Summary
Bacterial survival depends on the ability to switch between sessile and motile life styles in response to changing environmental conditions. In many species, this switch is governed by (3′-5′)-cyclic-diguanosine monophosphate (c-di-GMP) a signalling molecule, which is metabolized by proteins containing GGDEF and/or EAL domains. Salmonella Typhimurium contains 20 such proteins. Here, we show that the RNA-binding protein CsrA regulates the expression of eight genes encoding GGDEF, GGDEF-EAL and EAL domain proteins. CsrA bound directly to the mRNA leaders of five of these genes, suggesting that it may regulate these genes post-transcriptionally. The c-di-GMP specific phosphodiesterase STM3611, which reciprocally controls flagella function and production of biofilm matrix components, was regulated by CsrA binding to the mRNA, but was also indirectly regulated by CsrA through the FlhDC/FliA flagella cascade and STM1344. STM1344 is an unconventional (c-di-GMP-inactive) EAL domain protein, recently identified as a negative regulator of flagella gene expression. Here, we demonstrate that CsrA directly downregulates expression of STM1344, which in turn regulates STM3611 through fliA and thus reciprocally controls motility and biofilm factors. Altogether, our data reveal that the concerted and complex regulation of several genes encoding GGDEF/EAL domain proteins allows CsrA to control the motility-sessility switch in S. Typhimurium at multiple levels.
Introduction
The survival of bacteria in diverse environments largely depends on their ability to adjust their life style according to the surrounding conditions. An important factor that mediates the choice of an appropriate life style in various bacteria is the signalling molecule (3′-5′)-cyclic-diguanosine monophosphate (c-di-GMP) (recent reviews by Hengge, 2009; Jonas et al., 2009; Romling and Simm, 2009). In general, high intracellular levels of this second messenger promote sedentary biofilm-associated phenotypes, whereas low concentrations of c-di-GMP favour motility. In several bacteria, c-di-GMP has also been associated with the regulation of virulence and other phenotypes (Cotter and Stibitz, 2007; Tamayo et al., 2007). c-di-GMP is synthesized by diguanylate cyclases (DGCs), which contain catalytically active GGDEF domains, and degradation of the second messenger is mediated by phosphodiesterases (PDE), which harbour either EAL or HD-GYP domains (Paul et al., 2004; Simm et al., 2004; Christen et al., 2005; Ryjenkov et al., 2005; Schmidt et al., 2005; Ryan et al., 2006). Notably, individual bacterial genomes frequently encode numerous GGDEF and EAL/HD-GYP proteins (Galperin et al., 2001; Galperin, 2004), implying that the c-di-GMP network is a highly complex and tightly regulated intracellular signalling system (Jonas et al., 2009).
The ability to switch between different life styles is crucial for bacteria such as Salmonella that alternate between distinct niches in host organisms during infection as well as in the abiotic environment, in which they can persist for weeks in food, soil, water and other habitats. Salmonella enterica serovar Typhimurium contains a c-di-GMP system comprised of 5 GGDEF domain proteins, 7 GGDEF-EAL and 8 EAL domain proteins (Romling, 2005). The characterization of the phenotypes of these proteins has been the subject of several previous studies (Garcia et al., 2004; Kader et al., 2006; Simm et al., 2007; Solano et al., 2009). However, little is known about the regulation of the Salmonella c-di-GMP system by intra- and extracellular factors.
In the closely related species Escherichia coli, the carbon storage regulator CsrA controls the expression of at least seven of 29 genes encoding GGDEF/EAL domain proteins at the post-transcriptional level (Jonas et al., 2008). Members of the CsrA (RsmA) family are homodimeric RNA-binding proteins (Mercante et al., 2009) that are widely distributed among eubacteria and control various phenotypes including biofilm formation, motility, carbon flux, secondary metabolism, quorum sensing as well as interactions with animal and plant hosts (Romeo, 1998; Babitzke and Romeo, 2007; Lapouge et al., 2008; Lucchetti-Miganeh et al., 2008). In γ-proteobacteria CsrA proteins are antagonized by small non-coding RNAs (sRNAs) that bind and sequester multiple copies of CsrA (Babitzke and Romeo, 2007; Lapouge et al., 2008). Transcription of the sRNAs is controlled by a two-component system (BarA-UvrY in E. coli, BarA-SirA in S. Typhimurium), permitting the integration of environmental signals into the Csr system (Babitzke and Romeo, 2007; Lapouge et al., 2008).
By directly binding to target mRNAs CsrA can either down- or upregulate the expression of target genes (Babitzke and Romeo, 2007). CsrA binding to the mRNAs of the GGDEF proteins YcdT and YdeH in E. coli led to a strong downregulation in transcript levels (Jonas et al., 2008). Both proteins encode DGCs, which inhibit motility. YdeH is also involved in the positive regulation of biofilm formation (Boehm et al., 2009). Thus, by regulating the expression of these GGDEF domain proteins CsrA controls motility and biofilm behaviour in a c-di-GMP dependent manner. In addition, CsrA enhances motility and reciprocally inhibits biofilm formation in E. coli by binding to and stabilizing the transcript of the flagella master regulator FlhDC (Wei et al., 2001) and by blocking translation of pgaA and destabilizing pgaABCD mRNA (Wang et al., 2005), which encodes the synthesis and secretion apparatus of the PGA (poly-β-1,6-N-acetylglucosamine) biofilm polysaccharide adhesin (Itoh et al., 2008).
In contrast to E. coli, the role of CsrA in the regulation of the sessility-motility switch is less well understood in Salmonella. S. Typhimurium has a CsrA orthologue that is identical in its amino acid sequence to CsrA in E. coli. However, S. Typhimurium contains neither the pga operon nor orthologues of ycdT, ydeH and most of the other GGDEF/EAL genes that are regulated by CsrA in E. coli. Although CsrA regulates genes belonging to the flagella synthesis cascade as well as genes required for virulence (Altier et al., 2000a; Lawhon et al., 2003), no direct mRNA targets have been identified in Salmonella. Here, we show that CsrA controls the expression of eight genes encoding GGDEF/EAL domain proteins in S. Typhimurium by both direct and indirect mechanisms. The complex regulation of these genes enables CsrA to act at multiple levels in the signalling hierarchy mediating the switch between a motile and a sessile life style in S. Typhimurium.
Results
CsrA regulates steady state levels of transcripts encoding GGDEF, GGDEF-EAL and EAL domain proteins in S. Typhimurium
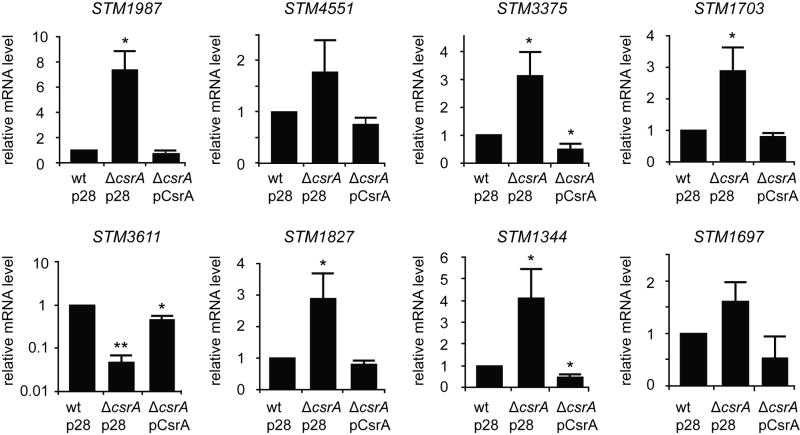
CsrA regulates GGDEF/EAL domain proteins in E. coli by affecting their mRNA steady state levels (Jonas et al., 2008). Since S. Typhimurium lacks orthologues of most of the CsrA regulated GGDEF/EAL genes in E. coli, we hypothesised that CsrA might control the expression of other GGDEF/EAL domain proteins in Salmonella. To identify such genes in S. Typhimurium UMR1, we analysed the effect of a csrA mutation on the mRNA steady state levels of all 20 genes that encode GGDEF, EAL or GGDEF-EAL domain proteins by quantitative Real-Time RT-PCR. This systematic screen led to the identification of eight genes whose mRNA levels were altered in the csrA mutant MAE125 compared to the wild type, when grown as liquid cultures at 37 °C to early stationary phase. Among these were two genes encoding GGDEF proteins (STM1987, STM4551), two genes encoding GGDEF-EAL domain proteins (STM1703, STM3375) as well as four genes encoding the EAL domain proteins STM3611, STM1827, STM1344 and STM1697 (Table 1). These genes have been phenotypically characterized in previous studies and most of them have been assigned to functions in the regulation of biofilm formation, motility and virulence (Table 1). Our data show that the csrA mutation caused a significant increase (3 to 7 fold) in the mRNA levels of STM1987, STM1703, STM3375, STM1827 and STM1344 (Fig. 1). Also the transcript levels of STM4551 and STM1697 were elevated in the csrA mutant compared to the wild type, albeit less strongly (1.6 to 1.8 fold). STM3611 (also known as yhjH), encoding an active PDE (Simm et al., 2004), was the only gene which showed a drastic decrease in mRNA steady state levels (>10 fold) in the csrA mutant. Ectopic csrA expression from pBADcsrA (pCsrA) fully complemented the effect of the csrA mutation for all genes that were upregulated in the csrA mutant. Expression of STM3611 was partially restored (to approx. 45 % of wt mRNA levels) by pCsrA. Altogether, these data demonstrate that CsrA down- or upregulates the mRNA steady state level of STM1987, STM4551, STM1703, STM3375, STM3611, STM1827, STM1344 and STM1697 in S. Typhimurium. Although we did not observe significant changes in the mRNA levels for the remaining 12 genes encoding GGDEF, GGDEF-EAL or EAL domain proteins in Salmonella (data not shown), we cannot rule out that CsrA affects the expression of these genes under different growth conditions or that potential changes in the half-lives of these mRNAs in the csrA mutant are compensated by unknown homeostatic mechanisms.
Table 1.
GGDEF/EAL proteins regulated by CsrA.
| Protein (synonym) | Regulation by CsrA | Motif | Enzymatic activity | Reported phenotype | Reference |
|---|---|---|---|---|---|
| STM1987 | negative, direct1 | GGEEF | DGC | Upregulation of cellulose production | (Garcia et al., 2004; Solano et al., 2009) |
| STM4551 | negative, indirect1? | GGDEF | DGC | Functions in motility, rdar morphotype expression, virulence | (Solano et al., 2009) |
| STM3375 (YhdA, CsrD) | negative, direct | HRSDF, ELM | no DGC, no PDE (in E. coli) | Regulation of sRNAs (in E. coli), upregulation of motility | (Suzuki et al., 2006; Simm et al., 2007) |
| STM1703 (YciR) | negative, indirect? | GGDEF, EAL | putative PDE, DGC activity not known | Downregulation of the rdar morphotype | (Garcia et al., 2004; Simm et al., 2007) |
| STM3611 (YhjH) | positive, direct and indirect | ELL | PDE | Upregulation of motility, downregulation of the rdar morphotype | (Simm et al., 2004; Simm et al., 2007) |
| STM1827 | negative, indirect? | EAL | putative PDE | Downregulation of the rdar morphotype | (Simm et al., 2007) |
| STM1344 (YdiV) | negative, direct | EII | no PDE | Downregulation of motility, upregulation of the rdar morphotype, role in virulence | (Hisert et al., 2005; Simm et al., 2007; Wozniak et al., 2008; Simm et al., 2009) |
| STM1697 | negative, direct | EIT | probably no PDE | Role in virulence (?) | (Lamprokostopoulou and Römling, unpublished) |
direct vs indirect regulation inferred from RNA-binding by CsrA to the respective transcripts as observed by gel-mobility shift assays.
Figure 1.
CsrA regulates the mRNA steady state level of genes encoding GGDEF/EAL domain proteins in S. Typhimurium. Relative mRNA levels of STM1987, STM4551, STM3375, STM1703, STM3611, STM1827, STM1344 and STM1697 were measured by quantitative Real-Time RT PCR in the wild type UMR1 (wt) and in the csrA::kan mutant MAE125 (ΔcsrA), carrying the empty vector pBAD28 (p28) or the CsrA vector pBADcsrA (pCsrA), respectively. Note that the data for STM3611 are displayed with a logarithmic scale. Total RNA was isolated from bacterial cultures grown at 37 °C in LB medium with 0.1 % arabinose to OD600 1.5. The data values represent means with standard deviations (** P < 0.01; * P < 0.05).
Potential CsrA binding sites in the 5′ untranslated leader sequences
CsrA is known to regulate gene expression by binding to specific sites in the 5′ untranslated regions (UTRs) of mRNA targets and thereby to affect mRNA steady state levels (Wei et al., 2001; Wang et al., 2005). Previous studies determined an optimal site [5′-(A/U)CA-GGA-G(U/A)-3′] for high affinity CsrA/RsmA binding (Valverde et al., 2004; Dubey et al., 2005; Schubert et al., 2007). However, the key feature that is recognized by CsrA in E. coli RNAs (pgaA, cstA, glgC, hfq, ydeH, ycdT, sepL, grlR, csrB and csrC) is the trinucleotide motif GGA, which is preferentially but not necessarily present in the loops of hairpin structures (Liu et al., 1997; Baker et al., 2002; Dubey et al., 2003; Weilbacher et al., 2003; Wang et al., 2005; Baker et al., 2007; Jonas et al., 2008; Bhatt et al., 2009). We determined the 5′ end of the transcripts, which were regulated by CsrA, and analysed the 5′UTRs for the presence of GGA triplets, the most invariant element of the CsrA binding site. The 5′UTRs of STM1987, STM3375, STM1703, STM3611, STM1344 and STM1697 contained at least one (STM1703) and at most 11 (STM1987) GGA motifs (Table 2), raising the possibility that CsrA directly interacts with the transcripts corresponding to these genes. No GGA triplets were present in the 5′UTRs of the transcripts of STM1827 and STM4551 (Table 2). Noticeably, STM1987, STM4551, STM1703, STM3375, STM3611, STM1827, STM1344 and STM1697 are not located in operons and, thus, possess their own promoters and 5′UTRs (S. 1). Likewise, most of the other genes encoding GGDEF/EAL proteins in Salmonella are stand-alone genes that are not clustered together with other functionally related genes (unpublished observation).
Table 2.
5′untranslated leader sequences of CsrA regulated genes.
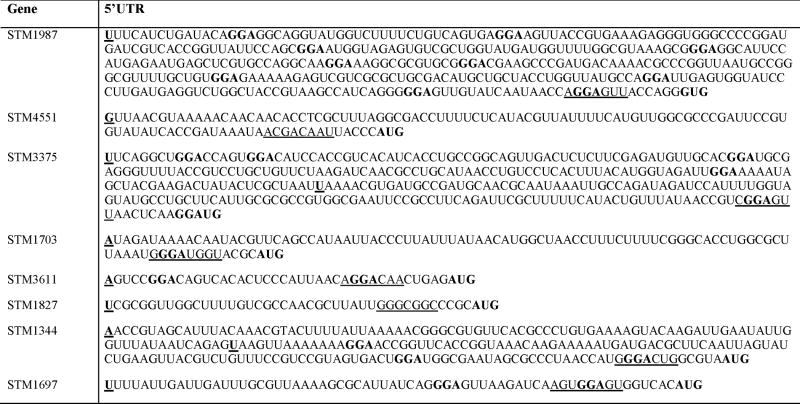 |
GGA - element of putative CsrA binding boxes; A, G or U – transcriptional start site; AUG or GUG – initiation codon; underlined – predicted Shine Dalgarno sequence
CsrA binds specifically to untranslated mRNA leaders of STM1987, STM3375, STM3611, STM1344 and STM1697
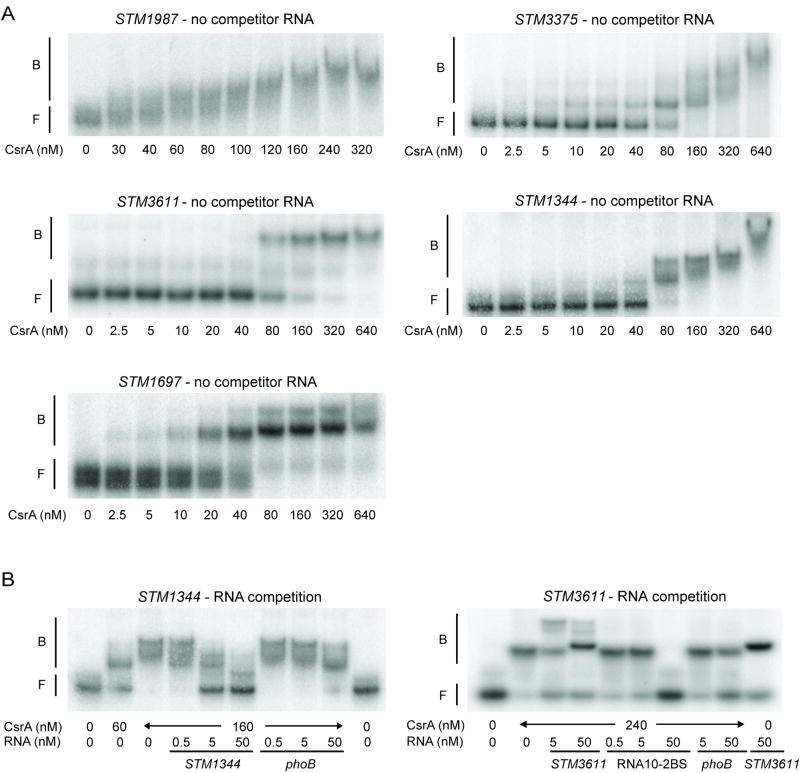
To determine whether CsrA directly interacts with the transcripts of the eight GGDEF/EAL genes found to be regulated by CsrA in Salmonella, gel-mobility shift assays were performed with CsrA protein, purified as previously described (Mercante et al., 2006), and in vitro synthesized transcripts, comprising the entire untranslated leader and a small part of the coding region, respectively: STM1987 (374 nt, 3 nt), STM4551 (109 nt, 3 nt), STM3375 (322 nt, 3 nt), STM1703 (95 nt, 3 nt), STM3611 (39 nt, 10 nt), STM1827 (42 nt, 38 nt), STM1344 (133 nt, 11 nt) and STM1697 (65 nt, 3 nt). CsrA binding to the transcripts of STM1987, STM3375, STM3611, STM1344 and STM1697 was detected as distinctly shifted bands in native gels (Fig. 2A). No shifts were detected for STM1703 and STM4551 (S. 2A), suggesting that CsrA regulates these genes by an indirect mechanism or requires additional factors that are necessary for an interaction. Consistent with the gel-shift data, no potential binding site for CsrA was detected in the 5′leader of STM4551 (Table 2). The leader of STM1703 contained one GGA motif. However, most previously identified direct CsrA targets in E. coli contain at least two binding sites (Mercante et al., 2009). Although a shifted band was seen for STM1827 at high CsrA concentrations, significant amounts of the free transcript remained unbound (S. 2A). In the 5′UTR of STM1827 no GGA motifs were found. Thus, it is possible that CsrA interaction with the STM1827 transcript may not be biologically relevant, although we cannot completely exclude this possibility to this date. We also cannot rule out that CsrA interacts with other regions than the 5′UTRs of the transcripts of STM4551, STM1703 and STM1827.
Figure 2.
Direct interaction between CsrA and the transcripts of STM1987, STM3375, STM3611, STM1344 and STM1697. A) Gel mobility shift analyses of CsrA–STM1987, CsrA–STM3375, CsrA–STM3611, CsrA–STM1344 and CsrA–STM1697 interactions in the absence of RNA competitor. The 5′ end-labelled respective transcript was incubated with CsrA at the indicated concentrations. The positions of free (F) and bound (B) RNA are shown. B) Competition reactions for STM1344 and STM3611 using specific (STM1344, STM3611 or RNA10-2BS) or non-specific (phoB) unlabelled RNA competitors. The concentration of competitor RNA is shown at the bottom of each lane.
More than one shifted complex was detected for STM1987, STM3375, STM1344 and STM1697 (Fig. 2A), suggesting that two or more CsrA proteins were bound to each transcript, although the stoichiometry of binding was not experimentally determined. In contrast, STM3611 formed only one shifted complex (Fig. 2A). A nonlinear least-squares analysis of these CsrA binding data yielded apparent Kd values of 53 ± 10 nM for STM1987, 34 ± 7 nM for STM3375, 119 ± 8 nM for STM3611, 58 ± 5 nM for STM1344 and 33 ± 3 nM for STM1697. These binding constants are comparable to the affinities previously measured for the interactions between CsrA and the E. coli mRNA targets glgC (39 nM), cstA (40 nM), pgaA (22 nM), hfq (38 nM) and sepL (23 nM) (Baker et al., 2002; Dubey et al., 2003; Wang et al., 2005; Baker et al., 2007; Bhatt et al., 2009). In contrast, CsrA binding to the transcripts of ycdT, ydeH (Kds ≈ 2.5 nM) and grlR (Kd ≈ 6 nM) was stronger (Jonas et al., 2008; Bhatt et al., 2009).
The specificity of the CsrA-RNA interactions was investigated by performing competition experiments with specific (STM1987, STM3375, STM3611, STM1344, STM1697 and RNA10-2BS) and non-specific (phoB 5′ untranslated region from E. coli K-12) unlabelled RNA competitors. Unlabelled STM1987, STM3375, STM1344 and STM1697 RNAs, respectively, were able to compete for CsrA binding with the corresponding labelled transcripts while phoB RNA did not compete (Fig. 2B, S. 2B). Unlabelled STM3611 RNA exhibited complex interactions (Fig. 2B). As the concentration of this RNA was increased, slower migrating bands were observed, the major species of which was also seen in the absence of CsrA (last lane), strongly suggesting the formation of complexes between the labelled STM3611 RNA, unlabelled STM3611 RNA and CsrA. Thus, to determine binding specificity of STM3611, unlabelled RNA10-2BS, which contains two optimal CsrA binding sites (Mercante et al., 2009) was used as a specific competitor for STM3611 RNA. RNA10-2BS was able to compete for CsrA binding without the formation of novel shifted species, while unlabelled phoB RNA did not compete with the STM3611-CsrA interaction (Fig. 2B). Altogether, these results indicate that CsrA binds specifically and with high affinity to STM1987, STM3375, STM1344, STM1697 and STM3611 RNA.
Indirect regulation of STM3611 through the FlhDC/FliA cascade
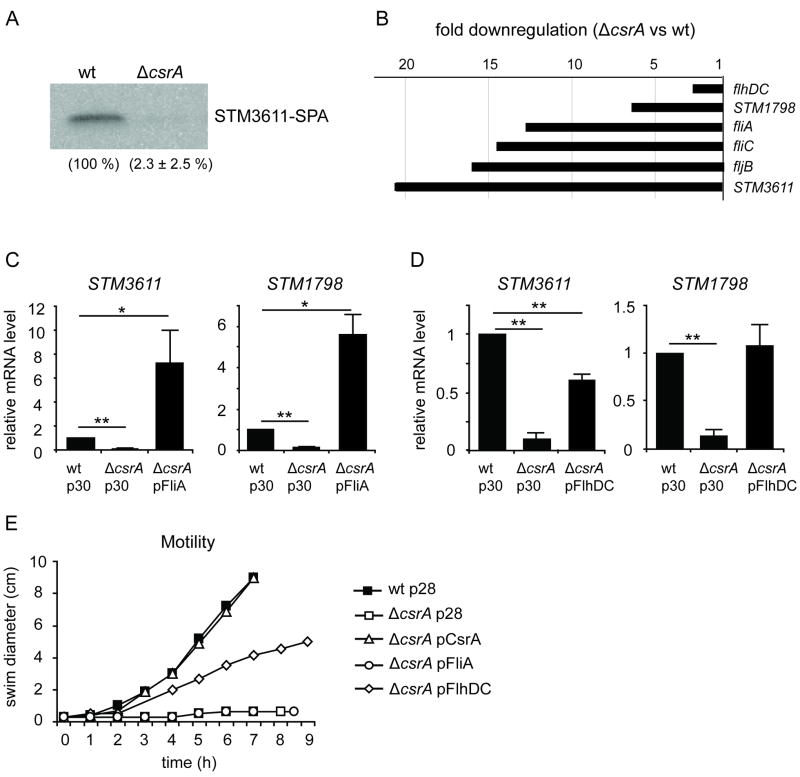
The mRNA level of STM3611 was drastically (>10 fold) downregulated in the csrA mutant compared to the wild type (Fig. 1). Consistent with this finding, the level of STM3611 protein was strongly decreased in the csrA mutant as observed by Western Blot analysis (Fig. 3A). STM3611 encodes a PDE, which positively controls flagella function and has a negative effect on the production of the Salmonella biofilm matrix components curli and cellulose (Simm et al., 2004; Simm et al., 2007). STM3611 is under the control of the flagella sigma factor FliA (Ko and Park, 2000; Wang et al., 2004; Frye et al., 2006; Claret et al., 2007). fliA, in turn is transcribed from a flagella gene class II promoter, which requires the master regulator of flagella synthesis, FlhDC (class I) for its activation (Chevance and Hughes, 2008).
Figure 3.
Indirect regulation of STM3611 by CsrA through the flagella cascade. A) Protein levels of SPA-tagged STM3611 in the wild type (wt) and the csrA deficient background (ΔcsrA). The bacteria were grown at 37 °C to OD600 1.5. B) A mutation in csrA results in a strong downregulation of flagella genes. mRNA levels were measured in the csrA mutant and the wild type by quantitative Real-Time RT PCR after growing the bacteria at 37 °C to OD600 1.5. C) Complementation of the effect of a csrA mutation on STM3611 and STM1798 mRNA levels by plasmid-borne expression of fliA from pIRF-2 (pFliA). p30 corresponds to the empty vector control pBAD30. The values represent means with standard deviations (** P < 0.01; * P < 0.05). D) Complementation with the plasmid pAS-0081 (pFlhDC). The values represent means with standard deviations (** P < 0.01; * P < 0.05). E) Restoration of swimming motility in the csrA mutant. Swimming motility of the wild type strain carrying the empty vector pBAD28 (wt p28) or the csrA mutant MAE125 (ΔcsrA) carrying pBAD28, pBADcsrA (pCsrA), pAS-0081 (pFlhDC) or pIRF-2 (pFliA), respectively, was analysed in 0.3 % LB agar with 0.1 % arabinose at 37 °C. Four μl of overnight cultures, which were grown in LB with 0.1 % arabinose, were used for the experiment. The swimming diameter vs. time is displayed.
E. coli and S. Typhimurium csrA mutants lack flagella and are non-motile (Wei et al., 2001; Lawhon et al., 2003). Furthermore, in E. coli CsrA was shown to control flagella synthesis by binding to and stabilizing flhDC mRNA (Wei et al., 2001). In S. Typhimurium, a microarray-based study revealed that genes belonging to the flhDC regulon were strongly downregulated in a csrA mutant (Lawhon et al., 2003). We found the mRNA levels of flhDC, fliA and genes belonging to the FliA regulon to be significantly decreased in the csrA mutant MAE125 compared to the wild type (Fig. 3B). Among these genes was also STM1798 (ycgR), encoding a PilZ domain containing c-di-GMP receptor protein (Ryjenkov et al., 2006), which has previously been shown to be co-regulated with STM3611 (Ko and Park, 2000; Wang et al., 2004; Frye et al., 2006; Claret et al., 2007).
As we considered it likely that the strong effect of CsrA on the mRNA levels of STM3611 was mediated, at least partly, by the indirect regulation of STM3611 through FlhDC and FliA, we attempted to complement the csrA mutation with plasmid-borne expression of fliA and flhDC, respectively. Ectopic expression of fliA from pBADfliA in the csrA mutant led to strongly elevated mRNA levels of both STM3611 and STM1798 (> 5 fold compared to wt) (Fig. 3C). We explain this result with the fact that the csrA mutant is deficient in the expression of the anti-sigma factor flgM, which is co-transcribed with fliA and counteracts FliA's activity in the wild type background (Chevance and Hughes, 2008). Ectopic expression of flhDC from pBADflhDC in the csrA mutant led only to a partial restoration of STM3611 mRNA levels (approx. 60 % of wt), but to a complete restoration of STM1798 transcript levels (approx. 105 % of wt) (Fig. 3D).
Furthermore, analysis of the swimming behaviour of the bacteria revealed that plasmid-borne expression of flhDC only partly restored the swimming defect of a csrA mutant (Fig. 3E). In contrast, expression of plasmid-borne csrA from pBADcsrA enabled the csrA mutant to swim in motility agar as the wild type. Overexpression of fliA did not restore the swimming ability in the csrA mutant, which is in agreement with the model that the other FlhDC regulated class II genes, which assemble the hook-basal body of flagella (Chevance and Hughes, 2008), are shut down in the csrA mutant.
Altogether, our data suggest that CsrA controls the flagella cascade by regulating flhDC, which results in the indirect upregulation of STM3611. Our data also indicate that CsrA does not exclusively regulate the flagella signalling hierarchy and STM3611 through flhDC, but also affects additional pathways.
Direct regulation of STM3611 by CsrA
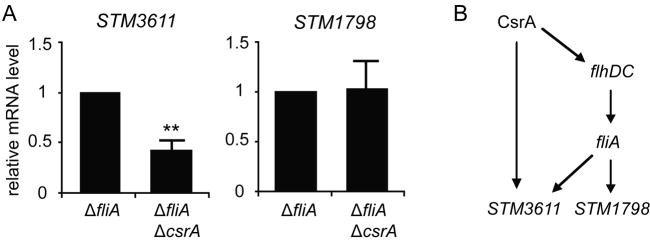
Gel-shift analysis revealed that CsrA specifically interacted with the transcript of STM3611 (Fig. 2A,B), suggesting that in addition to the indirect regulation through FlhDC/FliA CsrA might also regulate STM3611 mRNA stability. We noted that the leader of STM3611 contains two GGA motifs (Table 2), one of which seems to overlap the Shine-Dalgarno (SD) sequence, at which ribosome binding occurs. Previously identified CsrA mRNA targets in E. coli, which contain one CsrA binding site at the SD site and another one further upstream, have been demonstrated to be destabilized by CsrA (Mercante et al., 2009). However, our results show that in a fliA deficient background a mutation in csrA led to a further reduction in STM3611 mRNA levels (Fig. 4A), suggesting that binding of CsrA might have a stabilizing effect on STM3611 mRNA levels, independently of fliA. In contrast, the mRNA levels or STM1798, whose transcript did not interact with CsrA as determined by gel-mobility shift analysis (S. 2A), were the same in the fliA mutant and the csrA fliA double mutant (Fig. 4A).
Figure 4.
Regulation of STM3611 by CsrA. A) Effect of a csrA mutation on STM3611 and STM1798 mRNA levels in a fliA deficient background. RNA was isolated from bacterial cultures grown at 37 °C in LB medium with 0.1 % arabinose to OD600 1.5. mRNA levels of STM3611 and STM1798 were measured by quantitative Real-Time RT PCR in the csrA mutant MAE125 and the fliA csrA double mutant MAE1476 (fliA csrA). The values represent means with standard deviations (** P < 0.01). B) Schematic model depicting the inferred direct and indirect regulation of STM3611 by CsrA.
The regulation of flhDC by CsrA in E. coli has so far been the only example, in which binding of CsrA to a transcript resulted in mRNA stabilization (Wei et al., 2001). It is possible that CsrA-mediated activation processes require the formation of higher order RNA structural alterations that involve CsrA binding to additional uncharacterized sites in the transcript. However, to this date no molecular model exists for CsrA-mediated activation. Our data indicate that CsrA upregulates STM3611 expression by activating its transcription through the FlhDC/FliA cascade, as well as by directly interacting with STM3611 transcript (Fig. 4B).
Indirect regulation of STM3611 through STM1344
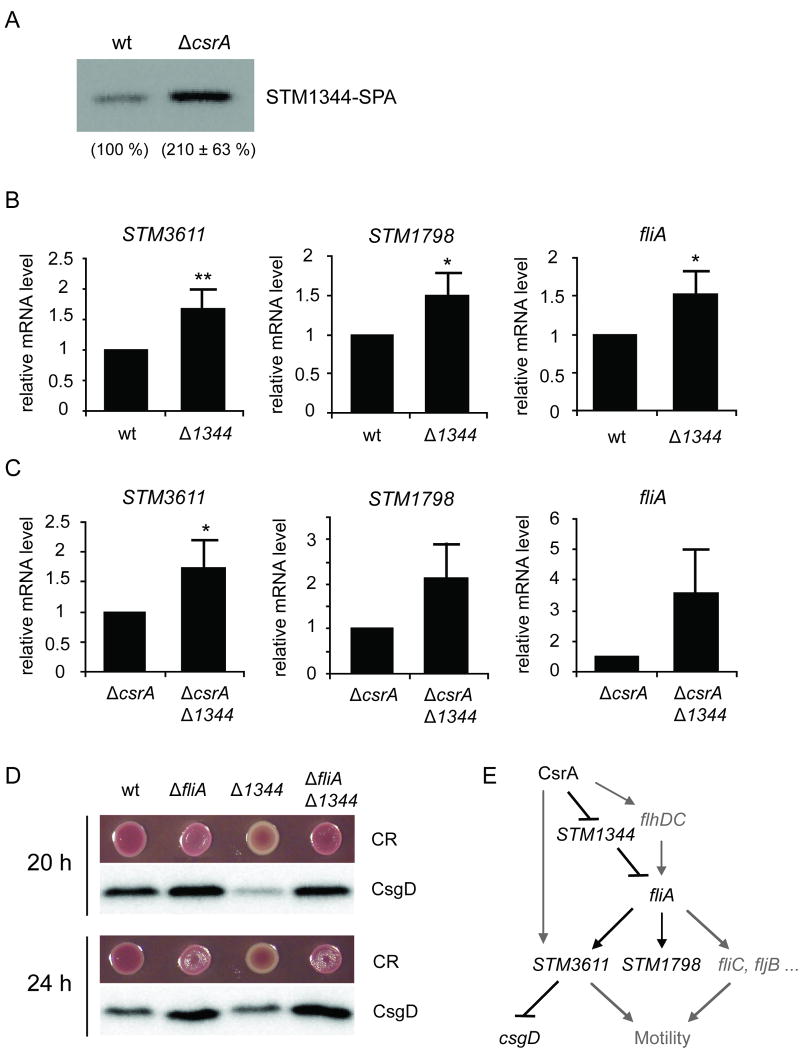
Besides STM3611, CsrA was found to strongly alter the mRNA and protein levels of another EAL domain protein, STM1344 (Fig. 1, Fig. 5A). In contrast to STM3611 and other conventional EAL domain proteins, STM1344 lacks activity as a c-di-GMP specific PDE and displays phenotypes, which are more typical for DGCs: the upregulation of biofilm behaviour and the downregulation of motility (Simm et al., 2009). Furthermore, STM1344 exerts its effect on biofilm behaviour through the PDEs STM3611 and STM1703, which are upregulated in the STM1344 mutant (Simm et al., 2009). In another parallel study STM1344 (also called YdiV) was identified as a negative regulator of flagella gene expression and it was suggested that STM1344 acts on post class I genes (Wozniak et al., 2008). In agreement with these previous studies, we found that a mutation in STM1344 caused an upregulation in STM3611 mRNA levels (Fig. 5B). Furthermore, the transcript levels of STM1798 and fliA were increased in the STM1344 mutant to similar extents, suggesting that STM1344 might regulate STM3611 expression through fliA. A similar upregulation of STM3611, STM1798 and fliA was detected in a csrA STM1344 double mutant (Fig. 5C), which is in agreement with our model that CsrA acts upstream of STM1344.
Figure 5.
CsrA-mediated regulation of STM1344 and its downstream effects. A) Protein levels of SPA-tagged STM1344 in the wild type (wt) and the csrA deficient background (ΔcsrA). The bacteria were grown at 37 °C to OD600 1.5. B) Effect of a mutation in STM1344 (Δ1344) on the mRNA levels of STM3611, STM1798 and fliA measured by quantitative Real-Time RT PCR, after growth of the bacteria at 37 °C to OD600 1.5. The values represent means with standard deviations (** P < 0.01; * P < 0.05). C) Effect of a double mutation in csrA and STM1344 (ΔcsrA Δ1344) on the mRNA levels of STM3611, STM1798 and fliA. The values represent means with standard deviations (* P < 0.05). D) STM1344 regulates rdar morphotype expression and CsgD levels through fliA. Rdar morphotype expression and CsgD protein levels were analysed in the wild type UMR1 and its isogenic mutants in fliA, STM1344 and fliA STM1344. The bacteria were grown for 20 h or 24 h, respectively, at 28 °C on Congo Red (CR) LB agar plates without salt. E) Schematic model depicting the regulation of STM3611 by STM1344 through fliA. The model illustrates that CsrA controls the flagella cascade at multiple levels, through FlhDC, STM1344 and STM3611.
The suggested sequential regulation of STM3611 by fliA and STM1344 was confirmed by additional studies, in which the ability of Salmonella to produce the biofilm matrix components curli and cellulose was used as a read-out. Curli and cellulose production can be conveniently visualized on Congo Red (CR) agar plates, on which colonies expressing these surface structures appear as red, dry and rough (rdar) and those that lack them as smooth and white (saw) (Romling, 2005). Under biofilm-inducing conditions (28 °C, LB agar plates without salt), the phosphodiesterase STM3611 was found to negatively affect rdar morphotype expression (Simm et al., 2007). Specifically, a mutant in STM3611 shows a more pronounced rdar phenotype on CR plates than the wild type and the protein level of CsgD, the major activator of curli and cellulose synthesis, is upregulated (Simm et al., 2007). Previous results also revealed that a mutation in STM1344 downregulates csgD expression through STM3611, resulting in a reduced production of curli and cellulose (Simm et al., 2009). Our data show that, similar to the STM3611 deletion strain, a mutant in fliA displayed elevated CsgD levels and enhanced rdar morphotype expression compared to the wild type (Fig. 5D). The same phenotype was also observed in a STM1344 fliA double mutant, confirming our model that STM1344 acts upstream of FliA and thereby regulates the PDE STM3611, which in turn positively controls motility behaviour and has a negative impact on the production of biofilm matrix components.
In summary, these data show that CsrA regulates the levels of STM3611 by an additional pathway, involving the direct regulation of STM1344, which in turn affects STM3611 by interfering with the flagella cascade upstream of fliA (Fig. 5E). Altogether these data indicate that CsrA controls the PDE STM3611 by at least three distinct pathways: the indirect regulation through flhDC, the direct regulation of its mRNA level and the indirect regulation through STM1344 (Fig. 5E). We suggest that this multi-layer control allows CsrA to precisely regulate the activity of STM3611 and, hence, the switch between motility and biofilm behaviour.
Discussion
The transition between a sessile and a motile life-style requires the complex integration of the pathways that regulate either bacterial behaviour. The present study describes the tight interplay between the Csr, c-di-GMP and motility systems in S. Typhimurium. In a recent study the regulation of c-di-GMP signalling by CsrA was investigated in E. coli (Jonas et al., 2008). However, the interconnection between Csr and c-di-GMP signalling differs substantially between E. coli and Salmonella. This let suggest that the utilization of conserved regulatory systems is highly adaptable and can vary between closely related species.
CsrA-mediated downregulation of the DGCs STM1987 and STM4551
Previous studies revealed that CsrA promotes motility and inhibits biofilm-associated phenotypes (Romeo, 1998; Wei et al., 2001; Jackson et al., 2002; Wang et al., 2005; Jonas et al., 2008). In agreement, CsrA was found to directly or indirectly downregulate the c-di-GMP producing enzymes STM1987 and STM4551, respectively, in S. Typhimurium (Figs. 1 and 2). Under certain growth conditions STM1987 promotes the synthesis of the biofilm polysaccharide cellulose (Garcia et al., 2004; Solano et al., 2009) and thus acts as an activator of biofilm formation. STM4551 has recently been shown to possess DGC activity, but it also appeared to act by a c-di-GMP-independent mechanism on diverse phenotypes (Solano et al., 2009). In E. coli CsrA downregulates the two DGCs YdeH and YcdT (Jonas et al., 2008), which are distinct from STM4551 and STM1987. Similar to STM1987, YdeH promotes the synthesis of a biofilm polysaccharide by a c-di-GMP dependent mechanism (Boehm et al., 2009). However, instead of cellulose, which is not produced by E. coli K12, YdeH controls production of the polysaccharide PGA (Boehm et al., 2009).
Complex regulation of the PDE STM3611 and the motility cascade
While CsrA inhibits c-di-GMP synthesis by downregulating STM1987 and STM4551, it upregulates the PDE STM3611 and thus promotes the degradation of c-di-GMP (Simm et al., 2004; Simm et al., 2007). STM3611 is suggested to influence the functionality of the flagella motor by degrading a local pool of c-di-GMP (Romling and Amikam, 2006; Wolfe and Visick, 2008). Under conditions when STM3611 is inactive, c-di-GMP accumulates and presumably binds to the receptor protein STM1798, containing a PilZ domain (Ryjenkov et al., 2006). The resulting c-di-GMP-STM1798 complex was suggested to negatively affect motor function (Romling and Amikam, 2006; Wolfe and Visick, 2008). In addition, STM3611 inhibits biofilm behaviour by negatively regulating the expression of csgD by a c-di-GMP dependent mechanism (Simm et al., 2007). Thus, by regulating STM3611 CsrA can reciprocally act on motility and biofilm behaviours.
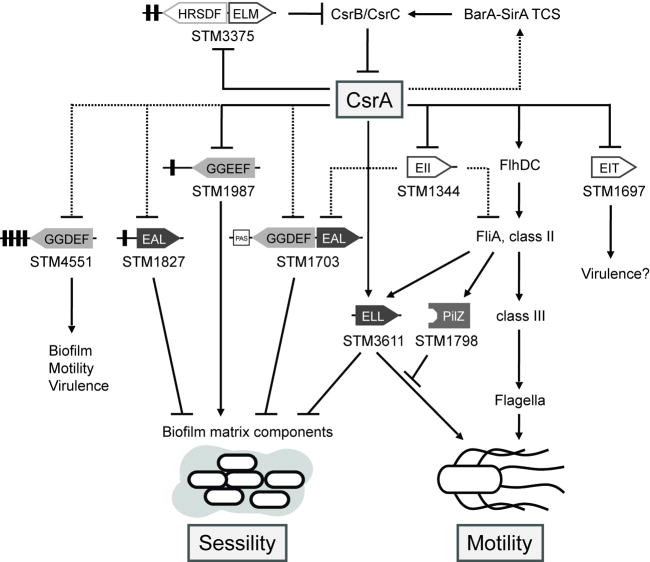
Notably, the regulation of STM3611 by CsrA occurred at multiple levels: by two indirect pathways involving FlhDC (Fig. 3) and STM1344 (Fig. 5), respectively, and by a direct, presumably post-transcriptional mechanism (Figs. 2 and 4). This three-tiered regulatory circuitry must permit precise control of the levels of the PDE STM3611. Furthermore, these results illustrate, for the first time, that CsrA controls the motility cascade at multiple levels (flhDC - class I, STM1344 - post-class I, STM3611 - class III) in the signalling hierarchy. We suggest that this complex control enables CsrA to coordinate flagella synthesis with flagella function. The feed-forward arrangement that directly regulates STM3611 might allow a rapid onset of the flagella motor, under conditions that favour motility. It is likely that CsrA also controls the expression of the orthologue of STM3611 in E. coli (YhjH), although this has not been experimentally demonstrated.
Noticeably, the mRNA levels of two other PDEs, STM1827 and STM1703, were downregulated by CsrA (Fig. 1), probably by indirect mechanisms. Both STM1827 and STM1703 have previously been shown to degrade c-di-GMP and thereby to downregulate csg expression (Simm et al., 2007).
Regulation of unconventional GGDEF/EAL domain proteins
CsrA was also found to control GGDEF and/or EAL domain proteins that have apparently lost the ability to metabolize c-di-GMP and have instead evolved alternative functions. The EAL-domain protein STM1344, which was directly downregulated by CsrA (Figs. 1, 2 and 5), contains a degenerate sequence motif in its EAL domain and is inactive in the degradation or binding of c-di-GMP (Simm et al., 2009). STM1344 negatively controls the flagella cascade by an unknown mechanism upstream of fliA (present study) and downstream of flhDC transcription (Wozniak et al., 2008). The indirect regulation of the PDE STM3611 by STM1344 results in the downregulation of motility and the upregulation of biofilm matrix production. STM1344 also controls the PDE STM1703 (Simm et al., 2009), which negatively regulates csg expression and, thus, biofilm behaviour (Simm et al., 2007) (Fig. 6). Hence, despite its degenerate EAL domain, STM1344 still maintains a function in the regulation of c-di-GMP metabolism.
Figure 6.
Schematic model illustrating the interplay between the Csr-, the c-di-GMP and the flagella regulatory system in Salmonella Typhimurium. Apparent direct (solid line) and indirect (dashed line) roles of CsrA in the expression of genes encoding GGDEF and EAL domain proteins resulting in the tight control of the sessility-motility switch in S. Typhimurium. STM4551 and STM1987 possess DGC activity (Garcia et al., 2004; Solano et al., 2009), whereas STM1703, STM1827 and STM3611 act as PDEs (Simm et al., 2004; Simm et al., 2007). The c-di-GMP metabolizing activities of these proteins control phenotypes in motility, biofilm formation or viruence. In contrast, STM1344, STM3375 and possibly STM1697 contain degenerate EAL/GGDEF domains (unshaded), which cannot synthesize or degrade c-di-GMP, but have apparently evolved alternative functions, e.g. regulatory functions (Simm et al., 2009). Notably, CsrA controls the flagella cascade at multiple levels in the hierarchy: by apparent direct regulation of the flagella master regulator FlhDC and STM1344, which influences the flagella cascade upstream of fliA, and by apparent direct and indirect regulation of STM3611. Presumably, this multi-layer control allows CsrA to coordinate flagella synthesis with motor function. By regulating STM3375 (CsrD), which, along with RNase E, destabilizes the CsrB and CsrC sRNAs in E. coli (Suzuki et al., 2006), CsrA seems to control its own activity by an autoregulatory loop. CsrB and CsrC are positively controlled by the two-component system (TCS) BarA-SirA (Altier et al., 2000b; Teplitski et al., 2003), which allows the integration of environmental signals into the regulatory network. In E. coli (Gudapaty et al., 2001) and in S. Typhimurium (our unpublished observations) transcription of csrB and csrC also requires upstream activation by CsrA, probably through the BarA-UvrY (BarA-SirA) TCS (Suzuki et al., 2002), indicative of an additional feedback loop.
Another EAL domain protein, STM1697, shows high sequence similarity to STM1344, and its mRNA level was directly regulated by CsrA (Figs. 1 and 2). Similar to STM1344, STM1697 does not contain the highly conserved sequence motifs (e.g. EXL, DDFGTG), which have previously been suggested to be critical for PDE activity (Schmidt et al., 2005; Rao et al., 2009), suggesting that STM1697 has no PDE activity. A mutant in STM1697 does not show a distinct phenotype in motility or biofilm behaviour (Simm et al., 2007). However, data from another on-going study suggest that the protein might be involved in the regulation of virulence phenotypes (Lamprokostopoulou and Römling, unpublished).
CsrA also regulated the expression of the unorthodox GGDEF-EAL protein STM3375 (Fig. 1, Fig. 2). The E. coli STM3375 orthologue CsrD (YhdA) is itself a component of the Csr system (Jonas et al., 2006; Suzuki et al., 2006), which facilitates RNaseE-dependent degradation of the sRNAs CsrB and CsrC, the molecular antagonists of CsrA (Suzuki et al., 2006). Thus, although not directly involved in the synthesis or degradation of c-di-GMP, CsrD controls the activity of CsrA, a global regulator of GGDEF and EAL domain proteins and c-di-GMP levels (Jonas et al., 2008). STM3375 and E. coli CsrD are highly similar proteins. They contain identical degenerate GGDEF and EAL motif signatures, as well as an identical sequence in the EAL domain that is unique to putative CsrD homologues (R579-TENQLLVQ-S588) and is required for activity of the E. coli protein (Suzuki et al., 2006). This suggests that these proteins function similarly. The finding that CsrA controls the expression of csrD (Suzuki et al., 2006; Jonas et al., 2008) and STM3375 indicates that it regulates its own activity through an autoregulatory loop in both of these species.
Signal integration into the Csr/c-di-GMP system
CsrA activity is also controlled by the BarA-SirA (BarA-UvrY in E. coli) two-component system (Fig. 6), which activates the transcription of csrB and csrC (Altier et al., 2000b; Suzuki et al., 2002; Teplitski et al., 2003; Weilbacher et al., 2003). The chemical nature of the signal acting on the BarA sensor and its orthologues in other bacteria has not been identified yet. However, studies in E. coli suggest that the metabolic status of the cells and the external pH regulate the activity of the two-component system (Pernestig et al., 2003; Mondragon et al., 2006; Jonas and Melefors, 2009). In Salmonella, the presence of bile salts seems to affect BarA-SirA dependent responses (Prouty and Gunn, 2000) and results from another study have shown that short chain fatty acids affect CsrA mediated phenotypes in a pathway involving SirA, but probably not BarA (Lawhon et al., 2002). Transcription of csrB and csrC somehow also requires upstream activation by CsrA, indicative of an additional feedback loop (Fig. 6) and strongly suggestive of a homeostatic mechanism controlling CsrA activity (Gudapaty et al., 2001; Suzuki et al., 2002; Weilbacher et al., 2003; Jonas and Melefors, 2009). The tight regulation of CsrA activity by BarA-SirA, CsrD and CsrA itself through the Csr sRNAs ensures that downstream targets of CsrA are precisely coordinated in response to multiple input signals.
Central role of CsrA in the mediation of life-style switches
Our data illustrate that the regulation of GGDEF and EAL domain proteins enables CsrA to tightly control the switch between sessility and motility at multiple levels (Fig. 6). In general, CsrA activates motility pathways and inhibits the production of biofilm factors. Notably however, in contrast to a csrA mutant in E. coli, in which biofilm formation is strongly enhanced (Jackson et al., 2002), a S. Typhimurium csrA mutant does not show increased adherence or rdar morphotype expression as would be expected (Teplitski et al., 2006)(our unpublished observations). Instead, colonies of the csrA mutant have a mucoid and smooth colony appearance (our unpublished observations). Although the molecular basis of this observation remains unknown, it indicates a complex role of CsrA in the regulation of cell surface / extracellular phenotypes in Salmonella.
Finally, CsrA plays an important role in the regulation of Salmonella virulence genes (Altier et al., 2000a; Lawhon et al., 2003). Likewise, c-di-GMP has been recently found to affect virulence properties in Salmonella (Lamprokostopoulou et al., 2009). In fact, several of the CsrA-regulated genes studied herein affect virulence phenotypes, including STM1344, STM4551 and STM1697 (Hisert et al., 2005; Solano et al., 2009). Thus it is likely, that the regulation of GGDEF and/or EAL domain proteins allows CsrA not only to control motility and sessility, but also a range of other bacterial behaviours that are important for Salmonella to adapt to changing environments.
Experimental Procedures
Bacterial strains and growth conditions
All strains used in this study are listed in Table 1. Chromosomal mutations were generated using the Datsenko method (Datsenko and Wanner, 2000). For the construction of the csrA::kan mutant allele, the kan gene was amplified from pKD4 by PCR using the primer pair CsrAKOup/CsrAKOdn (Table 3) and electroporated into arabinose-treated S. Typhimurium ATCC 14028 carrying pKD46. For the construction of the fliA mutant MAE1456 (UMR1 fliA::cat), the cat gene was amplified from pKD3 using the primer pair FliAKOfor/FliAKOrev and introduced by electroporation into arabinose-treated S. Typhimurium UMR1 carrying pKD46. Transformants were selected for the gain of kanamycin or chloramphenicol resistance, respectively, and the loss of ampicillin resistance and were verified by PCR using the appropriate control primer pairs (S. 3). To generate MAE125 (UMR1 csrA::kan) the csrA::kan allele was transferred by P22 transduction into Salmonella Typhimurium UMR1. For construction of the fliA csrA (MAE1476) and the STM1344 csrA (MAE1474) double mutants, the mutant alleles from MAE1456 (UMR1 fliA::cat) and MAE424 (UMR1 STM1344::cat), respectively, were transduced into the MAE125 background. For the construction of the fliA STM1344 double mutant MAE1481, the cat cassette was removed from MAE1456 followed by the transduction of STM1344::cat into the resulting strain (MAE1463). To generate MAE1493 (UMR1 ΔcsrA STM3611-SPA) and MAE1492 (UMR1 ΔcsrA STM1344-SPA), the kan cassette from MAE125 was flipped out using the the FLP recombinase followed by the transduction of the SPA-tagged STM1344 and STM3611 constructs from MAE132 (UMR1 STM1344-SPA kanr) and MAE130 (UMR1 STM3611-SPA kanr), respectively, into the resulting strain. Bacteria were routinely grown in LB medium at 37 °C with shaking at 200 r.p.m. If necessary, arabinose (0.1 %) or antibiotics were added: ampicillin 100 μg ml-1, kanamycin 50 μg ml-1 and chloramphenicol 30 μg ml-1.
Table 3.
Strains and plasmids used in this study.
| Strain/plasmid | Description or genotype | Reference |
|---|---|---|
| Strains | ||
| Salmonella enterica serovar Typhimurium ATCC 14028 | ||
| UMR1 | ATCC 14028-1s Nalr | (Romling et al., 1998) |
| MAE125 | UMR1 csrA∷kan | This study |
| MAE1491 | UMR1 ΔcsrA | This study |
| MAE130 | UMR1 STM3611-SPA kanr | (Simm et al., 2009) |
| MAE1493 | MAE1491 STM3611-SPA kanr | This study |
| MAE132 | UMR1 STM1344-SPA kanr | (Simm et al., 2009) |
| MAE1492 | MAE1491 STM1344-SPA kanr | This study |
| MAE1456 | UMR1 fliA∷cat | This study |
| MAE1463 | UMR1 ΔfliA:101 | This study |
| MAE1476 | MAE125 fliA∷cat | This study |
| MAE424 | UMR1 STM1344∷cat | (Simm et al., 2007) |
| MAE1481 | MAE1463 STM1344∷cat | This study |
| MAE1474 | MAE125 STM1344∷cat | This study |
| Plasmids | ||
| pKD46 | Temperature-sensitive λ red recombinase expression vector | (Datsenko and Wanner, 2000) |
| pKD4 | Template plasmid (kanr) for mutant construction | (Datsenko and Wanner, 2000) |
| pKD3 | Template plasmid (catr) for mutant construction | (Datsenko and Wanner, 2000) |
| pBAD28 | Arabinose inducible expression plasmid | (Guzman et al., 1995) |
| pBAD30 | Arabinose inducible expression plasmid | (Guzman et al., 1995) |
| pBADcsrA | csrA under control of the plasmid-borne PBAD promoter | (Jonas et al., 2008) |
| pIRF-2 | fliA under control of the plasmid-borne PBAD promoter | This study |
| pAS-0081 | flhDC under control of the plasmid-borne PBAD promoter | (Sittka et al., 2008) |
Plasmid construction
All plasmids used in this study are listed in Table 1. For construction of pIRF-2 the fliA gene was amplified from the S. Typhimurium UMR1 chromosome by PCR using the primers pBADfliAfor and pBADfliArev (S. 3). The resultant PCR product was cleaved with XbaI and HindIII and inserted between the corresponding sites of pBAD30, under the control of the arabinose inducible promoter PBAD. Sequencing verified the integrity of the fliA gene.
Quantitative real-time RT PCR
RNA was sampled, treated with RNAprotect Bacterial Reagent (Qiagen) and prepared using the RNeasy Mini Kit with on-column DNA digestion (Qiagen) according to the protocol. After determination of the RNA concentrations using the NanoDrop ND-1000 UV-Vis Spectrophotometer, 1 μg RNA was reverse transcribed in 20 μl reactions using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Primers were designed with the Primer Express Software v3.0 (Applied Biosystems). Twenty ng of template were used for the real-time PCR reaction using Power SYBR Green PCR Master Mix (Applied Biosystems). The cycling reaction was performed with an ABI 7500 Real Time PCR System (Applied Biosystems) using the standard run mode of the instrument. For detection of primer dimerization or other artefacts of amplification, dissociation curves and non-template controls were included in the real-time PCR analysis. Individual gene expression profiles were normalized against the recA gene or the rrnD gene (16S rRNA), serving as endogenous controls. All results were analysed using the 7500 SDS Software v1.3.1 (Applied Biosystems) and further prepared using Excel (Microsoft). In all experiments, the change in expression was measured relative to a calibrator, e.g. wild type, which was set to 1. The data values presented in all figures represent the mean values calculated from the results from at least three independent repetitions of the experiment. The error bars represent the standard deviations. For statistical evaluation p-values were calculated using the student t-test.
5′ Rapid amplification of cDNA ends (5′RACE)
The 5′ ends of the transcripts of STM1987, STM4551, STM3375, STM1703, STM3611, STM1827, STM1344, STM1697 and STM1798 were determined using the 5′RACE System for Rapid Amplification of cDNA ends (v2.0 Invitrogen) according to the protocol and as previously described (Jonas et al., 2008). The resulting RACE PCR products were sequenced using the Big Dye Terminator Cycle Sequencing Kit (v3.1).
RNA gel mobility shift assays
Quantitative gel mobility shift assays followed a previously published procedure (Yakhnin et al., 2000). CsrA-His6 protein was purified as described previously (Mercante et al., 2006). DNA templates for generating STM1697, STM1798, and phoB (non-specific competitor) RNA transcripts were produced by annealing primers STM1697-T7 and GC STM1697-T7, STM1798-T7 and GC STM1798-T7, or phoB-T7 and GC phoB-T7 in TES buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 100 mM NaCl). DNA templates for STM1344, STM1703, STM1827, STM1987, STM3375, and STM4551 were PCR-amplified from UMR1 genomic DNA using primers STM1344-F-T7 and STM1344-R-T7, STM1703-F-T7 and STM1703-R-T7, STM1827-F-T7 and STM1827-R-T7, STM1987-F-T7 and STM1987-R-T7, 3375-F1(P1)-T7 and 3375-R1(ATG)-T7 or STM4551-F-T7 and STM4551-R-T7 (S. 3). RNA was synthesized in vitro using the MEGAshortscript kit (Ambion, Austin, TX) using the annealed DNA primers (STM1697, STM1798 and phoB) or DNA templates (STM1344, STM1703, STM1827, STM1987, STM3375, and STM4551) as templates, and RNA was gel purified. STM3611 and RNA10-2BS RNA were synthesized by Integrated DNA Technologies (Coralville, IA). Transcripts were 5′ end-labelled using T4 polynucleotide kinase and [γ-32P]-ATP. Radiolabelled RNA was gel purified and resuspended in TE (10 mM Tris-HCl pH 8.0, 1 mM EDTA), heated to 85°C and chilled on ice. Increasing concentrations of purified CsrA-His6 recombinant protein were combined with 50 pM radiolabelled RNA in 10 μl of binding reactions [10 mM Tris-HCl pH 7.5, 10 mM MgCl2, 100 mM KCl, 3.25 ng total yeast RNA, 20 mM DTT, 7.5 % glycerol, 4 U SUPERasin (Ambion, Austin, TX)] for 30 min at 37 °C to allow for CsrA–RNA complex formation. Competition assays were performed in the absence or presence of unlabelled RNA specific and non-specific competitors. Binding reactions were separated using 8-12 % native polyacrylamide gels, and radioactive bands were visualized with a Molecular Dynamics phosphorimager. Free and bound RNA species were quantified with ImageQuant Software (Molecular Dynamics), and an apparent equilibrium binding constant (Kd) was calculated for CsrA–RNA complex formation according to a previously described cooperative binding equation (Mercante et al., 2006). The mean values and standard errors from at least two independent experiments were determined for each transcript. Graphpad Prism version 3.02 for Windows (San Diego, CA) software was used for calculations.
Western Blot analysis
For Western Blot analysis 5 μg of cells were harvested, resuspended in sample buffer, and heated to 95 °C for 10 min. The protein content was analysed by staining with a Coomassie blue solution (20 % methanol, 10 % acetic acid, 0.1 % Coomassie brilliant blue G). Equal amounts of protein were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis using 15 % (for SPA tagged proteins) or 12 % (for CsgD) resolving gels and 4 % stacking gels and were transferred onto a polyvinylidene difluoride membrane (Immobilon P, Millipore). The detection of the SPA tags was done using anti-FLAG antibody (1:2000) and anti-mouse immunoglobulin G conjugated with horseradish peroxidase (1:2000, Jackson ImmunoResearch Laboratories Inc.) as the secondary antibody. Detection of CsgD was performed as previously described (Romling et al., 2000) by using the polyclonal anti-CsgD peptide antibody (1:5000) as the primary antibody and goat anti-rabbit immunoglobulin G conjugated with horseradish peroxidase (1:2000) as the secondary antibody. Chemiluminescence from the Lumi-Light WB substrate (Roche) was recorded using the LAS-1000 system (Fujifilm). The intensity of the bands was quantified using Adobe Photoshop CS3. Each experiment was repeated at least three times and representative results were chosen for display.
Motility assay
To analyse the swimming behaviour of the bacteria, 0.3 % LB agar plates supplemented with 0.1 % arabinose were inoculated with 4 μl of overnight cultures, which were grown in LB with 0.1 % arabinose for 14 -16 h. The plates were incubated at 37 °C and the diameter of the swimming zone was measured over time. Each experiment was performed at least three times, and a representative result was chosen for display.
Congo Red binding assay
Samples of 5 μl of an overnight culture suspended in water (to an optical density at 600 nm [OD600] of 5) were spotted onto LB agar plates lacking NaCl and supplemented with Congo red (40 μg ml−1) and Coomassie brilliant blue (20 μg ml−1). Plates were incubated at 28°C and the development of the colony morphology and dye binding were analyzed over time.
Acknowledgments
We are grateful to Jörg Vogel for kindly providing the plasmid pAS-0081, to Fredrik Åslund for help with the construction of the csrA mutant MAE125, and to Karina Hentrich and Susanna Bächle for excellent technical assistance. This work was supported by grants from the Marie Curie Early Stage Research Training Fellowship of the European Community's Sixth Framework Program under Contract Number MEST-CT-2004-8475, the Swedish Research Council (621-2007-6509, to U.R.), the Carl Tryggers Stiftelse (CTS-07:306, to U.R.), the U.S. National Institutes of Health (GM059969, GM066794) and a PhD fellowship from the Higher Education Commission of Pakistan (HEC) to I.A.
References
- Altier C, Suyemoto M, Lawhon SD. Regulation of Salmonella enterica serovar typhimurium invasion genes by csrA. Infect Immun. 2000a;68:6790–6797. doi: 10.1128/iai.68.12.6790-6797.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altier C, Suyemoto M, Ruiz AI, Burnham KD, Maurer R. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol Microbiol. 2000b;35:635–646. doi: 10.1046/j.1365-2958.2000.01734.x. [DOI] [PubMed] [Google Scholar]
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol Microbiol. 2002;44:1599–1610. doi: 10.1046/j.1365-2958.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- Baker CS, Eory LA, Yakhnin H, Mercante J, Romeo T, Babitzke P. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J Bacteriol. 2007;189:5472–5481. doi: 10.1128/JB.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Edwards AN, Nguyen HT, Merlin D, Romeo T, Kalman D. The RNA binding protein CsrA is a pleiotropic regulator of the LEE pathogenicity island of enteropathogenic Escherichia coli. Infect Immun. 2009 doi: 10.1128/IAI.00418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, et al. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol. 2009;72:1500–1516. doi: 10.1111/j.1365-2958.2009.06739.x. [DOI] [PubMed] [Google Scholar]
- Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Christen B, Folcher M, Schauerte A, Jenal U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- Claret L, Miquel S, Vieille N, Ryjenkov DA, Gomelsky M, Darfeuille-Michaud A. The flagellar sigma factor FliA regulates adhesion and invasion of Crohn disease-associated Escherichia coli via a cyclic dimeric GMP-dependent pathway. J Biol Chem. 2007;282:33275–33283. doi: 10.1074/jbc.M702800200. [DOI] [PubMed] [Google Scholar]
- Cotter PA, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey AK, Baker CS, Romeo T, Babitzke P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. Rna. 2005;11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey AK, Baker CS, Suzuki K, Jones AD, Pandit P, Romeo T, Babitzke P. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J Bacteriol. 2003;185:4450–4460. doi: 10.1128/JB.185.15.4450-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M, Hughes KT. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:2233–2243. doi: 10.1128/JB.188.6.2233-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. Bacterial signal transduction network in a genomic perspective. Environ Microbiol. 2004;6:552–567. doi: 10.1111/j.1462-2920.2004.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- Garcia B, Latasa C, Solano C, Garcia-del Portillo F, Gamazo C, Lasa I. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol Microbiol. 2004;54:264–277. doi: 10.1111/j.1365-2958.2004.04269.x. [DOI] [PubMed] [Google Scholar]
- Gudapaty S, Suzuki K, Wang X, Babitzke P, Romeo T. Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J Bacteriol. 2001;183:6017–6027. doi: 10.1128/JB.183.20.6017-6027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Hisert KB, MacCoss M, Shiloh MU, Darwin KH, Singh S, Jones RA, et al. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: role of cyclic diGMP. Mol Microbiol. 2005;56:1234–1245. doi: 10.1111/j.1365-2958.2005.04632.x. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, et al. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2008;190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J Bacteriol. 2002;184:290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Melefors O. The Escherichia coli CsrB and CsrC small RNAs are strongly induced during growth in nutrient-poor medium. FEMS Microbiol Lett. 2009;297:80–86. doi: 10.1111/j.1574-6968.2009.01661.x. [DOI] [PubMed] [Google Scholar]
- Jonas K, Melefors O, Romling U. Regulation of c-di-GMP metabolism in biofilms. Future Microbiol. 2009;4:341–358. doi: 10.2217/fmb.09.7. [DOI] [PubMed] [Google Scholar]
- Jonas K, Tomenius H, Romling U, Georgellis D, Melefors O. Identification of YhdA as a regulator of the Escherichia coli carbon storage regulation system. FEMS Microbiol Lett. 2006;264:232–237. doi: 10.1111/j.1574-6968.2006.00457.x. [DOI] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Simm R, Romeo T, Romling U, Melefors O. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol. 2008;70:236–257. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader A, Simm R, Gerstel U, Morr M, Romling U. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;60:602–616. doi: 10.1111/j.1365-2958.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- Ko M, Park C. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol. 2000;303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- Lamprokostopoulou A, Monteiro C, Rhen M, Romling U. Cyclic di-GMP signalling controls virulence properties of Salmonella enterica serovar Typhimurium at the mucosal lining. Environ Microbiol. 2009 doi: 10.1111/j.1462-2920.2009.02032.x. [DOI] [PubMed] [Google Scholar]
- Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- Lawhon SD, Maurer R, Suyemoto M, Altier C. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol. 2002;46:1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- Lawhon SD, Frye JG, Suyemoto M, Porwollik S, McClelland M, Altier C. Global regulation by CsrA in Salmonella typhimurium. Mol Microbiol. 2003;48:1633–1645. doi: 10.1046/j.1365-2958.2003.03535.x. [DOI] [PubMed] [Google Scholar]
- Liu MY, Gui G, Wei B, Preston JF, 3rd, Oakford L, Yuksel U, et al. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- Lucchetti-Miganeh C, Burrowes E, Baysse C, Ermel G. The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology. 2008;154:16–29. doi: 10.1099/mic.0.2007/012286-0. [DOI] [PubMed] [Google Scholar]
- Mercante J, Suzuki K, Cheng X, Babitzke P, Romeo T. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J Biol Chem. 2006;281:31832–31842. doi: 10.1074/jbc.M606057200. [DOI] [PubMed] [Google Scholar]
- Mercante J, Edwards AN, Dubey AK, Babitzke P, Romeo T. Molecular geometry of CsrA (RsmA) binding to RNA and its implications for regulated expression. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon V, Franco B, Jonas K, Suzuki K, Romeo T, Melefors O, Georgellis D. pH-dependent activation of the BarA-UvrY two-component system in Escherichia coli. J Bacteriol. 2006;188:8303–8306. doi: 10.1128/JB.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernestig AK, Georgellis D, Romeo T, Suzuki K, Tomenius H, Normark S, Melefors O. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J Bacteriol. 2003;185:843–853. doi: 10.1128/JB.185.3.843-853.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty AM, Gunn JS. Salmonella enterica serovar typhimurium invasion is repressed in the presence of bile. Infect Immun. 2000;68:6763–6769. doi: 10.1128/iai.68.12.6763-6769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, Qi Y, Chong HS, Kotaka M, Li B, Li J, et al. The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J Bacteriol. 2009;191:4722–4731. doi: 10.1128/JB.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- Romling U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci. 2005;62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Amikam D. Cyclic di-GMP as a second messenger. Curr Opin Microbiol. 2006;9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Romling U, Simm R. Prevailing concepts of c-di-GMP signaling. Contrib Microbiol. 2009;16:161–181. doi: 10.1159/000219379. [DOI] [PubMed] [Google Scholar]
- Romling U, Sierralta WD, Eriksson K, Normark S. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol. 1998;28:249–264. doi: 10.1046/j.1365-2958.1998.00791.x. [DOI] [PubMed] [Google Scholar]
- Romling U, Rohde M, Olsen A, Normark S, Reinkoster J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol Microbiol. 2000;36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A. 2006;103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryjenkov DA, Simm R, Romling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Allain FH. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol. 2007;14:807–813. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Simm R, Lusch A, Kader A, Andersson M, Romling U. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J Bacteriol. 2007;189:3613–3623. doi: 10.1128/JB.01719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R, Remminghorst U, Ahmad I, Zakikhany K, Romling U. A role for the EAL-like protein STM1344 in regulation of CsgD expression and motility in Salmonella enterica serovar Typhimurium. J Bacteriol. 2009;191:3928–3937. doi: 10.1128/JB.00290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano C, Garcia B, Latasa C, Toledo-Arana A, Zorraquino V, Valle J, et al. Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc Natl Acad Sci U S A. 2009;106:7997–8002. doi: 10.1073/pnas.0812573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20:2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Wang X, Weilbacher T, Pernestig AK, Melefors O, Georgellis D, et al. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J Bacteriol. 2002;184:5130–5140. doi: 10.1128/JB.184.18.5130-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitski M, Goodier RI, Ahmer BM. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J Bacteriol. 2003;185:7257–7265. doi: 10.1128/JB.185.24.7257-7265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitski M, Al-Agely A, Ahmer BM. Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar Typhimurium. Microbiology. 2006;152:3411–3424. doi: 10.1099/mic.0.29118-0. [DOI] [PubMed] [Google Scholar]
- Valverde C, Lindell M, Wagner EG, Haas D. A repeated GGA motif is critical for the activity and stability of the riboregulator RsmY of Pseudomonas fluorescens. J Biol Chem. 2004;279:25066–25074. doi: 10.1074/jbc.M401870200. [DOI] [PubMed] [Google Scholar]
- Wang Q, Frye JG, McClelland M, Harshey RM. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol Microbiol. 2004;52:169–187. doi: 10.1111/j.1365-2958.2003.03977.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, et al. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- Wolfe AJ, Visick KL. Get the message out: cyclic-Di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol. 2008;190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak CE, Lee C, Hughes KT. T-POP array identifies EcnR and PefI-SrgD as novel regulators of flagellar gene expression. J Bacteriol. 2008 doi: 10.1128/JB.01177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Trimble JJ, Chiaro CR, Babitzke P. Effects of mutations in the L-tryptophan binding pocket of the Trp RNA-binding attenuation protein of Bacillus subtilis. J Biol Chem. 2000;275:4519–4524. doi: 10.1074/jbc.275.6.4519. [DOI] [PubMed] [Google Scholar]