Abstract
Proteins containing ubiquitin-like (UBL) and ubiquitin-associated (UBA) domains interact with various binding partners and function as hubs during ubiquitin-mediated protein degradation. A common interaction of the budding yeast UBL-UBA proteins Rad23 and Dsk2 with the E4 ubiquitin ligase Ufd2 has been described in endoplasmic reticulum-associated degradation among other pathways. The UBL domains of Rad23 and Dsk2 play a prominent role in this process by interacting with Ufd2 and different subunits of the 26 S proteasome. Here, we report crystal structures of Ufd2 in complex with the UBL domains of Rad23 and Dsk2. The N-terminal UBL-interacting region of Ufd2 exhibits a unique sequence pattern, which is distinct from any known ubiquitin- or UBL-binding domain identified so far. Residue-specific differences exist in the interactions of these UBL domains with Ufd2, which are coupled to subtle differences in their binding affinities. The molecular details of their differential interactions point to a role for adaptive evolution in shaping these interfaces.
Keywords: Protein Degradation, Protein Domains, Protein-Protein Interactions, Ubiquitin, Ubiquitin Ligase, Ubiquitylation, UBL-UBA Proteins, Ubiquitin Binding Domains, Ubiquitin-like Domains, Ufd2
Introduction
The ubiquitin proteasome system regulates diverse cellular processes including cell cycle progression, immune response, neurodegenerative diseases, and protein quality control (1–4). Ubiquitin-like (UBL)4 domains and ubiquitin- or UBL-binding domains (UBD) (5) are small and highly diversified domains that occur as integral parts of larger proteins (6–9). Integral UBLs display a similar fold as ubiquitin (Ub) and like Ub are described as protein-protein interaction modules without the modifier function of Ub (5, 10). So far more than 20 different classes of UBDs have been reported with a wide range of Ub binding specificities (11, 12). The ubiquitin-associated (UBA) domain was the first identified UBD, which exhibits the highest representation of all UBDs in the eukaryotic genome (13) with diverse Ub and Ub chain binding properties (14, 15). Although the source of this binding diversity in vivo remained elusive so far, remarkable structural studies have recently unraveled the unique poly-Ub binding mode of a few other UBDs (16–20) and contributed further to the understanding of how UBDs might have acquired their respective ligand specificity.
UBL-UBA proteins contain both a UBL domain and at least one UBA domain. Via these domains they interact simultaneously with ubiquitylated substrates and 26 S proteasome, thereby delivering substrates to the proteasome for degradation (21). Interestingly, UBL-UBA proteins are also binding partners of other proteins (22–25). For instance, the budding yeast UBL-UBA proteins Rad23 and Dsk2 can interact with the E4 ligase Ufd2 via their UBL domains (22, 26, 27). A common involvement of Ufd2, Rad23, and Dsk2 has been described in the endoplasmic reticulum-associated degradation, ubiquitin fusion degradation, and OLE-1 gene induction pathway (22, 28–30), where the UBL-Ufd2 interaction is indispensable. The association of UBL-UBA proteins with Ub ligases, their reported substrate specificity (31, 32), and the inhibitory effect of UBL-UBA proteins on Ub chain disassembly (33, 34) support the idea that UBL-UBA proteins might function as important regulatory and specificity factors in Ub-mediated cellular proteolysis (21). Therefore, understanding the binding behavior of the UBL domains of UBL-UBA proteins with their various interacting proteins will shed light on the regulatory role of these proteins. Despite the identification of a large number of UBDs, structural details of integral UBL-binding domains are limited. In some cases, the intra- and intermolecular interactions between these UBLs with known UBDs such as UBA or the ubiquitin-interacting motif (UIM) have been demonstrated by solution NMR (35–38).
Here, we are reporting crystal structures of budding yeast Ufd2 in complex with the UBL domains of Rad23 and Dsk2 and the molecular details of their interaction interfaces. We identify a novel sequence pattern in the N-terminal UBL-binding region of budding yeast Ufd2, which is conserved in lower eukaryotes and is distinct from any known UBD identified so far. Moreover, despite engaging the same binding region, residue-specific differences exist in the interactions of the UBL domains of Rad23 and Dsk2 with Ufd2, which are coupled to subtle differences in their overall binding affinities. Mutational analyses of the binding surface of the UBL domains and a closer inspection of the thermodynamic contributions of those residues point to adaptive evolution as a factor shaping these interfaces.
EXPERIMENTAL PROCEDURES
Cloning, site-directed mutagenesis, protein expression, and purification are described in the supplemental Experimental Procedures.
Crystallization of Ufd2·Rad23-UBL and Ufd2·Dsk2-UBL
For crystallization of the Ufd2·Rad23-UBL and Ufd2·Dsk2-UBL complexes, Ufd2 was incubated with Rad23-UBL or Dsk2-UBL at a molar ratio of 1:1.5 (77 μm Ufd2 and 115.5 μm UBL) for 1 h at 4 °C in the presence of 2 mm dithiothreitol. Crystals were grown by vapor diffusion in hanging drops containing equal volumes of protein in 50 mm HEPES, pH 7.4, 150 mm NaCl, and 2 mm dithiothreitol and a reservoir solution consisting of 16–18% (w/v) polyethylene glycol 3500 and 200 mm K3-citrate, pH 8.3, equilibrated against the reservoir solution. Crystals were cryo-protected by soaking in mother liquor containing 15–20% (v/v) glycerol. They belong to space group P212121 with approximate cell dimensions of a = 65 Å, b = 126 Å, and c = 181 Å with one complex per asymmetric unit.
Data Collection and Structure Determination
Crystals were flash-cooled in liquid nitrogen, and data collection was performed at 100 K. Data were collected at beamlines ID14–4 (European Synchrotron Radiation Facility (ESRF), Grenoble, France) and BL 14.1 (Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung (BESSY), Berlin, Germany) and processed using Mosflm and Scala (39, 40). Data collection statistics are summarized in supplemental Table S1. For subsequent calculations, the CCP4 suite was utilized (41) with exceptions as indicated. The Ufd2 structure was solved by molecular replacement using Phaser (42) with Protein Data Bank (PDB) entry 2QIZ as search model. Because Phaser could not find a solution for the UBL domain with different search models, this domain was fitted manually into the electron density using human ubiquilin 3 (PDB entry 1YQB) for the Ufd2·Rad23-UBL complex and the Dsk2-UBL domain (PDB entry 2BWF) for the Ufd2·Dsk2-UBL complex as a model. The structures were refined with Phenix (43) and REFMAC5 incorporating translation, libration, screw-rotation (TLS) refinement in all cycles (44, 45). Solvent molecules were automatically added with Coot (46). The figures were produced with PyMOL (65).
In Vitro Binding Assays
For pulldown assays, GST-tagged Ufd2 and variants were immobilized on glutathione (GSH) beads. In all experiments, 20 μl of GSH beads were incubated with 0.95 μm purified Ufd2 in 400 μl of phosphate-buffered saline buffer with 1 mm dithiothreitol and 0.1% (v/v) Triton X-100 at 4 °C for 1 h. WT-Ufd2 and GST alone were included as controls. After centrifugation (1250 × g, 30 s), beads were washed five times with 400 μl of binding buffer. Purified UBL proteins (0.95 μm) in a total volume of 400 μl of binding buffer were added to immobilized Ufd2 and treated in the same way as in the first step. Immobilized proteins were analyzed by 17% (v/v) SDS-PAGE or by immunoblotting with an anti-His antibody.
Isothermal Titration Calorimetry (ITC)
Proteins were extensively dialyzed against phosphate-buffered saline buffer (pH 7.4, 1 mm β-mercaptoethanol) followed by degassing. In all experiments, 75–150 μm Rad23- and Dsk2-UBL proteins were titrated as the ligand into the sample cell containing 5–10 μm Ufd2. A volume of 10 μl of ligand was added at a time with a total number of 30 injections, resulting in a final molar ratio of ligand-to-protein varying between 3:1 and 4:1. All experiments were performed using a VP-ITC instrument (MicroCal, GE Healthcare) at 25 °C. Buffer-to-buffer, buffer-to-Ufd2, as well as Rad23-UBL/Dsk2-UBL-to-buffer titrations were performed as described above. Corrected data were analyzed with a single-site binding model using software supplied by the ITC manufacturer and non-linear least squares fitting to calculate the dissociation constant (Kd).
Surface Plasmon Resonance (SPR) Measurements
SPR binding assays were performed alternatively on BIAcore X or BIAcore T100 instruments (GE Healthcare) at 25 °C in 10 mm HEPES, pH 7.4, 150 mm NaCl, 50 μm EDTA, 1 mm β-mercaptoethanol, and 0.005% (v/v) Surfactant P20. 100 response units of His-tagged Rad23- or Dsk2-UBL were captured on a nickel-nitrilotriacetic acid (Ni-NTA) sensor chip. GST-tagged Ufd2 for comparative binding assays and untagged Ufd2 for affinity analysis were applied to the UBL surfaces in random duplicates at a flow rate of 50 μl/min. After each cycle, the surface was regenerated using 350 mm EDTA in running buffer to remove bound Ni2+ and captured proteins. The BIAcore T100 evaluation software was used to calculate the steady state affinity constants. Data were plotted using GraphPad Prism. For comparative assays, the relative binding responses of the mutants to WT proteins were determined by obtaining the maximum response for each interaction at the end of each injection.
RESULTS
Ufd2 Binds the UBL Domains of Rad23 and Dsk2 with High Affinity
Although Rad23 and Dsk2 interact with Ufd2 via their UBL domains (22, 26), yeast two hybrid assays could only identify the isolated N-terminal fragment (residues 1–380) of Ufd2 as its UBL-interacting region (26). Additional details regarding the Ufd2-UBL interactions have not been unraveled so far. To further characterize the interactions of Ufd2 with the UBLs of Rad23 and Dsk2, we performed GST pulldown assays with GST-tagged full-length Ufd2 and C-terminally His-tagged UBLs (Fig. 1A). Both UBLs were readily captured using immobilized GST-Ufd2. In contrast, the UBL domain of Ddi1, a third UBL-UBA protein, does not interact with Ufd2 (Fig. 1A) (22). The differential binding of the Rad23- and Dsk2-UBLs to the proteasomal subunits Rpn1 and Rpn10 has been described (25, 47, 48). Hence, we used SPR interaction analysis to search for quantitative differences in their interactions. Steady state affinity analysis of Ufd2 on both Rad23-UBL (Fig. 1B, left panel, and 1C) and Dsk2-UBL surfaces (Fig. 1B, right panel, and 1C) provided a Kd of 55 ± 3 nm for the interaction of Rad23-UBL and a lower affinity for Dsk2-UBL with a Kd of 418 ± 56 nm.
FIGURE 1.
Interactions of Ufd2 with the UBL domains of Rad23 and Dsk2. A, GST-Ufd2 immobilized on GSH-beads was tested for binding to C-terminally His-tagged UBLs of Rad23, Dsk2, and Ddi1. Captured UBLs were visualized by immunoblotting (WB) with an anti-His antibody. 2% of the input and GST beads incubated with UBLs were loaded as controls. B, a series of 2-fold Ufd2 dilutions (233–3.6 nm) was applied on a Rad23- or Dsk2-UBL surface for 120 s (left and right panel, respectively). RU, response units. C, SPR binding isotherms of WT-Rad23- and WT-Dsk2-UBL and the quintuple and septuple Dsk2-UBL variants with Ufd2. conc., concentration. D, ITC analysis of Ufd2·Rad23-UBL (closed circles) and Ufd2·Dsk2-UBL (open circles) complexes.
The binding of the UBLs of Rad23 and Dsk2 to Ufd2 was also analyzed by ITC to allow for a complete thermodynamic characterization (Fig. 1D). These studies resulted in a Kd of 70 ± 6 nm for the interaction of Rad23-UBL with Ufd2 and for the binding of Dsk2-UBL to Ufd2 in a 2–3-fold higher Kd of 175 ± 19 nm. Although there is an excellent agreement between SPR and ITC for the Rad23-Ufd2 interaction, the two methods show an ∼2-fold difference for the Dsk2-Ufd2 interaction. More importantly, the enthalpic and entropic components to the free energy are highly different between the two UBLs. The interaction of Rad23-UBL and Ufd2 is more exothermic (ΔH = −17.3 kcal/mol) when compared with Dsk2-UBL (ΔH = −10.1 kcal/mol). However, this is offset by a substantial decrease in entropy for Rad23-UBL (−TΔS = 7.4 kcal/mol), whereas the entropic contribution is minimal for the Dsk2-UBL interaction (−TΔS = 0.8 kcal/mol).
Crystal Structures of Ufd2 in Complex with Rad23- and Dsk2-UBL
We solved the structures of Ufd2 in complex with Rad23-UBL carrying either an N-terminal or a C-terminal His tag, which showed no significant structural differences. Due to better data quality, the structure of Ufd2 with a C-terminal His-tagged UBL is presented here. The Ufd2·Rad23-UBL complex was refined at 2.4 Å resolution to a crystallographic R-factor of 20.3% and a free R-factor of 25.7% (Table 1). As described previously (49), Ufd2 is composed of an N-terminal variable domain, a core domain, and a C-terminal U-box domain with a fold similar to that of RING (really interesting new gene) domains, which are present in certain Ub ligases (Fig. 2A). Despite some conformational variability of the U-box domain, our Ufd2 structure in the complex is quite similar (1.5 Å root mean square (r.m.s.) deviation for 954 Cα atoms) to the published Ufd2 structure (49).
TABLE 1.
Refinement statistics
| Ufd2·Rad23-UBL | Ufd2·Dsk2-UBL | |
|---|---|---|
| Resolution limit (Å) | 45.2-2.4 | 73.5-2.4 |
| No. of reflections | 56,268 | 55,087 |
| No. of protein/ligand/solvent atoms | 8303/17/298 | 8288/17/182 |
| Rcryst (Rfree)a,b | 0.203 (0.257) | 0.210 (0.270) |
| r.m.s. deviations in: | ||
| Bond lengths (Å) | 0.016 | 0.015 |
| Bond angles (°) | 1.711 | 1.610 |
| Estimated coordinate error (Å) | 0.25 | 0.26 |
| Overall average B-factor (Å2) | 25.7 | 42.9 |
| Ramachandran statistics (%)c | 93.1/97.9/2.1 | 93.8/98.4/1.6 |
a Rcryst = Σhkl‖Fo| − |Fc‖/Σhkl|Fo| where Fo and Fc are the observed and calculated structure factor amplitudes.
b Rfree, same as Rcryst for 5% of the data randomly omitted from the refinement. The estimated coordinate error is based on Rfree.
c Ramachandran statistics indicate the fraction of residues in the favored (98%), allowed (>99.8%), and disallowed regions of the Ramachandran diagram, as defined by MolProbity (64).
FIGURE 2.
Structure of Ufd2 in complex with the UBL domain of Rad23. A, ribbon representation of the overall structure of the Ufd2·Rad23-UBL complex. The Rad23-UBL domain is shown in green, the N-terminal Ufd2 region is in orange, the Ufd2 core domain is in gray, and the Ufd2 U-box domain is in red. B, close-up view of the N-terminal Ufd2 domain in complex with Rad23-UBL with secondary structural elements labeled and color-coded as in A.
The N-terminal variable region of Ufd2 that binds to the UBL domain consists of eight α-helices. Helices α1 to α4 are arranged in a four-helix bundle, whereas helices α5 and α6 interact with α3 and α4 through hydrophobic contacts that are partly mediated by their connecting loops (Fig. 2B). The structure of Rad23-UBL is comprised of a five-stranded β-sheet, one α-helix, and one 310-helix (Fig. 2B). It displays a high degree of similarity with Ub (PDB entry 1UBQ, 1.1 Å r.m.s. deviation for 72 Cα atoms, z-score 14, 25% sequence identity) and the UBL domain of hHR23A (PDB entry 1P98, 1.6 Å r.m.s. deviation for 72 Cα atoms, z-score 11.3, 26% sequence identity), one of the two human homologs of budding yeast Rad23.
Subsequently, we solved the structure of Ufd2 with Dsk2-UBL by molecular replacement. The UBL domain in the Ufd2·Dsk2-UBL complex exhibits increased flexibility; in particular, with a C-terminally tagged UBL domain, the first 30 amino acids of this domain were largely disordered (data not shown). With an N-terminally tagged protein, the Ufd2·Dsk2-UBL structure was refined at 2.4 Å resolution to a crystallographic R-factor of 21.0% and a free R-factor of 27.0% (Table 1). Both Rad23-UBL and Dsk2-UBL structures can be superimposed with an r.m.s. deviation of 1.1 Å for 71 aligned residues (z-score 13.6, 30% sequence identity).
Analysis of the Ufd2·Rad23-UBL Interface
The Ufd2·UBL interface in the structure of the complex buries a total molecular surface of about 1260 Å2, which is comprised to ∼590 Å2 of the molecular surface of Ufd2 (∼1.3% of the total surface area) and ∼670 Å2 from UBL (∼14.6% of the total surface area). This interface is composed of almost equal parts of non-polar residues (38%), polar residues (33%), and charged residues (29%); however, there are only one salt bridge (UBL-Lys-10 Nζ–Ufd2-Glu-49 Oϵ1 with a distance of 2.6 Å) and two direct hydrogen bonds (UBL-Ser-47 Oγ–Ufd2-Arg-92 Nη2, UBL-Gln-52 Nϵ2–Ufd2-Glu-141 O at distances of 2.3 and 3.2 Å, respectively) present (Fig. 3A).
FIGURE 3.
The Ufd2·Rad23-UBL interface. A, residues involved in binding are shown in stick representation. Carbon atoms of Ufd2 residues are colored in orange and in green for Rad23-UBL. Dashed lines indicate H-bonds. B, structure-based sequence alignment of Rad23-UBL, Dsk2-UBL, hHR23A-UBL, and Ub. Secondary structure elements of Rad23-UBL were assigned using DSSP (61) and are labeled above the sequences. The alignment was performed using DaliLite (62), and the figure was prepared with ESPript (63). Strictly conserved amino acids are highlighted with a red background, and similar amino acids are shown as red letters. The three Ufd2-binding segments are indicated. Residues involved in Ufd2·Rad23-UBL interaction are labeled with green stars. C, superposition of the Ufd2·Rad23-UBL/Dsk2-UBL complex structures with the N-terminal binding domain of Ufd2 in orange (Rad23 complex) and gray (Dsk2 complex), with Rad23-UBL in green and Dsk2-UbL in yellow.
Three UBL segments are contacting Ufd2 (Fig. 3A). Segment I is located in the loop connecting β-strands one and two, segment II involves β-strands three and four, and segment III is located in β-strand five. Ufd2 residues from helix α2 and α4 as well as the loop connecting α4 with α5 contribute to the Ufd2·UBL interface. These residues contact the hydrophobic surface of the UBL β-sheet in the region of β-strands 3, 4, and 5. Participating residues from Ufd2 include Leu-44, Tyr-97, Val-100, and Trp-107, which are located in the hydrophobic UBL pocket formed by residues Phe-9, Ile-45, Val-50, Val-69, and Met-71 of Rad23 (Fig. 3, A and B).
For comparison, the principal recognition determinants in Ub are: 1) the hydrophobic pocket formed by the side chains of Leu-8 (Phe-9 in Rad23), Ile-44 (Ile-45 in Rad23), His-68 (Val-69 in Rad23), and Val-70 (Met-71 in Rad23) and 2) the main chain amide group of Gly-47 (Gly-48 in Rad23), which is involved in hydrogen bonding (50). Although the hydrophobic patch of Rad23-UBL is also crucial for its interaction with Ufd2, the main chain of Gly-48 does not form a hydrogen bond. Instead, the β-turn (Ser47–Gly48) connecting β-strands 3 and 4 is stabilized by the aforementioned strong hydrogen bond between Ufd2-Arg-92 and UBL-Ser-47, whereas Ufd2-Gly-96 and Ufd2-Tyr-97 contact UBL-Gly-48 (Fig. 3A). The aromatic ring of Ufd2-Tyr-97 is involved in a stacking interaction with the peptide bond between UBL residues 47 and 48 in this β-turn.
Probing the Ufd2·Rad23-UBL Interface
The importance of interface residues was analyzed by mutagenesis experiments. Eleven residues from Ufd2 and nine from Rad23-UBL were each replaced with Ala. With the exception of the Rad23-UBL-G48A variant that showed a reduced expression, all Ufd2 and Rad23-UBL variants behaved like the WT protein during and after purification, indicating that they were correctly folded (data not shown). Initially, the contribution of these residues was studied by GST pulldown and comparative SPR binding assays (Table 2, supplemental Figs. S1 and S2A). In SPR studies, the relative binding responses of mutants to WT proteins were determined and compared. The majority of Rad23-UBL single mutants revealed reduced binding to Ufd2 with Rad23-UBL-I45A displaying the most prominent binding defect. The contribution of the remaining residues to the interaction is augmented in double mutants (supplemental Fig. S1C). Analysis of the Ufd2 variants by SPR showed a largely reduced binding of the residues located in the hydrophobic region of the UBL-binding pocket (Leu-44, Tyr-97, Val-100, and Phe-107) and Asp-40 (Table 2 and supplemental Fig. S2A).
TABLE 2.
ITC and SPR parameters of Ufd2, Rad23-UBL, Dsk2-UBL, and variants
= indicates no change; ND indicates not detected (corresponding to at least a 104-fold decrease in binding affinity).
| Ufd2 | WT-UBL | ITC |
SPRa (% of relative response) | |
|---|---|---|---|---|
| Kd | Fold decrease | |||
| nm | ||||
| WT | Rad23 | 70 | 100 | |
| Dsk2 | 175 | 100 | ||
| E26A | Rad23 | 284 | 4 | 91 |
| Dsk2 | 521 | 3 | 83 | |
| D40A | Rad23 | 7900 | 110 | 20 |
| Dsk2 | 7600 | 40 | 0 | |
| L44A | Rad23 | 8300 | 120 | 31 |
| Dsk2 | 463 | 3 | 52 | |
| T48A | Rad23 | 72 | = | 70 |
| Dsk2 | 296 | 2 | 29 | |
| E49A | Rad23 | 413 | 6 | 69 |
| Dsk2 | 314 | 2 | 44 | |
| R92A | Rad23 | 265 | 4 | 76 |
| Dsk2 | 128 | = | 59 | |
| G96A | Rad23 | 592 | 8 | 51 |
| Dsk2 | 216 | = | 60 | |
| Y97A | Rad23 | 134,000 | 1900 | 3 |
| Dsk2 | 83,000 | 470 | 0 | |
| V100A | Rad23 | ND | >10,000 | 9 |
| Dsk2 | 3900 | 22 | 1 | |
| I104A | Rad23 | 1600 | 20 | 43 |
| Dsk2 | 1100 | 6 | 12 | |
| F107A | Rad23 | ND | >10,000 | 11 |
| Dsk2 | 3600 | 20 | 0 | |
| Ufd2 | Rad23-UBL | ITC |
SPRa (% of relative response) | |
|---|---|---|---|---|
| Kd | Fold decrease | |||
| nm | ||||
| WT | F9A | 376 | 5 | 80 |
| WT | K10A | 162 | 2 | 96 |
| WT | I45A | 9100 | 130 | 17 |
| WT | S47A | 606 | 9 | 62 |
| WT | V50A | 441 | 6 | 88 |
| WT | Q52A | 415 | 6 | 79 |
| WT | Q67A | 113 | 2 | 92 |
| WT | V69A | 478 | 7 | 70 |
| WT | M71A | 221 | 3 | 88 |
a For comparative SPR assays, the relative binding responses of the mutants to wt proteins were determined by obtaining the maximum response for each interaction at the end of injection.
ITC studies confirmed these results and allowed for a quantitative analysis (Table 2, supplemental Fig. S3 and supplemental Table S2). The most significant effect for Ufd2 was observed for all residues located in the hydrophobic UBL pocket. Mutation of Val-100 and Phe-107 to Ala completely abolished binding, the Y97A variant strongly reduced binding (1900-fold), and the I104A and L44A variants showed significantly decreased affinities (20- and 120-fold, respectively). Although not directly involved in complex formation (Fig. 3A), the Ufd2-D40A variant showed a 110-fold reduced affinity (Table 2), which probably is the result of the missing intramolecular hydrogen bond between Ufd2-Asp-40 and Ufd2-Tyr-97 (Oδ2–OH 2.5 Å). This hydrogen bond seems to be crucial for proper positioning of the aromatic side chain of Tyr-97 in the interface region and might be important to align helices α2 and α4 for interaction with the Rad23-UBL.
In Rad23-UBL, Ile-45 was shown to be integral for binding to Ufd2 by the detrimental effect (130-fold decrease) after exchange to Ala (Table 2). Mutation of Phe-9, Val-50, and Val-69, residues adjacent to Ile-45 in the hydrophobic patch, also decreased the affinity of Rad23-UBL 5–7-fold. Ser-47, which is in hydrogen-bonding distance to Ufd2-Arg-92 and next to UBL-Gly-48, showed a 9-fold reduced affinity. In Ub and in the human Rad23 homolog hHR23A, Ser-47 is replaced by Ala. Charged residues found in the interface (Ufd2, Glu-26, Glu-49, and Arg-92; UBL, Lys-10) do not contribute significantly to the interaction. In summary, our data indicate that the most prominent contact between Ufd2 and Rad23-UBL is the strong hydrophobic interaction between UBL-Ile-45 and Ufd2-Val-100 as well as Ufd2-Phe-107, which defines the core of the UBL-interacting region of Ufd2.
Molecular Discrimination between Rad23 and Dsk2
Despite a similar fold, the UBL domains of Rad23 and Dsk2 display only 30% sequence identity, which could give rise to differences in their interactions. A superposition of the bound Rad23-UBL and Dsk2-UBL in the two complex structures showed significant changes (Fig. 3C). Of the three UBL segments involved in the Ufd2 interaction (Fig. 3A), segment II including Ile-45 (Ile-44 in Ub) is highly conserved, and there are no conformational changes in both UBL structures, whereas segments I and III are not conserved and display structural changes (Fig. 3C). The loop, connecting β-strands one and two, adopts different conformations, and β-strand five shows a displacement that might affect binding (Fig. 3C).
Segment I includes Phe-9 in Rad23-UBL, corresponding to Leu-8 in Ub, where this residue is also involved in Ub recognition by UBDs (50, 51). Phe-9 is replaced by Gly-10 in Dsk2-UBL, and there is no corresponding hydrophobic interacting residue (supplemental Fig. S4A). Dsk2 residues Gly-10 and Gln-11 adopt different conformations when compared with Leu-8/Thr-9 of Ub and Phe-9/Lys-10 of Rad23-UBL. In the Ufd2·Dsk2-UBL structure, the Ufd2·Rad23-UBL salt bridge (Lys-10/Glu-49) is missing due to the Lys-10 to Gln-11 exchange, with the latter side chain no longer being located in the protein interface (supplemental Fig. S4A). The missing interaction from segment I in Dsk2 might be compensated by the displacement of β-strand five toward Ufd2 and a replacement of Val-69 to His-69 found in segment III resulting in a more pronounced interaction in this region when compared with Rad23-UBL (supplemental Fig. S4A). The presence of the salt bridge seems to be the reason for the more exothermic character of the Ufd2·Rad23-UBL interaction, a view that is also supported by the corresponding Ufd2-E49A and Rad23-K10A variants, which both display binding enthalpies similar to the Ufd2·Dsk2-UBL complex (supplemental Table S2). At the same time, the absence of the salt bridge in both mutants is accompanied by a more favorable entropic contribution, which is on a level similar to the Ufd2·Dsk2-UBL complex.
To identify residues important for the subtle molecular discrimination between the UBL domains of Rad23 and Dsk2, the interaction of Dsk2-UBL with Ufd2 mutants was analyzed by GST pulldown assays (data not shown), SPR, and ITC (Table 2, supplemental Figs. S2B and S3C). Quantitative ITC analysis showed reduced binding of Dsk2 to Ufd2 mutants Y97A (470-fold), V100A (22-fold), I104A (6-fold), and F107A (20-fold) (Table 2). However, binding of the V100A and F107A variants is not completely abolished, and when compared with Rad23-UBL, the binding affinities are less affected by a factor of about 3–7 in most of the mutants analyzed. In addition, the L44A mutant, which has a 120-fold reduced affinity with Rad23-UBL, is only three times reduced in the case of Dsk2-UBL.
In general agreement with the ITC affinity data, the comparative SPR binding assay revealed significant differences in the association of Ufd2 variants Y97A, V100A, I104A, and F107A with Rad23- and Dsk2-UBL surfaces (supplemental Fig. S4B). The observed SPR decrease for the binding of the T48A and E49A variants of Ufd2 to Dsk2-UBL seems to be compensated by slower dissociations, thus explaining why these mutants show no significant defect in the ITC analysis.
To further analyze the contribution of segments I and III to complex formation, a G10F/Q11K/S67Q/H69V/V71M quintuple Dsk2-UBL mutant was generated, where key residues in binding segments I and III were replaced with the corresponding residues from Rad23-UBL. Comparative binding as well as steady state affinity analysis by SPR revealed only a small increase (Kd = 348 nm) in binding affinity for Ufd2 when compared with WT-Dsk2-UBL (Kd = 418 nm) (data not shown and Fig. 1C). In addition, neither a crystal structure of the quintuple Ufd2·Dsk2-UBL complex (data not shown) nor the KD of 240 nm deduced by ITC revealed significant differences from WT-Dsk2-UBL (Kd = 175 nm). The ITC analysis did, however, reveal that the binding is now driven by an increase in entropy (−TΔS = −6.5 kcal/mol versus 0.8 and 7.4 kcal/mol for WT-Dsk2-UBL and -Rad23-UBL, respectively), whereas the binding enthalpy is reduced to only −2.5 kcal/mol when compared with −10.1 and −17.3 kcal/mol (supplemental Table S2). Interestingly, SPR and ITC analysis of a G10F/Q11K/I50V/K52Q/S67Q/H69V/V71M septuple Dsk2-UBL mutant, which has the additional I50V and K52Q substitutions in segment II, showed an even lower affinity (SPR, Kd = 648 nm; ITC, Kd = 875 nm) to Ufd2 when compared with WT-Dsk2-UBL (Fig. 1C).
The N Terminus of Ufd2 Represents a Unique and Conserved UBL-binding Domain
A multiple sequence alignment of Ufd2 from different yeast species displays a distinct pattern of conserved residues involved in UBL binding (Fig. 4A). Among the available yeast genomes, the Schizosaccharomyces pombe sequence is most similar to those from higher eukaryotes; thus we isolated cDNA fragments for the coding region of the UBL domains of Rad23 and Dsk2 and full-length Ufd2 from this organism and examined their interactions by GST pulldown assays (Fig. 4B) as well as SPR (data not shown). We could show that SpUfd2 interacts strongly with the UBL domains of SpRad23 and SpDsk2 as well as with the UBL domains of ScRad23 and ScDsk2 and vice versa. This cross species interaction, despite the diversified UBL and Ufd2 amino acid sequences, indicates that the identified sequence pattern represents a real UBL-interacting domain. A surface representation of this motif is shown in Fig. 4C.
FIGURE 4.
The N terminus of Ufd2 represents a conserved UBL-interacting domain in lower eukaryotes. A, alignment of the N-terminal sequences of fungal Ufd2s. Invariant or conserved residues with surface access are colored in dark blue, buried ones are in light blue. Residues labeled with red stars represent the core region of the binding domain, which is essential for UBL interaction, whereas residues labeled with yellow stars contribute moderately to the interaction. K. lactis, Kluyveromyces lactis; C. glabrata, Candida glabrata; Z. rouxii, Zygosaccharomyces rouxii; L. thermotolerans, Lachancea thermotolerans; C. tropicalis, Candida tropicalis; C. dubliniensis, Candida dubliniensis; P. guilliermondii, Pichia guilliermondii; D. hansenii, Debaryomyces hansenii. B, GST pulldown assay demonstrates the cross interactions of S. pombe and S. cerevisiae proteins. 5% of inputs and GST beads incubated with UBLs were loaded as controls. WB, Western blot. C, surface representation of the N-terminal UBL-binding domain of Ufd2, color-coded as in A.
The N terminus of budding yeast Ufd2 displays only limited sequence homology with the human Ufd2s, E4A and E4B (supplemental Fig. S5) and other Ufd2s from higher eukaryotes. In agreement with this finding, there are no reports that hHR23A/B interacts with either of the human homologs of Ufd2. Interestingly, our SPR studies showed that the UBL domain of hHR23A interacts with ScUfd2, albeit with lower affinity (data not shown). Apparently, the high affinity interaction of the UBL domains of Rad23 and Dsk2 has been lost during the evolution of this domain. The absence of conservation of the Ufd2-UBL interface could potentially be used for therapeutic interventions against pathogenic yeasts such as Candida albicans by designing low molecular weight compounds that disrupt this interface. However, further functional studies in pathogenic yeasts are required to examine the suitability of this surface as a drug target.
DISCUSSION
Families and superfamilies of protein domains or folds have evolved through a process of homologous recombination and gene duplication (52) followed by sequence divergence. Members of different classes of UBDs such as UBA or UBL domains result from these processes. For instance, the UBL domains of Rad23 and Dsk2 display only 30% sequence identity but adopt the same fold and utilize the same binding surface to recognize a common UBL-binding domain of Ufd2 to form complexes that display similarly high affinity. Nevertheless, not all interacting residues are conserved; in particular, there is sequence diversity in binding segments I and III of UBLs. Our attempts to interconvert the UBL domains by altering non-conserved interfacial residues were not successful, thus suggesting that additional elements exist and play a role in the respective Ufd2-UBL interaction. Interestingly, these results resemble earlier studies on WW domains (53, 54), where a statistical analysis of multiple sequence alignments was utilized to identify co-evolving residues. The authors demonstrated that not only interfacial residues but also buried residues distal to the interface co-evolved with interfacial residues and contribute significantly to the interactions. They concluded that certain sequence patterns in interacting domains are due to adaptive evolution. In agreement with these findings, our data prove that substitution of key interfacial residues of Dsk2-UBL has no significant effect on its overall binding affinity to Ufd2. In case of the septuple mutant, we even observed a decrease in binding affinity, which could be due to the imposed disorder into the evolutionary inter-residue relations within the UBL fold. This is supported by the fact that when compared with Dsk2-UBL and in particular Rad23-UBL, the binding of the quintuple Dsk2-UBL mutant is driven strongly by entropy. These findings indicate that binding interfaces can be modulated by changes in residues that affect either the binding enthalpy or the entropy, thus providing additional freedom to maintain an interaction during the course of evolution, an effect that has been described previously as entropy/enthalpy compensation (55, 56).
Our studies suggest that UBL domains have co-evolved with Ufd2 to reach optimal binding affinities by altering specific residue-to-residue interactions (co-evolution at the residue level) (57), while at the same time, all functional aspects of Rad23 or Dsk2 are preserved. Therefore, the primary sequence degeneracy of protein domains such as UBAs or UBLs has been tolerated and established in favor of the cooperative nature of the interactions and their functionality within their respective protein complexes. This further suggests that differential binding properties observed for the interactions of Ufd2 with UBLs (this study) or for the interactions of UBAs with Ub and Ub chains (14) can arise not necessarily due to their interaction with different ligands but can also result from the adaptive co-evolution of these domains with the same interacting partners. Seemingly, these interfacial domains have evolved to hold protein-protein interactions in a suitable form within multicomponent complexes until they are challenged by downstream events.
Numerous structures of Ub receptors in complex with their respective Ub/UBL-binding domains have been reported. The so far characterized Ub receptors of the 26 S proteasome in budding yeast encompass the two proteasomal subunits Rpn10 (S5a in humans) and Rpn13 and the three UBL-UBA proteins Rad23, Dsk2, and Ddi1, which associate with the proteasome and function as shuttle factors (21). Experimental evidence for the existence of additional candidates exist (21, 58). Rad23 and Dsk2 interact with the proteasomal subunit Rpn1 via their UBL domains (21, 47). Aside from their known interactions with Ub, Rpn13 and Rpn10/S5a alternatively interact with UBL-UBA proteins (21, 35, 37, 38, 48, 51, 59). For instance, the preferential association of Rpn1 with Rad23 and Rpn10 with Dsk2 has been reported (25, 38, 47, 48). Based on the binding of hpLIC2 (Dsk2 homolog) with Rpn13, an interaction of Dsk2 with Rpn13 has been proposed (51, 59).
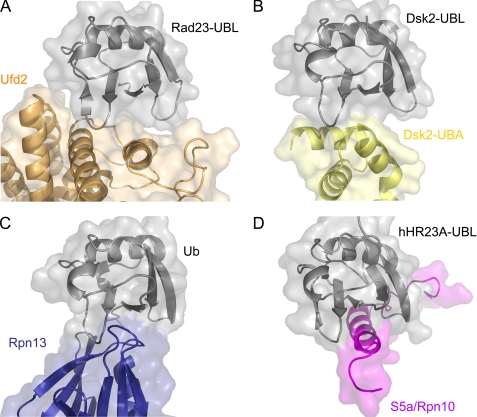
Although the aforementioned examples engage essentially the same surface of Ub/UBL, they diverge in both structure and patterns of Ub/UBL recognition (Fig. 5). For instance, hRpn10/S5a recognizes the UBL domain of hHR23A, one of the two human homologs of Rad23, via a Ub-interacting motif, which consists of a single α-helix (35, 37). Rpn13 binds Ub via a pleckstrin homology domain, which is a seven-stranded β-sandwich capped by an α-helix (51). The Ub-binding surface of Rpn13 is formed by three loops that bridge β-strands. Another Ub-binding element is the UBA domain found for example in Dsk2 (60). The UBA domain is composed of a three-helix bundle. With the exception of Rpn13, which exclusively binds via loops, it seems that the majority of Ub/UBL-binding domains fold into α-helical structures including the known UBDs, UIM, and UBA, and the UBL-binding domain of Ufd2 identified in this study. Despite the predominant interaction involving α-helices as Ub/UBL-binding elements, the three-dimensional structure of the UBL-binding domain of Ufd2 differs from other known examples, hence providing the first structural description for how Ufd2 acts as a UBL receptor while at the same time further enhancing the diversity of UBDs in general.
FIGURE 5.
Mode of Ub/UBL recognition by different Ub/UBL-binding domains. In each panel, ribbon representations together with the molecular surfaces of both binding partners are shown with Ub/UBL in the same orientation. A, Ufd2·Rad23-UBL. B, Dsk2-UBA·Dsk2-UBL (PDB entry 2BWE). C, Rpn13·Ub (PDB entry 2Z59). D, S5a/Rpn10·hHR23A-UBL (PDB entry 1P9D).
Supplementary Material
Acknowledgments
We thank Martin Scheffner and Keith Wilkinson for critical reading of the manuscript. We thank Stefan Jentsch for providing the original plasmids for the expression of Rad23, Dsk2, and Ufd2 and for Ufd2-specific antibodies used in the initial phase of this study. We also thank David Fischer and Rodrigo Villaseñor for the contribution to this study and Sven Eiselein for providing us with C-terminal GST-tagging plasmid.
This work was supported by Deutsche Forschungsgemeinschaft Grant RA1643/2-1 (to S. R.) and Rudolf Virchow Center for Experimental Biomedicine Grant FZ 82 (to H. S.).
The atomic coordinates and structure factors (codes 3M62 and 3M63) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Tables S1 and S2, and Figs. S1–S5.
- UBL
- ubiquitin-like
- UBA
- ubiquitin-associated
- UIM
- ubiquitin-interacting motif
- UBD
- ubiquitin-binding or ubiquitin-like binding domain
- UFD
- ubiquitin fusion degradation
- Ub
- ubiquitin
- GST
- glutathione S-transferase
- ITC
- isothermal titration calorimetry
- SPR
- surface plasmon resonance
- r.m.s.
- root mean square
- WT
- wild type
- PDB
- Protein Data Bank
- h
- human
- Sc
- S. cerevisiae
- Sp
- S. pombe.
REFERENCES
- 1.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2.Ross C. A., Pickart C. M. (2004) Trends Cell Biol. 14, 703–711 [DOI] [PubMed] [Google Scholar]
- 3.Haglund K., Dikic I. (2005) EMBO J. 24, 3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukhopadhyay D., Riezman H. (2007) Science 315, 201–205 [DOI] [PubMed] [Google Scholar]
- 5.Grabbe C., Dikic I. (2009) Chem. Rev. 109, 1481–1494 [DOI] [PubMed] [Google Scholar]
- 6.Buchberger A. (2002) Trends Cell Biol. 12, 216–221 [DOI] [PubMed] [Google Scholar]
- 7.Hicke L., Schubert H. L., Hill C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 8.Hurley J. H., Lee S., Prag G. (2006) Biochem. J. 399, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harper J. W., Schulman B. A. (2006) Cell 124, 1133–1136 [DOI] [PubMed] [Google Scholar]
- 10.Hochstrasser M. (2009) Nature 458, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dikic I., Wakatsuki S., Walters K. J. (2009) Nat. Rev. Mol. Cell Biol. 10, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofmann K. (2009) DNA Repair 8, 544–556 [DOI] [PubMed] [Google Scholar]
- 13.Hofmann K., Bucher P. (1996) Trends Biochem. Sci. 21, 172–173 [PubMed] [Google Scholar]
- 14.Raasi S., Varadan R., Fushman D., Pickart C. M. (2005) Nat. Struct. Mol. Biol. 12, 708–714 [DOI] [PubMed] [Google Scholar]
- 15.Varadan R., Assfalg M., Raasi S., Pickart C., Fushman D. (2005) Mol. Cell 18, 687–698 [DOI] [PubMed] [Google Scholar]
- 16.Rahighi S., Ikeda F., Kawasaki M., Akutsu M., Suzuki N., Kato R., Kensche T., Uejima T., Bloor S., Komander D., Randow F., Wakatsuki S., Dikic I. (2009) Cell 136, 1098–1109 [DOI] [PubMed] [Google Scholar]
- 17.Lo Y. C., Lin S. C., Rospigliosi C. C., Conze D. B., Wu C. J., Ashwell J. D., Eliezer D., Wu H. (2009) Mol. Cell 33, 602–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato Y., Yoshikawa A., Mimura H., Yamashita M., Yamagata A., Fukai S. (2009) EMBO J. 28, 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato Y., Yoshikawa A., Yamashita M., Yamagata A., Fukai S. (2009) EMBO J. 28, 3903–3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulathu Y., Akutsu M., Bremm A., Hofmann K., Komander D. (2009) Nat. Struct. Mol. Biol. 16, 1328–1330 [DOI] [PubMed] [Google Scholar]
- 21.Finley D. (2009) Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim I., Mi K., Rao H. (2004) Mol. Biol. Cell 15, 3357–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara T., Kamura T., Kotoshiba S., Takahashi H., Fujiwara K., Onoyama I., Shirakawa M., Mizushima N., Nakayama K. I. (2005) Mol. Cell. Biol. 25, 9292–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivantsiv Y., Kaplun L., Tzirkin-Goldin R., Shabek N., Raveh D. (2006) Mol. Cell. Biol. 26, 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishii T., Funakoshi M., Kobayashi H. (2006) EMBO J. 25, 5492–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richly H., Rape M., Braun S., Rumpf S., Hoege C., Jentsch S. (2005) Cell 120, 73–84 [DOI] [PubMed] [Google Scholar]
- 27.Hoppe T. (2005) Trends Biochem. Sci. 30, 183–187 [DOI] [PubMed] [Google Scholar]
- 28.Rape M., Jentsch S. (2004) Biochim. Biophys. Acta 1695, 209–213 [DOI] [PubMed] [Google Scholar]
- 29.Medicherla B., Kostova Z., Schaefer A., Wolf D. H. (2004) EMBO Rep. 5, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raasi S., Wolf D. H. (2007) Semin. Cell Dev. Biol. 18, 780–791 [DOI] [PubMed] [Google Scholar]
- 31.Verma R., Oania R., Graumann J., Deshaies R. J. (2004) Cell 118, 99–110 [DOI] [PubMed] [Google Scholar]
- 32.Liu C., van Dyk D., Li Y., Andrews B., Rao H. (2009) BMC Biol. 7, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raasi S., Pickart C. M. (2003) J. Biol. Chem. 278, 8951–8959 [DOI] [PubMed] [Google Scholar]
- 34.Hartmann-Petersen R., Hendil K. B., Gordon C. (2003) FEBS Lett. 535, 77–81 [DOI] [PubMed] [Google Scholar]
- 35.Walters K. J., Kleijnen M. F., Goh A. M., Wagner G., Howley P. M. (2002) Biochemistry 41, 1767–1777 [DOI] [PubMed] [Google Scholar]
- 36.Walters K. J., Lech P. J., Goh A. M., Wang Q., Howley P. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12694–12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller T. D., Feigon J. (2003) EMBO J. 22, 4634–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang D., Chen T., Ziv I., Rosenzweig R., Matiuhin Y., Bronner V., Glickman M. H., Fushman D. (2009) Mol Cell. 36, 1018–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leslie A. G. W. (1992) Joint CCP4+ESF-EAMCB Newsletter on Protein Crystallography, Vol. 26, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 40.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 41.Bailey S. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 42.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 44.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 45.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 46.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 47.Elsasser S., Gali R. R., Schwickart M., Larsen C. N., Leggett D. S., Müller B., Feng M. T., Tübing F., Dittmar G. A., Finley D. (2002) Nat. Cell Biol. 4, 725–730 [DOI] [PubMed] [Google Scholar]
- 48.Matiuhin Y., Kirkpatrick D. S., Ziv I., Kim W., Dakshinamurthy A., Kleifeld O., Gygi S. P., Reis N., Glickman M. H. (2008) Mol. Cell. 32, 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tu D., Li W., Ye Y., Brunger A. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15599–15606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohno A., Jee J., Fujiwara K., Tenno T., Goda N., Tochio H., Kobayashi H., Hiroaki H., Shirakawa M. (2005) Structure 13, 521–532 [DOI] [PubMed] [Google Scholar]
- 51.Schreiner P., Chen X., Husnjak K., Randles L., Zhang N., Elsasser S., Finley D., Dikic I., Walters K. J., Groll M. (2008) Nature 453, 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Te Velthuis A. J., Bagowski C. P. (2008) Curr. Genomics. 9, 88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Russ W. P., Lowery D. M., Mishra P., Yaffe M. B., Ranganathan R. (2005) Nature 437, 579–583 [DOI] [PubMed] [Google Scholar]
- 54.Socolich M., Lockless S. W., Russ W. P., Lee H., Gardner K. H., Ranganathan R. (2005) Nature 437, 512–518 [DOI] [PubMed] [Google Scholar]
- 55.Reyes-Turcu F. E., Shanks J. R., Komander D., Wilkinson K. D. (2008) J. Biol. Chem. 283, 19581–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunter C. A., Tomas S. (2003) Chem. Biol. 10, 1023–1032 [DOI] [PubMed] [Google Scholar]
- 57.Pazos F., Valencia A. (2008) EMBO J. 27, 2648–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lam Y. A., Lawson T. G., Velayutham M., Zweier J. L., Pickart C. M. (2002) Nature 416, 763–767 [DOI] [PubMed] [Google Scholar]
- 59.Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., Dikic I. (2008) Nature 453, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lowe E. D., Hasan N., Trempe J. F., Fonso L., Noble M. E., Endicott J. A., Johnson L. N., Brown N. R. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 177–188 [DOI] [PubMed] [Google Scholar]
- 61.Kabsch W., Sander C. (1983) Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 62.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 64.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.