Summary
Dehydrophos is a vinyl phosphonate tripeptide produced by Streptomyces luridus with demonstrated broad spectrum antibiotic activity. To identify genes necessary for biosynthesis of this unusual compound we screened a fosmid library of S. luridus for the presence of the phosphoenolpyruvate mutase gene, which is required for biosynthesis of most phosphonates. Integration of one such fosmid clone into the chromosome of Streptomyces lividans led to heterologous production of dehydrophos. Deletion analysis of this clone allowed identification of the minimal contiguous dehydrophos cluster, which contained 17 open reading frames (ORFs). Bioinformatic analyses of these ORFs are consistent with a proposed biosynthetic pathway that generates dehydrophos from phosphoenolpyruvate. The early steps of this pathway are supported by analysis of intermediates accumulated by blocked mutants and in vitro biochemical experiments.
Keywords: phosphonate, antibiotic, Streptomyces, biosynthetic, gene cluster
Introduction
Dehydrophos, originally designated A53868, was first described by Eli Lilly in 1984 as a broad spectrum antibiotic present in culture supernatant of Streptomyces luridus (Johnson et al., 1984). Dehydrophos has in vitro activity against gram negative and positive bacteria and demonstrated in vivo activity in a Salmonella/chicken model system (Johnson et al., 1984). The chemical structure of dehydrophos has been revised multiple times, most recently in 2007 (Whitteck et al., 2007). The revised structure of dehydrophos reveals a unique O-methylated vinylphosphonate connected to a glycine-leucine dipeptide by an amide bond. Based on similarity of the vinylphosphonate moiety to dehydroalanine, it was proposed that the compound be renamed dehydrophos (Whitteck et al., 2007).
Phosphonic and phosphinic acids are characterized by a C-P bond and contain phosphorus in a reduced form (+3, +1 respectively). Interest in these compounds as potential antibiotics lies in the structural similarity shared with phosphate esters and carboxylate-containing compounds, ubiquitous participants in cellular biology. Unlike phosphate esters, reduced phosphorus compounds are resistant to hydrolysis and enzymatic cleavage by phosphatases. These properties afford phosphonates the potential to inhibit a wide variety of metabolic processes (Metcalf and van der Donk, 2009). Indeed, known naturally occurring phosphonates such as K-26, phosphinothricin tripeptide (PTT), and fosfomycin (Seto and Kuzuyama, 1999) are structurally diverse and affect diverse biological processes. Despite widespread use in medicine and agriculture, understanding the biosynthesis of these compounds is still in a preliminary stage.
We have previously used a gene-targeted approach to identify gene clusters required for the biosynthesis of PTT (Blodgett et al., 2005), fosfomycin (Woodyer et al., 2006), and FR900098 (Eliot et al., 2008). This approach relies on the observation that biosynthesis of most known phosphonates requires the enzyme phosphoenolpyruvate (PEP) mutase (Seidel et al., 1988), with the notable exception of the phosphonic acid K-26 (Ntai et al., 2005). The conservation of the gene encoding PEP mutase allowed for design of degenerate PCR primers that have proven useful in the identification of phosphonic acid biosynthetic gene clusters from uncharacterized soil isolates and known phosphonic acid producers. Following identification and sequencing of the gene clusters required for the biosynthesis of these reduced phosphorus compounds, bioinformatic analysis has allowed hypothetical biosynthetic pathways to be tested by genetic and biochemical means (Blodgett et al., 2007; Eliot et al., 2008; Lee et al., 2009; Schwartz et al., 2004; Schwartz et al., 2005; Shao et al., 2008).
Previous studies of phosphonate biosynthesis have revealed a wealth of unprecedented and interesting biochemistry (Blodgett et al., 2007; Cicchillo et al., 2009; Liu et al., 2001). The unique structure of dehydrophos suggested that unusual transformations would also be present in its biosynthetic pathway. Here we report the cloning, sequencing, heterologous expression, and mutant analysis of the dehydrophos biosynthetic gene cluster from S. luridus and detail the early steps of the biosynthetic pathway.
Results
Cloning of the dehydrophos gene cluster and heterologous production in S. lividans
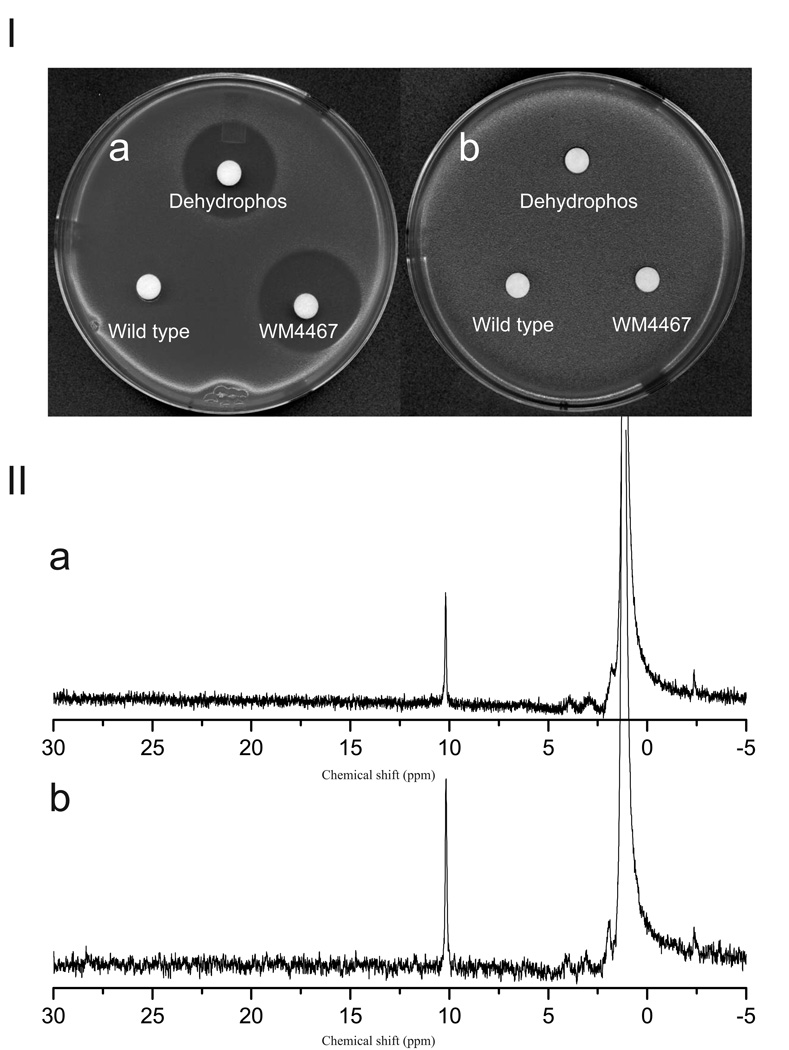
To identify genes necessary for dehydrophos biosynthesis, a fosmid library of S. luridus NRRL 15101 was assembled in E. coli and individual clones were screened by PCR for the PEP mutase gene. Of 2496 fosmid clones that were screened, ten were PEP mutase positive. To test whether these clones contained all required genes for dehydrophos production, they were modified with helper plasmid pAE4 to allow their transfer into the chromosome of S. lividans (Eliot et al., 2008). One such recombinant, S. lividans WM4467, produced a compound that inhibited the growth of E. coli DH5α, but not of an isogenic dehydrophos resistant mutant, suggesting the strain acquired the ability to synthesize dehydrophos. No growth inhibition was detected using supernatant from unmodified S. lividans (Figure 1). Further evidence that the compound produced by the recombinant strain was dehydrophos was provided by 31P NMR analysis. Supernatant from the recombinant strain exhibits a peak in the 31P NMR spectrum with a chemical shift of ~10.2 ppm, which corresponds closely to the chemical shift of dehydrophos from the native producer, S. luridus. Addition of a synthetic dehydrophos standard resulted in increased intensity of the 10.2 ppm peak and lacked the appearance of new peaks, corroborating the identity of this phosphonate as dehydrophos (Figure 1 II). An additional 31P NMR peak was observed in supernatant from the heterologous producer during longer incubations. Located at 24.5 ppm, this peak is also present in the supernatant of the native producer and increases in intensity over time, possibly indicating a breakdown product of the tripeptide. Control experiments revealed that unmodified S. lividans converts dehydrophos into a compound with the same 31P NMR spectrum, supporting this contention (data not shown).
FIG. 1.
(I) Bioassay demonstrating heterologous production of dehydrophos. Production of dehydrophos was assayed against sensitive (a) and resistant (b) strains of E. coli. Filter disks were spotted with supernatant from WM4467, synthetic standard of dehydrophos, or supernatant from wild type S. lividans, as indicated. (II) 31P NMR demonstrating heterologous production of dehydrophos. (a) Supernatant collected from WM4467, (b) As (a) but containing synthetic dehydrophos added as an internal standard.
Sequencing, delineation of boundaries, and bioinformatic analysis of the gene cluster
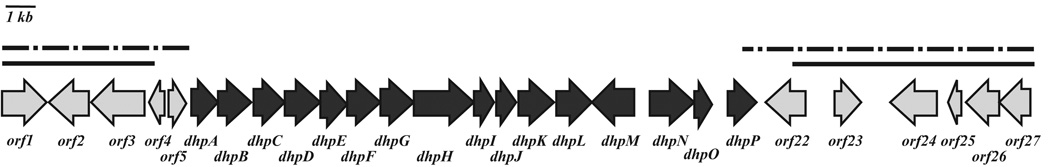
The fosmid clone that conferred dehydrophos production to S. lividans was sequenced and shown to carry a 35.9 kb insert of S. luridus genomic DNA (Figure 2). In silico sequence analyses identified 27 potential genes within the insert. To determine the minimal contiguous cluster required for dehydrophos production, deletion derivatives that truncated the fosmid from either the 5′ or 3′ end of the insert were constructed and transferred to the chromosome of S. lividans. The effects of these deletions on dehydrophos production were monitored by 31P NMR spectroscopy and bioassay. A recombinant strain lacking ORFs 1–4 retained the ability to produce dehydrophos, whereas a strain in which ORFs 1–5 and a portion of dhpA were removed abolished production of dehydrophos (Figure S1). Likewise, a strain in which ORFs 22–27 were deleted continued to produce dehydrophos; whereas a strain in which a portion of dhpP was deleted in addition to ORFs 22–27 was unable to produce dehydrophos (Figure S1). These experiments revealed that a contiguous segment of DNA, comprised of ORFs orf5 to orf21, is needed to confer dehydrophos production on a heterologous host. Accordingly, ORFs orf6 to orf21 were designated dhpA-dhpP.
FIG. 2.
Gene organization of fosmid 17E11-4. Dark arrows indicate ORFs thought to be involved in dehydrophos biosynthesis. The solid lines indicate DNA fragments whose deletion has no effect on production of dehydrophos, while the dotted lines indicate deletions that abolish production.
The organization of the dhpA-dhpL genes suggests they are co-transcribed (Figure 2). Many of these genes share overlapping start/stop codons (dhpA-dhpB, dhpC-dhpD, dhpE-dhpF-dhpG-dhpH). Three of these genes, dhpE (PEP mutase), dhpF (phosphonopyruvate decarboxylase), and dhpG (Fe-dependent alcohol dehydrogenase) have homologs present in multiple phosphonate biosynthetic clusters (Shao et al., 2008). Bioinformatic analysis, consisting of BLAST searches followed by alignment to representative proteins and identification of conserved active site residues allowed a proposed function to be assigned to each open reading frame (Table 1).
TABLE 1.
Summary of 17E11 open reading frames
| ORF | No. of amino acids |
Protein homology1 | Amino acid identity (%) |
|---|---|---|---|
| orf1 | 509 | Streptomyces lividans d-Ala-d-Ala carboxypeptidase (YP_002193425) | 215/366 (59%) |
| orf2 | 469 |
Thermomonospora curvata FAD dependent oxidoreductase (ZP_04033692) |
336/465 (72%) |
| orf3 | 624 |
Salinispora arenicola polysaccharide pyruvyl transferase (YP_001538394) |
228/366 (62%) |
| orf4 | 170 | Bacillus sp. SG-1steroid Δ-isomerase domain protein (ZP_01861119) | 51/109 (47%) |
| orf5 | 86 |
Streptomyces ghanaensis putative transglycosylase associated protein (ZP_04690162) |
75/81(92%) |
| dhpA | 299 | uncultured microorganism putative γ-butyrobetaine hydroxylase (ABZ07908) |
72/254 (28%) |
| dhpB | 383 | Vibrio splendidus glycerate kinase (YP_002394749) | 115/332 (35%) |
| dhpC | 371 |
Aciduliprofundum boonei malate/l-lactate dehydrogenase (YP_002578914) |
130/367(35%) |
| dhpD | 397 |
Candidatus Pelagibacter ubique aspartate transaminase (ZP_01264754) |
112/403 (28%) |
| dhpE | 296 | Frankia alni putative PEP phosphomutase (YP_716511) | 147/275 (53%) |
| dhpF | 381 |
Amycolatopsis orientalis putative phosphonopyruvate decarboxylase (CAB45023) |
182/371(49%) |
| dhpG | 396 | Amycolatopsis orientalis putative alcohol dehydrogenase (CAB45024) | 152/367(41%) |
| dhpH | N′ terminal domain (355 AA) Amycolatopsis orientalis putative aminotransferase (CAB45025) |
169/355 (48%) | |
| C′ terminal domain (343 AA) Yersinia pseudotuberculosis FAD linked oxidase domain-containing protein (YP_001721344) |
101/348 (29%) | ||
| dhpI | 218 |
Streptomyces clavuligerus putative S-adenosylmethionine-dependent methyltransferase (YP_002193190) |
89/211 (42%) |
| dhpJ | 230 | Streptomyces albus peptide aspartate β-dioxygenase (ZP_04702360) | 113/230 (49%) |
| dhpK | 422 | marine γ proteobacterium hypothetical protein (YP_002651317) | 72/271 (27%) |
| dhpL | 422 |
Bradyrhizobium japonicum putative integral membrane transporter (NP_774016) |
160/352 (45%) |
| dhpM | 503 | Frankia sp. CcI3 Na+/solute symporter (YP_481567) | 274/465 (59%) |
| dhpN | 520 | Frankia sp. CcI3 Na+/solute symporter (YP_481567) | 307/507(61%) |
| dhpO | 207 | Frankia sp. CcI3 σ-24 (YP_481568) | 105/172 (61%) |
| dhpP | 339 |
Streptosporangium roseum lacI family transcription regulator (ZP_04472611) |
219/336 (65%) |
| Orf22 | 452 |
Actinosynnema mirum cellulase (glycosyl hydrolase family 5) ZP_03814915 |
193/322 (60%) |
| Orf23 | 294 | Streptomyces ambofaciens hypothetical protein (CAI78012) | 125/228 (55%) |
| Orf24 | 519 | Streptomyces ambofaciens putative export protein (CAJ90162) | 420/527 (80%) |
| Orf25 | 163 | Streptosporangium roseum transcriptional regulator (ZP_04478568) | 112/156 (72%) |
| Orf26 | 392 | Streptomyces sp. Mg1 aspartate aminotransferase (YP_002180528) | 305/384 (79%) |
| Orf27 | 350 | Streotomvces sp. Ma1 hvoothetical protein (YP_0021805291) | 251/306 (82%) |
Results generated by BLAST analysis of deduced open reading frames. Accession numbers are listed in parentheses.
Mutant analysis of dhpA-dhpP
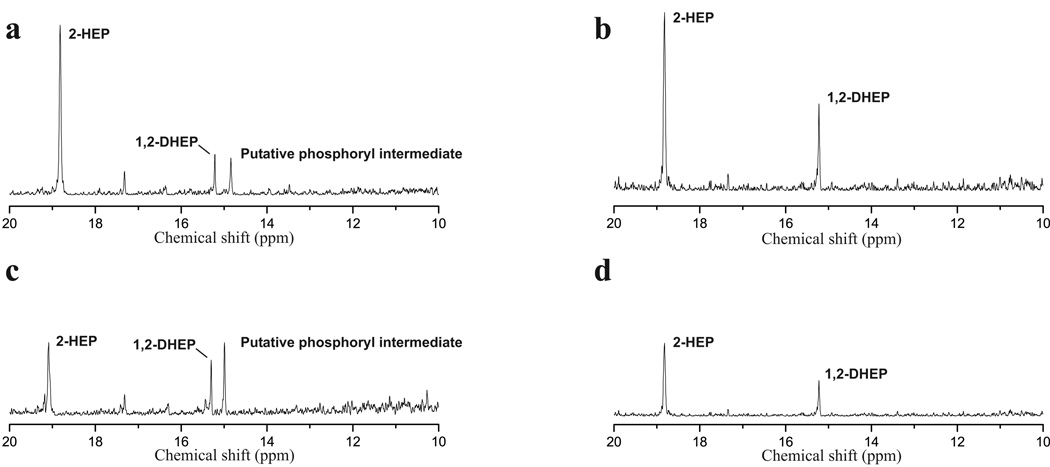
To define the role of individual dhp genes in dehydrophos biosynthesis we constructed a series of plasmids with deletions or disruptions of each gene in the biosynthetic cluster using λ red based recombination (Datsenko and Wanner, 2000). To prevent polar effects, these cassettes were designed to preserve the original gene spacing in the operon, with the exception of a 6 bp AvrII scar. The resultant plasmids were then transferred into the heterologous host S. lividans and dehydrophos production was queried by bioassay and 31P NMR spectroscopy. Mutations in dhpA, dhpB, dhpC, dhpD, dhpE, dhpF, dhpG, dhpH, dhpI, dhpJ, dhpK, dhpM and dhpP abolished antibiotic production (Figure S2) and led to the disappearance of the dehydrophos peak from the 31P NMR spectra (Figure S3). Most of these strains also show accumulation of new peaks in the 10–25 ppm range (Figure S3), suggesting that intermediates are accumulating in these blocked mutants (see below). Mutations affecting dhpL and dhpN have no discernible effect on bioactivity (Figure S2). In sum, these results indicate that all genes examined, with the exceptions of dhpL and dhpN are required for dehydrophos biosynthesis.
Genetic complementation experiments were carried out on select mutants to examine the possibility that their phenotypes were mediated by transcriptional polarity. To do this, a functional copy of the deleted gene was introduced at a distant site in the S. lividans chromosome under the control of the constitutive Perm* promoter. Restoration of dehydrophos biosynthesis after complementation of the dhpB, dhpH, and dhpI mutations demonstrates that these particular mutations were not affecting expression of downstream genes (Figure S2). Other mutations were not tested for polar effects due to their position in the gene cluster (i.e. they are not upstream of any other dhp genes), or because the mutation did not effect bioactivity. Mutant gene clusters of dhpC and dhpJ were not tested for complementation and therefore the formal possibility of polar effects being responsible for their observed phenotypes exists. We view this as unlikely in the case of dhpC because the products of the downstream genes (dhpE, dhpF, and dhpG) are needed to produce the intermediates observed to accumulate in the dhpC mutant (see below).
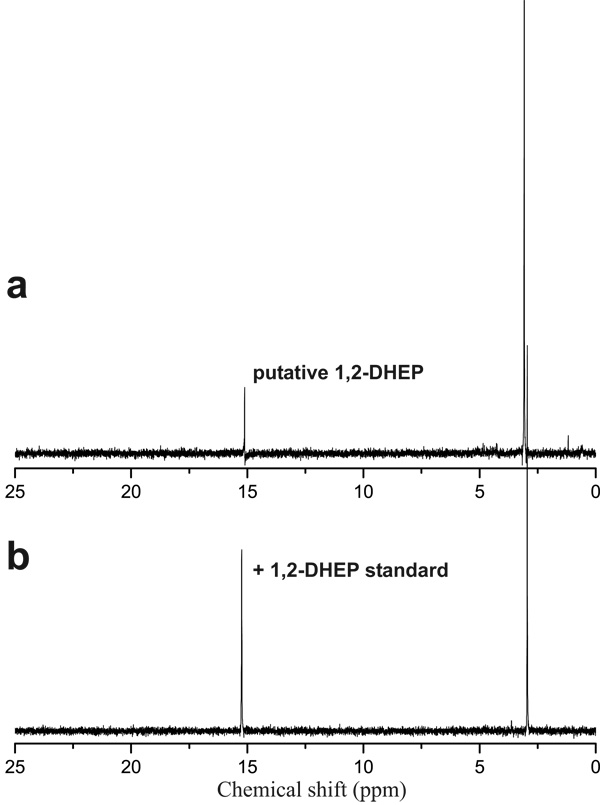
As described above, a number of the mutants accumulated putative phosphonate intermediates in spent media. In an attempt to identify these compounds, authentic standards of commercially available putative intermediates were directly added to NMR samples containing concentrated culture supernatant and new spectra were acquired using the same parameters. In many cases, putative intermediates were unavailable commercially, which necessitated chemical synthesis (see Materials and Methods). The accumulation of 2-hydroxyethylphosphonate (HEP, 31P chemical shift at 19 ppm), and 1,2 dihydroxyethylphosphonate (DHEP, 31P chemical shift at 15.2 ppm) in dhpA and dhpB mutants respectively, were confirmed in this manner (Figure 3, Figure S4). Mutations in dhpK, dhpP, and dhpD abolished dehydrophos production based on NMR data and bioassays (Figure S1, S2); however, supernatants from these strains produce a 31P NMR spectrum that contains a phosphonate with a chemical shift of 24.5 ppm, similar to the break-down product witnessed in both the wild-type and heterologous producers of dehydrophos.
FIG. 3.
dhpB mutants accumulate DHEP. 31P NMR spectra of concentrated culture supernatant from a dhpB mutant (a), and that same sample spiked with a synthetic standard of DHEP (b).
In vitro activity of DhpA
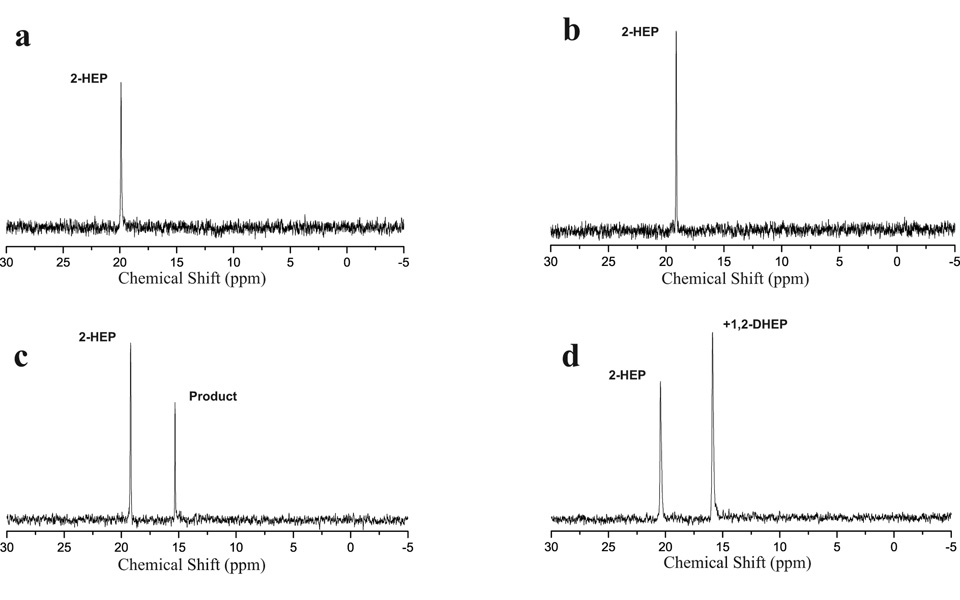
The homology of DhpA to known 2-oxoglutarate-dependent dioxygenases, and accumulation of HEP in the supernatant of a ΔdhpA mutant led to the expectation that DhpA converted HEP to DHEP. To test this hypothesis, the protein was overproduced in E. coli as a 6X-His-tagged protein, purified, and assayed for 2-oxoglutarate-dependent dioxygenase activity with synthetic HEP. The reaction was monitored via 31P NMR spectroscopy. After a 30 minute incubation, the presence of a second phosphonate peak was observed in the 31P NMR spectrum (Figure 4, panel C). Addition of authentic DHEP into the sample confirmed that DhpA converted HEP into DHEP (Figure 4, panel D). This reaction occurred in a 2-oxoglutarate dependent fashion, but did not proceed to completion in the reaction time. Additional experiments indicated DhpA could also convert HEP O-phosphomonomethylester into DHEP O-phosphomonomethylester in a 2-oxoglutarate dependent fashion (data not shown)
FIG. 4.
31P NMR spectra of DhpA assay. HEP was incubated with purified DhpA and α-ketoglutarate (c), incubated without enzyme (a), or incubated without α-ketoglutarate(b). The reaction shown in panel c was then spiked with DHEP to verify the product (d).
Identification of a CIAP labile phosphonate intermediate
Based on 31P NMR spectroscopy, culture supernatant from dhpC and dhpH mutants was known to contain DHEP (Figure 5 31P chemical shift at 15.2 ppm) and hypothesized to contain a phosphorylated form of DHEP (Figure 5, 31P chemical shift at 14.9 ppm). To test this, the supernatant was treated with calf intestinal alkaline phosphatase (CIAP), which is expected to hydrolyze most phosphate esters. Phosphatase treatment eliminated the resonance at 14.9 ppm, with a concomitant increase in the intensity of the DHEP peak at chemical shift of 15.2 ppm (Figure 5). The identity of DHEP was confirmed prior to and following phosphatase treatment by addition of authentic standard. Thus the phosphonate intermediates in these mutants contain a phosphate ester modification. This intermediate fails to demonstrate P-P splitting in the NMR spectrum, and therefore we feel this modification occurs at the more distally located 2-hydroxy position.
FIG. 5.
dhpC and dhpH mutants accumulate a phosphate ester. 31P NMR spectra of concentrated culture supernatants from a dhpC mutant (a), a dhpH mutant,(c) and those same supernatants after treatment with calf intestinal alkaline phosphatase (b and d, respectively).
Discussion
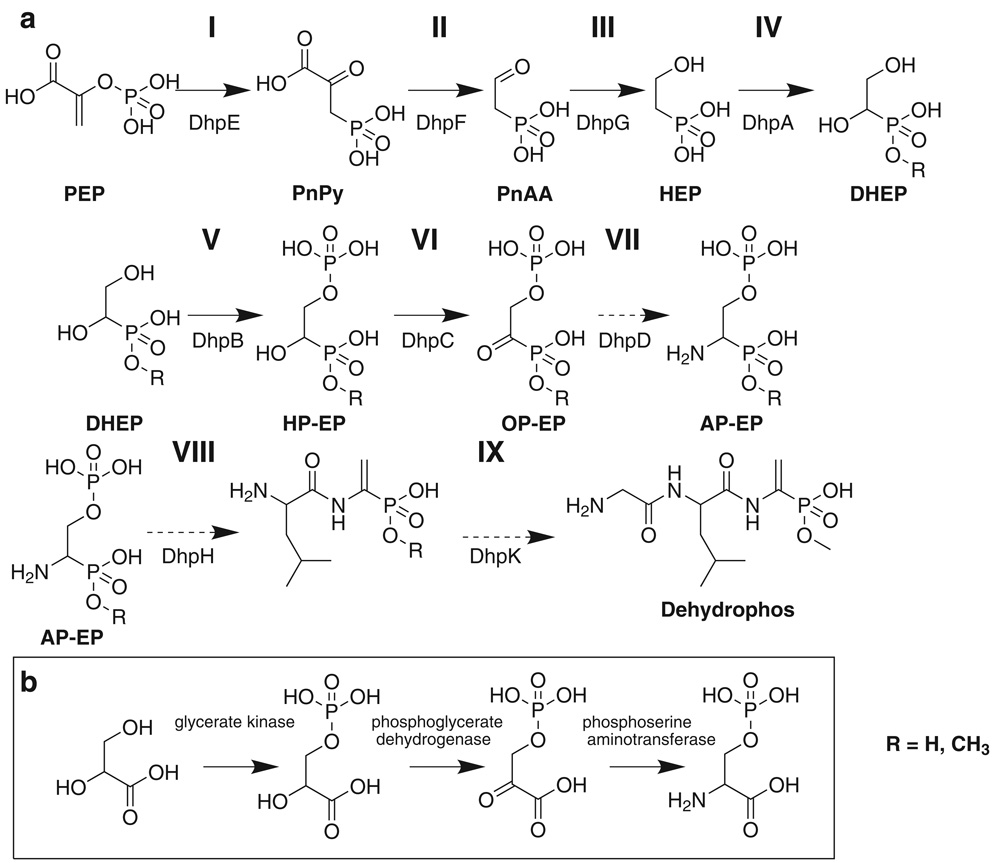
Dehydrophos is an unusual natural product with several features that are unique among known natural products. The data presented here led to a proposed biosynthetic pathway that accounts for the synthesis of some, but not all of these features (Figure 6). This pathway begins with conversion of PEP to phosphonopyruvate (PnPy) (Step I), subsequent decarboxylation (Step II) to generate phosphonoacetaldehyde (PnAA), and reduction of PnAA to yield HEP (Step III). A hydroxy group is then introduced at C1 of HEP (Step IV). Following this, the C2 hydroxy group undergoes phosphorylation (Step V), and the C1 hydroxy group is oxidized (Step VI), and transaminated (Step VII). At this point generation of the vinyl group is coupled to the first peptide bond formation (Step VIII). Addition of the glycine residue and O-methylation round out completion of the dehydrophos molecule (Step IX).
FIG 6.
Proposed pathway for dehydrophos biosynthesis (a), and the portion of the serine biosynthetic pathway that bears structural similarity to step V, VI, and VII of the proposed pathway (b). Broken arrows indicate hypothetical reactions. OP-EP = 1-oxo-2-phosphorylethylphosphonate.
The first two steps proposed in the pathway mirror those of all other known phosphonate biosynthetic pathways. The PEP mutase reaction is thermodynamically unfavorable (Bowman et al., 1988) and often coupled to PnPy decarboxylase to yield net synthesis of phosphonates. Based on significant amino acid homology to biochemically characterized examples, DhpE and DhpF catalyze the PEP mutase and PnPy decarboxylase reactions, respectively. Both of these enzymes also possess complete conservation of residues known to be important for catalytic activity (Jia et al., 1999; Zhang et al., 2003).
The next proposed reaction is a metal-dependent dehydrogenase catalyzed reduction of PnAA to HEP. This reaction is common to several unrelated phosphonate biosynthetic pathways. We previously overexpressed and purified DhpG. The enzyme was shown to be a Group III Fe-dependent PnAA dehydrogenase which consumes NAD(P)H during the reduction reaction (Shao et al., 2008).
Given the final structure of dehydrophos, biochemical logic suggests that a free primary amine be available at C1 for peptide bond formation. The hypothetical pathway achieves this placement by hydroxylation of HEP at C1, followed by oxidation and transamination reactions (Fig 7, Steps VI, VII). The accumulation of HEP in blocked mutants implicated DhpA as a 2-oxoglutarate dependent dioxygenase responsible for the hydroxylation (step IV). Subsequent in vitro experiments confirmed this function.
DHEP is a phosphonic acid analog of glyceric acid. Thus, it was interesting to note that DhpC has significant homology to malate and lactate dehydrogenases, which catalyze oxidation of similar substrates. Further, comparison of DhpC to known malate dehydrogenases revealed conservation of all important active site residues in DhpC (Goward and Nicholls, 1994). Based on these observations we expected DhpC to catalyze oxidation of DHEP to 1-oxo, 2-hydroxyethylphosphonate (OHEP). As such, a deletion of the dhpC gene was expected to result in the accumulation of DHEP, which we showed has a chemical shift of approximately 15 ppm in 31P NMR. Surprisingly, the dhpC mutant accumulates two phosphonate intermediates with 31P NMR peaks near 15 ppm, one of which was shown to be DHEP. Treatment of dhpC culture supernatant with alkaline phosphatase clearly shows that the second compound is a phosphate ester of DHEP. Consistent with this idea, the protein product of dhpB (located directly upstream of dhpC) is homologous to glycerate kinase, including conservation of seven out of nine important active site residues (Cheek et al., 2005). Thus, DhpB could be responsible for phosphorylation of DHEP producing 1-hydroxy-2-phosphorylethylphosphonate (HP-EP), the phosphonate analog of 3-phosphoglycerate, which would represent the second phosphonate compound seen in culture supernatants of the dhpC mutant (Figure 6, Step V). Supernatant from mutants containing deletions of dhpB accumulate only DHEP, suggesting that the DhpB reaction occurs before the DhpC reaction. Therefore, we have assigned DhpB to step V and DhpC to stepVI (Figure 6).
The function that phosphorylation of DHEP serves is uncertain. During lantibiotic biosynthesis, phosphorylation precedes the dehydration reaction involved in the formation of dehydroalanine and dehydrobutyrine residues (Willey and van der Donk, 2007). A similar reaction could generate the vinyl group in dehydrophos. However, the dehydrophos gene cluster lacks ORFs with recognizable homology to the enzymes that perform these dehydrations in lantibiotic biosynthesis. Alternatively, phosphorylation could provide protection from an otherwise toxic intermediate, in a manner similar to acetylation of demethylphosphinothricin (Blodgett et al., 2005).
Step VII is predicted to be a transamination of OHEP. DhpD contains sequence homology to aspartate aminotransferases and residues known to be involved in PLP binding (Inoue, et al., 1991) are conserved. Mutants lacking dhpD fail to produce dehydrophos, as observed by 31P NMR spectroscopy and bioassay. However, these mutants also fail to accumulate the expected intermediates from this blockage (DHEP and HP-EP). We suspect that endogenous aminotransferases present within S. lividans may be able to act in a promiscuous fashion and bypass this step in the pathway in an inefficient manner. Similar promiscuous transaminations have been witnessed in research on other phosphonate clusters (Blodgett et al., 2007). Pending further investigation, we have tentatively assigned DhpD to step VII.
The reactions putatively catalyzed by DhpB, DhpC, and DhpD lend remarkable structural similarity between the intermediates and transformations found in steps V, VI, VII of the proposed dehydrophos pathway and those found in serine biosynthesis (Figure 6, panel b). Interestingly, several other phosphonate biosynthetic pathways contain reactions ‘borrowed’ from central metabolic pathways (Blodgett et al., 2007; Eliot et al., 2008; Metcalf and van der Donk, 2009). This suggests the intriguing possibility that dehydrophos biosynthesis evolved from enzymes of the related serine biosynthetic pathway.
The compound resulting from these reactions, 1-amino-2-phosphorylethylphosphonate (AP-EP) is proposed to be four steps away from dehydrophos: addition of leucine and glycine residues, dehydration to form the vinyl group, and O-methylation. Although dehydrophos contains two peptide bonds, the minimal cluster lacks identifiable non-ribosomal peptide synthetases. It seems unlikely that dehydrophos is ribosomally synthesized, because of its small size and the need for the specialized genetic code required to identify, charge, and load a phosphonate precursor. The protein products of dhpH and dhpK contain putative Acyl-CoA N-acyltransferase domains. Similar domains are found in FemXAB family proteins responsible for peptide bond formation during cross-linking of bacterial cell wall peptidoglycan (Biarrotte-Sorin et al., 2004). FemXAB proteins utilize activated aminoacyl-tRNAs to generate peptide bonds. Additional examples of aminoacyl-tRNA based peptide bond formation in small molecule biosynthesis (Gondry et al., 2009) make the potential use of an analogous mechanism by DhpH and DhpK intriguing.
The mechanism of vinyl group formation also remains uncertain. It seems likely that this step occurs either simultaneously with, or after, peptide bond formation because dehydration of the primary amine would form an eneamine, which would tautomerize to the imine and undergo hydrolysis. An attractive solution to this problem is present in the ORF encoding DhpH. The ORF encodes a large protein (698 AA) that consists of two distinct domains. The N′ terminal domain possesses homology to serine-glyoxylate aminotransferases and contains conserved residues known to bind pyridoxal phosphate (PLP). The C′ terminal domain displays homology to proteins with oxidase activity, but further analysis by InterProScan reveals the aforementioned N-acyl CoA acyltransferase functionality presumably responsible for peptide bond formation. PLP containing enzymes are capable of catalyzing β-eliminations (Eliot and Kirsch, 2004) and therefore, DhpH could eliminate the phosphate from intermediate VIII, generating 1-aminovinylphosphonate. If so, this intermediate would have to be protected from hydrolysis to allow peptide bond formation by the second domain. Alternatively, the vinyl group could be formed after peptide bond formation, driven by hydrolysis of the phosphate ester on the 1-amino-2-phosphorylethylphosphonate moiety.
Dehydrophos contains a unique O-methylation of the phosphonate group. A likely candidate to perform this reaction is DhpI, which is predicted to be a SAM-dependent methyltransferase. The point at which this reaction takes place remains to be determined, because methylated derivatives of HEP and AEP can be seen accumulating to low amounts in dhpA and dhpJ mutants (31P NMR peaks at approximately 25.5 ppm and 23.5 ppm respectively). However, the concentration of methylated early intermediates is much lower than that of the corresponding unesterified compounds, suggesting early methylation may be an artifact of mutations which block the pathway. Preliminary data indicate that purified DhpA is capable of utilizing HEP O-phosphomonomethylester as a substrate (unpublished data). Additional enzymatic studies may clarify the preferred substrate of DhpA.
BLAST homology analysis of the protein product of dhpL reveals similarity to membrane transport proteins, suggesting that DhpL is involved in self-resistance and/or export of the finished compound. Similarly DhpM and DhpN show homology to Na+/solute symporters and are predicted to contain the 13 transmembrane domains characteristic of these proteins. DhpM and DhpN show remarkable homology to each other (68% identity, 318/471 residues) suggesting they may function as a dimer.
Finally, DhpP is homologous to σ-24 transcriptional factors, while DhpO is a member of the LacI repressor family of transcriptional regulators. Both of these may be involved in regulation of the cluster, although the specific roles they play in the heterologous host are unknown. Orf5 falls within the segment of DNA necessary to confer production to S. lividans. This ORF contains a small peptide with weak homology to signal peptides and transglycosylases. It is uncertain what role, if any, this ORF plays in the biosynthesis of dehydrophos.
Still unaddressed are questions regarding the mode of dehydrophos activity. PTT and rhizocticin (an antifungal phosphonate produced by Bacillus subtilis ATCC 6633) promote uptake by synthesis as tripeptides which are imported by non-specific oligopeptide permeases (Diddens et al., 1976; Kugler et al., 1990). Once inside the cell, these compounds are hydrolyzed to release biologically active phosphonate. Hydrolysis of the analogous dehydrophos tripeptide would release 1-aminovinylphosphonate (a dehydroalanine analogue) and result in an eneamine expected to tautomerize to the imine and undergo hydrolysis to produce methyl acetylphosphonate. This compound is a potent inhibitor of pyruvate dehydrogenase, with a Ki (5 × 10−8 M) 125 times lower than unesterified acetylphosphonate (O'Brien et al., 1980). This increased activity may provide a rationale for the O-methylation present on the dehydrophos phosphonate moiety. Methylation reduces the negative charge on the phosphonate moiety by one, causing the charge distribution to be more similar to a carboxylate group. This strategy of mimicking carboxylates is similar to the C-P-C arrangement found in PTT, but is constructed by a much simpler biosynthetic route. Identification and manipulation of the enzyme responsible for this reaction could lead to facile methylation of existing phosphonates, potentially increasing their efficacy or altering their targets by allowing these compounds to more closely resemble carboxylate containing compounds.
Significance
This work addresses the issue of bacterial resistance by exploring the biosynthesis of the antibiotic dehydrophos. Research focusing on the biosynthesis of phosphonates as secondary metabolites is rather limited and the data presented here may be of general use in deciphering future phosphonate gene clusters. The data raise specific questions about peptide bond formation and vinyl group formation. Although dehydrophos is a tripeptide, no identifiable non-ribosomal peptide synthetases are present within the minimal cluster, indicating an alternate mode of peptide bond formation. Similarly, dehydrophos contains an unusual vinylphosphonate moiety with no obvious mechanism for formation within the gene cluster. Understanding these transformations could reveal unique and useful chemistry in its own right, and aid in the combinatorial biosynthesis of phosphonates and other small molecules. Identification of the protein responsible for the novel methylation of the dehydrophos molecule is particularly attractive as a means of altering the charge distribution of the phosphonate to resemble a carboxylate, which could prove invaluable in downstream engineering applications. Finally this work provides insight into the evolutionary origin of dehydrophos biosynthesis, suggesting that portions of the pathway originated in serine biosynthesis.
Materials and Methods
Bacterial strains, plasmids and culture conditions
The strains and plasmids used in this study are listed in Supplementary Table S1. Streptomyces strains were propagated on ISP2 medium (Difco, Becton Dickinson Microbiology Systems, Sparks, MY) at 30 °C. Heterologous production cultures were inoculated with 50 µl of spore stock (109 colony forming units) of the appropriate strain and incubated in 500 mL baffled flasks containing 100 mL of nutrient broth (Difco) agitated at 250 rpm for 48 hours. E. coli was routinely grown in Luria-Bertani broth (LB) or on 1.5% agar LB plates supplemented appropriately. Antibiotics were used at the following concentrations to select for plasmid propagation or integration events: ampicillin 100 µg/mL, chloramphenicol 10 µg/mL, apramycin 37.5 µg/mL, kanamycin 50 µg/mL. Diaminopimelic acid was added to media at a concentration of 1 mM for growth of dapA strains. For dehydrophos bioassays E. coli BW13711 was grown to an OD600 of 0.1, and then 100 µL was mixed with 2 mL of molten ‘E’ media 0.5 % top agar (Vogel and Bonner, 1956) and pipetted onto ‘E’ media 1.5% agar plates and allowed to solidify. These plates were incubated at 37 °C for 12 hours. Succinate (40 mM) was provided as the carbon source during bioassays.
DNA isolation, manipulation, cloning and sequencing
Cloning was accomplished by routine methods (Sambrook et al., 1989). Restriction endonucleases and T4 DNA ligase were purchased from New England Biolands (Ipswich, MA) or Invitrogen (Carlsbad, CA). KOD HotStart polymerase (Novagen, EMD Chemicals Inc., Gibbstown, NJ) was used for PCR amplification in combination with Failsafe PCR Premix buffers (Epicentre, Madison, WI). Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). DNA fragments used in cloning experiments were isolated by gel purification using the Promega Wizard PCR cleanup kit (Madison, WI). Plasmid isolation was accomplished by Qiagen Maxiprep or Miniprep kits (Valencia, CA). Sequencing reactions were performed at the W.M. Keck Center for Biotechnology at the University of Illinois Urbana-Champaign utilizing primers T7-for/T7-rev or primers pBTC034-For/pBTC034-Rev. Sequence data were compiled using Sequencer (Gene Codes Co., Ann Arbor, MI) and deduced ORFs were subjected to BLAST (Altschul et al., 1990), InterProScan (Zdobnov and Apweiler, 2001), and ClustalW alignments with proteins of determined function.
Generation of a S. luridus genomic library, library screening and sequencing of fosmid 17E11-4
Construction of a S. luridus genomic library was accomplished as described previously (Eliot et al., 2008) except WM4489 was used as the cloning host. This library was screened by PCR for the presence of the PEP mutase gene as previously described (Eliot et al., 2008). PCR reactions contained 1 µL of culture broth as template, 500 nM of primers luridusppm-for/luridusppm-rev, and Taq polymerase in Failsafe buffer G. To provide the functions necessary for downstream genetic manipulation, the PEP mutase positive fosmid clones were recombined with plasmid pAE4 (Eliot et al., 2008) by in vitro recombination using BP clonase as directed by the manufacturer (Invitrogen, Carlsbad, CA). WM4489 cells containing fosmid/pAE4 cointegrants were selected for on LB + kanamycin following transformation. Following in vitro transposon mutagenesis with the mini-Mu containing BglII fragment of pAE5 as described (Eliot et al., 2008), sequencing of fosmid 17E11-4 was performed at the W. M. Keck Center for Biotechnology with primers SeqAETnR/SeqAETnL. The insert sequence has been deposited into GenBank as GU199252.
Delineation of the minimal contiguous dehydrophos biosynthetic cluster
Deletion constructs were generated by in vitro recombination between FRT or loxP sites present on the fosmid backbone and corresponding sites on individual mini-Mu transposon insertions within the cluster DNA as described (Eliot et al., 2008). Following site-specific recombination catalyzed by the appropriate recombinase (FLP or Cre), all intervening DNA between the transposon insertion and the fosmid backbone was deleted. Conjugal transfer of fosmids was performed as described elsewhere (Martinez et al., 2004) with the modifications described below. The donor strain (WM6026) containing the fosmid to be transferred, was mixed with recipient S. lividans and the entire solution was spotted onto R2 (no sucrose) plates (Kieser et al., 2000) in 10 µl aliquots and incubated at 37 °C. After 12 hours the plates were flooded with 2 mls of 1mg/mL apramycin or neomycin and returned to the incubator. After 48 hours, exconjugants were picked and restreaked twice onto selective ISP2 before PCR verification.
Generation of additional mutant clusters
Three types of individual gene mutants were generated for analysis of dehydrophos biosynthesis; transposon inactivated mutants, gene replacements, and gene deletions. Gene replacements and gene deletions were constructed in WM7264 by utilizing the λ Red mediated recombination method of Datsenko and Wanner (Datsenko and Wanner, 2000). Briefly, regions of homology upstream and downstream of the target are included on PCR primers designed to amplify a chosen resistance cassette. Following recombination between the PCR product and fosmid 17E11-4, the target gene is replaced by the kanamycin resistance gene of pAE5. Gene replacements of dhpA, dhpD, dhpM, dhpN, and dhpO were made in this fashion and retain the kanamycin resistance cassette. This cassette has a synthetic, outward-directed promoter designed to express downstream genes to minimize polar effects. The primers used in gene replacement construction also introduce an AvrII site not present on fosmid 17E11-4, which allows for excision of the antibiotic cassette following digestion and self-ligation of the fosmid. Digestion and self-ligation results in deletion of the target gene and the resistance cassette, leaving only a 6 base pair AvrII scar. Gene deletions of dhpB, dhpC, dhpE, dhpF, dhpG, dhpH, and dhpI were constructed using the aforementioned procedure. The remainder of the genes analyzed (dhpJ, dhpK, dhpL) are insertional mutants obtained following the saturation transposon mutagenesis with pAE5 generated for sequencing of the cluster. After PCR verification, the mutant clusters were electroporated into WM6026 and conjugally transferred to S. lividans as described above.
31P NMR detection and identification of dehydrophos and phosphonate metabolites
Culture supernatant was passed through a 0.2 micron filter, concentrated twenty fold by evaporation, and D2O was added to 25% as a lock solvent. 31P NMR spectra were acquired on a Varian Unity Inova 600 spectrophotometer equipped with a 5 mm AutoTuneX probe at the Varian Oxford Instrument Center for Excellence at the University of Illinois Urbana-Champaign. An external standard of 85% phosphoric acid was defined as 0 ppm. Supernatant obtained following growth on liquid ISP4 (Difco) media was utilized to confirm heterologous dehydrophos production. All other production experiments were conducted in nutrient broth. To verify heterologous production of dehydrophos, an authentic standard of dehydrophos was introduced into the NMR sample to a final concentration of 1 mM and the spectrum of the sample was retaken using identical parameters. To demonstrate that intermediates in the dehydrophos pathway contained a phosphate ester, 100 µl of calf intestinal alkaline phosphatase (CIAP) (Promega, Madison, Wisconsin) was added directly to the NMR tube, mixed by inversion, and allowed to incubate at 37 °C for one hour before reacquiring the NMR spectra under identical conditions. Experiments to confirm the identity of pathway intermediates were routinely performed by adding 1–5 mM of standard to the sample and then reacquiring the spectra.
Bioassay of dehydrophos production
Streptomyces lividans WM4467 and the collection of isogenic dhp mutants were grown on ISP4 plates at 30 °C for 48 hours. Agar cores were then taken from these plates and placed onto bioassay medium containing either E. coli DH5α or WM4553 (a spontaneous dehydrophos resistant mutant of DH5α). Alternatively, filter disks spotted with 10 µl authentic standard or concentrated culture supernatant were subjected to the same procedure as the agar cores. These plates were incubated at 37 ° for 16 hours and exam ined for growth inhibition of the indicator strain.
Overexpression, purification, and activity assay of His6-DhpA
WM7284 was grown at 37 °C to an OD 600 of 0.6, then cooled to 22 °C for ten minutes prior to the addition of 250 µM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added. The culture was incubated for an additional 18 h at 22 °C before harvesting by centrifugation at 5000 × g for ten minutes. The cell pellet was resuspended in 15 mL of ice cold Buffer 1 (50 mM HEPES pH 7.5, 250 mM NaCl, 25 mM imidazole, and 0.5% glycerol) and cells were disrupted by a single passage through a chilled French press cell at 20,000 psi. Solid debris was removed by centrifugation at 13,000 × g for 30 minutes at 4 °C. Cleared supernatant was passed over a column containing 5 mL of Ni-NTA affinity resin (Qiagen, Valencia, California) equilibrated with Buffer 1. Unbound proteins were washed from the column with 50 mL wash Buffer 2 (Buffer 1 with 50 mM imidazole). His6-DhpA was eluted from the column with 5 mL Buffer 3 (Buffer 1 with 250 mM imidazole). The eluant was desalted by passage over a PD-10 column (GE Healthcare), following the manufacturer’s instructions and eluted with Buffer 4 (Buffer 1 lacking imidazole). His6-DhpA purity was evaluated by visual inspection of an SDS-PAGE gel stained with Coomassie Brilliant Blue (estimated to be at least 95% pure). His6-DhpA activity was assayed at 30°C for 30 min in 50 mM HEPES buffer pH 7.5, 250 mM NaCl, 0.5% glycerol. Each 1 mL reaction also included: 5 mM HEP or HEP O-phosphomonomethylester, approximately 100 µg purified protein, 5 mM 2-oxoglutarate, and 10 µM ferrous ammonium sulfate. The reactions were stopped by incubation at 100 °C for 5 minutes, and 100 µL of 0.5M EDTA was added to chelate excess Fe(II) prior to 31P NMR analysis.
Construction of complementation vectors
Complementation experiments were performed using ΦBT1 integrating vectors developed by Smith et al (Gregory et al., 2003). However, these vectors did not include a streptomycete promoter for expression purposes. This function was included by PCR amplification and subsequent cloning of the strong, constitutive streptomycete promoter pERM* from pANT841 into the pRT802 vector. This resulted in the creation of pBTC034, which was used as the vector for cloning and conjugal transfer of genes being tested for complementation.
Chemical Syntheses of Phosphonates
All NMR spectra were recorded on Varian Unity 400 or Varian Unity 500 spectrometer. 1H NMR spectra are referenced to TMS at 0 ppm or H2O at 4.67 ppm, and 13C NMR spectra are referenced to CHCl3 at 77.0 ppm. For 31P NMR spectra 85% phosphoric acid in D2O was used as an external reference to 0 ppm. Fractions collected during silica gel column chromatography were analyzed by thin layer chromatography (TLC). Unless otherwise specified, all compounds and solvents were obtained from Fisher or Aldrich. CH2Cl2 was freshly distilled over CaH2 prior to use.
Dehydrophos (1)
Dehydrophos was synthesized and purified according to literature procedure (Whitteck et al., 2001).
Dimethyl 1,2-dihydroxyethylphosphonate ester (2) (Wang et al., 1994)
To a suspension of AD mix β* (6.02 g) in 50 mL of 1:1 tBuOH/H2O which had been stirred for 15 min at rt dimethyl vinylphosphonate was added dropwise (4.3 mmol, Alfa Aesar, USA) and the solution was stirred for 4 d at rt. The yellow suspension was quenched by addition of Na2SO3 (5.0 g) while on ice bath upon which the solution turned to green. After 20 min the ice bath was removed, and the suspension was further stirred at room temperature for another 20 min. The solution was extracted with EtOAc (3 × 50 mL), and the combined organic layers were dried over MgSO4. After evaporating excess solvents and reagents, the crude product was purified by a silica gel column chromatography (EtOAc:MeOH = 4:1) to afford the desired product in 29% yield as a colorless oil. 1H NMR (500 MHz, CDCl3) δ 4.13-4.10 (1H, m, PCH), 3.91-3.85 (2H, m, CH2), 3.83 (3H, d, J = 10.5 Hz, OCH3), 3.82 (3H, d, J = 10.5 Hz, OCH3); 13C (125.6 MHz, CDCl3) δ 68.8 (CH, d, J = 160 Hz), 62.3, 53.3 (OCH3, d, J = 24.9 Hz); 31P (202.3 MHz, CDCl3) δ 26.9. (*Stereospecific dihydroxylation by AD mix β yields enantiomerically enriched product, however, NMR experiments described in this report do not differentiate between enantiomers, and therefore it was not necessary for this report to determine the enantiomeric excess of the product.)
1,2-Dihydroxyethylphosphonate methyl hydrogen ester (3)
To a 10 mL round-bottomed flask charged with ester 2 (167 mg, 0.982 mmol) was added 4.5 mL acetone and stirred at rt. To this suspension was added p-toluene sulfonic acid (9.3 mg, 5 mol%) and further stirred overnight. The temperature was raised on an oil bath, and the reaction mixture was refluxed for 2 h. After cooling, the crude product was extracted with CH2Cl2 (3 × 40 mL), and the combined organic layers were dried over MgSO4. The purification was achieved by silica gel column chromatography (EtOAc:MeOH = 10:1) to isolate a colorless oil. To this was added 10% NaOH solution and the reaction was heated to 100 °C. After refluxing for 3 h, the reaction was acidified using 1 M HCl, and passed through a Dowex (H+) column to yield the desired acid in a 36.1% overall yield. 31P NMR (202.3 MHz, D2O) δ 20.8.
1, 2-Dihydroxyethylphosphonic acid (4)
To a 50 mL round-bottomed flask were added 91.8 mg (0.54 mmol) of ester 2 and 5 mL of dry CH2Cl2. While stirring at rt, iodotrimethylsilane (380 mg, 3.5 equiv) was added dropwise. After stirring at rt for 2 h, methanolic water was added. The excess solvents and reagents were removed under reduced pressure and after diluting with water, the product was mixed with charcoal and filtered through celite. 1H NMR (500 MHz, CDCl3) δ 3.68 (1H, ddd, J = 10, 9, 3 Hz, PCH), 3.60 (1H, ddd, J = 12, 7, 3 Hz, CH2), 3.43 (1H, ddd, J = 12, 9, 5.5 Hz, CH2); 31P NMR (202.3 MHz, D2O) δ 21.3.
2-Hydroxyethylphosphonate methyl hydrogen ester (5)
To a solution of 10% sodium hydroxide (10 mL) was added dimethyl 2-hydroxyethylphosphonate ester (6.49 mmol), and refluxed for 1 h. After the reaction mixture was cooled to rt, it was acidified with 2 M HCl, concentrated, and redissolved in 20 mL methanol and filtered through celite. 1H NMR (400 MHz, CDCl3) δ 3.70 (2H, dt, J = 14.4, 7.2 Hz, CH2OH), 3.35 (3H, d, J = 11.2 Hz, OCH3), 2.01 (2H, dt, J = 18.4, 7.2 Hz, PCH2); 31P NMR (202.3 MHz) δ 28.6.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (GM PO1 GM077596 and R01 GM059334). BTC was supported by a National Institutes of Health Chemistry-Biology Interface Training Program (GM070421). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS or NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Biarrotte-Sorin S, Maillard A, Delettré J, Sougakoff W, Arthur M, Mayer C. Crystal structures of Weissella viridescens FemX and its complex with UDP-MurNAc-pentapeptide: insights into FemABX family substrates recognition. Structure. 2004;12:257–267. doi: 10.1016/j.str.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Blodgett J, Thomas P, Li G, Velasquez J, van der Donk W, Kelleher N, Metcalf W. Unusual transformations in the biosynthesis of the antibiotic phosphinothricin tripeptide. Nat Chem Biol. 2007;3:480–485. doi: 10.1038/nchembio.2007.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blodgett J, Zhang J, Metcalf W. Molecular cloning, sequence analysis, and heterologous expression of the phosphinothricin tripeptide biosynthetic gene cluster from Streptomyces viridochromogenes DSM 40736. Antimicrob Agents Chemother. 2005;49:230–240. doi: 10.1128/AAC.49.1.230-240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman E, McQueney M, Barry RJ, Dunawaymariano D. Catalysis and thermodynamics of the phosphoenolpyruvate phosphonopyruvate rearrangement - entry into the phosphonate class of naturally-occurring organophosphorus compounds. Journal of the American Chemical Society. 1988;110:5575–5576. [Google Scholar]
- Cheek S, Ginalski K, Zhang H, Grishin N. A comprehensive update of the sequence and structure classification of kinases. BMC Struct Biol. 2005;5:6. doi: 10.1186/1472-6807-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchillo R, Zhang H, Blodgett J, Whitteck J, Li G, Nair S, van der Donk W, Metcalf W. An unusual carbon-carbon bond cleavage reaction during phosphinothricin biosynthesis. Nature. 2009;459:871–874. doi: 10.1038/nature07972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K, Wanner B. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diddens H, Zahner H, Kraas E, Gohring W, Jung G. Transport of tripeptide antibiotics in bacteria. European Journal of Biochemistry. 1976;66:11–23. doi: 10.1111/j.1432-1033.1976.tb10420.x. [DOI] [PubMed] [Google Scholar]
- Eliot A, Griffin B, Thomas P, Johannes T, Kelleher N, Zhao H, Metcalf W. Cloning, expression, and biochemical characterization of Streptomyces rubellomurinus genes required for biosynthesis of antimalarial compound FR900098. Chem Biol. 2008;15:765–770. doi: 10.1016/j.chembiol.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliot A, Kirsch J. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- Gondry M, Sauguet L, Belin P, Thai R, Amouroux R, Tellier C, Tuphile K, Jacquet M, Braud S, Courçon M, et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat Chem Biol. 2009;5:414–420. doi: 10.1038/nchembio.175. [DOI] [PubMed] [Google Scholar]
- Goward C, Nicholls D. Malate dehydrogenase: a model for structure, evolution, and catalysis. Protein Sci. 1994;3:1883–1888. doi: 10.1002/pro.5560031027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory M, Till R, Smith M. Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J Bacteriol. 2003;185:5320–5323. doi: 10.1128/JB.185.17.5320-5323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Kuramitsu S, Okamoto A, Hirotsu K, Higuchi T, Kagamiyama H. Site-directed mutagenesis of Escherichia coli aspartate aminotransferase: role of Tyr70 in the catalytic processes. Biochemistry. 1991;30:7796–7801. doi: 10.1021/bi00245a019. [DOI] [PubMed] [Google Scholar]
- Jia Y, Lu Z, Huang K, Herzberg O, Dunaway-Mariano D. Insight into the mechanism of phosphoenolpyruvate mutase catalysis derived from site-directed mutagenesis studies of active site residues. Biochemistry. 1999;38:14165–14173. doi: 10.1021/bi990771j. [DOI] [PubMed] [Google Scholar]
- Johnson R, Kastner R, Larsen S, Ose E. Antibiotic A53868 and process for production thereof (USPTO, ed.) U.S.A.: Eli Lilly and Company; 1984. p. 14. [Google Scholar]
- Kieser T, Bibb M, Buttner M, Chater K, Hopwood D. Practical Streptomyces Genetics. Norwich, England: The John Innes Foundation; 2000. [Google Scholar]
- Kugler M, Loeffler W, Rapp C, Kern A, Jung G. Rhizocticin A, an antifungal phosphono-oligopeptide of Bacillus subtilis ATCC 6633: biological properties. Arch Microbiol. 1990;153:276–281. doi: 10.1007/BF00249082. [DOI] [PubMed] [Google Scholar]
- Lee J, Evans B, Li G, Kelleher N, van der Donk W. In vitro characterization of a heterologously expressed nonribosomal peptide synthetase involved in phosphinothricin tripeptide biosynthesis. Biochemistry. 2009;48:5054–5056. doi: 10.1021/bi900164d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Murakami K, Seki T, He X, Yeung S, Kuzuyama T, Seto H, Liu H. Protein purification and function assignment of the epoxidase catalyzing the formation of fosfomycin. J Am Chem Soc. 2001;123:4619–4620. doi: 10.1021/ja004153y. [DOI] [PubMed] [Google Scholar]
- Martinez A, Kolvek S, Yip C, Hopke J, Brown K, MacNeil I, Osburne M. Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl Environ Microbiol. 2004;70:2452–2463. doi: 10.1128/AEM.70.4.2452-2463.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf W, van der Donk W. Biosynthesis of phosphonic and phosphinic acid natural products. Annu Rev Biochem. 2009;78:65–94. doi: 10.1146/annurev.biochem.78.091707.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntai I, Manier M, Hachey D, Bachmann B. Biosynthetic origins of C-P bond containing tripeptide K-26. Org Lett. 2005;7:2763–2765. doi: 10.1021/ol051091d. [DOI] [PubMed] [Google Scholar]
- O'Brien T, Kluger R, Pike D, Gennis R. Phosphonate analogues of pyruvate. Probes of substrate binding to pyruvate oxidase and other thiamin pyrophosphate-dependent decarboxylases. Biochim Biophys Acta. 1980;613:10–17. doi: 10.1016/0005-2744(80)90186-2. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schwartz D, Berger S, Heinzelmann E, Muschko K, Welzel K, Wohlleben W. Biosynthetic gene cluster of the herbicide phosphinothricin tripeptide from Streptomyces viridochromogenes Tu494. Applied and Environmental Microbiology. 2004;70:7093–7102. doi: 10.1128/AEM.70.12.7093-7102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Grammel N, Heinzelmann E, Keller U, Wohlleben W. Phosphinothricin tripeptide synthetases in Streptomyces viridochromogenes Tu494. Antimicrobial Agents and Chemotherapy. 2005;49:4598–4607. doi: 10.1128/AAC.49.11.4598-4607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H, Freeman S, Seto H, Knowles J. Phosphonate biosynthesis: isolation of the enzyme responsible for the formation of a carbon-phosphorus bond. Nature. 1988;335:457–458. doi: 10.1038/335457a0. [DOI] [PubMed] [Google Scholar]
- Seto H, Kuzuyama T. Bioactive natural products with carbon-phosphorus bonds and their biosynthesis. Nat Prod Rep. 1999;16:589–596. doi: 10.1039/a809398i. [DOI] [PubMed] [Google Scholar]
- Shao Z, Blodgett J, Circello B, Eliot A, Woodyer R, Li G, van der Donk W, Metcalf W, Zhao H. Biosynthesis of 2-hydroxyethylphosphonate, an unexpected intermediate common to multiple phosphonate biosynthetic pathways. J Biol Chem. 2008;283:23161–23168. doi: 10.1074/jbc.M801788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H, Bonner D. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- Wang ZM, Kakiuchi K, Sharpless KB. Osmium-catalyzed asymmetric dihydroxylation of cyclic cis-disubstituted olefins. Journal of Organic Chemistry. 1994;59:6895–6897. [Google Scholar]
- Whitteck J, Ni W, Griffin B, Eliot A, Thomas P, Kelleher N, Metcalf W, van der Donk W. Reassignment of the structure of the antibiotic A53868 reveals an unusual amino dehydrophosphonic acid. Angew Chem Int Ed Engl. 2007;46:9089–9092. doi: 10.1002/anie.200703810. [DOI] [PubMed] [Google Scholar]
- Willey J, van der Donk W. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- Woodyer R, Shao Z, Thomas P, Kelleher N, Blodgett J, Metcalf W, van der Donk W, Zhao H. Heterologous production of fosfomycin and identification of the minimal biosynthetic gene cluster. Chem Biol. 2006;13:1171–1182. doi: 10.1016/j.chembiol.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Zdobnov E, Apweiler R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Zhang G, Dai J, Lu Z, Dunaway-Mariano D. The phosphonopyruvate decarboxylase from Bacteroides fragilis. J Biol Chem. 2003;278:41302–41308. doi: 10.1074/jbc.M305976200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.