. SC-8409, Santa Cruz Biotechnology), and tubulin (1:1000, catalogue no. CP06, Calbiochem).
For p75NTR ICD Western blotting, neurons were also treated with 10 μm proteasome inhibitor ZLLLH (Peptide Institute Inc., Osaka, Japan), which was required to detect the ICD. The proteasome inhibitor was added with or without 200 ng/ml BDNF, and the cells were harvested and lysed in Laemmli buffer and subjected to p75NTR ICD Western blotting (1:2000, p75NTR antiserum was generated using a fusion protein of glutathione S-transferase and the full-length intracellular domain) as performed before (18). To detect the p75NTR cleavage in vivo, whole SCG from wild-type or jnk3−/− mice were isolated at P4, lysed in Laemmli buffer, and subjected to Western blotting using p75ICD and tubulin antibodies.
In some of the experiments, 293 cells stably expressing p75NTR (293-p75NTR cells) were transfected with JNK1, JNK2, or JNK3, and 36 h later treated with 10 μm ZLLLH for 12 h. The cells were then harvested, lysed in Nonidet P-40 lysis buffer, and subjected to p75NTR ICD, TACE/ADAM-17, and tubulin Western blotting.
Adenoviral Infection of Sympathetic Neurons
Sympathetic neurons from P4 rat SCGs were cultured as described above in NGF (20 ng/ml) for 48 h, then incubated with adenovirus expressing GFP, MEKK1, p75NTR ICD, or DN-JNK at dilutions sufficient to give similar levels of infection, based on immunofluorescent detection of the expressed protein. 24 h after infection, the neurons were harvested for phospho-JNK, total-JNK, p75NTR ICD, TACE/ADAM17, and tubulin Western blotting or fixed in 4% paraformaldehyde for counting apoptotic nuclei. In some of the experiments the neurons were treated with 200 ng/ml BDNF as described above, then lysed in Nonidet P-40 lysis buffer and subjected to phospho-JNK and total-JNK Western blotting. For p75NTR ICD Western blotting, the cells were lysed in Laemmli buffer at the indicated time after BDNF and ZLLLH treatment. For treatment with the γ-secretase inhibitor Compound E (100 nm) or JNK inhibitor SP600125 (10 μm), the neurons were pretreated 1 h before infection with adenovirus expressing MEKK1 and also treated during incubation with adenovirus.
Transfection of Sympathetic Neurons
Sympathetic neurons from P4 rats were isolated and cultured in 20 ng/ml NGF as described above, then 24 h later transfected with RNAi to TACE (TACE: duplex RNA with following sequence: duplex-01, sense sequence (CGUCAGAGCCGAGUUGAUAUU) and antisense sequence (5′-PUAUCAACUCGGCUCUGACGUU); duplex-02, sense sequence (UAUGGGAACUCUUGGAUUAUU) and antisense sequence (5′-PUAAUCCAAGAGUUCCCAUAUU); neurons were co-transfected with duplex-01 and -02) or a control, scrambled RNAi by using a transfection reagent (catalogue no. T-2003, Dharmacon RNA Technologies, Chicago, IL). 24 h after transfection, the neurons were treated with 200 ng/ml BDNF, as indicated. The cells were then lysed and subjected to TACE, p75NTR ICD, and tubulin Western blotting.
For some experiments, the sympathetic neurons were transfected immediately after dissociation with wild-type or catalytically inactive mutant TACE (generously provided by Dr. Peter Dempsey, University of Michigan), wild-type p75NTR, γ-secretase-resistant p75NTR (p75FasTM), or an empty vector by electroporation using a rat neuron Nucleofector kit (cat no. VGP-1003, Amaxa Biosystems, Walkersville, MD) and program O-003 on an Amaxa Nucleofector device. 36 h after transfection, the neurons were left untreated or treated with 10 μm of proteasome inhibitor ZLLLH with or without 200 ng/ml BDNF, and then lysed and subjected to Western blotting. In some experiments neurons were electroporated as described above with GFP together with RNAi to TACE or control RNAi (RNAi raised to GFP had no effect on GFP we used) or GFP alone. 36 h later cells were left untreated or treated with 200 ng/ml BDNF for 48 h, and then fixed in 4% paraformaldehyde, and GFP-positive neurons were scored for apoptosis. Approximately 150 neurons were counted for each condition.
JNK Assay
Cultured sympathetic neurons from wild-type P4 rats were treated with or without 200 ng/ml BDNF, as described above, and after the indicated time harvested and lysed in Nonidet P-40 lysis buffer. The immunoprecipitation kinase assays for JNK1, -2, and -3 were performed as described previously (14, 21).
Neuronal Apoptosis Assay
Sympathetic neurons from P4 wild-type and jnk3−/− mice were cultured in 20 ng/ml NGF for 2 days, then NGF was removed, and the cells were rinsed and switched to medium containing anti-NGF (0.1 μg/ml) together with 12.5 mm KCl, to promote survival, with or without the addition of 200 ng/ml BDNF. After 48 h the neurons were fixed in 4% paraformaldehyde and mounted using Vectashield with 4′,6-diamidino-2-phenylindole to score the nuclei as apoptotic or non-apoptotic. Approximately 150 neurons were counted for each condition in every experiment.
Total RNA Isolation, First Strand cDNA Synthesis, Primer Validation, and Quantitative PCR
Sympathetic neurons from P4 rats were cultured and treated with 200 ng/ml BDNF, as described above, and 4 h later total RNA was isolated using TRIzol (Invitrogen). Total RNA (200 ng) from each sample was reverse transcribed to cDNA using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). Real-time PCR was performed with an ABI Prism 7300 System (Applied Biosystems) using 2 ng of cDNA per 50-μl reaction volume, 2× SYBR green master mix, and gene-specific primers. The primers were as follows: rat Adam17 (NM_020306, 89-bp PCR) forward (5′-actggatcaccagaggatgg-3′) and reverse (5′-ggaaggggtccttctcaaag-3′); rat Pgk1 (phosphoglycerate kinase 1 NM_053291, 98 bp PCR) forward (5′-ggctcgagctaagcagattg-3′) and reverse (5′-gctttcaccacctcatccat-3′); rat Actb (beta actin NM_031144, 104-bp PCR) forward (5′-gtagccatccaggctgtgtt-3′) and reverse (5′-ccctcatagatgggcacagt-3′). All samples were run in quadruplicate. Data from the PCR reactions were analyzed using the comparative cycle number determined as threshold (Ct) method (User Bulletin No. 2, PerkinElmer Life Sciences) (22). Differential expression was calculated as ΔΔCt against expression of Actb and Pgk1 as normalizers.
NGF Withdrawal
Sympathetic neurons from P4 rats were cultured and maintained in NGF for 48 h, then rinsed with medium containing anti-NGF (0.1 μg/ml, Chemicon International), and maintained in medium lacking NGF and containing ZLLLH for various times as indicated. For some experiments, the neurons were kept in medium lacking NGF in the presence of the pan-Caspase inhibitor boc-aspartyl-(OMe)-fluoromethyl ketone (40 μm, MP Biomedicals) then lysed and subjected to Western blot analysis, as indicated.
RESULTS
Activation of JNK Is Necessary and Sufficient to Induce p75NTR Proteolysis in Sympathetic Neurons
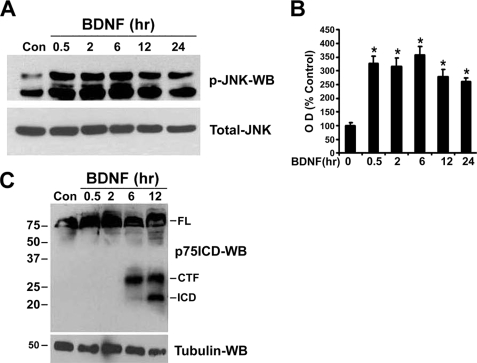
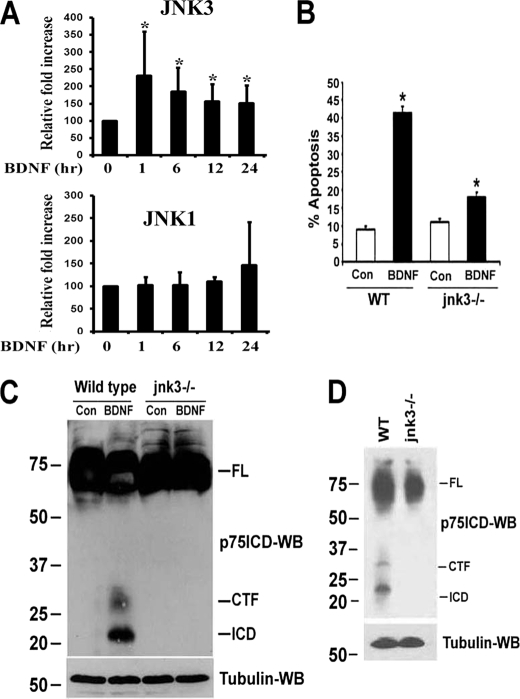
Recent studies from our laboratory revealed that p75NTR-mediated apoptosis in sympathetic neurons requires regulated proteolysis of the receptor (18). Because previous studies demonstrated that the activation of the stress kinase JNK was necessary for p75NTR to induce cell death (21–24), we investigated whether these signals are in a common pathway. To selectively activate p75NTR without any influence from TrkA, sympathetic neurons cultured with NGF were switched to medium lacking NGF and containing 12.5 mm KCl to promote survival. BDNF treatment activated JNK within 30 min and the kinase remained active for at least 24 h (Fig. 1, A and B). In contrast, BDNF-induced p75NTR cleavage by TACE, and γ-secretase was not observed until ∼6 and 12 h, respectively (Fig. 1C) (18). Therefore, we hypothesized that JNK activation was upstream of receptor processing.
FIGURE 1.
Kinetics of JNK activation and p75NTR cleavage by BDNF in sympathetic neurons. Sympathetic neurons from P4 rats cultured for 2 days with 20 ng/ml NGF were rinsed to remove the NGF and refed with medium containing 12.5 mm KCl, to promote survival, and 200 ng/ml BDNF for various times (A). To detect p75NTR cleavage, the neurons were treated with 10 μm proteasome inhibitor ZLLLH with or without the addition of 200 ng/ml BDNF for various times (C). After the indicated times, the neurons were lysed and subjected to Western blot analysis using antibodies to phospho-JNK (p-JNK), total-JNK (A), p75NTR ICD (C), or tubulin. B, p-JNK Western blots were quantified and expressed as percent of control. Data are means ± S.D., n = 3 independent experiments (*, p < 0.009).
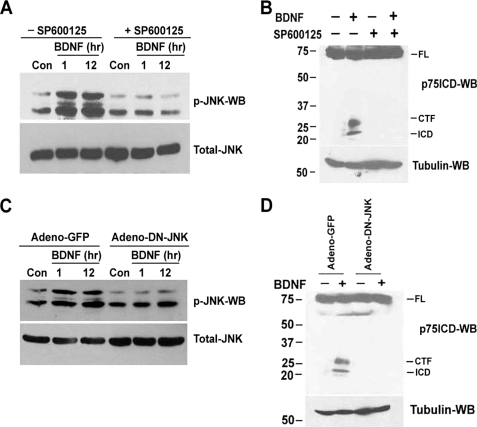
To determine whether inhibition of JNK could block ligand-induced p75NTR cleavage, sympathetic neurons were pretreated with the selective JNK inhibitor SP600125 for 1 h before treating with BDNF. The proteasome inhibitor ZLLLH was also added to detect the intracellular domain fragment, because it is rapidly degraded following cleavage (16–18). SP600125 completely blocked BDNF-induced JNK phosphorylation (Fig. 2A) as well as the release of carboxyl-terminal fragment (CTF) and ICD of p75NTR receptor (Fig. 2B). To confirm a requirement for JNK activation prior to ligand-dependent p75NTR cleavage, a DN-JNK (DN-JNK), which was previously shown to inhibit p75NTR-mediated apoptosis (21, 23), was ectopically expressed in the neurons using adenovirus. In Adeno-GFP-infected neurons, BDNF activated JNK and induced p75NTR cleavage; however, Adeno-DN-JNK completely blocked both activation of the kinase and release of the CTF and ICD of p75NTR (Fig. 2, C and D). These results demonstrate that p75NTR-mediated activation of JNK is required for receptor proteolysis.
FIGURE 2.
Inhibition of JNK activation blocks BDNF-induced p75NTR cleavage in sympathetic neurons. A and B, sympathetic neurons from P4 rats cultured and maintained for 2 days in 20 ng/ml NGF, were rinsed to remove the NGF and refed with medium containing 12.5 mm KCl, to promote survival. The neurons were treated for 1 h with 10 μm of the JNK inhibitor SP600125, and then treated with 200 ng/ml BDNF for 1 or 12 h (A). For p75NTR cleavage, the neurons were treated with 10 μm ZLLLH in the presence and absence of JNK inhibitor SP600125 (10 μm) with or without the addition of 200 ng/ml BDNF for 12 h (B). Then neurons were lysed and subjected to Western blot analysis using antibodies to phospho-JNK (p-JNK), total-JNK (A), p75NTR ICD, or tubulin (B). C and D, sympathetic neurons were infected with an adenovirus expressing GFP (Adeno-GFP) or DN-JNK (DN-JNK). 24 h later the neurons were rinsed to remove NGF and refed with medium containing 12.5 mm KCl with or without the addition of 200 ng/ml BDNF for 1 or 12 h (C) or treated with 10 μm ZLLLH with or without the addition of 200 ng/ml BDNF for 12 h (D). Lysates were subjected to Western blot analysis using phospho-JNK, total-JNK (C), and p75NTR ICD and tubulin (D) antibodies. The full-length receptor (FL), CTF, and the ICD are indicated. Data are the representative of three independent experiments.
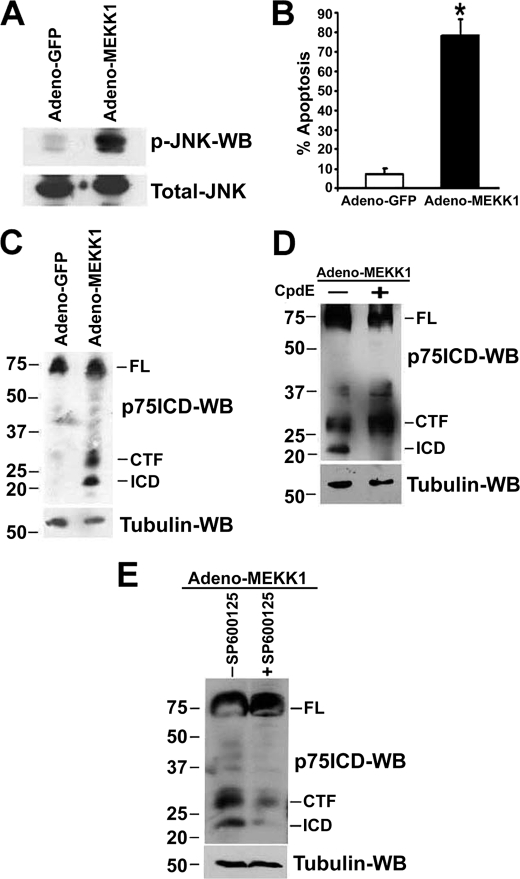
We next investigated whether activation of JNK, independent of p75NTR, was sufficient to promote receptor proteolysis. MEKK1, the upstream activator of JNK, or GFP was overexpressed in sympathetic neurons using adenovirus. As was expected, overexpression of MEKK1 was sufficient to activate JNK and induce apoptotic death in sympathetic neurons (Fig. 3, A and B). In addition, receptor fragments were generated corresponding to the molecular weights of the metalloprotease and γ-secretase cleavage products (Fig. 3C). The generation of the smaller MEKK1-induced p75NTR fragment was γ-secretase- and JNK-dependent, because it was not detected in the presence of the γ-secretase inhibitor Compound E and JNK inhibitor SP600125 (Fig. 3, D and E). Taken together, these results demonstrate that JNK activation precedes and is necessary for ligand-dependent p75NTR cleavage in sympathetic neurons.
FIGURE 3.
Overexpression of MEKK1 induces JNK activation, apoptosis, and p75NTR cleavage in sympathetic neurons. Sympathetic neurons, cultured for 2 days with 20 ng/ml NGF, were infected with an adenovirus expressing GFP or MEKK1. After 24 h, the neurons were lysed and Western blotted using phospho-JNK antibody (A). Some neurons were fixed 24 h after adenoviral exposure and stained with 4′,6-diamidino-2-phenylindole, and the apoptotic nuclei were quantified (B). Shown are the means ± S.D., n = 3 (*, p < 0.008). C–E, 12 h after infection with adenovirus expressing GFP or MEKK1, the neurons were treated with 10 μm proteasome inhibitor ZLLLH alone (C) and/or 100 nm of the γ-secretase inhibitor Compound E (CpdE, D) or 10 μm of the JNK inhibitor SP600125 (E). After 12 h the neurons were lysed and subjected to Western blotting using p75NTR ICD antibody. Data are the representative of three independent experiments.
p75NTR Activates JNK in a Biphasic Manner, First Prior to Receptor Proteolysis, Then through a Cleavage-dependent Process
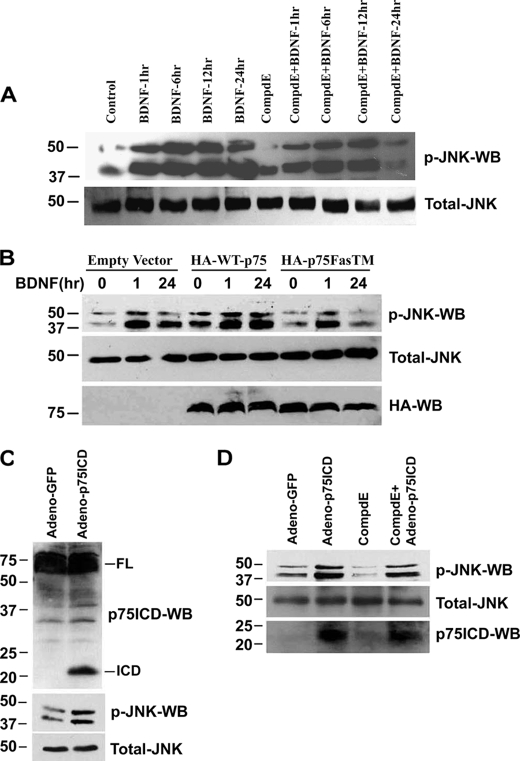
Treatment of sympathetic neurons with BDNF leads to activation of JNK within 30 min, but the kinase remained active for at least 24 h (Fig. 1, A and B). In contrast, p75NTR cleavage by TACE and γ-secretase occurred at 6 and 12 h, respectively (Fig. 1C), which was prevented by JNK inhibition (Fig. 2). Because JNK remained active after receptor cleavage, we investigated whether this long term activation was dependent on p75NTR proteolysis. Therefore, neurons were treated with the γ-secretase inhibitor Compound E for 1 h before treating with BDNF for 1, 6, 12, and 24 h, and the effect on JNK activation was measured. Compound E had no effect on BDNF-induced JNK activation at 1, 6 and 12 h; however, it completely blocked the stimulation of JNK at 24 h (Fig. 4A), suggesting that the later activation of JNK was dependent on p75NTR cleavage.
FIGURE 4.
p75NTR activates JNK in a biphasic manner, initially independent of cleavage, then in a manner dependent on receptor proteolysis. A, sympathetic neurons derived from P4 rats were cultured in NGF for 2 days. The NGF was then removed and the neurons were cultured with medium containing 12.5 mm KCl, and the cells were pretreated for 1 h with 100 nm Compound E (CompdE) or left untreated or treated with 200 ng/ml BDNF for 1, 6, 12, and 24 h. Cells were then lysed and Western blotted using phospho-JNK and total-JNK antibodies. Results are representative of three independent experiments. B, sympathetic neurons were electroporated with pcDNA3 (empty vector), wild-type p75NTR, or γ-secretase-resistant p75NTR (p75FasTM). 36 h later, the neurons were left untreated or treated with 200 ng/ml BDNF for 1 or 24 h, and then cells were lysed and Western blotted using p-JNK, total-JNK, and HA antibodies. C and D, sympathetic neurons were infected with an adenovirus expressing GFP or p75NTR ICD. 24 h after infection the neurons were lysed and Western blotted using p75NTR ICD, phospho-JNK, and total-JNK antibodies (C). In some experiments, 12 h after infection with p75NTR ICD-expressing adenovirus, the neurons were treated with 100 nm of the γ-secretase inhibitor Compound E (CompdE) and 12 h later lysed and subjected to Western blotting using phospho-JNK, total-JNK, and p75NTR ICD antibodies (D). Data are representative of three independent experiments.
To confirm that it is the specific cleavage of p75NTR by γ-secretase that is required for long term JNK activation, we used a mutant p75NTR (p75FasTM) resistant to γ-secretase. This mutant acts as a dominant negative, preventing the cleavage of the endogenous receptor (25, 26), and preventing receptor-mediated cell death (18). We transfected the neurons with empty vector, wild-type p75NTR, or p75FasTM by electroporation, and 36 h later the cells were treated with BDNF for 1 or 24 h. As anticipated, BDNF induced JNK activation in neurons transfected with empty vector and wild-type p75NTR at 1 and 24 h; however, in neurons expressing p75FasTM the activation of JNK was only detected at 1 h and was completely blocked at 24 h (Fig. 4B). Taken together these results indicate that JNK is initially activated independently of p75NTR cleavage but later, long term activation is dependent on p75NTR proteolysis.
To determine whether the release of p75NTR ICD by γ-secretase is sufficient for JNK activation, we expressed the free ICD in sympathetic neurons using adenovirus. The expression of this construct in PC12 cells (27) and in sympathetic neurons (18) was previously shown to induce apoptosis. We found that expression of the p75NTR ICD in sympathetic neurons activated JNK (Fig. 4C). Compound E had no effect on this activation (Fig. 4D), suggesting that the release of the p75NTR ICD is sufficient to stimulate the kinase and to induce cell death without contribution from other γ-secretase substrates.
JNK3 Is Preferentially Activated by p75NTR and Is Required for Receptor Cleavage and Apoptosis
There are three mammalian genes for JNK1, -2, and -3; therefore, to investigate which JNK is required for p75NTR cleavage, we initially transfected JNK1, -2, or -3 into HEK293 cells stably expressing p75NTR and assessed receptor processing by Western blot. Overexpression of each of the JNKs resulted in their activation (supplemental Fig. 1A), and all three forms of the kinase were able to induce p75NTR cleavage (supplemental Fig. 1B). However, JNK3 was reported to be the specific isoform mediating the p75NTR apoptotic signal in oligodendrocytes (21). Therefore, we investigated which JNK was activated by p75NTR in sympathetic neurons. The neurons were treated with BDNF for various times, the lysates were immunoprecipitated using selective antibodies to different JNKs, and an in vitro kinase assay was performed. In concordance with the results in oligodendrocytes (21), BDNF selectively activated JNK3 at 1, 6, 12, and 24 h (Fig. 5A). Some marginal JNK1 activation was also detected; however, this was not significant. There was never any activation of JNK2 observed (data not shown). Taken together with the preceding results (Fig. 4), these findings indicate that the full-length receptor stimulates JNK3 at early time points and the released ICD leads to prolonged JNK3 activation.
FIGURE 5.
JNK3 is selectively activated by p75NTR and is required for receptor cleavage and apoptosis. A, P4 rat sympathetic neurons were cultured in NGF for 48 h, then NGF was washed off, and the cells were treated with medium containing 12.5 mm KCl alone or with 200 ng/ml BDNF for the times indicated. The cells were then lysed and subjected to immunoprecipitation using antibodies to JNK1 or -3, and an in vitro kinase assay was performed using glutathione S-transferase-c-Jun as a substrate. The blots were quantitated, and data are expressed as relative -fold increase over control, untreated neurons. Shown are the means ± S.D., n = 4–5 independent experiments (*, p < 0.05). B, sympathetic neurons from P4 wild-type (WT) and jnk3−/− mice were cultured for 2 days in 20 ng/ml NGF, and then the NGF was removed and the cells refed with medium containing 12.5 mm KCl alone (Con), or 200 ng/ml BDNF (BDNF). 48 h later, the cells were fixed and stained with 4′,6-diamidino-2-phenylindole, and apoptotic nuclei were quantified. Shown are the means ± S.D., n = 3 (*, p < 0.004). C, sympathetic neurons from P4 wild-type (WT) and jnk3 −/− mice cultured in medium containing 12.5 mm KCl were treated with 10 μm ZLLLH alone (Con) or in combination with 200 ng/ml BDNF for 12 h (BDNF). Then the neurons were lysed and immunoblotted using anti-p75NTR ICD. D, SCG were isolated at P4 from wild-type and jnk3−/− mice and Western blotted with antibody to the p75NTR ICD. Data are representative of three independent experiments.
To determine whether JNK3 was specifically required for the apoptotic signal of p75NTR, sympathetic neurons were cultured from jnk3−/− mice and treated with BDNF. The cell death induced by this neurotrophin was significantly decreased in the null neurons, demonstrating the requirement for this signaling component in the apoptotic pathway (Fig. 5B). The preceding findings suggested that p75NTR cleavage would also be attenuated in the absence of JNK3. To test this hypothesis, we measured receptor processing in neurons from wild-type and jnk3−/− mice following BDNF treatment. Although both the CTF and ICD of p75NTR were detected in wild-type neurons, no proteolytic fragments were found in jnk3−/− neurons (Fig. 5C). Given that p75NTR contributes to developmental apoptosis in the SCG in vivo (11, 28, 23), and receptor cleavage was previously observed in the SCG during the period of normal cell death (18), we predicted that receptor proteolysis would be reduced in the ganglia from the JNK3 null mice. Indeed, neither the free CTF nor the ICD were detectable in SCG from jnk3 −/− animals, unlike wild-type (Fig. 5D). All together, these results demonstrate that p75NTR activation of JNK3 is required for receptor cleavage and apoptosis in sympathetic neurons.
p75NTR Activation of JNK3 Results in the Up-regulation of TACE/ADAM17 in Sympathetic Neurons
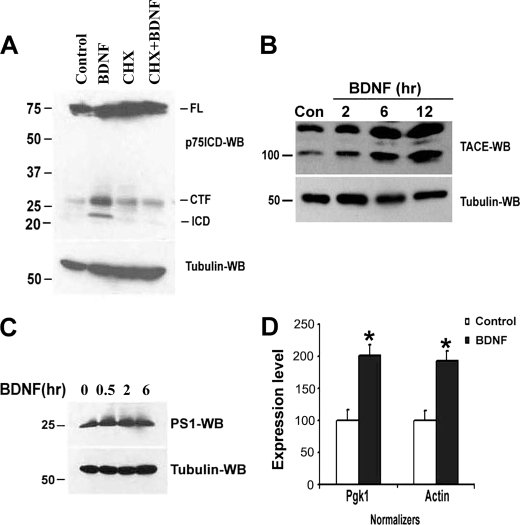
JNK is well characterized as an activator of several AP-1 transcription factors, which suggested that one role of the early JNK activation in p75NTR signaling may be to transcriptionally up regulate a component necessary for cleaving the receptor. A requirement for macromolecular synthesis could also explain why ligand-induced p75NTR cleavage takes 6 h in sympathetic neurons (Fig. 1C). We previously demonstrated that p75NTR-mediated apoptosis in sympathetic neurons could be blocked by the protein synthesis inhibitor cycloheximide (20); therefore, we tested whether p75NTR processing was also sensitive to this inhibitor. When neurons were treated with cycloheximide and BDNF for 12 h no p75NTR cleavage was detected (Fig. 6A), indicating that BDNF-mediated receptor proteolysis requires new protein synthesis.
FIGURE 6.
BDNF-induced p75NTR cleavage in sympathetic neurons requires protein synthesis and correlates with up-regulation of TACE. A, sympathetic neurons were cultured in 20 ng/ml NGF for 2 days, then the NGF was rinsed off and the neurons refed with medium containing 12.5 mm KCl and 10 μm ZLLLH in the presence or absence of 200 ng/ml BDNF for 12 h, with or without the protein synthesis inhibitor cycloheximide (CHX, 100 ng/ml). Cells were then lysed and Western blotted using p75NTR ICD and tubulin antibodies. B and C, sympathetic neurons cultured in medium containing 12.5 mm KCl were treated with or without BDNF (200 ng/ml) for various times, as indicated. The neurons were then lysed and subjected to Western blot analysis using TACE (B), presenilin-1 (PS1) (C), or tubulin antibodies. D, neurons in medium containing 12.5 mm KCl were left untreated (empty bar, Control) or treated with 200 ng/ml BDNF for 4 h (black bar, BDNF). Total RNA was isolated and reverse-transcribed, and quantitative PCR was performed for TACE, Pgk1, and actin using an ABI Prism 7300 system. Differential expression of TACE message was calculated as ΔΔCt against the expression of normalizers Pgk1 and actin. Depicted is the relative expression level of TACE with Pgk1 or actin set as 100%, as indicated. Data are representative of three independent experiments.
Cleavage of the p75NTR has been suggested to occur in two steps: first the metalloprotease TACE/ADAM17 sheds the extracellular domain of the receptor, producing the CTF (29, 25), then the γ-secretase complex cleaves in the transmembrane domain to release the ICD (16–18). Therefore, we evaluated the effects of BDNF on the levels of TACE and presenilin 1, the catalytic subunit of the γ-secretase complex, in sympathetic neurons. Following BDNF treatment, there was a 323% ± 88% (n = 3, mean ± S.D.) increase in TACE protein (Fig. 6B), while the level of presenilin 1 remained unchanged (Fig. 6C). Note that the level of both the mature, active form of TACE (lower band) and the precursor (upper band) increased. This up-regulation of TACE protein was also reflected by an increase in mRNA (Fig. 6D), further supporting a transcriptional induction of the protease upon p75NTR activation.
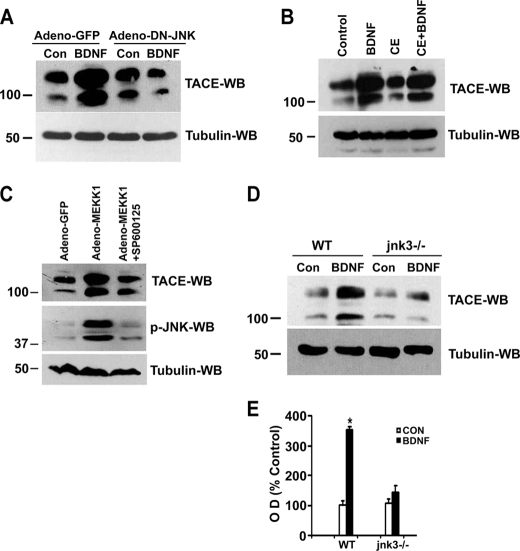
To determine whether BDNF activation of JNK was responsible for the up-regulation of TACE in sympathetic neurons we first blocked the kinase using DN-JNK. As shown in Fig. 7A, expression of DN-JNK prevented BDNF-mediated TACE up-regulation (Fig. 7A), indicating activation of the kinase was required for the increase in TACE expression. In contrast, inhibition of γ-secretase did not prevent p75NTR-mediated TACE up-regulation (Fig. 7B). We then asked whether JNK activation, in the absence of receptor stimulation, was sufficient to induce TACE. The neurons were infected with adenovirus expressing MEKK1 or GFP, as a control, in the presence and absence of JNK inhibitor SP600125 and TACE levels measured 24 h later. As depicted in Fig. 7C, MEKK1 substantially up-regulated the level of this protease, and the JNK inhibitor blocked MEKK1-induced JNK activation and TACE up-regulation. This confirms that JNK activation is sufficient to increase the expression of TACE. The ability of JNK activation to up-regulate TACE was not specific to neurons; overexpression of JNK in HEK293 cells was also able to increase the amount of the protease (supplemental Fig. 1).
FIGURE 7.
p75NTR-mediated TACE up-regulation is dependent on JNK3 activation not on p75NTR cleavage. A, sympathetic neurons were infected with an adenovirus expressing GFP (Adeno-GFP) or DN-JNK (Adeno-DN-JNK). 24 h later the neurons were rinsed to remove the NGF and refed with medium containing 12.5 mm KCl with or without the addition of 200 ng/ml BDNF for 6 h. The cells were then lysed and subjected to Western blot analysis using TACE and tubulin antibodies. B, sympathetic neurons were left untreated or treated with 200 ng/ml BDNF for 12 h in the presence or absence of 100 nm Compound E (CE). The cells were then lysed and Western blotted using TACE or tubulin antibodies. C, sympathetic neurons were infected with adenovirus expressing GFP (Adeno-GFP) or MEKK1 (Adeno-MEKK1) in the presence or absence of JNK inhibitor SP600125, and 24 h later lysed and subjected to Western blotting using TACE, phospho-JNK, or tubulin antibodies. D, sympathetic neurons from P4 wild-type (WT) and jnk3 −/− mice were cultured in NGF for 2 days, and then NGF was removed, and the neurons were refed with medium containing 12.5 mm KCl alone (Con) or in combination with 200 ng/ml BDNF (BDNF). After 6 h, the neurons were lysed and Western blotted using TACE or tubulin antibodies. E, quantitation of TACE Western blots, expressed as a percentage of control. Results are means ± S.D. of three independent experiments (*, p < 0.037).
Because our data indicated that BDNF binding to p75NTR results in the activation of JNK3, which leads to receptor cleavage (Fig. 5), we hypothesized that the up-regulation of TACE should be dependent on this isoform of the kinase. To this aim, we treated jnk3−/− or +/+ sympathetic neurons with BDNF and, upon analysis of TACE levels, found up-regulation only in wild-type neurons (Fig. 7, D and E). Together, these results indicate that BDNF activates JNK3, resulting in increased TACE expression, which then cuts off the extracellular domain of the receptor, leaving the CTF to be cleaved by γ-secretase.
BDNF Binding to p75NTR Induces TACE-dependent Cleavage of the Receptor, Which Is Necessary for Apoptosis
Although shedding of the extracellular domain of p75NTR has long been known to be mediated by a metalloprotease (30, 31), only recently was TACE suggested to be the specific enzyme responsible for this cleavage (29). However, this result was based on fibroblasts ectopically expressing the receptor, and TACE was shown to shed the ECD in a constitutive fashion or following treatment with pervanadate or phorbol ester. It has not been determined whether this metalloprotease cleaves p75NTR in response to ligand binding in neurons. Because our data demonstrated that BDNF increased the expression of this protease in sympathetic neurons, we reasoned that TACE was carrying out the observed proteolysis. To determine whether TACE up-regulation is sufficient to induce cleavage of p75NTR, we overexpressed the wild-type enzyme or a catalytically inactive mutant (32) in the neurons. Interestingly, expression of wild-type TACE was sufficient to release the CTF of p75; however, it did not result in the release of the γ-secretase product (supplemental Fig. 2). Thus, although JNK activation alone was sufficient to stimulate both proteolytic events (Fig. 3), p75NTR cleavage by TACE alone is not enough to induce γ-secretase-mediated processing of the receptor in sympathetic neurons.
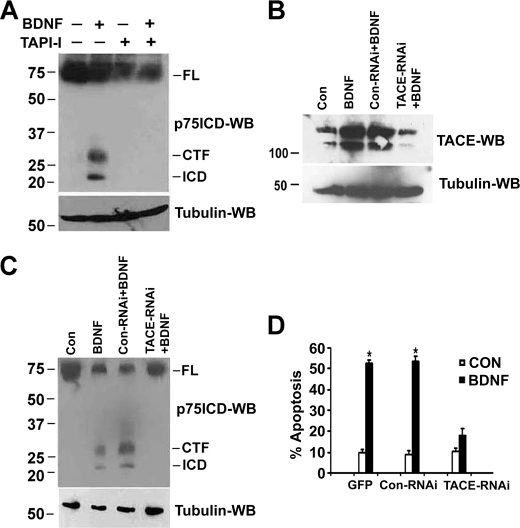
Kanning et al. (17) previously demonstrated that phorbol ester-induced cleavage of p75NTR in fibroblasts could be prevented by TAPI-1, an inhibitor of several metalloproteases, including TACE. Therefore, we tested the effects of TAPI-1 on BDNF-induced proteolysis in sympathetic neurons. Pretreatment of the neurons with TAPI-1 for 1 h before adding BDNF for 12 h completely blocked the generation of p75NTR CTF and ICD (Fig. 8A). To specifically address the requirement for TACE, we used RNAi to down-regulate its expression. Neurons were transfected with control RNAi or TACE RNAi and after 24 h, treated with BDNF. After confirming TACE knockdown (Fig. 8B), we assessed the effects on p75NTR cleavage and receptor-induced apoptosis. As shown in Fig. 8 (C and D), reducing TACE levels completely blocked BDNF-mediated p75NTR proteolysis and receptor-mediated apoptosis, indicating that TACE up-regulation is essential for ligand-dependent receptor processing and apoptosis.
FIGURE 8.
Inhibition or down-regulation of TACE blocks ligand-dependent p75NTR cleavage and apoptosis in sympathetic neurons. A, sympathetic neurons were cultured in NGF for 2 days, and then the NGF was rinsed off and the neurons were treated with medium containing 12.5 mm KCl and 10 μm ZLLLH alone or together with 200 ng/ml BDNF in the presence or absence of the metalloprotease inhibitor TAPI-1 (10 μm) for 12 h. The cells were then lysed and subjected to Western blot analysis using p75NTR ICD and tubulin antibodies. B, sympathetic neurons were cultured in 20 ng/ml NGF for 2 days, and then transfected with control RNAi or TACE RNAi using Dharmafect. 24 h after transfection, NGF was rinsed off, and the cells were cultured with medium containing 12.5 mm KCl alone (Control) or in combination with 200 ng/ml BDNF. 6 h later the cells were lysed and subjected to TACE and tubulin Western blotting. C, 24 h after RNAi transfection, the neurons were cultured in 12.5 mm KCl and treated with 10 μm ZLLLH alone (Control) or together with 200 ng/ml BDNF for 12 h. The cells were then lysed and subjected to p75NTR ICD and tubulin Western blotting. Data are representative of three independent experiments. D, sympathetic neurons were transfected by electroporation with GFP alone or co-transfected with control RNAi or TACE RNAi and 36 h later treated with 200 ng/ml BDNF for 48 h. The cells were then fixed and stained with 4′,6-diamidino-2-phenylindole, and apoptotic nuclei were quantified. Shown are the means ± S.D., n = 3 (*, p < 0.0031).
DISCUSSION
The p75NTR is a critical regulator of sympathetic neuron survival and target tissue innervation during development. Gene deletion of p75NTR in mice causes a reduction in the normal programmed cell death in sympathetic ganglia, resulting in more neurons (11, 12, 23) and altered innervation of some target tissues (10, 33–35). Unfortunately, the molecular mechanisms by which p75NTR signals cell death have yet to be fully elucidated. In this study, we demonstrated that BDNF binding to p75NTR in sympathetic neurons first activates JNK, predominantly JNK3, which then results in the transcriptional up-regulation of the metalloprotease TACE/ADAM17, leading to receptor cleavage. The freed ICD prolongs the activation of JNK, ultimately resulting in apoptosis.
The stress kinase JNK plays an essential role in the apoptotic pathway activated by the p75NTR (13–15). Of the three JNK genes, we demonstrated that p75NTR preferentially stimulates JNK3, and this gene was required for the up-regulation of TACE (Fig. 7) and the apoptotic response (Fig. 5). A role for JNK3 in p75NTR signaling is in agreement with a previous report that cell death in oligodendrocytes by p75NTR involved the activation of JNK3 (21).
Cell death induced by JNK activation has been shown to occur through both transcription-dependent and -independent mechanisms; for example, JNK activates members of the AP-1 family of transcription factors, leading to up-regulation of pro-apoptotic genes such as Bim (36, 37), Bak (38), and Fas ligand (39), but it also promotes apoptosis through phosphorylation of cell death factors such as p53 (40), BAD (41), and Bim (42, 43). Similarly, p75NTR-mediated cell death was demonstrated to involve JNK phosphorylation of Bim (43), Bad (41), and p53 (44). However, protein synthesis inhibitors blocked p75NTR-dependent apoptosis in sympathetic neurons, suggesting that transcription may have a role (20), although the genes required for inducing cell death have not been identified nor have the specific isoforms of JNK involved.
In our efforts to understand the mechanisms of p75NTR-mediated apoptosis in sympathetic neurons, we recently demonstrated that the receptor undergoes proteolytic cleavage in response to apoptotic ligands (18). The cleavage facilitated ubiquitination of the DNA binding protein NRIF by the E3 ligase TRAF6, resulting in nuclear translocation of NRIF, which was necessary for cell death (18, 19). How receptor processing, NRIF nuclear shuttling, and JNK activation were related was not known.
Here we demonstrate that JNK activation by p75NTR transcriptionally up-regulates TACE/ADAM17, which is necessary for cleavage to occur. Surprisingly, in addition to the initial activation of the kinase prior to receptor proteolysis, there is also a subsequent, cleavage-dependent activation of JNK. The molecular mechanisms underlying JNK activation by p75NTR are not well understood. Numerous p75NTR-interacting proteins have been linked to JNK activation, including Rac (21), NRAGE (45, 24), NRIF (46), and TRAF6 (23, 47). Recently, the activation of JNK by BDNF was shown to be significantly reduced at the 1-h time point in sympathetic neurons from nrage−/− mice (45), implicating this interactor in the early, pre-cleavage phase of JNK stimulation. In contrast, BDNF treatment of traf6−/− sympathetic neurons activates JNK at the 1-h time point (supplemental Fig. 4), but, at 24 h BDNF failed to activate JNK in traf6−/− mice (23). However, p75NTR still underwent proteolysis by TACE and γ-secretase in TRAF6 null neurons (18), suggesting that TRAF6 is critical for the post-cleavage activation of JNK. Similarly, in sympathetic neurons from nrif−/− mice, there was no activation of JNK at the 24-h time point (46). Because TRAF6 and NRIF directly interact (48), it is likely that a complex forms involving these two proteins and the released p75NTR ICD, which is then responsible for the later phase JNK activation. In summary, we propose the model shown in Fig. 9 for p75NTR-mediated apoptosis in sympathetic neurons. Pre-existing dimers of p75NTR become activated upon ligand binding, leading to recruitment of various interactors (49); notably, NRAGE associates with the receptor, which we suggest is required for JNK3 activation, leading to up-regulation of TACE. This metalloprotease then cleaves p75NTR allowing γ-secretase-mediated cleavage. Upon release of the ICD, NRIF and TRAF6 promote JNK3 activation and NRIF nuclear translocation, all of which lead to apoptosis.
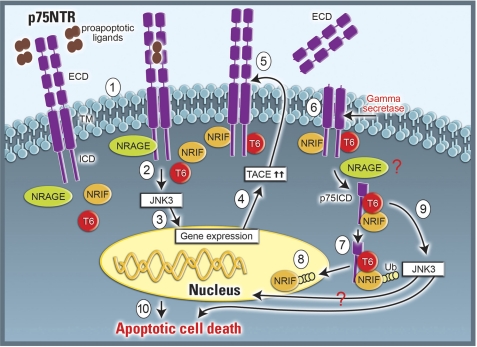
FIGURE 9.
Schematic illustration of p75NTR-mediated apoptotic signaling. In sympathetic neurons binding of pro-apoptotic ligands, such as pro-BDNF and mature-BDNF, to pre-existing p75NTR dimers (1) recruits cytosolic interactors like NRAGE to the intracellular domain (ICD) of the receptor, resulting in activation of JNK3 (2). Stimulation of the kinase (3) leads to up-regulation of TACE (4), which cleaves the extracellular domain of p75NTR (5) and allows γ-secretase mediated release of the receptor's intracellular domain (p75NTR ICD) (6). The liberated p75NTR ICD forms a ternary complex with NRIF and TRAF6 (p75NTR ICD-NRIF-TRAF6) to promote TRAF6-dependent ubiquitination of NRIF (7) and nuclear translocation (8); in addition, the p75NTR ICD-NRIF-TRAF6 complex also activates JNK3 (9), all of which lead to p75NTR-mediated apoptosis (10).
Previous studies suggested that p75NTR is a substrate of TACE/ADAM17 (29, 50); however, none have shown this specific enzyme responsible for the ligand-induced proteolysis. Hence, this is the first demonstration that TACE cleaves the receptor in response to ligand activation. Moreover, we found that TACE is transcriptionally up-regulated through p75NTR activation of JNK. Several other cytokines and growth factors have been shown to increase TACE transcription (51, 52), and the mitogen-activated protein kinases p38 and ERK have been implicated in transducing the signal from some of these ligands (53), although the mechanisms remain largely unknown. Interestingly, JNK overexpression in 293 cells was also sufficient to induce the protein (supplemental Fig. 1), suggesting that this kinase could mediate the response to other receptors as well.
The requirement for TACE transcription prior to p75NTR cleavage accounts, at least in part, for the long time between ligand binding to the receptor and the subsequent proteolysis by TACE and γ-secretase in sympathetic neurons (Fig. 1C) (18). Typically, receptor activation leads to rapid cleavage; for example, neuregulin binding to ErbB4 induces cleavage by TACE and γ-secretase within 30 min (54). The relative basal levels of TACE may also explain why p75NTR cleavage was detected within 1 h of myelin-associated glycoprotein binding to cerebellar neurons (26) and neurotrophin treatment of PC12 cells (55) or cortical neurons (56), but in sympathetic neurons required 6 h of BDNF exposure. However, the mechanisms of cleavage must also be somewhat different in PC12 cells compared with sympathetic neurons, because NGF binding to TrkA could induce proteolysis of p75NTR by both metalloprotease and γ-secretase, whereas in sympathetic neurons p75NTR processing was not detected in response to NGF (18). Furthermore, NGF treatment of PC12 cells leads to survival and differentiation, whereas our results indicate that p75NTR cleavage is only associated with apoptosis in the sympathetic neurons. Indeed, proteolytic processing of the receptor was detected not only in response to BDNF, but also following NGF withdrawal (supplemental Fig. 3).
Previously, we did not observe p75NTR processing 24 h after NGF withdrawal from sympathetic neurons (18); however, when we looked 30 h after the trophic factor removal (keeping the cells alive with a pan-caspase inhibitor), both the CTF and ICD fragments were observed (supplemental Fig. 3B). This correlates well with the kinetics of JNK activation: BDNF activates JNK more rapidly than removing NGF (compare Fig. 1A to supplemental Fig. 3A). Hence, activation of JNK by p75NTR or by NGF withdrawal can induce p75NTR proteolysis, which promotes apoptosis.
As discussed above, JNK activation promotes cell death through multiple mechanisms. Moreover, our results suggest that, even in regulating p75NTR cleavage, there is more than one function required of this kinase. Despite the fact that JNK stimulation was sufficient for cleavage of p75NTR by both TACE and γ-secretase, overexpression of TACE alone was not enough to prompt γ-secretase-mediated release of the ICD (supplemental Fig. 2), indicating that additional signaling mediated by JNK is necessary. In this context, it is notable that JNK can induce γ-secretase cleavage of amyloid precursor protein through phosphorylation of its CTF (57, 58). Because amyloid precursor protein is thought to undergo cleavage by γ-secretase following endocytosis (59), it has been suggested that the phosphorylation promotes amyloid precursor protein internalization, thereby facilitating the proteolysis (60). Interestingly, Urra and colleagues recently provided evidence that p75NTR internalization after ligand binding is necessary for γ-secretase to cleave the receptor (55); therefore, JNK activation may also promote receptor internalization. Further elucidation of the mechanisms regulating p75NTR processing during apoptotic signaling will be the focus of future studies.
Supplementary Material
Acknowledgments
We thank Regeneron for providing BDNF, Dr. Peter Dempsey, University of Michigan, for plasmids expressing wild-type and mutant TACE, and Dr. Philip Barker, McGill University, for adenovirus expressing the p75NTR-ICD. We also thank Ana Perdigoto and Cedric Jones for help with the Traf6 animals.
This work was supported, in whole or in part, by National Institutes of Health Grants NS038220 (to B. D. C. and R. S. K.) and NS039472 (to S. O. Y.). This work was also supported by the Fondo Nacional de Desarrollo Científico y Tecnológico (1085273) and Conicyt PFB12/2007 (to F. C. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- p75NTR
- p75 neurotrophin receptor
- CTF
- C-terminal fragment
- ICD
- intracellular domain
- BDNF
- brain-derived neurotrophic factor
- NGF
- nerve growth factor
- JNK
- c-Jun N-terminal kinase
- TACE
- tumor necrosis factor-α-converting enzyme
- ADAM17
- a disintegrin and metallopeptidase domain 17
- MEKK1
- mitogen-activated protein kinase kinase 1
- NRIF
- neurotrophin receptor-interacting factor
- TRAF6
- TNF receptor-associated factor 6
- SCG
- superior cervical ganglia
- P
- postnatal day
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- RNAi
- RNA interference
- DN
- dominant negative
- ERK
- extracellular signal-regulated kinase
- TAP-I
- tumor necrosis factor-α protease inhibitor 1
- ZLLH
- Z-Leu-Leu-Leu-H (aldehyde).
REFERENCES
- 1.Glebova N. O., Ginty D. D. (2005) Annu. Rev. Neurosci. 28, 191–222 [DOI] [PubMed] [Google Scholar]
- 2.Deppmann C. D., Mihalas S., Sharma N., Lonze B. E., Niebur E., Ginty D. D. (2008) Science 320, 369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levi-Montalcini R., Cohen S. (1960) Ann. N.Y. Acad. Sci. 85, 324–341 [DOI] [PubMed] [Google Scholar]
- 4.Korsching S., Thoenen H. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 3513–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shelton D. L., Reichardt L. F. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 7951–7955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hempstead B. L., Martin-Zanca D., Kaplan D. R., Parada L. F., Chao M. V. (1991) Nature 350, 678–683 [DOI] [PubMed] [Google Scholar]
- 7.Barker P. A., Shooter E. M. (1994) Neuron 13, 203–215 [DOI] [PubMed] [Google Scholar]
- 8.Epa W. R., Markovaska K., Barrett G. L. (2004) J. Neurochem. 89, 344–353 [DOI] [PubMed] [Google Scholar]
- 9.Kuruvilla R., Zweifel L. S., Glebova N. O., Lonze B. E., Valdez G., Ye H., Ginty D. D. (2004) Cell 118, 243–255 [DOI] [PubMed] [Google Scholar]
- 10.Singh K. K., Park K. J., Hong E. J., Kramer B. M., Greenberg M. E., Kaplan D. R., Miller F. D. (2008) Nat. Neurosci. 11, 649–658 [DOI] [PubMed] [Google Scholar]
- 11.Bamji S. X., Majdan M., Pozniak C. D., Belliveau D. J., Aloyz R., Kohn J., Causing C. G., Miller F. D. (1998) J. Cell Biol. 140, 911–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansen P., Giehl K., Nyengaard J. R., Teng K., Lioubinski O., Sjoegaard S. S., Breiderhoff T., Gotthardt M., Lin F., Eilers A., Petersen C. M., Lewin G. R., Hempstead B. L., Willnow T. E., Nykjaer A. (2007) Nat. Neurosci. 10, 1449–1457 [DOI] [PubMed] [Google Scholar]
- 13.Casaccia-Bonnefil P., Carter B. D., Dobrowsky R. T., Chao M. V. (1996) Nature 383, 716–719 [DOI] [PubMed] [Google Scholar]
- 14.Yoon S. O., Casaccia-Bonnefil P., Carter B., Chao M. V. (1998) J. Neurosci. 18, 3273–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman W. J. (2000) J. Neurosci. 20, 6340–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung K. M., Tan S., Landman N., Petrova K., Murray S., Lewis R., Kim P. K., Kim D. S., Ryu S. H., Chao M. V., Kim T. W. (2003) J. Biol. Chem. 278, 42161–42169 [DOI] [PubMed] [Google Scholar]
- 17.Kanning K. C., Hudson M., Amieux P. S., Wilney J. C., Bothwell M., Schecterson L. C. (2003) J. Neurosci. 23, 5425–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenchappa R. S., Zampieri N., Chao M. V., Barker P. A., Teng H. K., Hempstaed B. L., Carter B. D. (2006) Neuron 50, 219–232 [DOI] [PubMed] [Google Scholar]
- 19.Geetha T., Kenchappa R. S., Wooten W. M., Carter B. D. (2005) EMBO J. 24, 3859–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmada M., Kanwal S., Rutkoski N. J., Gustafson-Brown C., Johnson R. S., Wisdom R., Carter B. D., Gufstafson-Brown C. (2002) J. Cell Biol. 158, 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrington A. W., Kim J. Y., Yoon S. O. (2002) J. Neurosci. 22, 156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurrasch D. M., Huang J., Wilkie T. M., Repa J. J. (2004) Methods Enzymol. 389, 3–15 [DOI] [PubMed] [Google Scholar]
- 23.Yeiser E. C., Rutkoski N. J., Naito A., Inoue J., Carter B. D. (2004) J. Neurosci. 24, 10521–10529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salehi A. H., Xanthoudakis S., Barker P. A. (2002) J. Biol. Chem. 277, 48043–48050 [DOI] [PubMed] [Google Scholar]
- 25.Zampieri N., Xu C. F., Neubert T. A., Chao M. V. (2005) J. Biol. Chem. 280, 14563–14571 [DOI] [PubMed] [Google Scholar]
- 26.Domeniconi M., Zampieri N., Spencer T., Hilaire M., Mellado W., Chao M. V., Filbin M. T. (2005) Neuron 46, 849–855 [DOI] [PubMed] [Google Scholar]
- 27.Roux P. P., Bhakar A. L., Kennedy T. E., Barker P. A. (2001) J. Biol. Chem. 276, 23097–23104 [DOI] [PubMed] [Google Scholar]
- 28.Majdan M., Walsh G. S., Aloyz R., Miller F. D. (2001) J. Cell Biol. 155, 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weskamp G., Schlöndorff J., Lum L., Becherer J. D., Kim T. W., Saftig P., Hartmann D., Murphy G., Blobel C. P. (2004) J. Biol. Chem. 279, 4241–4249 [DOI] [PubMed] [Google Scholar]
- 30.DiStefano P. S., Chelsea D. M. (1990) Brain Res. 534, 340–344 [DOI] [PubMed] [Google Scholar]
- 31.DiStefano P. S., Chelsea D. M., Schick C. M., McKelvy J. F. (1993) J. Neurosci. 13, 2405–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garton K. J., Gough P. J., Philalay J., Wille P. T., Blobel C. P., Whitehead R. H., Dempsey P. J., Raines E. W. (2003) J. Biol. Chem. 278, 37459–37464 [DOI] [PubMed] [Google Scholar]
- 33.Kohn J., Aloyz R. S., Toma J. G., Haak-Frendscho M., Miller F. D. (1999) J. Neurosci. 19, 5393–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jahed A., Kawaja M. D. (2005) Auton Neurosci. 118, 32–42 [DOI] [PubMed] [Google Scholar]
- 35.Habecker B. A., Bilimoria P., Linick C., Gritman K., Lorentz C. U., Woodward W., Birren S. J. (2008) Auton Neurosci. 140, 40–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ham J., Eilers A., Whitfield J., Neame S. J., Shan B. (2000) Biochem. Pharmacol. 60, 1015–1021 [DOI] [PubMed] [Google Scholar]
- 37.Putcha G. V., Moulder K. L., Golden J. P., Bouillet P., Adams J. A., Strasser A., Johnson E. M., Jr (2001) Neuron 29, 615–628 [DOI] [PubMed] [Google Scholar]
- 38.Jin H. O., Park I. C., An S., Lee H. C., Woo S. H., Hong Y. J., Lee S. J., Park M. J., Yoo D. H., Rhee C. H., Hong S. I. (2006) J. Cell. Physiol. 206, 477–486 [DOI] [PubMed] [Google Scholar]
- 39.Le-Niculescu H., Bonfoco E., Kasuya Y., Claret F. X., Green D. R., Karin M. (1999) Mol. Cell. Biol. 19, 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuchs S. Y., Adler V., Pincus M. R., Ronai Z. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10541–10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhakar A. L., Howell J. L., Paul C. E., Salehi A. H., Becker E. B., Said F., Bonni A., Barker P. A. (2003) J. Neurosci. 23, 11373–11381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Putcha G. V., Le S., Frank S., Besirli C. G., Clark K., Chu B., Alix S., Youle R. J., LaMarche A., Maroney A. C., Johnsom E. M., Jr. (2003) Neuron 38, 899–914 [DOI] [PubMed] [Google Scholar]
- 43.Becker E. B., Howell J., Kodama Y., Barker P. A., Bonni A. (2004) J. Neurosci. 24, 8762–8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aloyz R. S., Bamji S. X., Pozniak C. D., Toma J. G., Atwal J., Kaplan D. R., Miller F. D. (1998) J. Cell Biol. 143, 1691–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertrand M. J., Kenchappa R. S., Andrieu D., Leclerca-Smekens M., Nyugen H. N., Carter B. D., Muscatelli F., Barker P. A., De Backer O. (2008) Cell Death Differ 15, 1921–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linggi M. S., Burke T. L., Williams B. B., Harrington A., Kraemer R., Hempstead B. L., Yoon S. O., Carter B. D. (2005) J. Biol. Chem. 280, 13801–13808 [DOI] [PubMed] [Google Scholar]
- 47.Khursigara G., Orlinick J. R., Chao M. V. (1999) J. Biol. Chem. 274, 2597–2600 [DOI] [PubMed] [Google Scholar]
- 48.Gentry J. J., Rutkoski N. J., Burke T. L., Carter B. D. (2004) J. Biol. Chem. 279, 16646–16656 [DOI] [PubMed] [Google Scholar]
- 49.Vilar M., Charalampopoulos I., Kenchappa R. S., Simi A., Karaca E., Reversi A., Choi S., Bothwell M., Mingaroo I., Freidman W. J., Schiavo G., Bastiaens P. I., Verveer P. J., Carter B. D., Ibáñez C. F. (2009) Neuron 62, 72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srinivasan B., Wang Z., Brun-Zinkernagel A. M., Collier R. J., Black R. A., Frank S. J., Barker P. A., Roque R. S. (2007) Mol. Cell Neurosci. 36, 449–461 [DOI] [PubMed] [Google Scholar]
- 51.Bzowska M., Jura N., Lassak A., Black R. A., Bereta J. (2004) Eur. J. Biochem. 271, 2808–2820 [DOI] [PubMed] [Google Scholar]
- 52.Worley J. R., Baugh M. D., Hughes D. A., Edwards D. R., Hogan A., Sampson M. J., Gavrilovic J. (2003) J. Biol. Chem. 278, 51340–51346 [DOI] [PubMed] [Google Scholar]
- 53.Heidinger M., Kolb H., Krell H. W., Jochum M., Ries C. (2006) Biol. Chem. 387, 69–78 [DOI] [PubMed] [Google Scholar]
- 54.Ni C. Y., Murphy M. P., Golde T. E., Carpenter G. (2001) Science 294, 2179–2181 [DOI] [PubMed] [Google Scholar]
- 55.Urra S., Escudero C. A., Ramos P., Lisbona F., Allende E., Covarrubias P., Parraguez J. I., Zampieri N., Chao M. V., Annaert W., Bronfman F. C. (2007) J. Biol. Chem. 282, 7606–7615 [DOI] [PubMed] [Google Scholar]
- 56.Volosin M., Trotter C., Cragnolini A., Kenchappa R. S., Light M., Hempstead B. L., Carter B. D., Friedman W. J. (2008) J. Neurosci. 28, 9870–9879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y. F., Wang B. J., Cheng H. T., Kuo L. H., Wolfe M. S. (2004) J. Biol. Chem. 279, 49523–49532 [DOI] [PubMed] [Google Scholar]
- 58.Shen C., Chen Y., Liu H., Zhang K., Zhang T., Lin A., Jing N. (2008) J. Biol. Chem. 283, 17721–17730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Small S. A., Gandy S. (2006) Neuron 52, 15–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sodhi C. P., Perez R. G., Gottardi-Littell N. R. (2008) Brain Res. 1198, 204–212 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.