Abstract
A priori global identifiability is a structural property of biological and physiological models. It is considered a prerequisite for well-posed estimation, since it concerns the possibility of recovering uniquely the unknown model parameters from measured input-output data, under ideal conditions (noise-free observations and error-free model structure). Of course, determining if the parameters can be uniquely recovered from observed data is essential before investing resources, time and effort in performing actual biomedical experiments. Many interesting biological models are nonlinear but identifiability analysis for nonlinear system turns out to be a difficult mathematical problem. Different methods have been proposed in the literature to test identifiability of nonlinear models but, to the best of our knowledge, so far no software tools have been proposed for automatically checking identifiability of nonlinear models. In this paper, we describe a software tool implementing a differential algebra algorithm to perform parameter identifiability analysis for (linear and) nonlinear dynamic models described by polynomial or rational equations. Our goal is to provide the biological investigator a completely automatized software, requiring minimum prior knowledge of mathematical modelling and no in-depth understanding of the mathematical tools. The DAISY (Differential Algebra for Identifiability of SYstems) software will potentially be useful in biological modelling studies, especially in physiology and clinical medicine, where research experiments are particularly expensive and/or difficult to perform. Practical examples of use of the software tool DAISY are presented. DAISY is available at the web site http://www.dei.unipd.it/~pia/.
Keywords: Identifiability, Linear and nonlinear dynamical systems, A priori global identifiability, Differential algebra, Model identification, Identifiability software
1 INTRODUCTION
Parameters characterizing the behavior of unobservable features of biological and physiological systems, e.g. the effect of a drug on its target organ, are usually not amenable to direct measurement. Their measurement is thus usually approached indirectly as a parameter estimation problem [1, 13]. A dynamic model describing the internal structure of the system is formulated theoretically, based on physical, chemical, conservation and transport laws e.g. of enzyme kinetics and pharmacokinetics. In general, these physiological models take the mathematical form of linear or nonlinear dynamic state-space models, depending on unknown parameters. Usually these parameters are unknown and cannot be pre-specified, and need to be estimated from data collected experimentally by measuring the observable model variables (inputs and outputs). In order to solve the estimation problem, input-output (I/O) experiments are designed. A fundamental prerequisite for parameter estimation to be well posed is global identifiability of the parametric model (see e.g. [1, 5, 8, 13]). This property states that, under ideal conditions of noise-free observations and error-free model structure, the unknown parameters of the postulated model can be uniquely (and exactly) recovered from the knowledge of the input-output variables of the designed input-output experiment.
Note that the property of a priori identifiability regards an ideal context of error-free model structure and noise-free measurements and thus it is a necessary, but not a sufficient condition to ensure that an accurate identification of the model parameters from real input/output data is possible (e.g. data may be too noisy or the problem too badly conditioned, etc.). However, if the parameters of the postulated model are not uniquely identifiable, even in the theoretical most favorable situation, they will never be identifiable in a practical experiment where model structure misspecification and noise in the measurements are inevitably present. Without a guarantee of a priori identifiability, the estimates of the parameters which could, nevertheless, sometimes be obtained by some numerical optimization algorithms, will be totally unreliable and random. Unfortunately, despite its essential role in parameter identification, identifiability analysis has often been neglected by many researchers. Use of a non uniquely identifiable model in a clinical setting, may possibly compromise the distinction of the normal vs. pathological state, ultimately leading the researcher physician to draw potentially erroneous conclusions.
Identifiability also impacts on the design of experiments, by providing guidelines on the selection of input and output sites to allow unique identifiability [12]. This is particularly useful when dealing with intact physiological systems, where both the number and the location of possible inputs and outputs is often very limited. It has been shown that a priori identifiability results can be used to achieve the formulation of a minimal, i.e. necessary and sufficient, input-output configuration for complex experimental design.
Identifiability analysis can be helpful also to provide guidelines to deal with non-identifiability, either providing hints on how to simplify the model structure or indicating when more information (measured data) are needed for the specific experiment.
Identifiability analysis of nonlinear systems is in general very difficult. The need for such a theory is unquestionable as dealing with nonlinear models, i.e. the Michaelis-Menten equation, is very common in modelling say enzyme kinetics and drug metabolism. Unfortunately its applicability has been seriously hampered by the heavy computational burden of the available techniques. Specifically, the problem translates mathematically into checking solvability of an unusually large system of nonlinear algebraic equations. The number of equations and their degree generally increases with the model order. Note that identifiability of the linearized model can provide information on the identifiability of the original nonlinear model only under certain restrictions [4] but, in general, this is not true and the identifiability of nonlinear models has to be tackled directly.
Different approaches have been proposed in literature [1, 2, 5, 4, 8, 9, 14] but, to the best of our knowledge, no general software tools currently exist to perform the identifiability analysis for a nonlinear dynamic model, despite the crucial importance of this step in the modelling process.
As our contribution to the solution of this problem, we have developed a new differential algebra algorithm [2, 11] which integrates the different strategies proposed in [8, 9] and broadens their domain of applicability to models described by (linear and) nonlinear models involving polynomial or rational functions, and are initialized at either unknown or known initial conditions. We refer the reader to [2, 11] where this algorithm is described in detail. In this paper we shall present a new software tool, DAISY (Differential Algebra for Identifiability of SYstems) implementing the algorithm in the symbolic language REDUCE version 3.8. The goal of this new software tool, which we propose to make broadly available, is to bring to the field of biomedical applications a piece of software which, although being based on a rather sophisticated set of mathematical tools, will not require knowledge of higher mathematics and computer algebra by the user and yet will allow them to tackle problems which are hard and computationally intensive in a transparent way, without requiring any knowledge of high-level programming languages.
The layout of the paper is the following:
In Section 2, the basic dynamic structure of the models under test is introduced. Some definitions regarding parameter identifiability are recalled and the identifiability test based on the characteristic set is briefly illustrated.
In Section 3, a simple example to illustrate the differential algebra method is presented.
In Section 4, the algorithm and its features are described.
In Section 5, the software tool DAISY is presented.
In Section 6, a case of usage of the software DAISY for the analysis of a priori global identifiability of a biomedical model is presented in some detail. In particular, the input file that the user has to provide to DAISY and the corresponding output file with the identifiability results are reported.
Some basic concepts of differential algebra useful for identifiability analysis test are recalled in the Appendix. In particular, differential ideals and the characteristic set are defined together with their principal properties.
2 CHECKING A PRIORI IDENTIFIABILITY
This section provides the reader with a brief description of the theory behind our software tool. Such an overview is not meant to be complete. A detailed documentation of the theory is reported in [2, 11].
2.1 Definitions
Many dynamic models of biological and physiological systems can be described by a general nonlinear system Σp in state space form, depending on an unknown parameter vector p:
| (1) |
| (2) |
where x is the n-dimensional state variable, representing e.g. masses, concentrations etc.; u the m-dimensional input vector made of smooth functions 1; y is the r-dimensional output, e.g. time histories of drug concentrations; p ∈ 𝒫 is a ν-dimensional vector of unknown parameters. The entries of f, G = [g1, …, gm] and h are assumed to be polynomial or rational functions of their arguments.
Note that (1,2) describe the system together with a certain choice of input-output variables which is designed for the identification experiment. In biological/medical applications, identification experiments are often performed on systems at rest or started from known (equilibrium) initial conditions. If initial conditions are specified, the relevant equation x(t0) = x0 (or x(t0) = x0(p) if initial conditions are parameter dependent), is added to the system. The essential assumption here is that the system is accessible from its initial conditions, which is a “generic” controllability-like assumption. See [11]2.
Equality constraints (linear or nonlinear) on p may be also present. We shall denote them
| (3) |
where E is a polynomial or a rational vector function.
A priori global identifiability of the system-experiment model (1,2), is a condition guaranteeing theoretical uniqueness of solution to the problem of recovering the model parameters from input-output data.
In more precise terms, a priori global identifiability addresses the following question: given arbitrary parameters p1, p2 ∈ 𝒫, p1 ≠ p2 and assuming the systems Σp1 and Σp2 are initialized at the same initial state; does there exist (at least) an input function u for which systems Σp1 and Σp2 yield different outputs? If so, Σp1 and Σp2 are distinguishable from the experiment and the system Σp is said to be a priori globally (or uniquely) identifiable from input-output data. In this case, at least in principle, the parameter values p1 and p2 can be distinguished (from at least one input-output experiment). If instead systems Σp1 and Σp2 yield the same output no matter which input function is applied and no matter what initial state the system is started at, the parameter values p1 and p2 can never be distinguished and are said to be indistinguishable from input-output experiments.
If there are at most a finite number of indistinguishable parameter values, the system is called locally (or nonuniquely) identifiable from input-output data.
Finally, if there is an infinite number of different indistinguishable parameter values, the system is commonly called nonidentifiable or unidentifiable [1, 5, 8, 14].
One should point out that the definition above does not exactly apply to many concrete identification problems of biological/biomedical systems. In these applications, most of the time there is very little (if none at all) freedom to choose a suitable input function guaranteeing identifiability. See for example the model describing the control of insulin on glucose utilization in humans and the model of glucose metabolism in the brain from PET data (Examples 2 and 4, respectively, in [2]). Often one has to deal with experiments where the input function is fixed (typically an impulse-like function of time). The condition of a priori global identifiability from input-output data has to be replaced with the (more stringent) condition of a priori global identifiability with a fixed input function. The approach presented in this paper also applies to this notion of identifiability, provided of course, one keeps in mind that the input function is fixed.
2.2 The differential algebra approach to identifiability
In order to see how differential algebra techniques play a role in system identifiability analysis, we shall look upon the set of n + r equations defining the dynamic system (1,2) just as a system of algebraic (in fact, polynomial or, more generally, rational) equations in the relevant state, input, output variables and their derivatives. The algebraic apparatus of differential algebra permits to deal with polynomials which may involve derivatives of some variables (typically the state), and allows polynomials themselves to be differentiated, still keeping a formal similarity with classical commutative algebra. See the Appendix for a very succinct review of basic concepts. A basic idea is that of characteristic set of a differential ideal. The choice of which particular (differential) polynomial ring one wants to work is nontrivial. In [11, 2] it is shown that the ring of differential polynomials with rational (in p) coefficients, denoted R(p)[u, y, x], is a particularly convenient choice for identifiability analysis. Here the variables u, y, x represent inputs, outputs, states, and, possibly, their derivatives of some finite order.
A characteristic set is a special basis (i.e. a “minimal” set of differential polynomials) which generates the same differential ideal generated by an arbitrary given set of differential polynomials. Practically speaking, knowledge of a characteristic set in our base ring, allows one to eliminate the state variables and to find the input-output relation of the dynamic system: a set of r polynomial differential equations involving only the variables (u, y).
It can be shown that the characteristic set of a dynamic system in state space form has a special structure which easily allows one to test parameter identifiability. The computation of the characteristic set is performed via the famous Ritt’s pseudodivision algorithm [10], a far-reaching generalization of the Euclidian algorithm for polynomials.
As described in the Appendix, in order to calculate the characteristic set of the differential ideal generated by the polynomials defining the dynamic system (1,2), we need to apply the pseudodivision algorithm. This requires the introduction of a ranking among the model variables (inputs, outputs and states and their derivatives)3. The standard ranking used in system identification declares the input and the output components, which are known variables in this context, as the lowest ranked variables, and the highest rank is given to the state variable components:
| (4) |
In principle, given the rank among the variables, their derivatives can be ranked in different ways. In system identifiability it has turned out convenient to choose the following ranking:
| (5) |
The pseudodivision algorithm to calculate the characteristic set is then applied with respect to the declared ranking (5). The result is a family of differential polynomials belonging to the differential ring R(p)[u, y, x] of the following form:
| (6) |
where the explicit dependence on p is not shown. Equating these polynomials to zero provides a (finite) set of n + r nonlinear differential equations which generate exactly the same totality of functions (u, y, x) which satisfy the equations of the original system (1,2).
Note that the first r differential polynomials Ai, i = 1, …, r of (6) do not depend on x. In fact, they are obtained after elimination of the state variables x from the system (1,2). In principle, these differential polynomials are identically known hence all their coefficients are known. The corresponding polynomial differential equations are called the input-output relation of the system. They describe all input-output pairs which satisfy the system equations (1,2). These r input-output differential equations are the basic tool used in input-output identifiability analysis.
To fix uniquely the coefficients of the polynomials listed in (6), one has to normalize each of the above polynomials to make it monic. Denote with cij ∈ R(p), i = 1, …, r, j = 1, …, νi, (here j is an index running over the monomial indices of the polynomial Ai, the monomials being ordered, say, in a lexicographic ordering) the coefficients of the first r polynomials listed in (6). It follows that the functions cij(p) are uniquely attached to the input-output relation of the system. Let ν := ∑i νi and let c : 𝒫 → ℝν be the map defined by stacking the scalar components, {cij}, in some prescribed order. The map c is called the exhaustive summary of the model [9], since c embodies the parameter dependence of the input-output model completely.
After the exhaustive summary is found, in order to test global input-output identifiability of the system (1,2) the injectivity of the map c from the parameter space to its range, a subset of the ν-dimensional Euclidean space, has to be checked. To do this, one has to solve the system of algebraic nonlinear equations
| (7) |
for arbitrary right-hand members in the range of c. Equality constraints (3), if present, can be added to (7).
The resulting system of nonlinear equations may be solved by a suitable computer algebra method, using e.g. the Buchberger algorithm [3]. This algorithm calculates the Groebner basis, which is a set of polynomials with specific properties which make it a powerful tool for solving systems of nonlinear equations. In particular, from the structure of the Groebner basis it is possible to see by inspection if the system admits one solution, a finite number of solutions or infinitely many solutions for each parameter, thus allowing one to distinguish between global or local identifiability or unidentifiability. This is particularly useful in biological and biomedical modelling applications, where, due to the possibility of imposing physiological plausibility constraints, in some cases, models may still be useful even if they are only locally identifiable.
2.3 Identifiability with known initial conditions
It has been shown that the construction of the characteristic set ignores the initial conditions. However, in the fortunate circumstances of known initial conditions, the data available to solve the identifiability problem should include also this knowledge. In this case the additional assumption of algebraic observability 4 is required.
In particular, assume that l, (l ≤ n), initial conditions of a dynamical model of order n are known. To check global identifiability, first we have to check if the corresponding states are algebraically observable. To do this one has to verify if the derivatives of these state components appear or not in the corresponding l polynomials of the characteristic set. If not, the above states are algebraically observable and we can proceed with the identifiability test as follows. The l polynomials are evaluated at time t = 0 and set equal to their corresponding known values. Note that the right hand members of these equations, evaluated at t = 0 become known polynomial functions of p with coefficients in the known data x0, u(0), u̇(0), ü(0), …, y(0), ẏ(0), ÿ(0),…. The identifiability test should be based on the new exhaustive summary of the model obtained by adding these l equations to the previous exhaustive summary (calculated in absence of known initial conditions).
For example we have shown, by applying our software DAISY, that the follow mamillary model of four compartments
| (8) |
when all the initial conditions are unknown is locally identifiable, while when at least two initial conditions between x2(0), x3(0), x4(0) are known it becomes globally identifiable.
3 ANALYSIS OF AN EXAMPLE
Here we shall analyze in mathematical notation, a nonlinear model for which the calculations can be done by hand. This is meant to illustrate the basic steps of the differential algebra identifiability algorithm described in the previous section. Another example analyzed via the software tool, will be presented in Section 6.
Example 1
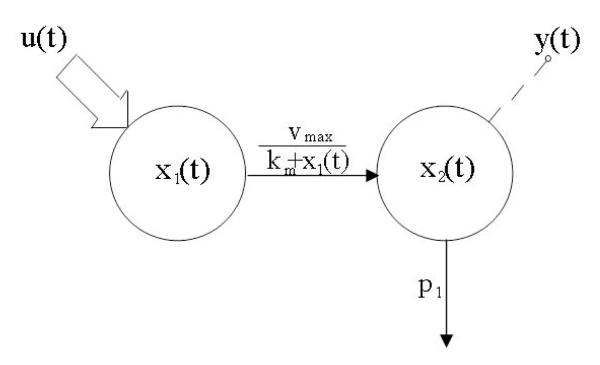
Consider a two-compartment model with Michaelis-Menten kinetics. The model structure, together with its input-output configuration, is shown in Fig. 1. The model is mathematically described by the following rational differential equations:
| (9) |
where x1, x2 are drug masses in blood and tissues respectively, u is the drug input, y the measured drug output in the blood, p1 is a constant rate parameter, vmax and km are the classical Michaelis-Menten parameters.
LEGEND FIG.1.
A nonlinear two-compartment model with Michaelis-Menten kinetics. The input-output experiment configuration is also shown (the large arrow denotes the input and the dashed line ending with a bullet the output).
The question is whether the unknown parameter vector p = [p1, vmax, km] is globally identifiable from the input-output experiment.
The following (standard) ranking of the input, output and state variables is declared
| (10) |
The differential polynomials of the system are ranked according to the above chosen rank:
| (11) |
With this information, the pseudodivision algorithm to calculate the characteristic set can start. Since the third polynomial contains the leader of the first polynomial, we have to reduce it with respect to the first polynomial. The remainder of this first pseudodivision is calculated:
| (12) |
The new system is:
| (13) |
Now the second polynomial is already reduced with respect the first one, while the third polynomial has to be reduced with respect to the first. The result of the pseudodivision is:
| (14) |
The new system is:
| (15) |
Now the system is autoreduced, thus it is the characteristic set for the model.
The first polynomial has been obtained after elimination of the state variable x and represents the input-output relation of the model (1,2). As explained in Section 2.2, to fix uniquely the coefficients of the input-output relation, we have to make it monic. To do this, the first polynomial in (15) is divided by the factor km vmax, obtaining
| (16) |
To construct the exhaustive summary of the model, the coefficients of the above input-output polynomial are extracted:
| (17) |
To calculate the range set of this function, the coefficients (17) are evaluated at a symbolic parameter value p = [α, β, γ].
This gives the following set of algebraic nonlinear equations:
| (18) |
Solving this system (18) by the Buchberger algorithm provides the following Groebner basis:
| (19) |
showing that the parameters have one and only one solution, i. e. the model is globally identifiable.
In practice, to speed up the process, instead of choosing a symbolic parameter value, we can use a set of pseudo-randomly chosen numerical points in the range set. For example, choose a numerical value for the parameter vector, say
| (20) |
and calculate the corresponding value of the coefficients of the input-output relation, as we did before with the symbolic value.
This provides the following system of algebraic nonlinear equations:
| (21) |
Solve this system by the Buchberger algorithm, which provides a unique value for the Groebner basis:
| (22) |
This shows that the model is identifiable about the parameter value (20). Repeating this procedure for a “reasonably” chosen set of parameter values provides a “practical” test of global identifiability 5. Here, by repeating the test, one obtains that the model is globally identifiable, as obtained with the symbolic point.
4 THE DAISY (Differential Algebra for Identifiability of SYstems) ALGORITHM
The input of the algorithm is provided by the differential polynomials defining the dynamic system (1, 2), the number of inputs and outputs, the ranked list of input, output and state variables, the list of the unknown parameters and, if present, the equality constraints among the parameters (3).
The algorithm consists of the following sequence of steps:
If one or more polynomials are rational, they are reduced to the same denominator (this can be easily accomplished using the built-in REDUCE commands).
A binary matrix is assembled, where all the information on the structure of the dynamical system can be summarized. This matrix has as many rows as model equations, and as many columns as there are variables (inputs, outputs and states) and their corresponding derivatives. In particular, the matrix is constructed by considering one equation, i.e. row, at a time, and by writing 1 (one) if the corresponding variable or derivative is present in the equation, 0 (zero) if that element is absent.
- In the input file the specific components of the input, output and state variables are assigned in the standard ranking (inputs<outputs<states) to a list. These variables are automatically ranked with their derivatives as follows:
This implies a rank between the polynomials. Thus, the polynomials defining the system are ranked by increasing rank.(23) Each polynomial is compared with the previous ones. If it is of equal or higher rank, it is reduced with respect to the preceding ones by applying the pseudodivision algorithm.
Steps 2 - 4 are repeated until no further reductions can be performed. At the end, the autoreduced set of minimum rank is reached. This constitutes the characteristic set.
An observability test is performed: if in the characteristic set a state component appears without derivatives then it is algebraically observable [8].
Extract from the characteristic set the polynomials without the x variable, i.e. containing only y, u and their derivatives. These polynomials must be as many as the model outputs and constitute the input-output relation of the dynamic model (1,2). Their coefficients are polynomial functions of p. If some input-output relation does not contain a monomial term with coefficient equal to one, it is suitably normalized to make it monic.
The coefficients, functions of the unknown parameter p, of the monomials in y, u and their derivatives, are extracted from the input-output relation and provide the exhaustive summary of the original dynamic model. If parameter equality constraints (3) are present, they are included in the exhaustive summary.
Each function of the unknown p in the exhaustive summary is evaluated at a pseudo-randomly chosen numerical value for the vector p and set equal to the obtained numerical value. Thus, a system of algebraic nonlinear equations in the unknown p is constructed 6.
These algebraic nonlinear equations are submitted to the REDUCE module “groebner”, which solves the system with the routine “groesolve” and returns the solutions for each unknown parameter thus providing the model identifiability results, i.e. global or local identifiability or nonidentifiability.
If j initial conditions of algebraically observable states are known, the corresponding j polynomials of the characteristic set (6) are evaluated at time t = 0. These known algebraic polynomial functions of p are added to the polynomials calculated in step 7. The algorithm proceeds with step 8 where, in particular, the added polynomials are set equal to their corresponding known values, and step 9 which provides the identifiability results.
The computation of the identifiability results is usually very fast (few seconds) also for complex nonlinear models of order 4 or 5 even in a computer with only 128 M of RAM. Note that the most computationally demanding step of the whole algorithm is represented by the Buchberger algorithm (embedded Reduce function) for solving the system of algebraic nonlinear equations provided by the exhaustive summary. Thus, if the model is very complex, for example with more than one nonlinearity, the algorithm may not successfully terminate due to a lack of memory of the system running the application. Very likely, this limit will be substantially improve in the future due to the rapid progress in hardware.
5 THE SOFTWARE TOOL DAISY
The software tool implementing the above algorithm is written in REDUCE version 3.8. REDUCE 3.8 is an interactive program designed for general algebraic computations of interest to mathematicians, scientists and engineers. The main aim of REDUCE is to support algebra calculations that are not feasible by hand. The REDUCE computer algebra system is not public domain, but it is available at a small fee. The identifiability software tool DAISY is available at the web site http://www.dei.unipd.it/~pia/.
The identifiability test program involves a number of subroutines. Below is a list of the components of DAISY:
DAISY.B is the REDUCE compiled module which loads all the subroutines into memory for all the successive elaborations.
- The subroutine DAISY computes the characteristic set and the exhaustive summary of the model and calls the REDUCE module “groebner” which solves the exhaustive summary system with the routine “groesolve” and returns the solutions for each unknown parameter thus providing the model identifiability results. In particular DAISY invokes the following subroutines:
- — The subroutine CREATEIO assembles a vector with all variables and their derivatives in the given ranking.
- — The subroutine CREATE assembles one suitable matrix where all the information on the dynamic system is present and writes all the input data to a file.
- — The surboutine SORTG sorts the system equations according to the particular ranking given as input.
- — The subroutine SORTEQ exchanges the matrix rows according to the given ranking.
- — The subroutine SORTV exchanges the elements of the equation leaders vector according to the given ranking.
- — The subroutine REDUCTION simplifies the equations according to the presence of common factors.
- — The subroutine PSEUDOREM calculates the rest of the pseudodivision among polynomials.
- — The subroutine CONDINIZ, if called by the user in the input file, deals with the known initial conditions. In particular, the subroutine implements step 11 of the algorithm.
To run the identifiability program, one must submit an input file that follows a specific format, which is described in the next Section, where also the format of the output file provided by the software tool is reported.
The program is available together with a README file where detailed instructions about its usage are reported and a folder with the input and the output files of some representative biological case studies is available to the user.
6 A CASE STUDY
DAISY has been tested and evaluated “in house” on a representative set of biological and biomedical cases. To check global identifiability with DAISY, the dynamical model should be provided in a separate file according to a specific scheme. We shall illustrate this scheme by giving an example.
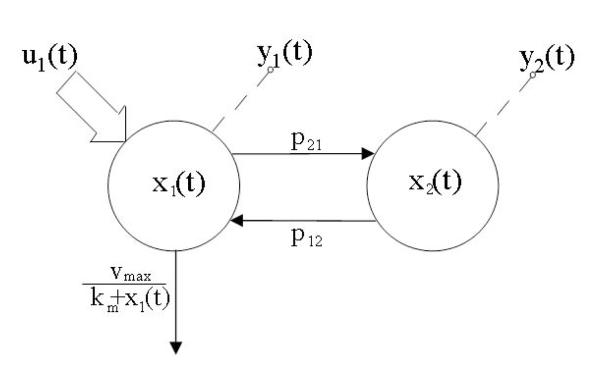
We will use a very common model in enzyme kinetics and drug metabolism, i.e. the two-compartment open model with Michaelis-Menten elimination. The model structure, together with its input-output configuration, is shown in Fig. 2. The model is mathematically described by the following rational differential equations:
| (24) |
where x1, x2 are drug masses in blood and tissues respectively, u1 is the input, y1 and y2 the measured outputs, p = [p21, p12, vmax, km] is the unknown parameter vector.
LEGEND FIG.2.
A nonlinear two-compartmental model. The same notation of Fig.1 is used.
The question is whether the unknown parameter vector p is globally identifiable from the input-output experiment.
To solve this identifiability problem with DAISY, the standard ranking of the input, output and state variables is assigned
| (25) |
In the following, the input file that the user has to provide to DAISY to check the global identifiability of model (24) and the corresponding output file with all the identifiability results are reported.
6.1 The input file
Here we write in lower letters the portion to be inserted by the user in order to distinguish it from the fixed structure, written in upper letters, of the file. Note that in REDUCE comments to the code are preceded by symbol %.
WRITE “Case study 1. A two-compartment Michaelis-Menten model”$
% B_ is the vector of all the input, output and state variables
B_:={u1,y1,y2,x1,x2}$
FOR EACH EL_ IN B_ DO DEPEND EL_,T$
% B1_ is the vector of the unknown parameters
B1_:={p21,p12,vmax,km}$
% NY_ and NU_ indicate the number of inputs and outputs respectively
NU_:=1$
NY_:=2$
% C_ is the system’s differential equations describing the dynamic model
C_:={df(x1,t)+(p21+vmax/(km+x1))*x1-p12*x2-u1,
df(x2,t)-p21*x1+p12*x2,
y1-x1,
y2-x2}$
% main program and subroutines
DAISY() $
% If the initial conditions are equal to given values, the user should write these values and call the CONDINIZ subroutine, for example:
% LET x10=1,x20=2$
% CONDINIZ()$
6.2 The output file
The results provided by DAISY are shown below. To be as brief as possible, we do not report the whole results file, but just the relevant results for identifiability testing:
Case study 1. A two-compartment Michaelis-Menten model$
NUMBER OF EQUATIONS$
n_:= 4$
VARIABLE VECTOR$
b_ := {u1, y1, y2, x1, x2}$
PARAMETER VECTOR$
b1_ := {p21,p12,vmax,km}$
RANKING AMONG THE VARIABLES$
bb_ := {u1, y1, y2, df(u1,t), df(u1,t,2), df(y1,t),df(y1,t,2),df(y2,t),df(y2,t,2), x1,x2,df(x1,t),df(x2,t)}$
NUMBER OF INPUTS$
nu_ := 1$
NUMBER OF OUTPUTS$
ny_ := 2$
MODEL EQUATIONS$
c_ := {df(x1,t)*(km + x1) + km*(- p12*x2 + p21*x1
- u1) - p12*x1*x2 + p21*x1**2 - u1*x1 + vmax*x1,
df(x2,t) + p12*x2 - p21*x1,
- x1 + y1,
- x2 + y2}$
CHARACTERISTIC SET$
% aa(1)_ and aa(2)_ constitute the input-output relation of the system
aa(1)_ :=df(y1,t)*(km + y1) + km*(- p12*y2 + p21*y1 - u1) + - p12*y1*y2 + p21*y1**2 - u1* y1 + vmax*y1$
aa(2)_ :=df(y2,t) + p12*y2 - p21*y1$
aa(3)_ := - x1+ y1$
aa(4)_ := - x2 + y2$
THE SYSTEM IS ALGEBRAICALLY OBSERVABLE$
PSEUDO-RANDOM NUMERICAL PARAMETER VECTOR$
b2_ := {14,28,15,17}$
EXHAUSTIVE SUMMARY$
flist_:= {km - 17, - km + 17, p21 - 14, - p12 + 28, - km*p12 + 476, - p21 + 14, p12 - 28, km*p21 + vmax - 253}$
MODEL PARAMETER SOLUTION(S)$
g := {{vmax=15,p12=28,p21=14,km=17}}$
The results show that the model is globally identifiable, since the Groebner basis g provides one and only one solution for each model parameter.
7 CONCLUSIONS
In this paper we have described DAISY (Differential Algebra for Identifiability of SYstems), a general software tool allowing biomedical researchers to perform global identifiability analysis for linear and nonlinear dynamic models. In particular, DAISY effectively facilitates the solution to the underappreciated problem of determining if unique parameter estimation from the experimental data is theoretically possible. Although DAISY is a computer-algebra code implementing a differential algebra algorithm, high-level programming languages, mathematical and computer algebra skills are not prerequisites for using the software. The design of an intuitive graphical user interface is under study, in particular, we are planning to develop a graphical interface by translating the REDUCE identifiability algorithm in C++. We are confident that this will also improve the execution speed of the program. The software tool can be downloaded from the website http://www.dei.unipd.it/~pia/.
8 Acknowledgments
This work was partially supported by National Institute of Health grant R01 GM070635.
APPENDIX
Background on differential algebra
For a formal treatment of differential algebra, the reader is referred to [7, 6, 10]. Here we shall just recall the definitions and notions which are necessary to follow the notations used in the rest of the paper.
Let z := [z1, …, zn] be a vector of smooth functions of the variable t (time); the totality of polynomials in the variables zi and their derivatives with coefficients in a field K, is a differential polynomial ring which will be denoted K[z].
Consider a set S of differential polynomials belonging to K[z]. The differential ideal I = IS, sometimes also denoted I(S), generated by S, is the smallest subset of K[z] containing S, which is closed with respect to addition, multiplication by arbitrary elements of K[z] and with respect to differentiation. The elements of S are generators of the ideal.
A differential ideal I is called prime if AiAj ∈ I implies that Ai ∈ I or Aj ∈ I and perfect if A ∈ I whenever Ak does (i.e. a perfect ideal coincides with its own radical).
In order to handle differential ideals, a ranking, i.e. a total ordering, denoted “<”, among the variables and their derivatives, must be introduced. Let and be arbitrary derivatives. Then the ranking should be such that, for arbitrary positive integer k:
| (26) |
The leader uj of a polynomial Aj is the highest ranking derivative of the variables appearing in that polynomial (in particular it can be a derivative of order zero).
The polynomial Ai is said to be of lower rank than Aj if ui < uj or, whenever ui = uj and degui(Ai) < deguj(Aj), where degu(A) denotes the algebraic degree.
A polynomial Ai will be said to be reduced with respect to a polynomial Aj if Ai contains neither the leader of Aj with equal or greater algebraic degree, nor its derivatives.
If Ai is not reduced with respect to Aj it can be reduced by using the pseudodivision algorithm described below.
if Ai contains the kth-derivative, (possibly k = 0), of the leader of Aj, Aj is differentiated k times so its leader becomes ;
multiply the polynomial Ai by the coefficient of the highest power of ; let R be the remainder of the division of this new polynomial by with respect to the variable . Then R is reduced with respect to . The polynomial R is called the pseudoremainder of the pseudodivision;
the polynomial Ai is replaced by the pseudo-remainder R and the process is iterated using in place of and so on, until the pseudoremainder is reduced with respect to Aj.
A set of differential polynomials A := {A1, A2, …, Ar} that are all reduced with respect to each other, is called an autoreduced set.
Let π be a differential polynomial. If we apply the pseudodivision algorithm to reduce π with respect to all Aj, j = 1, …, r, the final pseudoreminder is called the pseudoremainder of π with respect to the autoreduced set A. Such a pseudoremainder is said to be reduced with respect to A (compare [10] where an autoreduced set is called a chain).
Two autoreduced sets, A = {A1, A2, …, Ar} and B = {B1, B2, …, Bs} ordered in increasing rank so that A1 < A2 < … < Ar, B1 < B2 < … < Bs, are ranked according to the following principle.
If there is an integer k, k ≤ min(s, r) such that rank Ai = rank Bi, i = 1, …k - 1, rank Ak < rank Bk then A is said to be of lower rank than B.
If r < s and rank Ai = rank Bi, i = 1,…r, then A is also said to be of lower rank than B.
Definition 1 A lowest rank autoreduced set that can be formed with polynomials from a given set S of differential polynomials, is called a characteristic set of S.
The concept of characteristic set of a differential ideal has been introduced by Ritt [10] who also proposed the pseudodivision algorithm to construct it. The important property of a characteristic set is that it can be used to generate a differential ideal by means of a finite number of polynomials [6].
In principle, the characteristic set is not unique. It can however be normalized in such a way as to render it unique.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Sometimes in biological/medical applications the input functions are assumed to be impulsive. Clearly nonlinear operations on distributions are in general not defined and mathematically impulsive inputs cannot in general be applied to nonlinear systems. The difficulty is however purely formal, and can be circumvented by noting that in reality the input is always a smooth function of time. “Impulsive inputs” are a mathematical idealization to describe signals which have a support of duration negligible with respect to the slowest time constant of the system.
For systems satisfying this assumption, the software presented in this paper performs global identifiability analysis in a completely automatized way. The software works also for systems which are not accessible from initial conditions, but in this case requires an external intervention by the user.
For checking identifiability of a dynamic system, it is sufficient to consider the derivative up to n (model order) of the input and output variables and only the first derivative of the state variables.
We remember that a state of a dynamical model of order n is algebraically observable if its derivatives do not appear in the last n polynomials of the characteristic set of the model [8]
Since a priori identifiability is a structural property, i.e. it holds almost everywhere, the numerical point strategy gives the right result with probability one. To be sure that the pseudo-randomly chosen numerical point does not belong to a set of zero measure, the user is required to repeat several times the identifiability test.
The use of a pseudo-random numerical point allows one to significantly reduce the symbolic computation and hence broaden the class of testable models. When the system dimension increases, in fact, the use of a symbolic point affects the complexity of the Groebner basis calculation, thus severely limiting the class of testable models. In this case, to test global identifiability, the user is required to repeat the test several times (see last paragraph of Section 3).
References
- [1].Audoly S, D’Angiò L, Saccomani MP, Cobelli C. Global identifiability of linear compartmental models. IEEE Trans. Biomed. Eng. 1998;vol. 45:36–47. doi: 10.1109/10.650350. [DOI] [PubMed] [Google Scholar]
- [2].Audoly S, Bellu G, D’Angiò L, Saccomani MP, Cobelli C. Global identifiability of nonlinear models of biological systems. IEEE Trans. Biomed. Eng. 2001;vol. 48(no. 1):55–65. doi: 10.1109/10.900248. [DOI] [PubMed] [Google Scholar]
- [3].Buchberger B. An algorithmical criterion for the solvability of algebraic system of equation. Aequationes Mathematicae. 1988;vol. 4(no. 3):45–50. [Google Scholar]
- [4].Chapman MJ, Godfrey KR, Chappell MJ, Evans ND. Structural identifiability of non-linear systems using linear/non-linear splitting. Int. J. Control. 2003;vol. 76(no. 3):209–216. [Google Scholar]
- [5].Chappell MJ, Godfrey KR. Structural identifiability of the parameters of a nonlinear batch reactor model. Math. Biosci. 1992;vol. 108:245–251. doi: 10.1016/0025-5564(92)90058-5. [DOI] [PubMed] [Google Scholar]
- [6].Forsman K. Constructive Commutative Algebra in Nonlinear Control Theory. Linköping University; Sweden: 1991. Linköping Studies in Science and Technology. Dissertation No. 261. [Google Scholar]
- [7].Kolchin E. Differential Algebra and Algebraic Groups. Academic Press; New York: 1973. [Google Scholar]
- [8].Ljung L, Glad ST. On global identifiability for arbitrary model parameterizations. Automatica. 1994;vol. 30(no. 2):265–276. [Google Scholar]
- [9].Ollivier F. Le problème de l’identifiabilité structurelle globale: étude théorique, méthodes effectives et bornes de complexité, Thèse de Doctorat en Science. École Polytéchnique; Paris, France: 1990. [Google Scholar]
- [10].Ritt JF. Differential Algebra. American Mathematical Society; Providence, RI: 1950. [Google Scholar]
- [11].Saccomani MP, Audoly S, D’Angiò L. Parameter identifiability of nonlinear systems: the role of initial conditions. Automatica. 2003;vol. 39:619–632. [Google Scholar]
- [12].Saccomani MP, Cobelli C. A minimal input-output configuration for a priori identifiability of a compartmental model of leucine metabolism. IEEE Trans Biomed Eng. 1993;vol. 40:797–803. doi: 10.1109/10.238464. [DOI] [PubMed] [Google Scholar]
- [13].Walter E. Identifiability of State Space Models. Springer-Verlag; Berlin: 1982. [Google Scholar]
- [14].Walter E, Lecourtier Y. Global approaches to identifiability testing for linear and nonlinear state space models. Math. and Comput. in Simul. 1982;vol. 24:472–482. [Google Scholar]