Abstract
Cyclooxygenase (COX)-2 enzyme catalyzes the rate-limiting step of prostaglandin formation in pathogenic states, and overexpression of COX-2 occurs at multiple stages of colon carcinogenesis, allowing elevated prostaglandin synthesis to occur in the tumor microenvironment. In normal cells, COX-2 expression levels are potently regulated at the posttranscriptional level through various RNA sequence elements present within the mRNA 3′ untranslated region (3′UTR). A conserved AU-rich element functions to target COX-2 mRNA for rapid decay and translational inhibition through association with various RNA-binding proteins to influence the fate of COX-2 mRNA. The 3′UTR contains alternative polyadenylation signals that result in a shortened 3′UTR and loss of regulatory elements. Specific microRNAs have been identified to bind regions within the COX-2 3′UTR and control COX-2 expression. Recent evidence demonstrates the functional significance of the COX-2 3′UTR and how improper recognition of the 3′UTR can contribute to COX-2 overexpression in colorectal cancer.
Keywords: COX-2, Colon cancer, Posttranscriptional regulation, AU-rich element
Introduction
Cyclooxygenase (COX) enzymes perform the rate-limiting step in the conversion of free arachidonic acid into prostaglandins and facilitate normal homeostasis in the gastrointestinal tract [1]. Two primary COX enzyme isoforms (COX-1 and COX-2) have been identified to play distinct roles in physiologic and pathologic conditions. COX-1 (PTGS1) is constitutively expressed in most cell types, and COX-1—derived prostaglandins are necessary for protecting gastric mucosa and maintaining vascular tone. In contrast to the expression pattern of COX-1 is COX-2 (PTGS2). Normally absent in most cells, COX-2 is an immediate-early response gene whose expression is rapidly induced by a variety of proinflammatory and growth-associated stimuli. Elevated COX-2 expression often is associated with many chronic inflammatory diseases and cancers, making it a clinically important target for pharmacologic intervention.
The relationship between COX-2 expression and colorectal cancer was first suggested by studies demonstrating the efficacy of aspirin and nonsteroidal anti-inflammatory drugs in reducing the relative risk of colon cancer and promoting tumor regression in both humans and experimental animal models of colon cancer [1]. Investigation of the molecular basis of these observations showed that high levels of COX-2 protein were present in both human and animal colorectal tumors, whereas the normal intestinal mucosa had low to undetectable COX-2 expression [1]. This association between COX-2 overexpression and carcinogenesis was established further in genetic studies demonstrating a significant reduction in intestinal polyposis in mice deficient in the PTGS2 gene [2]. These findings clearly indicate that chronic elevation of COX-2 is pathologic and suggest that inhibition of COX-2 via pharmacologic means or regulation of its expression can limit the development or progression of colorectal cancer.
A variety of evidence gathered from epidemiologic, experimental animal, and cellular studies indicates that unregulated COX-2 expression is an important step in colorectal cancer, and it is generally well accepted that transcriptional activation of COX-2 may occur early in tumorigenesis [3]. Current efforts now indicate the significance of COX-2 regulation at the level of mRNA decay and translational control, and demonstrate that loss of posttranscriptional regulation promotes COX-2 overexpression in colorectal tumors. This review summarizes the current advances in the area of COX-2 posttranscriptional regulation in colorectal cancer, focusing largely on the mRNA 3′ untranslated region (3′UTR) and associated regulatory factors that play a role in controlling COX-2 expression at the posttranscriptional level.
The COX-2 mRNA 3′ Untranslated Region Mediates Posttranscriptional Regulation
A characteristic feature regulating the expression of inducible genes in response to growth stimulation, inflammation, and cellular stress signaling occurs through mechanisms of posttranscriptional regulation that control mRNA stability and subsequent translation [4]. The impact of this level of regulation is evident, as microarray analysis has determined that 40% to 50% of the changes in inducible gene expression occur at the level of mRNA stability [5]. Eukaryotic mRNAs contain two characteristic features integral to their function, a 5′ 7-methylguanosine cap and a 3′ poly(A) tail. In mammalian cells, most mRNA decay is initiated by shortening of the poly(A) tail (deadenylation). At this point, degradation initiated at the 5′ end involves removal of the 5′ cap by decapping enzymes, followed by 5′-to-3′ exonucleolytic decay. Alternatively, the mRNA can be degraded by 3′-to-5′ exonucleolytic degradation through a complex of exonucleases known as the exosome [4]. These two decay pathways are not mutually exclusive, and there appears to be overlap between the pathways, although the relative contribution of each mechanism is under debate [4]. Many of these mRNAs targeted for degradation are localized to processing (P)-bodies, which are small cytoplasmic foci containing components of both 3′-to-5′ and 5′-to-3′ decay machinery, suggesting these decay pathways converge at P-bodies [4]. An alternative fate of mRNAs observed under situations of cellular stress is their trafficking to cytoplasmic stress granules, where they are translationally silenced, and current work now indicates a functional interaction between P-bodies and stress granules, suggesting that mRNAs destined for decay are sorted at stress granules and delivered to P-bodies for degradation [6].
mRNA regulatory elements that play a critical role in identifying specific transcripts for posttranscriptional regulation typically reside within the 3′UTR of the transcript [4]. Identification of multiple mRNA regulatory elements present within the COX-2 3′UTR was the first evidence suggesting that COX-2 might be regulated at a posttranscriptional level [3]. Exon 10 of the human PTGS2 gene contains the entire 3′UTR, and within this region are multiple polyadenylation signals, two of which are used primarily to yield transcripts approximately 4.6 and 2.8 kb in length [7]. Observed in most cells is the larger 4.6-kb COX-2 transcript that results from processing at a distal canonical (AAUAAA) polyadenylation site. However, a proximal noncanonical polyadenylation signal (AUUAAA) can yield transcripts with shortened 3′UTRs [7]. The significance of this alternative mRNA processing event has been shown in colorectal cancer cells, as a polyadenylation variant of COX-2 mRNA lacking the distal region of the 3′UTR was selectively stabilized upon cell growth to confluence [8]. These findings suggest that COX-2 mRNA can escape rapid decay through alternative polyadenylation site use, resulting in deletion of potential 3′UTR regulatory elements, and this phenomenon of shortening of mRNA transcripts appears to be a widespread feature occurring in cancer cells [9••].
A characteristic feature controlling the expression of many inflammatory cytokines, growth factors, and protooncogenes is their inherent ability to be targeted for rapid mRNA decay. These growth-related immediate-early gene transcripts are unstable because of the presence of a common cis-acting element known as the AU-rich element (ARE) [10]. The importance of this particular RNA element is evident within the human genome, because it is estimated that approximately 8% of transcripts contain putative AREs [11]. AREs are organized into several classes and often are composed of multiple copies of an AUUUA sequence motif [11]. Within the 3′UTR of COX-2 mRNA, the ARE is composed of a 116-nucleotide region containing a cluster of six AUUUA sequence elements located near the stop codon of COX-2 [12]. Posttranscriptional regulation has been shown to depend on this conserved ARE, because its presence confers rapid decay of a normally stable reporter mRNA [12]. Furthermore, this AU-rich region is highly conserved in both sequence and location among various species of COX-2, implying that ARE function has been evolutionarily conserved [3].
Normal cellular growth is associated with rapid decay of ARE-containing mRNAs, and targeted mRNA decay is an essential way to control their pathogenic overexpression. This aspect of COX-2 regulation is observed in non-transformed intestinal epithelial cells, where rapid degradation of COX-2 mRNA occurs (half-life, ~13 minutes) [13]. However, several observations have implicated the loss of ARE-mediated posttranscriptional regulation in the neo-plastic transformation of cells [10], and recent findings have demonstrated that this subset of transcripts is enriched during colon tumorigenesis. Gene expression profiling comparing adenomas with late-stage adenocarcinomas showed a three-to four-fold enrichment in ARE-containing genes compared with the genome as a whole, and a similar enrichment is observed in tumors as early as stage I [14•]. With regard to COX-2 regulation, similar findings have been observed in human colon carcinoma cells [13, 15-17]. As a result of the inability of the COX-2 ARE to function properly, enhanced stability of COX-2 mRNA was detected, and increased expression of a reporter gene containing the COX-2 3′UTR also was observed. Based on the inherent genetic instability of tumor cells, it might be expected that mutations in AREs occur that allow for dysregulated protein expression. However, few naturally occurring mutations in AREs have been described, and the ARE region of the PTGS2 gene is intact in healthy individuals as well as in colon tumor cells [3]. This implies that loss of ARE function in colon tumors is caused primarily by altered ARE recognition by cellular trans-acting regulatory factors.
COX-2 ARE-Binding Proteins
AREs mediate their regulatory function through the association of trans-acting RNA-binding proteins that display high affinity for AREs. The best-studied ARE-binding proteins can promote rapid mRNA decay, mRNA stabilization, or translational silencing [4]. Through these mechanisms, ARE-binding proteins exhibit pleiotropic effects on gene expression, as a single ARE-binding protein can bind to multiple mRNAs [10].
Various cytoplasmic proteins have been observed to bind the COX-2 ARE, and similar RNA/protein complexes are seen between the AREs from COX-2 and granulocyte-macrophage colony-stimulating factor [12]. When cross-linked to the COX-2 ARE, this complex is composed of several distinct proteins, with sizes ranging from 35 to 90 kD. To date, 16 different RNA-binding proteins have been reported to bind the COX-2 3′UTR (Table 1). This review focuses on the most well-documented RNA-binding proteins that regulate COX-2 expression in the gastrointestinal tract and their potential impact on colorectal cancer.
Table 1.
Cyclooxygenase 2 mRNA 3′ untranslated region–binding proteins
| Studies | Protein | Gene | Function |
|---|---|---|---|
| Dixon et al. [15], Young et al. [24••] | HuR | ELAVL1 | mRNA stabilization Translational enhancer |
| Sawaoka et al. [8], Young et al. [24••], Carballo et al. [26] | TTP | ZFP36 | mRNA destabilization |
| Mukhopadhyay et al. [32], Murmu et al. [33] | CUGBP2 | CUGBP2 | mRNA stabilization Translational suppression |
| Dixon et al. [16], Lopez de Silanes et al. [35] | TIA-1 | TIA-1 | Translational suppression |
| Cok et al. [44] | TIAR | TIAL1 | Translational suppression |
| Anant et al. [36], Blanc et al. [37] | Apobec-1 | APOBEC1 | mRNA stabilization |
| Cok et al. [45] | AUF1 | HNRNPD | mRNA destabilization |
| Cok et al. [45], Sureban et al. [46] | RBM3 | RBM3 | mRNA stabilization Translational enhancer |
| Lee and Jeong [47] | β-Catenin | CTNNB1 | mRNA stabilization |
| Winzen et al. [48] | KSRP | KHSRP | mRNA destabilization |
| Cok et al. [45], Dean et al. [49] | CBF-A | HNRNPAB | mRNA stabilization |
| Rousseau et al. [50] | hnRNP A0 | HNRNPA0 | mRNA stabilization |
| Cok et al. [45] | hnRNP A2/B1 | HNRNPA2B1 | mRNA transport |
| Cok et al. [45] | hnRNP A3 | HNRNPA3 | mRNA transport |
| Shanmugam et al. [41] | hnRNP K | HNRNPK | mRNA stabilization |
| Cok et al. [44] | hnRNP U | HNRNPU | ND |
ND—not determined
HuR
The HuR protein is a ubiquitously expressed member of the ELAV (Embryonic-Lethal Abnormal Vision in Drosophila) family of RNA-binding proteins [18]. The human Hu proteins (ubiquitously expressed HuR and neuronal-specific HuB, HuC, and HuD) originally were discovered as antigens in patients displaying paraneo-plastic disorders, such as cerebrellar carcinoma. The cloning and characterization of HuR demonstrated that it contains three RNA recognition motifs with a high affinity and specificity for AREs and that its overexpression stabilizes ARE-containing transcripts and promotes their translation [18].
The ability of HuR to function as an ARE-stability factor appears to be linked to its subcellular localization [19]. HuR is localized predominantly in the nucleus (>90%) and can shuttle between the nucleus and cytoplasm. It is hypothesized that the ability of HuR to promote mRNA stabilization requires its translocation to the cytoplasm, and overexpression of HuR promotes localization to the cytoplasm, where it binds target ARE-containing mRNAs and interferes with their rapid decay [18, 19]. A variety of cellular signals known to activate MAPK pathways involving p38 and ERK kinases, the PI-3 kinase pathway, and the Wnt signaling pathway have been shown to trigger cytoplasmic HuR localization and promote ARE-containing mRNA stabilization [20-22]. Insight into the mechanism of HuR-mediated mRNA stabilization has advanced with the identification of low molecular weight inhibitors of HuR [23••]. These compounds interfere with the formation of HuR dimers and RNA binding, indicating that HuR homodimerizes before ARE binding. Furthermore, these compounds inhibit HuR cytoplasmic localization, suggesting that an HuR dimer is the active species involved in nucleocytoplasmic trafficking.
Based on its ability to bind the COX-2 ARE, HuR has been identified as a trans-acting factor involved in regulating COX-2 expression [15]. The enhanced stabilization of COX-2 mRNA observed in colon cancer cells is partly the result of elevated HuR levels [15, 24••]. Recent work evaluating HuR expression in colon tissue demonstrates that in normal tissue, HuR is expressed at low levels and is located in the nucleus, whereas HuR overexpression and cytoplasmic localization have been observed in colon adenomas, adenocarcinomas, and metastases; consistent with these observations, overexpression of COX-2 colocalized with elevated HuR [24••]. Furthermore, several studies indicate that HuR overexpression and cytoplasmic localization are a marker for elevated COX-2 that is correlated with advancing stages of malignancy and poor clinical outcome [3]. Based on its recognized role as a regulator of COX-2 and other genes promoting cell proliferation, angiogenesis, metastasis, and cell survival, elevated HuR has been indicated to contribute to the malignant phenotype.
Tristetraprolin
Tristetraprolin (TTP, ZFP36, TIS11) is a member of a small family of tandem Cys3His zinc finger proteins consisting of TTP, ZFP36L1, and ZFP36L2 [25]. TTP originally was identified as an immediate-early response gene whose expression was transiently induced by extracellular stimuli such as serum or phorbol ester. These early studies implicated a role for TTP as a transcription factor; however, it now is known that TTP acts on a posttranscriptional level to promote rapid decay of ARE-mRNAs by direct ARE binding [26]. The binding of TTP to AREs targets the transcript for rapid degradation through association with various decay enzymes [27, 28].
The ability of TTP to promote rapid ARE-mRNA decay has been shown to be controlled by several mechanisms. The subcellular localization of TTP has been observed to occur in both nuclear and cytoplasmic regions; however, treatment of cells with growth factor or mitogen results in enhanced translocation of TTP to the cytoplasm, presumably through increased nucleocytoplasmic shuttling of TTP [25]. When present in the cytoplasm, TTP localizes to P-bodies [29], suggesting a critical role in ARE-mRNA delivery to the decay machinery residing within P-bodies [30]. TTP also is a target of phosphorylation by the ERK and p38 MAPK pathways both in vitro and in vivo, and the phosphorylation state likely plays a role in regulating TTP function [25]. Phosphorylation via the p38 MAPK pathway allows TTP to interact with 14-3-3 adaptor proteins, which in turn promotes TTP cytoplasmic localization and protein stabilization; however, phosphorylation of TTP compromises its ability to promote ARE-mediated decay.
Efforts to characterize the function of TTP have focused primarily on its regulation of inflammatory mediators, such as tumor necrosis factor (TNF)-α [26]. The physiologic consequences of TTP loss are evident, as TTP knockout mice develop multiple inflammatory syndromes resulting from increased inflammatory factors, including COX-2, due to defects in rapid mRNA turnover [31], and recent work has shown COX-2 to be a target of TTP [8, 24••]. With regard to its role in controlling COX-2 expression in colon cancer, TTP expression is low or not apparent in colon cancer cell lines and adenoma and adenocarcinoma tissue [24••]. This is in contrast to normal tissue, in which TTP expression is highest in normal epithelium and localized predominantly to the cytoplasm. Furthermore, adenoviral-mediated delivery of TTP to colon cancer cells resulted in downregulation of COX-2 expression coupled with a dramatic reduction in cell growth and proliferation [24••]. These findings indicate that the presence of TTP in normal colon epithelium serves a protective role by controlling expression of various inflammatory mediators, including COX-2, whereas the loss of TTP expression in tumors allows HuR to promote the stabilization and translation of COX-2 mRNA.
CUGBP2
CUGBP2 is an RNA-binding protein containing three RNA recognition motifs and is a member of the CELF (CUGBPETR-3—like factors) family [32, 33]. Work investigating COX-2 regulation in colon cancer cells and intestinal epithelium subjected to ionizing radiation elucidated a novel role for CUGBP2 in regulating COX-2 expression on a posttranscriptional level [32, 33]. CUGBP2 resides predominately in the nucleus; however, its expression is rapidly induced in response to radiation, leading to increased cytoplasmic trafficking and CUGBP2 inhibition after radiation treatment leads to increased cellular survival associated with increased COX-2 expression. Further work demonstrated that CUGPB2 has a high affinity for the COX-2 ARE, and this interaction serves to repress COX-2 translation through the disruption of polysomal loading of the COX-2 mRNA [32]. Interestingly, the radiation-induced overexpression of CUGBP2 led to COX-2 mRNA stabilization similar to overexpression of HuR protein. Given that HuR enhances and CUGBP2 inhibits COX-2 protein expression, this indicates that these two ARE-binding proteins differ in their regulation of COX-2 mRNA translation. Although both proteins have similar affinities for the COX-2 ARE, CUGBP2 was effective in competing with HuR for ARE binding, leading to a translational block in COX-2 expression [34]. While demonstrating the ability of CUGBP2 to regulate the opposing functions of COX-2 mRNA stabilization and translational repression, these findings suggest a possible role for CUGBP2 in the early stages of tumorigenesis, counteracting the effects of HuR overexpression to repress COX-2 protein synthesis.
T-Cell Intracellular Antigen 1
T-cell intracellular antigen 1 (TIA-1), originally identified in activated T lymphocytes, is an RNA-binding protein containing three RNA recognition motifs with specificity to mRNAs containing short sections of uridylate repeats [35]. Under normal cellular conditions, this protein is predominantly nuclear and in response to cellular stress, trans-locates to the cytoplasm, where it is associated with untranslated mRNAs in discrete cytoplasmic stress granules, implicating a role in translational regulation [6, 29]. Consistent with these observations, TIA-1 has been shown to specifically bind the COX-2 ARE and regulate its expression through translational inhibition without altering COX-2 mRNA turnover [16]. However, in colon cancer cells, deficiencies in TIA-1 binding to the COX-2 ARE were observed, allowing for increased polysome association with the COX-2 mRNA. The significance of TIA-1—mediated regulation of COX-2 expression was observed in TIA-1—deficient fibroblasts that produced significantly more COX-2 protein and prostaglandins than wild-type fibroblasts. Similarly, deletion of TIA-1 in mice led to elevated COX-2 levels and the development of arthritis [31]. These findings implicate TIA-1 as a translational silencer of COX-2 expression and suggest that loss of TIA-1 function may be a contributing factor promoting enhanced expression of COX-2 in neoplasia.
Apobec-1
Apobec-1 is a sequence-specific cytidine deaminase named for its ability to catalyze C-to-U editing of apolipoprotein B mRNA. In the context of ARE-binding proteins, Apobec-1 binds the AREs of COX-2 in addition to other inflammatory mediator and oncogene mRNAs and promotes their respective mRNA stabilization [36]. The importance of Apobec-1 expression in intestinal epithelium is seen in the acute response to radiation injury; in Apobec-1—deficient mice, COX-2 expression is suppressed, leading to increased levels of radiation-induced apoptosis [36]. With regard to colon cancer, Apobec-1 overexpression occurs in human colorectal cancer, and current results using the Apcmin/+ mouse model of gastrointestinal tumorigenesis demonstrate that Apobec-1 deletion in the Apcmin/+ background is associated with reduced COX-2 expression in adenomas of Apcmin/+ apobec-1-/-mice, leading to an overall reduction in polyp number in these compound mice [37]. These findings identify Apobec-1 as a critical regulator of COX-2 through ARE-mediated mRNA stabilization in different pathologic situations.
MicroRNA-Mediated Regulation of COX-2
MicroRNAs (miRNAs) are small noncoding RNAs that have emerged as fundamental global regulators of gene expression via posttranscriptional mechanisms. Within the genome, miRNAs may reside within gene exons or introns and may be transcribed as part of a protein-coding gene, or their transcription may be regulated independently [38•]. Once transcribed, the primary miRNA transcript is processed in the nucleus by the RNase III endonuclease Drosha into a precursor miRNA stem-loop. The precursor miRNA is actively transported by exportin-5 to the cytoplasm, where it is processed a second time by another endonuclease, Dicer, to form a double-stranded miRNA duplex. The miRNA duplex is incorporated into the RNA-induced silencing complex (RISC), and the mature miRNA strand is preferentially retained in the RISC complex, where it can negatively regulate target mRNA expression [38•].
In humans, miRNAs alter most gene expression by binding to complementary sites in the 3′UTR of its target mRNA and inhibiting its translation or promoting mRNA cleavage. Although similar in nature to short interfering RNAs, which bind target mRNA regions with 100% complementarity and promote mRNA degradation, miRNAs can bind imperfectly to the 3′UTR of targeted transcripts to attenuate target gene expression. Based on this characteristic, a single miRNA can potentially affect expression of many proteins with various cellular functions. To date, 721 human miRNAs have been identified, and they are proposed to regulate an estimated 30% of human mRNAs [38•]. Because of the substantial amount of control they have over several putative mRNA targets, it is of considerable interest how alterations in miRNA expression in cancer can contribute to tumorigenesis. In colorectal cancer, differential expression of several miRNAs has been observed, and current efforts have demonstrated that specific miRNA loss or overexpression may have an impact on various cellular pathways associated with colon tumorigenesis [38•, 39]. Currently, three miRNAs are reported to target COX-2 mRNA and attenuate its expression.
miR-16
miR-16, part of an miRNA cluster that also includes miR-15a, was discovered via its loss of expression in chronic lymphocytic leukemia resulting from genomic deletion, and this miRNA can serve in a tumor suppressor capacity through targeting of the antiapoptotic gene Bcl-2 [39]. Based on its RNA sequence, miR-16 displays complementarity to AU-rich regions, and it has been demonstrated that miR-16 can target AREs, particularly those of TNF-α, interleukin (IL)-6, IL-8, and COX-2, to alter their mRNA stability [40]. Interestingly, the same studies determined that miR-16 works in conjunction with TTP to promote decay of ARE-containing mRNAs, through interactions between TTP and components of the RISC complex. More recent work in inflammatory cells has determined that miR-16 and the RNA-binding protein hnRNP K play opposing roles in COX-2 mRNA stabilization and that hnRNP K can compete with miR-16 for binding to the COX-2 3′UTR [41]. Taken together, these findings emphasize the significance of this ARE-targeting miRNA and highlight the dynamic nature of miRNA-mediated posttranscriptional regulation.
miR-101 and miR-199
In work investigating the role of COX-2 during embryo implantation, the murine miRNAs miR-101a and miR-199a were identified to posttranscriptionally regulate murine COX-2, and expression of these miRNAs is inversely correlated with uterine COX-2 protein levels [42]. Further work in human cells established the ability of miR-101a and miR-199a to target human COX-2 [42]. In the context of colon cancer, an inverse correlation between human miR-101 and COX-2 expression in colon cancer cell lines and colon tumors has been shown, along with the ability of miR-101 to inhibit COX-2 translation when ectopically expressed in cells [43]. Along the same lines, COX-2—expressing colon cancer cells do express miR-199 [42], indicating that limited expression of these miRNAs in colon cancer may contribute to observed COX-2 overexpression.
Conclusions
Posttranscriptional gene regulatory events involving mRNA decay and translational silencing is emerging as a fundamental and effective way to alter the expression of COX-2 and other cancer-associated genes that are functionally related on the mRNA level (Fig. 1). The identification and characterization of RNA-binding proteins that interact with the COX-2 3′UTR are fundamental for understanding the mechanisms allowing COX-2 overexpression to occur in colorectal cancer. By virtue of their altered expression and function in colon tumors, allowing for enhanced mRNA stabilization and translation, these RNA-binding proteins hold great promise as candidates for therapeutic targeting to allow the nascent cellular RNA degradation machinery to counteract the oncogenic effects of mRNA stabilization. Furthermore, continued investigation in the area of miRNA-mediated posttranscriptional regulation undoubtedly will elucidate the role of miRNAs in colorectal tumor development and identify new COX-2—targeting miRNAs whose expression may be altered during tumorigenesis.
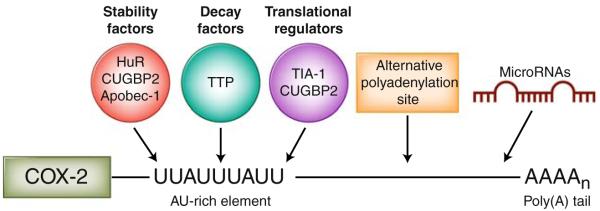
Fig. 1.
Posttranscriptional regulation of cyclooxygenase 2 (COX-2) within the 3′ untranslated region (3′UTR). COX-2 expression is tightly regulated at the posttranscriptional level by RNA-binding proteins that promote mRNA stability (HuR, CUGBP2, and Apobec-1), mRNA decay (tristetraprolin [TTP]), and translational inhibition (T-cell intracellular antigen 1 [TIA-1] and CUGBP2) through their binding of the COX-2 AU-rich element. The presence of alternative polyadenylation signals may lead to a shortened 3′UTR and subsequent loss of regulatory elements and microRNA binding sites within the 3′UTR. Specific microRNAs have been determined to bind the COX-2 3′UTR and attenuate COX-2 expression
Acknowledgments
The authors thank David Young for graphics support. The authors were funded by grants from the National Institutes of Health (R01CA134609) and American Cancer Society (RSG-06-122-01-CNE). They apologize to their colleagues for not being able to reference all primary work owing to space limitations.
Footnotes
Disclosure No potential conflicts of interest relevant to this article were reported.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Wang D, Mann JR, DuBois RN. The role of prostaglandins and other eicosanoids in the gastrointestinal tract. Gastroenterology. 2005;128:1445–1461. doi: 10.1053/j.gastro.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 2.Oshima M, Dinchuk JE, Kargman SL, et al. Suppression of intestinal polyposis in APCΔ716 knockout mice by inhibition of cyclooxygenase 2 (COX-2) Cell. 1996;87:803–809. doi: 10.1016/s0092-8674(00)81988-1. [DOI] [PubMed] [Google Scholar]
- 3.Dixon DA. Regulation of COX-2 expression in human cancer. Prog Exp Tumor Res. 2003;37:52–71. doi: 10.1159/000071363. [DOI] [PubMed] [Google Scholar]
- 4.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 5.Cheadle C, Fan J, Cho-Chung YS, et al. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics. 2005;6:75. doi: 10.1186/1471-2164-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Hall-Pogar T, Zhang H, Tian B, et al. Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res. 2005;33:2565–2579. doi: 10.1093/nar/gki544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sawaoka H, Dixon DA, Oates JA, et al. Tristetrapolin binds to the 3′ untranslated region of cyclooxygenase-2 mRNA: a polyadenylation variant in a cancer cell line lacks the binding site. J Biol Chem. 2003;278:13928–13935. doi: 10.1074/jbc.M300016200. [DOI] [PubMed] [Google Scholar]
- 9••.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. This article highlights the significance of alternative polyadenylation in cancer.
- 10.de Silanes I Lopez, Quesada MP, Esteller M. Aberrant regulation of messenger RNA 3′-untranslated region in human cancer. Cell Oncol. 2007;29:1–17. doi: 10.1155/2007/586139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon DA, Kaplan CD, McIntyre TM, et al. Post-transcriptional control of cyclooxygenase-2 gene expression. The role of the 3′-untranslated region. J Biol Chem. 2000;275:11750–11757. doi: 10.1074/jbc.275.16.11750. [DOI] [PubMed] [Google Scholar]
- 13.Sheng H, Shao J, Dixon DA, et al. Transforming growth factor-beta1 enhances Ha-ras-induced expression of cyclooxygenase-2 in intestinal epithelial cells via stabilization of mRNA. J Biol Chem. 2000;275:6628–6635. doi: 10.1074/jbc.275.9.6628. [DOI] [PubMed] [Google Scholar]
- 14•.Kanies CL, Smith JJ, Kis C, et al. Oncogenic Ras and transforming growth factor-beta synergistically regulate AU-rich element-containing mRNAs during epithelial to mesenchymal transition. Mol Cancer Res. 2008;6:1124–1136. doi: 10.1158/1541-7786.MCR-07-2095. This article reports that selective ARE-containing mRNA stabilization occurs during colon tumorigenesis.
- 15.Dixon DA, Tolley ND, King PH, et al. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon DA, Balch GC, Kedersha N, et al. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J Exp Med. 2003;198:475–481. doi: 10.1084/jem.20030616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao J, Sheng H, Inoue H, et al. Regulation of constitutive cyclooxygenase-2 expression in colon carcinoma cells. J Biol Chem. 2000;43:33951–33956. doi: 10.1074/jbc.M002324200. [DOI] [PubMed] [Google Scholar]
- 18.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keene J. Why is Hu where? Shuttling of early-response messenger RNA subsets. Proc Natl Acad Sci USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briata P, Ilengo C, Corte G, et al. The Wnt/beta-catenin—>Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol Cell. 2003;12:1201–1211. doi: 10.1016/s1097-2765(03)00407-6. [DOI] [PubMed] [Google Scholar]
- 21.Ming XF, Stoecklin G, Lu M, et al. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol Cell Biol. 2001;21:5778–5789. doi: 10.1128/MCB.21.17.5778-5789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Wang W, Fan J, et al. Prostaglandin A2-mediated stabilization of p21 mRNA through an ERK-dependent pathway requiring the RNA-binding protein HuR. J Biol Chem. 2004;279:49298–49306. doi: 10.1074/jbc.M407535200. [DOI] [PubMed] [Google Scholar]
- 23••.Meisner NC, Hintersteiner M, Mueller K, et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat Chem Biol. 2007;3:508–515. doi: 10.1038/nchembio.2007.14. This article identifies, for the first time, small molecule inhibitors of HuR and characterizes their function in inflammatory cells.
- 24••.Young LE, Sanduja S, Bemis-Standoli K, et al. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136:1669–1679. doi: 10.1053/j.gastro.2009.01.010. This article reports on how loss of TTP expression coupled with HuR overexpression occurs during colon tumorigenesis and results in COX-2 overexpression.
- 25.Baou M, Jewell A, Murphy JJ. TIS11 family proteins and their roles in posttranscriptional gene regulation. J Biomed Biotechnol. 2009:634520. doi: 10.1155/2009/634520. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 27.Chen CY, Gherzi R, Ong SE, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 28.Fenger-Gron M, Fillman C, Norrild B, et al. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 29.Kedersha N, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips K, Kedersha N, Shen L, et al. Arthritis suppressor genes TIA-1 and TTP dampen the expression of tumor necrosis factor alpha, cyclooxygenase 2, and inflammatory arthritis. Proc Natl Acad Sci U S A. 2004;101:2011–2016. doi: 10.1073/pnas.0400148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukhopadhyay D, Houchen CW, Kennedy S, et al. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 33.Murmu N, Jung J, Mukhopadhyay D, et al. Dynamic antagonism between RNA-binding protein CUGBP2 and cyclooxygenase-2-mediated prostaglandin E2 in radiation damage. Proc Natl Acad Sci U S A. 2004;101:13873–13878. doi: 10.1073/pnas.0406066101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sureban SM, Murmu N, Rodriguez P, et al. Functional antagonism between RNA binding proteins HuR and CUGBP2 determines the fate of COX-2 mRNA translation. Gastroenterology. 2007;132:1055–1065. doi: 10.1053/j.gastro.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 35.de Silanes I Lopez, Galban S, Martindale JL, et al. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anant S, Murmu N, Houchen CW, et al. Apobec-1 protects intestine from radiation injury through posttranscriptional regulation of cyclooxygenase-2 expression. Gastroenterology. 2004;127:1139–1149. doi: 10.1053/j.gastro.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 37.Blanc V, Henderson JO, Newberry RD, et al. Deletion of the AU-rich RNA binding protein Apobec-1 reduces intestinal tumor burden in Apcmin mice. Cancer Res. 2007;67:8565–8573. doi: 10.1158/0008-5472.CAN-07-1593. [DOI] [PubMed] [Google Scholar]
- 38•.O’Hara SP, Mott JL, Splinter PL, et al. MicroRNAs: key modulators of posttranscriptional gene expression. Gastroenterology. 2009;136:17–25. doi: 10.1053/j.gastro.2008.11.028. This thorough review discusses the role of miRNAs in the gastrointestinal tract.
- 39.Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 40.Jing Q, Huang S, Guth S, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 41.Shanmugam N, Reddy MA, Natarajan R. Distinct roles of heterogeneous nuclear ribonuclear protein K and microRNA-16 in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products. J Biol Chem. 2008;283:36221–36233. doi: 10.1074/jbc.M806322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakrabarty A, Tranguch S, Daikoku T, et al. MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc Natl Acad Sci U S A. 2007;104:15144–15149. doi: 10.1073/pnas.0705917104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strillacci A, Griffoni C, Sansone P, et al. MiR-101 down-regulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp Cell Res. 2009;315:1439–1447. doi: 10.1016/j.yexcr.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 44.Cok SJ, Acton SJ, Morrison AR. The proximal region of the 3′-untranslated region of cyclooxygenase-2 is recognized by a multimeric protein complex containing HuR, TIA-1, TIAR, and the heterogeneous nuclear ribonucleoprotein U. J Biol Chem. 2003;278:36157–36162. doi: 10.1074/jbc.M302547200. [DOI] [PubMed] [Google Scholar]
- 45.Cok SJ, Acton SJ, Sexton AE, et al. Identification of RNA-binding proteins in RAW 264.7 cells that recognize a lipopolysaccharide-responsive element in the 3-untranslated region of the murine cyclooxygenase-2 mRNA. J Biol Chem. 2004;279:8196–8205. doi: 10.1074/jbc.M308475200. [DOI] [PubMed] [Google Scholar]
- 46.Sureban SM, Ramalingam S, Natarajan G, et al. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene. 2008;27:4544–4556. doi: 10.1038/onc.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee HK, Jeong S. Beta-Catenin stabilizes cyclooxygenase-2 mRNA by interacting with AU-rich elements of 3′-UTR. Nucleic Acids Res. 2006;34:5705–5714. doi: 10.1093/nar/gkl698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winzen R, Thakur BK, Dittrich-Breiholz O, et al. Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol Cell Biol. 2007;27:8388–8400. doi: 10.1128/MCB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dean JL, Sully G, Wait R, et al. Identification of a novel AU-rich-element-binding protein which is related to AUF1. Biochem J. 2002;366:709–719. doi: 10.1042/BJ20020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rousseau S, Morrice N, Peggie M, et al. Inhibition of SAPK2a/p38 prevents hnRNP A0 phosphorylation by MAPKAP-K2 and its interaction with cytokine mRNAs. EMBO J. 2002;21:6505–6514. doi: 10.1093/emboj/cdf639. [DOI] [PMC free article] [PubMed] [Google Scholar]